Abstract
Transcriptional regulation of transgenes depends upon genomic localization in higher eukaryotes. For the applied use of transgenic organisms as producers of pharmaceutically relevant proteins or as pest population control agents, a method to make transgene expression predictable is highly desirable. A targeting method that allows precise cassette replacement comprising solely genes of interest (without extraneous donor vector sequences) would be highly advantageous for insects and other multicellular organisms. In this report, we describe a method for transgene targeting to predefined chromosomal sites in Drosophila by using a transposon vector that, once integrated in the germ line, acts as an acceptor site for donor vectors. To make recombinational insertions irreversible, a FLP recombinase-mediated cassette exchange strategy was used, and to enhance donor-target pairing, a homing sequence from the linotte locus was used. Site-specific recombinants were screened by interconvertible eye fluorescence marker phenotypes yielding, on average, targeted insertions at a frequency of 23%. The cassette exchange system provides for repetitive integrations into the same locus, allowing comparative analysis of true transgenic alleles. Furthermore, this method was used to stabilize a targeted transgene by the postintegration excision of putatively mobile transposon sequences. The genomic targeting and stabilization strategy described for Drosophila should be applicable to other insects, specifically for the goals of optimizing heterologous protein expression and enhancing ecological safety of transgenic strains intended for release in biocontrol programs.
Keywords: homing sequence, insect transformation, recombinase-mediated cassette exchange, transgene stabilization
Site-specific recombinases such as FLP, Cre, and ΦC31 have emerged as powerful tools to manipulate genomes of eukaryotic model organisms (1). In mice, site-specific recombinase technology is being applied to modify chromosomal DNA in a spatially and temporally controlled manner, thereby bypassing embryonic lethality associated with many germ-line null alleles (2). In Drosophila, the FLP/FRT (FRT, FLP recombinase target site) (3) has been extensively used to generate genetic mosaics in soma and germ-line (4), and chromosome rearrangements (5). Recently, systematic collections of molecularly defined deletions spanning >50% of the Drosophila melanogaster genome have been engineered by using FLP/FRT (6, 7).
Site-specific recombinase proteins catalyze the cutting and rejoining of two different DNA segments at specific target sequences. Depending on the relative orientation of the recombinase target sites, the outcome of a recombination reaction is excision and insertion of a circular DNA molecule, respectively, or inversion of intervening DNA. Excision of genomic DNA located between two equally oriented target sites is effectively irreversible due to the creation and, in dividing cells, loss of an episomal reaction product. In contrast, integration of a plasmid at a genomic target site is considered by nature an inefficient process, because the integration product is kinetically and thermodynamically disfavored and prone to reexcision (8). A strategy termed recombinase-mediated cassette exchange (RMCE) has been developed to increase the efficiency of site-specific integration (9). RMCE based on the FLP recombinase system utilizes a set of two 48-bp recombinase target sites, FRT and FRT3, engineered to be heterospecific. Although both sites are recognized by FLP, crossreactivity between FRT and FRT3 has been eliminated by the substitution of four of eight base pairs in the FRT3 spacer element (9). Heterospecificity of FRT and FRT3 is the basis for targeted replacement of a genomic target cassette by an incoming plasmid donor cassette, with the exchange product being stable even under the permanent influence of FLP recombinase (10). FLP-RMCE methodology has been applied for the purpose of targeting and modifying tagged loci in mammalian genomes (11). However, inherent to current RMCE technology is the dependence on identifying recombinant cell clones by a chemical selection scheme limiting the application to cell culture and organisms with an established embryonic stem cell manipulation technique.
For Drosophila and other insect systems, site-specific targeting would be a valuable means to eliminate position effect variegation that often confounds direct comparison of allelic transgenes. As an interchromosomal targeting strategy, mobilization of FRT-flanked DNA to target a FRT site elsewhere in the Drosophila genome has been accomplished (12). However, this approach depends on both the donor and target FRT containing constructs being genomically integrated before the targeting experiment. On the other hand, gene-targeting methodologies using P element-dependent gene conversion (13) or double-strand break-induced homologous recombination (14) have been established for Drosophila. Although valuable to “knock out” or “knock in” a defined gene function of interest, the creation of multiple transgenic strains and execution of homologous targeting experiments require considerable effort (15, 16).
In this report, we introduce FLP-mediated cassette exchange as a tool for site-specific gene targeting in Drosophila. The objective of this concept is to maximize the efficiency of cassette exchange to enable targeting in vivo without the need for chemical selection. A sequence from the Drosophila linotte locus has been reported to mediate homing of P element vectors to the endogenous linotte locus with high precision and efficiency (17). We reasoned that this linotte sequence would also support site-specific targeting if included in both the plasmid RMCE donor and the genomic RMCE target. Following this concept, genomic loci were targeted that were tagged previously by a transgene construct carrying heterospecific FRT sites, the linotte homing sequence and a gene for a visible marker phenotype. Furthermore, we demonstrate the use of targeting to facilitate transposon terminus deletion, which has been shown to stabilize transgenic insertions (18). The application of site-specific targeting will not only further basic research in Drosophila but also aid in the development of insect strains bearing stabilized transgenic insertions for applied purposes, such as heterologous protein expression and biological control of pest species.
