Abstract
UV-light-induced cyclobutane pyrimidine dimers (CPDs) present a severe block to synthesis by replicative DNA polymerases (Pols), whereas Polη promotes proficient and error-free replication through CPDs. Although the archael Dpo4, which, like Polη, belongs to the Y family of DNA Pols, can also replicate through a CPD, it is much less efficient than Polη. The x-ray crystal structure of Dpo4 complexed with either the 3′-thymine (T) or the 5′ T of a cis-syn TT dimer has indicated that, whereas the 3′ T of the dimer forms a Watson–Crick base pair with the incoming dideoxy ATP, the 5′ T forms a Hoogsteen base pair with the dideoxy ATP in syn conformation. Based upon these observations, a similar mechanism involving Hoogsteen base pairing of the 5′ T of the dimer with the incoming A has been proposed for Polη. Here we examine the mechanisms of CPD bypass by Dpo4 and Polη using nucleotide analogs that specifically disrupt the Hoogsteen or Watson–Crick base pairing. Our results show that both Dpo4 and Polη incorporate dATP opposite the 5′ T of the CPD via Watson–Crick base pairing and not by Hoogsteen base pairing. Furthermore, opposite the 3′ T of the dimer, the two Pols differ strikingly in the mechanisms of dATP incorporation, with Dpo4 incorporating opposite an abasic-like intermediate and Polη using the normal Watson–Crick base pairing. These observations have important implications for the mechanisms used for the inefficient vs. efficient bypass of CPDs by DNA Pols.
Keywords: cyclobutane pyrimidine dimer, translesion DNA synthesis, Watson–Crick base pairing, Hoogsteen base pairing
UV light (UV) causes the formation of DNA lesions, among which cyclobutane pyrimidine dimers (CPDs) are the most prevalent form, and which, if not removed, lead to mutations and cancer formation. Although a cis-syn thymine-thymine (TT) dimer introduces only a modest deformation of the DNA helix, bending it by ≈30° and unwinding it by ≈9°, and the ability of the two Ts in the dimer to form Watson–Crick base pairs with the As is not significantly altered (1–4), a CPD still presents a strong block to synthesis by most DNA polymerases (Pols), because the covalent linkage of the two Ts in the CPD prevents the kinking of the DNA backbone so that the 5′ T of the dimer cannot be pushed out of the active site. This then prevents the classical replicative and repair Pols from replicating through this lesion, because they can accommodate only a single templating nucleotide in their active site (5–7), which provides for a tight fit of the template base with the correct incoming nucleotide and thereby imposes a high degree of geometric selectivity.
Because of the steric constraints imposed by the active site, the binding pocket of high-fidelity replicative and repair DNA Pols accommodates only the base pairs with correct Watson–Crick geometry (8). Consequently, these Pols are inhibited from replicating through DNA lesions that distort the Watson–Crick base-pairing geometry. The Y family DNA Pols, on the other hand, can synthesize past DNA lesions, but they synthesize DNA with much lower fidelity and processivity than the replicative/repair Pols. The eukaryotic Y family Pols, however, exhibit a high degree of specificity in their lesion bypass abilities (9), and of these, Polη has the unique ability to replicate through CPDs proficiently and accurately (10, 11). Consequently, inactivation of Polη in both yeast and humans results in increased UV light mutagenesis (12–15), and defects in Polη in humans cause the skin cancer-prone syndrome, the variant form of xeroderma pigmentosum (11, 16). Hence, Polη prevents skin cancers by reducing the incidence of mutations resulting from the mutagenic bypass of CPDs formed at TT, TC, and CC sites in DNA (17, 18).
Steady-state kinetic analyses with yeast and human Polη have indicated that these enzymes incorporate an A opposite both the 3′ T and the 5′ T with nearly the same efficiency and fidelity as opposite the two undamaged Ts (19, 20). Also, from presteadystate kinetic analyses, it has been determined that yeast Polη incorporates As opposite the two Ts of a CPD with only a slight decrease in the affinity (Kd) for dATP, and the rate of nucleotide incorporation (kPol) remains the same as opposite undamaged Ts (21). All these biochemical studies have supported the inference that Polη can accommodate both the residues of a CPD in its active site and can carry out proficient nucleotide incorporation opposite each residue of the CPD by using the intrinsic Watson–Crick base-pairing ability of the lesion. Although the structure of a ternary complex of Polη has not been determined, modeling of the yeast Polη apo structure with a cis-syn TT dimer in template DNA and with an incoming dATP opposite the 3′ Tof the dimer shows that the 5′ T of the dimer is also accommodated in the Polη active site without any steric hindrance (22).
Although Dpo4, an archael Y family Pol, can replicate through a cis-syn TT dimer, it does so with a much more reduced efficiency than does Polη (23). However, it has been suggested that in its lesion bypass properties, including its ability to bypass CPDs, it is more akin to Polη than to the other Y family Pols (23). Interestingly, the x-ray crystal structure of Dpo4 complexed with a cis-syn TT dimer has shown that, whereas the 3′ T of the CPD forms a Watson–Crick base pair with the incoming dideoxy ATP, the 5′ T forms a Hoogsteen base pair with the dideoxy ATP in syn conformation (24). Based upon these structures, a similar mechanism for nucleotide incorporation opposite a TT dimer has been proposed for Polη (24) and, moreover, the somewhat higher fidelity of Dpo4 and Polη at the 5′ T of the CPD than at the 3′ T has been suggested to arise because of the involvement of the 5′ T in Hoogsteen base pairing (25).
Dpo4 is phylogenetically related to the DinB group of Y family Pols, members of which, like Escherichia coli Pol IV and human Polκ, are highly inefficient at replicating through CPDs; moreover, in its other properties, such as the propensity for frameshifting, Dpo4 resembles other DinB members (see ref. 26 for discussion and references). Here we have carried out biochemical studies with the aim of determining whether Dpo4, in fact, uses the mechanisms for CPD bypass indicated from the structural studies, and we have also done comparable studies with Polη to determine whether the two Pols use the same mechanisms for CPD bypass. We find, however, that Dpo4 differs strikingly from Polη in the mechanisms it utilizes for replicating through the two Ts of a CPD, and moreover, our results show that neither Dpo4 or Polη engages in Hoogsteen base pairing for incorporating an A opposite the 5′ T of the CPD. Based upon our observations, we propose that Pols use two very distinct mechanisms for CPD bypass: one is highly inefficient and has been shown previously to be used by T7 Pol, which we infer to also be the mechanism used by Dpo4, and the other which is highly efficient and also accurate and is used uniquely by Polη.
Materials and Methods
Protein and DNA Substrates. Human DNA Polη and Solfolobus solfactaricus Dpo4 were expressed as GST fusion proteins and affinity-purified from yeast as described (27, 28). GST tags were removed by treatment with PreScission protease (Amersham Pharmacia), which resulted in an N-terminal 7-aa leader peptide attached to each protein. Primer/template DNA substrates consisted of an oligonucleotide primer, which was 5′-32P-end-labeled using polynucleotide kinase (Roche Molecular Biochemicals) and [γ-32P]ATP (Amersham Pharmacia Biotech), annealed to an oligonucleotide DNA template by heating a mixture of primer/template at a 1:1.5 molar ratio to 95°C and allowing it to cool to room temperature over several hours. The template 75-mer oligonucleotide contained the sequence 5′-AGCAA GTCACCAATG TCTAAGAGTT CGTATTATGC CTACACTGGA GTACCGGAGC ATCGTCGTGA CTGGG AAAAC-3′ and was either undamaged or contained a cis-syn TT dimer (CPD) at the underlined position. The AP site containing the template was 5′-AGCTACCATG CCTGCCTCAA GAATTCGTAA oATGCCTACA CTGGAGTACC GGAGCATCGT CGTGACTGGG AAAAC-3′, where the o indicates the position of the tetrahydrofuran moiety. For steady-state kinetic analyses of nucleotide insertion opposite the 3′ T of the CPD, its undamaged counterpart or the AP site, the primer 5′-GTTTTCCCAG TCACGACGAT GCTCCGGTAC TCCAGTGTAG GCAT-3′ was annealed to the appropriate 75-nt template. For steady-state kinetic analyses of nucleotide insertion opposite the 5′ T of the CPD or its undamaged counterpart, the primer 5′-GTTTTCCCAG TCACGACGAT GCTCCGGTAC TCCAGTGTAG GCATA-3′ was used. The running start primer was identical in sequence to the first 32 nucleotides of the primers described above.