Methods
Plasmid Construction. Details on plasmid construction are described in Supporting Text, which is published as supporting information on the PNAS web site. In brief, pBac{3xP3-FRT-ECFP-linotte-FRT3} (ECFP, enhanced cyan fluorescent protein) harbors (i) a 3xP3-ECFPaf (19) marker gene interrupted by the wild-type FRT sequence (20) [derived from pSL>AB> (21)]; (ii) nucleotides 1-1603 from the Drosophila linotte gene (17); and (iii) the FRT spacer mutant “F3” (9), assembled in the piggyBac p3E1.2 vector (22). pSL-FRT-EYFP-linotte-FRT3 (EYFP, enhanced yellow fluorescent protein) was derived from pSL-3xP3-EYFPaf (19) by replacing the 3xP3 promoter by the wild-type FRT sequence and inserting linotte and “F3.” Further RMCE donor vectors were derived from pSL-FRT-EYFP-linotte-FRT3 by placing 3xP3-DsRed (23) downstream to the FRT-FRT3 cassette (pSL-FRT-EYFP-linotte-FRT3-3xP3-DsRed) and both pBacR2 [from pBac{3xP3-DsRedaf} (23)] and 3xP3-DsRed downstream to the promoter-free eyfp gene (pSL-FRT-EYFP-pBacR2-3xP3-DsRed-linotte-FRT3), respectively. pKhsp82-FLP contains the FLP ORF positioned downstream to the hsp82 promoter in pKhsp82 (24).
D. melanogaster Culture, Transformation, and RMCE. Fly strains were reared under standard laboratory conditions (25). Genetic mapping of transgenes was carried out by segregation analysis by using the balancer chromosomes CyO and TM2, Ubx. In our strain nomenclature, “M” refers to stocks established initially from male individuals.
piggyBac Transformation and FLP-RMCE. piggyBac-mediated Drosophila germ-line transformation to integrate the RMCE target vector was performed as described (26). FLP-RMCE was achieved by transformation protocols (26) where RMCE donor plasmids (500 ng/μl) and pKhsp82-FLP helper plasmid (300 ng/μl) were coinjected into RMCE target line embryos. At 12-16 h after microinjection, a 90-min 37°C heat shock was applied. For convenience of rearing, only male G0 adult survivors were mated individually to four virgin w females, with their progeny screened for the presence of fluorescing eyes by epifluorescence microscopy (23). Recombinant G1 individuals were bred to homozygosity and subjected to Southern analysis.
piggyBac Terminus Remobilization. Remobilization of the piggyBacL1-R2 transposon was performed by recombining the target line M4 ECFP by RMCE using the donor plasmid pSL-FRT-EYFP-pBacR2-3xP3-DsRed-linotte-FRT3. Recombinant G1 individuals were crossed to homozygous piggyBac-expressing “jumpstarter” strains [Mi{3xP3-DsRed, hsp70-piggyBac} M5.II and Her{3xP3-ECFP, αtub-piggyBacK10} M6.II and M10.III (27)]. Male G2 progeny carrying both the jumpstarter and the recombinant RMCE transgenes were mated to w virgins. For each jumpstarter line used, G3 progeny from 20 fertile matings were scored for individuals exhibiting either EYFP eye fluorescence (indicative for piggyBacL1-R2 deletion) or both EYFP and DsRed eye fluorescence.
Inverse PCR. Inverse PCR was performed with primers and protocols described (27) by using genomic DNA from the target lines M4 ECFP, M7 ECFP, M8 ECFP, M9 ECFP, and recombinant lines thereof (donor pSL-FRT-EYFP-linotte-FRT3).
Southern Hybridization Analysis. Genomic DNA (3-5 μg) was digested with the indicated restriction enzymes, separated on 0.8% agarose gels, blotted to nylon filters, and immobilized by UV irradiation. Probes were radiolabeled with [32P]-dCTP by random priming (Invitrogen) and hybridized to blots in phosphate buffer pH 7.5; 1% BSA/7% SDS at 65°C with an initial wash in 2× SSC/0.2% SDS at 25°C and two washes in 1× SSC/0.1% SDS at 55°C for 30 min. Autoradiography was performed by exposure on Kodak X-Omat film at -70°C.
Results
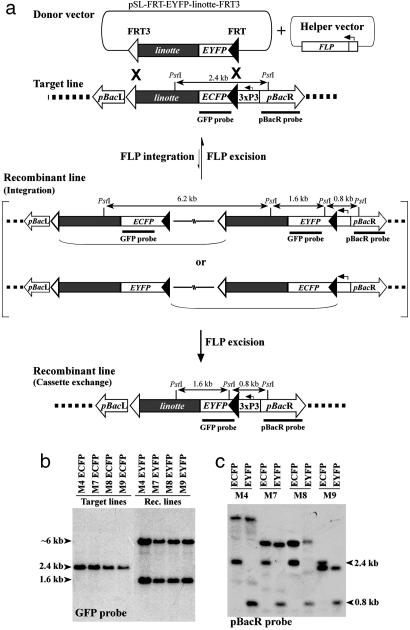
Site-Specific Targeting in Drosophila with High Efficiency. The concept of site-specific targeting involves the following steps illustrated in Fig. 1a:(i) genomic loci are tagged by target vector insertions by using piggyBac (or other transposon)-mediated germ-line transformation, thereby providing the docking sites for subsequent donor cassette recombinations. (ii) The donor vector is microinjected in conjunction with a FLP recombinase-expressing helper vector into the target line. Because both the donor and the target constructs contain a cassette bounded by FRT and FRT3 sites, recombination products may form with the donor vector by single crossover at either the FRT or FRT3 site, whereas cassette exchange is the consequence of a double crossover using both recognition sites. Because of the heterospecific nature of FRT and FRT3 sites, the cassette exchange product, once formed, is stable. A linotte sequence was included as a genomic homing device directing plasmid donor DNA to genomic target DNA. (iii) Recombinant individuals are identified by the change in fluorescent marker phenotype. The donor vector contains a promoter-free eyfp ORF, whereas the target is marked by an ecfp gene placed under control of the eye-specific 3xP3 promoter (28). Therefore, loss of cyan eye fluorescence coinciding with gain of yellow eye fluorescence indicates recombination by either integration or cassette exchange (Fig. 1a).
Fig. 1.
FLP recombinase-mediated cassette exchange in vivo. (a) The targeting concept. For this strategy, the donor vector harbors a cassette bounded by a wild-type FRT site (filled arrowhead) and a mutant FRT3 site (open arrowhead) in parallel orientation. A promoter-free eyfp gene (open box) and a 1.6-kb homing sequence (shaded box) from the Drosophila linotte locus (17) are placed internally to the FLP recombinase recognition sites. The target line is transgenic for a cassette identical to the donor cassette except for the ecfp gene that is placed under control of the 3xP3 promoter (28). Recombination at either pair of FRT or FRT3 sites, mediated by FLP-recombinase-expressing helper vector, results in intermediate configurations containing an integration of the entire donor vector plasmid. Intermediates can be resolved to the original configuration by FLP excision reusing the identical target site pairs. Alternatively, FLP excision using the different target site pairs leads to cassette exchange. Due to the heterospecificity of FRT and FRT3 sites, the formation of the cassette exchange product is regarded as an irreversible reaction (symbolized by an unidirectional arrow). PstI fragments and probes indicative for Southern analysis are denoted. (b and c) Southern analysis of four D. melanogaster target and corresponding recombinant lines. For each line, PstI-digested genomic DNA was hybridized to GFP (b) and pBacR (c) probes [calculated PstI-fragment sizes: 11.7 (M4), 4.2 (M7), 4.6 (M8), and 1.2 kb (M9)], respectively.
To analyze the functionality of this concept, we performed a targeting experiment in the model insect D. melanogaster. The target plasmid pBac{3xP3-FRT-ECFP-linotte-FRT3} was genomically integrated using piggyBac-mediated germ-line transformation. Four target lines carrying the transgene in a homozygous state were established: M4 ECFP, M8 ECFP, M9 ECFP (genetically mapped to the second chromosome), and M7 ECFP (mapped to the third chromosome). In these lines, ECFP eye fluorescence was observed as described (19), indicating that positioning of the FRT site between the 3xP3 promoter and the ecfp start codon does not interfere significantly with ecfp gene expression.
For site-specific targeting, the donor vector pSL-FRT-EYFP-linotte-FRT3 was coinjected with the FLP recombinase-expressing helper vector pKhsp82-FLP into preblastoderm embryos of each of the four target lines. In total, 2,850 embryos were injected. To screen for recombinants, the G1 generation was analyzed for eye fluorescence phenotypes. Depending on the target line used, between 22% and 31% of fertile male injection survivors produced offspring exhibiting an EYFP eye fluorescence phenotype (Table 1). Those individuals consistently lacked ECFP fluorescence that was observed in their siblings. Segregation analysis of EYFP-positive male offspring indicated, for all four lines, localization of the eyfp marker gene to the chromosome to which the target transgene was previously genetically mapped. These results suggested that recombination of the donor plasmid with a target docking site occurred at four independent genomic loci. Notably, recombinants were obtained with an efficiency comparable to well established P element-based Drosophila transformation protocols (25), demonstrating that site-specific recombination is achievable with a similar experimental effort as transposon-mediated germ-line transformation.
Table 1. Targeting experiments with the donor vector pSL-FRT-EYFP-Iinotte-FRT3.
| Target lines* | Eggs injected | Fertile F1 crosses | EYFP-pos. F1 crosses | Recombination frequency, %† |
|---|---|---|---|---|
| M4 | 750 | 70 | 22 | 31 |
| M7 | 750 | 72 | 17 | 24 |
| M8 | 600 | 54 | 12 | 22 |
| M9 | 750 | 109 | 27 | 25 |
Drosophila lines transgenic for the target vector pBac {3xP3-FRT-ECFP-linotte-FRT3} with the following molecular locations: AE003662.3, 2046 (M4); AE003558.3, 1710 (M7); AE003618.2, 1541 (M8); and AE003662.3, 1580 (M9). Sequence number and nucleotide positions refer to the Release 3.2 sequence of the Drosophila genome (www.fruitfly.org/annot/release3.html).
Percentage of (F1 crosses with at least one EYFP-fluorescing offspring)/(fertile F1 crosses).
To examine whether recombination mechanistically occurred as an integration of the donor vector or as cassette exchange (Fig. 1a), Southern blot analysis was performed on PstI-digested genomic DNA, because an additional PstI site is introduced by the eyfp gene that allows a straightforward diagnostic test for donor vector integration. A probe hybridizing to both gfp-derivative genes ecfp and eyfp recognized a 2.4-kb PstI fragment in four target lines and a 1.6-kb PstI fragment in four recombined lines (Fig. 1b). This pattern is in accordance with replacement of ecfp by eyfp under 3xP3 promoter control. A PstI fragment of ≈6 kb hybridizing in recombinant lines, however, suggested integration of the entire donor vector by a single FRT recombination, rather than double recombination cassette exchange, in the four randomly selected recombinant lines.
It was expected that site-specific targeting would result in genomic integration loci of the transgenes to remain identical prior and subsequent to the recombination reaction. Therefore, we compared the genomic insertion site sequences flanking the transgenes in target and recombinant lines. Using inverse PCR, molecular localization of the target vector could be determined for all four target lines, and moreover, sequence identity for corresponding pairs of target and recombinant lines was determined for either or both the 5′ and 3′ piggyBac junctions to the D. melanogaster genome (Table 1). Interestingly, lines M9 ECFP and M9 EYFP were found to carry the transgene integrated at intron 1 of the linotte gene. We interpret this result as a confirmation of the homing properties previously assigned to the 1.6-kb linotte sequence (17).
To further confirm identical transgene location in pairs of target and recombination lines, Southern analysis was performed by using a probe hybridizing to the transposon terminus pBacR. The predicted result of a crossover event at FRT is a PstI fragment spanning the junction between pBacR and the Drosophila genome to be of identical size, whereas the internal 2.4-kb PstI fragment would be digested to a 0.8-kb fragment (Fig. 1a). A hybridization pattern consistent with these predictions was observed for all four corresponding pairs; however, the external PstI fragment size in line M9 differed from sequence-based calculation (Fig. 1c). Furthermore, for all four corresponding pairs, two distinct hybridization signals were obtained, consistent with single-copy integrations of the target transgene.
In summary, three lines of evidence indicate that four independent genomic docking sites were targeted by the donor plasmid pSL-FRT-EYFP-linotte-FRT3: (i) the change from ECFP to EYFP eye color fluorescence; (ii) ecfp- and eyfp-indicative hybridization fragments from target and recombinant lines, respectively; and (iii) the identical size of genomic fragments hybridizing to pBacR probe.
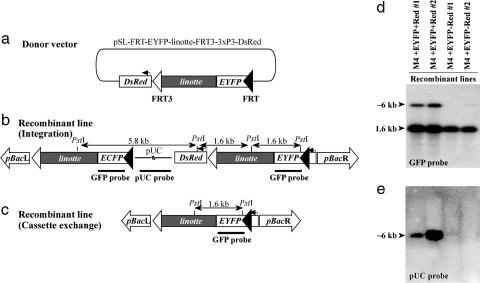
Double-Reciprocal Cassette Exchange Recombination Products Can Be Selected For. The mechanism of RMCE relies on double-reciprocal crossover at both recombinase target sites, FRT and FRT3 (Fig. 1a). However, the four recombinant lines analyzed putatively harbored a fully integrated donor vector (Fig. 1b). One possible explanation is a preference of FLP recombinase for the wild-type FRT over the mutant FRT3 site. To examine whether both FRT and FRT3 sites are active FLP targets in Drosophila in vivo, a donor plasmid was constructed that allows discrimination of recombination products resulting from integration or cassette exchange. The donor plasmid pSL-FRT-EYFP-linotte-FRT3-3xP3-DsRed encodes an additional marker gene, 3xP3-DsRed, which was placed external to the FRT-EYFP-linotte-FRT3 cassette (Fig. 2a). Upon recombination with the target vector mediated by single crossover at FRT, an integration product is formed that can be visualized by yellow and red eye fluorescence (Fig. 2b). In contrast, recombinant individuals resulting from a double crossover at both FRT and FRT3 sites are expected to exhibit solely yellow eye fluorescence (Fig. 2c). A targeting experiment using pSL-FRT-EYFP-linotte-FRT3-3xP3-DsRed and the target line M4 ECFP revealed that both mechanisms occur at about an equivalent frequency. Exclusively EYFP fluorescent recombinant offspring were recovered from five male founder individuals, whereas a combination of EYFP and DsRed double and EYFP single fluorescent flies were observed in progeny from six founder individuals. In total, 41 single and 48 double EYFP and DsRed fluorescent recombinants were obtained. Recombinant G1 offspring were observed in 11 of 84 fertile G1 crosses, corresponding to a recombination frequency of 13%.
Fig. 2.
Mechanism of FLP-mediated recombination in Drosophila. Targeting of the donor vector pSL-FRT-EYFP-linotte-FRT3-3xP3-DsRed (a) into line M4 ECFP results in either of two recombinant configurations referred to as integration (b) or cassette exchange (c). These configurations can be differentiated by an additional 3xP3-DsRed marker gene (expressed exclusively in integrations). To molecularly discriminate integration from cassette exchange, PstI-digested genomic DNA from recombinant lines were hybridized to probes complementary to both eyfp and ecfp (GFP probe, d) or to the donor vector backbone (pUC probe, e).
To correlate the fluorescent phenotypes to the proposed recombinant configurations, Southern analysis was performed on two independent EYFP and DsRed fluorescing (referred to as M4 +EYFP+Red) and two solely EYFP fluorescing recombinant lines (referred to as M4 +EYFP-Red). The 1.6-kb PstI fragment indicative for eyfp (Fig. 2 b and c) was recognized by a GFP probe in all recombinant lines (Fig. 2d). Exclusively in M4 +EYFP+Red lines, an additional PstI fragment of ≈6-kb size was detected. To demonstrate that this fragment contains the pUC-derived backbone of the donor vector, we used a pUC hybridization probe. This probe recognized genomic DNA from M4 +EYFP+Red lines but not from M4 +EYFP-Red lines (Fig. 2e). These hybridization patterns are in accordance with integration of the entire donor plasmid in M4 +EYFP+Red recombinants (vector configuration shown in Fig. 2b) and cassette exchange in M4 +EYFP-Red recombinants (configuration shown in Fig. 2c).
In conclusion, we have shown that RMCE is mechanistically practicable and efficient in D. melanogaster. The application of an additional phenotypic marker, placed external to the RMCE cassette, allows the discrimination between single crossover recombinants carrying an integrated vector plasmid and true double crossover cassette exchange recombinants.
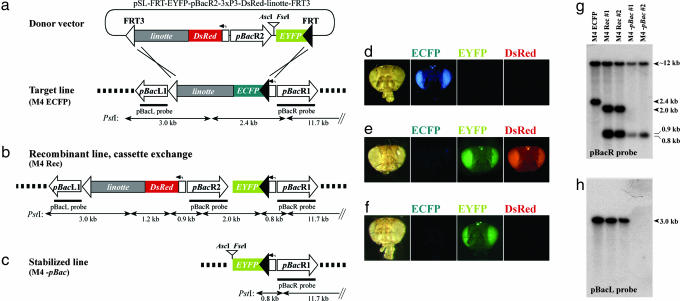
Transposon Deletion Subsequent to Site-Specific Targeting. Postintegrational stabilization of transgenes is considered an important precondition for engineering genetically modified insects for prospective use in biological control programs (29). Recently, stabilization of a terminus-deleted piggyBac vector was demonstrated (18) and, to investigate whether terminus deletion is also feasible in concert with site-specific targeting, we designed the donor vector pSL-FRT-EYFP-pBacR2-3xP3-DsRed-linotte-FRT3 (Fig. 3a). As a main feature, this vector contains a 3′ piggyBac terminus sequence, pBacR2, placed in a configuration so that a head-to-tail tandem duplication of piggyBacR termini will be created upon RMCE with the target line (Fig. 3b). To visualize the presence and loss of the internal pBacR2 terminus, a 3xP3-DsRed marker gene was introduced between the L1 and R2 termini. If the internal piggyBacL1-R2 transposon is mobilizable by the presence of piggyBac transposase, only a single pBacR1 terminus at the target site will remain (Fig. 3c).
Fig. 3.
Application of FLP RMCE for transgene stabilization. (a-c) The transposon terminus deletion concept. The donor plasmid pSL-FRT-EYFP-pBacR2-3xP3-DsRed-linotte-FRT3 (a) carries a 2.6-kb insertion of the piggyBacR2 terminus and the 3xP3-DsRed marker gene. Unique AscI-FseI cloning sites facilitate introduction of gene(s) of interest. FLP-mediated targeting of this donor vector to target line M4 ECFP results in recombinant lines, referred to as M4 Rec, with a head-to-tail tandem duplication of piggyBacR termini (b). Excision of the internal piggyBacL1-R2 transposon is accompanied by loss of the marker gene 3xP3-DsRed (c). Subsequent to excision, only a single piggyBacR terminus remains in the insect genome, resulting in stabilization of DNA insertions placed at the AscI-FseI cloning sites. The expected genomic PstI fragmentation pattern is depicted. (d-f) Eye fluorescence phenotypes. Although the target line M4 ECFP shows ECFP fluorescence (d), and the recombinant line M4 Rec #1 exhibits both EYFP and DsRed fluorescence (e), only EYFP fluorescence expression is detected in the stabilized line M4-pBac #1 (f), indicating piggyBacL1-R2 excision. (g and h) Comparative Southern analysis of Drosophila lines M4 ECFP, M4 Rec (#1 and #2), and M4-pBac (#1 and #2). PstI-digested genomic DNAs isolated from indicated Drosophila lines were hybridized by using pBacR (g) and pBacL (h) probes, respectively.
To examine the functionality of this concept, the line M4 ECFP (phenotype shown in Fig. 3d) was targeted with the donor vector pSL-FRT-EYFP-pBacR2-3xP3-DsRed-linotte-FRT3. G1 individuals lacking ECFP but showing both EYFP and DsRed eye fluorescence (Fig. 3e) were obtained at a frequency of 22% (34 of 158 fertile G1 crosses), consistent with the recombination frequency observed previously (Table 1). In particular, extension of the donor cassette size by 2.6 kb did not significantly influence targeting efficiency, portending that donor derivatives carrying additional gene(s) of interest can be recombined at a similar frequency.
To remobilize the internal piggyBacL1-R2 transposon (Fig. 3b), male individuals of two recombinant lines (referred to as M4 Rec #1 and #2) established from EYFP and DsRed double-positive recombinants (Fig. 3e) were mated to different piggyBac transposase-expressing “jumpstarter” strains. Subsequent to outcrossing to w flies, progeny were analyzed for eye fluorescence phenotypes (see Methods for details). Loss of DsRed fluorescence associated with maintenance of EYFP fluorescence indicated remobilization by excision of piggyBacL1-R2 (Fig. 3 c and f). From mobilization with the jumpstarter Mi{3xP3-DsRed, hsp70-piggyBac} (27), 25 EYFP+/DsRed- and 1,106 EYFP+/DsRed+ individuals resulted. Two independent EYFP+/DsRed- individuals were used to establish lines M4-pBac #1 and #2. From mobilization with the jumpstarter Her{3xP3-ECFP, αtub-piggyBacK10} (27), sole EYFP fluorescence was observed at frequencies of 0.4% (4/996 individuals) and 0.9% (10/1106 individuals), respectively.
To substantiate that jumpstarter activity resulted in physical excision of piggyBacL1-R2, PstI-digested genomic DNA from lines M4 ECFP, M4 Rec (#1 and #2) and M4-pBac (#1 and #2) was hybridized with probes against both piggyBac termini. The pBacR probe recognized both the external pBacR1 and the internal pBacR2 fragments in M4 Rec lines (Fig. 3 b and g, lanes 2 and 3). In contrast, the pBacR probe hybridized only to the external pBacR1 fragments in M4-pBac lines (Fig. 3 c and g, lanes 4 and 5). The pBacL probe recognized a single 3.0-kb PstI fragment in the target line M4 ECFP and in two M4 Rec lines but failed to detect this PstI fragment in M4-pBac lines (Fig. 3h). Together, the Southern analysis data prove physical deletion of the piggyBacL and piggyBacR2 sequences from the Drosophila genome in M4-pBac lines. Because the linotte and FRT3 sequences are excised together with piggyBacL1-R2, this technique also removes the genomic docking site and (except for targeting the linotte locus itself) the linotte mutation introduced by the target transgene.
In summary, piggyBac deletion subsequent to site-specific introduction of an appropriately oriented transposon terminus is technically feasible and can be easily monitored by a combination of three separable and widely functional fluorescent marker genes (23). This will allow a step-wise integration of gene(s) of interest by site-specific targeting followed by stabilization of the transgene by transposon deletion in many insects and potentially other organisms.
Discussion
To devise an in vivo method for site-specific targeting in Drosophila, we took advantage of (i) an RMCE strategy with heterospecific FLP-recombinase target sites (9) and (ii) genomic homing abilities mediated by the linotte sequence (17). In six independent targeting experiments with four target lines and three donor vectors, an average recombination frequency of 23% was obtained. Notably, at this frequency, the experimental effort for site-specific recombinase-mediated genomic targeting and transposon-mediated germ-line transformation is comparable. Because not all recombinant individuals were produced by double-crossover events at the FRT and FRT3 sites, a strategy was developed that allows for straightforward identification of RMCE recombinants by eye fluorescence phenotypes. Alternatively, intermediate configurations resulting from a single crossover may be converted to cassette exchange products by crossing to FLP-expressing Drosophila strains. The phenomenon of linotte homing has been shown to occur with high frequency and precision (up to 20% of linotte-containing P elements inserted within 1 kb of the endogenous locus) (17); however, the requirement for linotte to increase the targeting frequency in the RMCE system described here is unknown. If linotte is required for efficient RMCE, the possible caveat of unwanted targeting of the endogenous linotte locus may be circumvented readily in Drosophila by using target lines created in the lio deficiency strain having a 4-kb deletion that includes the 5′ linotte region, and that is homozygous-viable (30).
RMCE expands the tools available for reverse genetics in Drosophila and complements current transposon-based germ-line transformation technology. Site-specific targeting with multiple heterospecific recombination sites should allow one to repetitively anchor different transgenes, successively, at an identical position within the host genome and thus generate series of true allelic transgenes. Different position effects influencing the expression profile of transgenes randomly integrated within the genome can thus be eliminated. By using RMCE, allelic transgenes can be comparatively analyzed without restriction on their numbers, and this advancement should greatly facilitate the study of regulatory elements such as enhancers, silencers, and insulators. Moreover, with varying position effects excluded, the effects of different mutations on gene expression or expression of interspecific homologous genes can be more exactly compared.
A different approach for site-specific targeting of the Drosophila genome based on the integrase system from phage ΦC31 has recently been reported (31). In this study, a transformation efficiency comparable to our targeting strategy was achieved by using the property of ΦC31 integrase to catalyze integration of an entire donor plasmid in a unidirectional manner. An advantage of RMCE relative to a ΦC31-mediated integration is that extraneous DNA sequences of prokaryotic origin, such as vector sequences, antibiotic resistance genes, or reporter genes, can be excluded from the insect host genome. The RMCE strategy therefore precludes apriori the unintentional but possible influence of such vector backbone sequences on the regulation of the modified genome. The integrase system may be modified, however, by addition of homotypic recombination sites (such as loxP or FRT) or transposon termini (and an additional marker) to mediate postintegration excision of unwanted sequences.
Transposase-mediated germ-line transformation is typically limited by the vector insert size, with shorter vectors being more transpositionally efficient (shown for P elements; ref. 32). In contrast, the efficiency of site-specific recombination should not depend on the total size of the donor molecule, because the reaction catalyzed by FLP recombinase relies only on an interaction with two 48-bp FRT (or mutant FRT) sites. Therefore, we anticipate that genomic integration of very large vectors, such as bacterial artificial chromosomes, might be feasible by using RMCE.
RMCE might also be a valuable tool for forward genetics in Drosophila. A variety of docking sites can be produced, tagged, and analyzed with respect to various parameters, such as tissue-, cell type-, or developmental stage-specific enhancer activity, strength of reporter gene expression, and stringency of conditional gene expression. We anticipate that FRT/FRT3 flanked cassettes will add a novel degree of flexibility for large-scale forward insertional mutagenesis approaches, including those that use the piggyBac transposon, among others, as a mutagen (27, 33, 34).
The site-specific targeting system was designed with components that allow general application in insects where recent progress has resulted in transposon-mediated germ-line transformation of nearly 20 species from four different insect orders (35, 36). In particular, the functionality of piggyBac-based transformation systems and fluorescent transformation marker genes has been demonstrated in a broad spectrum of insect species (23, 37). Although FLP activity in embryos of the silkworm Bombyx mori and the mosquito Aedes aegypti could be demonstrated (38, 39), FLP-mediated excision of a FRT-flanked marker gene from the mosquito chromosome could not be detected (40). Thus, the activity of FLP recombinase, as well as the behavior of Drosophila linotte or linotte-homologous sequences in insect species of interest, remains to be investigated.
What benefits from RMCE and vector stabilization can be envisaged for nonmodel insects? In general, the establishment of homozygous transgenic lines will be facilitated, mapping of transgenes will be dispensable after initial genomic localization of the target vector insertion site, and genomic docking sites that do not interfere with vital gene functions when mutated can be selected for and repetitively used. For transgenic insect strains intended for biocontrol programs, site-specific targeting will allow a controlled assessment of different transgene constructs on the fitness and host competitiveness in a truly comparative approach (41, 42). Taken with defined target sites having minimal position-effect suppression, the efficient and repetitive creation of highly fit transgenic strains optimized for field release will be greatly facilitated. Controlling for position effects by genomic targeting will also allow optimization of heterologous protein expression in beneficial insects used as protein factories, such as the silkworm B. mori (43).
To stabilize the genomically integrated gene of interest and enhance the ecological safety of such strains, removal of at least one transposon terminus has been proposed (29) and conceptually proven in Drosophila (18). As demonstrated in the present study, transposon deletion is also feasible by using an RMCE donor that provides a transposon terminus in a correct orientation. Taken with the advantages of RMCE genomic targeting for creation and analysis of transgenic strains for biological control programs, the ability to stabilize integrated transgenes should further enhance the development and utilization of transgenic insects for applied use.
Supplementary Material
Acknowledgments
We thank Gary Struhl (Columbia University, New York), Peter Atkinson (University of California, Riverside), and Malcolm Fraser, Jr. (University of Notre Dame, Notre Dame, IN), for providing plasmids. C.H. thanks Ernst A. Wimmer for supervising the generation of progenitor vectors pSL-3xP3-FRT-ECFP, pSL-FRT-ECFP, pSL-3xP3-FRT-EYFP, and pBac{3xP3-FRT-ECFP} and the FLP helper vector pKhsp82-FLP. C.H. further acknowledges support from the Fonds der Chemischen Industrie and the Robert Bosch Foundation (by a junior research group to Ernst A. Wimmer) and thanks Christian F. Lehner and the members of the Lehrstuhl für Genetik for support and encouragement. A.M.H. acknowledges support from the U.S. Department of Agriculture National Research Initiative Competitive Grants Program.
Author contributions: C.H. designed research; C.H. and A.M.H. performed research; C.H. contributed new reagents/analytic tools; C.H. and A.M.H. analyzed data; and C.H. and A.M.H. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: FRT, FLP recombinase target site; RMCE, recombinase-mediated cassette exchange; EYFP, enhanced yellow fluorescent protein; ECFP, enhanced cyan fluorescent protein.
References
- 1.Branda, C. S. & Dymecki, S. M. (2004) Dev. Cell 6, 7-28. [DOI] [PubMed] [Google Scholar]
- 2.Metzger, D. & Feil, R. (1999) Curr. Opin. Biotechnol. 10, 470-476. [DOI] [PubMed] [Google Scholar]
- 3.Golic, K. G. & Lindquist, S. (1989) Cell 59, 499-509. [DOI] [PubMed] [Google Scholar]
- 4.Theodosiou, N. A. & Xu, T. (1998) Methods Enzymol. 14, 355-365. [DOI] [PubMed] [Google Scholar]
- 5.Golic, K. G. & Golic, M. M. (1996) Genetics 144, 1693-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parks, A. L., Cook, K. R., Belvin, M., Dompe, N. A., Fawcett, R., Huppert, K., Tan, L. R., Winter, C. G., Bogart, K. P., Deal, J. E., et al. (2004) Nat. Genet. 36, 288-292. [DOI] [PubMed] [Google Scholar]
- 7.Ryder, E., Blows, F., Ashburner, M., Bautista-Llacer, R., Coulson, D., Drummond, J., Webster, J., Gubb, D., Gunton, N., Johnson, G., et al. (2004) Genetics 167, 797-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baer, A. & Bode, J. (2001) Curr. Opin. Biotechnol. 12, 473-480. [DOI] [PubMed] [Google Scholar]
- 9.Schlake, T. & Bode, J. (1994) Biochemistry 33, 12746-12751. [DOI] [PubMed] [Google Scholar]
- 10.Seibler, J. & Bode, J. (1997) Biochemistry 36, 1740-1747. [DOI] [PubMed] [Google Scholar]
- 11.Seibler, J., Schübeler, D., Fiering, S., Groudine, M. & Bode, J. (1998) Biochemistry 37, 6229-6234. [DOI] [PubMed] [Google Scholar]
- 12.Golic, M. M., Rong, Y. S., Peterson, R. B., Lindquist, S. L. & Golic, K. G. (1997) Nucleic Acids Res. 25, 3665-3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gloor, G. B., Nassif, N. A., Johnson-Schlitz, D. M., Preston, C. R. & Engels, W. R. (1991) Science 253, 1110-1117. [DOI] [PubMed] [Google Scholar]
- 14.Rong, Y. S. & Golic, K. G. (2000) Science 288, 2013-2018. [DOI] [PubMed] [Google Scholar]
- 15.Gloor, G. B. (2004) Methods Mol. Biol. 260, 97-114. [DOI] [PubMed] [Google Scholar]
- 16.Rong, Y. S. (2002) Biochem. Biophys. Res. Commun. 297, 1-5. [DOI] [PubMed] [Google Scholar]
- 17.Taillebourg, E. & Dura, J. M. (1999) Proc. Natl. Acad. Sci. USA 96, 6856-6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Handler, A. M., Zimowska, G. J. & Horn, C. (2004) Nat. Biotechnol. 22, 1150-1154. [DOI] [PubMed] [Google Scholar]
- 19.Horn, C. & Wimmer, E. A. (2000) Dev. Genes Evol. 210, 630-637. [DOI] [PubMed] [Google Scholar]
- 20.Jayaram, M. (1985) Proc. Natl. Acad. Sci. USA 82, 5875-5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wimmer, E. A., Cohen, S. M., Jäckle, H. & Desplan, C. (1997) Development (Cambridge, U.K.) 124, 1509-1517. [DOI] [PubMed] [Google Scholar]
- 22.Cary, L. C., Goebel M., Corsaro B. G., Wang H. G., Rosen, E. & Fraser, M. J. (1989) Virology 172, 156-169. [DOI] [PubMed] [Google Scholar]
- 23.Horn, C., Schmid, B. G., Pogoda, F. S. & Wimmer, E. A. (2002) Insect Biochem. Mol. Biol. 32, 1221-1235. [DOI] [PubMed] [Google Scholar]
- 24.Coates, C. J., Howells, A. J., O'Brochta, D. A. & Atkinson, P. W. (1996) Gene 175, 199-201. [DOI] [PubMed] [Google Scholar]
- 25.Roberts, D. B., ed. (1998) Drosophila: A Practical Approach (IRL, Oxford).
- 26.Horn, C., Jaunich, B. & Wimmer, E. A. (2000) Dev. Genes Evol. 210, 623-629. [DOI] [PubMed] [Google Scholar]
- 27.Horn, C., Offen, N., Nystedt, S., Häcker, U. & Wimmer, E. A. (2003) Genetics 163, 647-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berghammer, A. J., Klingler, M. & Wimmer, E. A. (1999) Nature 402, 370-371. [DOI] [PubMed] [Google Scholar]
- 29.Handler, A. M. (2004) Insect Biochem. Mol. Biol. 34, 121-130. [DOI] [PubMed] [Google Scholar]
- 30.Dura, J. M., Taillebourg, E. & Preat, T. (1995) FEBS Lett. 370, 250-254. [DOI] [PubMed] [Google Scholar]
- 31.Groth, A. C., Fish, M., Nusse, R. & Calos, M. P. (2004) Genetics 166, 1775-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spradling, A. C. (1986) in Drosophila: A Practical Approach, ed. Roberts, D. B. (IRL, Oxford).
- 33.Häcker, U., Nystedt, S., Barmchi, M. P., Horn, C. & Wimmer, E. A. (2003) Proc. Natl. Acad. Sci. USA 100, 7720-7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thibault, S. T., Singer, M. A., Miyazaki, W. Y., Milash, B., Dompe, N. A., Singh, C. M., Buchholz, R., Demsky, M., Fawcett, R., Francis-Lang, H. L., et al. (2004) Nat. Genet. 36, 283-287. [DOI] [PubMed] [Google Scholar]
- 35.Handler, A. M. (2001) Insect Biochem. Mol. Biol. 31, 111-128. [DOI] [PubMed] [Google Scholar]
- 36.O'Brochta, D. A. & Atkinson, P. W. (2004) Methods Mol. Biol. 260, 227-254. [DOI] [PubMed] [Google Scholar]
- 37.Handler, A. M. (2002) Insect Biochem. Mol. Biol. 32, 1211-1220. [DOI] [PubMed] [Google Scholar]
- 38.Tomita, S., Kanda, T., Imanishi, S. & Tamura, T. (1999) Appl. Entomol. Zool. 34, 371-377. [Google Scholar]
- 39.Morris, A. C., Schaub, T. L. & James, A. A. (1991) Nucleic Acids Res. 19, 5895-5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jasinskiene, N., Coates, C. J., Ashikyan, A. & James, A. A. (2003) Nucleic Acids Res. 31, e147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Catteruccia, F., Godfray, H. C. J. & Crisanti, A. (2003) Science 299, 1225-1227. [DOI] [PubMed] [Google Scholar]
- 42.Irvin, N., Hoddle, M. S., O'Brochta, D. A., Carey, B. & Atkinson, P. W. (2004) Proc. Natl. Acad. Sci. USA 101, 891-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tomita, M., Munetsuna, H., Sato, T., Adachi, T., Hino, R., Hayashi, M., Shimizu, K., Nakamura, N., Tamura, T. & Yoshizato, K. (2003) Nat. Biotechnol. 21, 52-56. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.