DNA Pol Assays. The standard DNA Pol reaction (5 μl) contained 25 mM Tris·HCl (pH 7.5), 5 mM MgCl2, 1 mM dithiolthreitol, 100 μg/ml BSA, 10% glycerol, 10 nM DNA substrate, and the indicated deoxynucleotide triphosphates. dNTPs were purchased from Roche Biochemicals (Indianapolis). The 7-deaza-2′-deoxyadenosine-5′-triphosphate (7dzA) and 2-aminopurine-2′-deoxyribose-5′-triphosphate (2AP) were purchased from TriLink BioTechnologies (San Diego). N1-methyl-2′-deoxyadenosine-5′-triphosphate (1meA) was generated by treatment of dATP with DMS (Sigma) and purified by HPLC as described (29). The identity and purity of the N1-methyl nucleotide triphosphate thus generated was confirmed by acid hydrolysis and by comparing the HPLC elution profile of the free base to N1-methyl, N3-methyl, and N7-methyladenine standards. Reactions containing Polη (0.17 or 0.34 nM) were carried out at 37°C for 5 min. Reactions containing Dpo4 (0.025, 0.1, or 0.2 nM) were carried out at 60°C for 5 or 10 min. Deoxynucleotides were present at the concentrations indicated in the legend to Fig. 1. Reactions were terminated by the addition of 6 volumes of loading buffer (95% formamide/0.05% cyanol blue/0.05% bromophenol blue) before resolving on 10% polyacrylamide gels containing 8 M urea. Gels were dried before autoradiography at –70°C or PhosphorImage analysis.
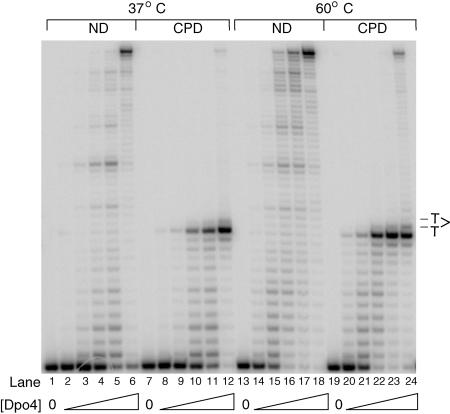
Fig. 1.
Inefficient bypass of a cis-syn TT dimer (CPD) by Dpo4. DNA synthetic activity of Dpo4 was assayed at either 37°C or 60°C. The running start primer–template DNA substrate contained either two undamaged T residues (ND) or a CPD. The position of the CPD is indicated on the right. Assays contained 10 nM DNA substrate; 10 μM dNTP; and 0.01 nM, 0.05 nM, 0.2 nM, 0.5 nM, or 5 nM Dpo4, and were carried out for 10 min.
Steady-State Kinetic Analysis. Steady-state kinetic analyses for deoxynucleotide incorporation were done as described (30). Gel band intensities of the substrate and products of the deoxynucleotide incorporation reactions were quantitated by using a PhosphorImager and the imagequant software (Molecular Dynamics). The observed rate of deoxynucleotide incorporation, vobs, was determined by dividing the amount of product formed by the reaction time and protein concentration. The vobs was graphed as a function of the deoxynucleotide concentration, and the data were fit to the Michaelis–Menten equation describing a hyperbola: vobs = (kcat[E] × [dNTP])/(Km + [dNTP]). From the best fit curve, the apparent Km and kcat steady-state kinetics parameters were obtained for the incorporation of dATP and the modified deoxynucleotides by the various DNA Pols and the efficiencies of nucleotide incorporation (kcat/Km) determined.
Results
CPD Bypass by Dpo4. To assess the proficiency of Dpo4 in replicating through a CPD, we used a running start assay in which the Pol must synthesize 12 nucleotides before encountering the lesion. As is shown in Fig. 1, Dpo4 is severely blocked by the presence of a cis-syn TT dimer in the template. Even at 60°C, excessive amounts of Dpo4 are needed to carry out minimal bypass of the CPD (maximally 8% complete bypass, lane 24). This is in contrast to the highly efficient bypass of a CPD by Polη, where extensive steady-state (19, 20) and presteady-state kinetic analyses (21) of nucleotide incorporation opposite both the Ts of the cis-syn TT dimer have shown that the efficiency of A incorporation opposite the two Ts of a CPD is nearly identical to that opposite the undamaged T.
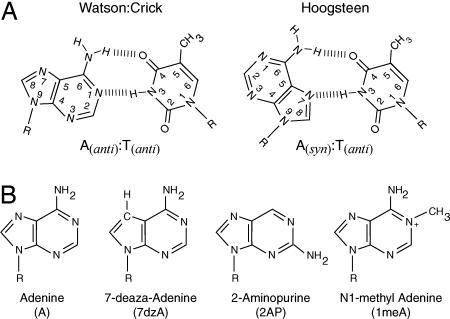
Base Analogs. To determine the mechanisms used by Dpo4 vs. Polη for replicating through a cis-syn TT dimer, we examined the nucleotide incorporation efficiency of these Pols by using dATP analogs that specifically disrupt the Hoogsteen or Watson–Crick base pairing (Fig. 2). To disrupt the Hoogsteen base pairing, we used the adenine analogs, 7dzA and 2AP (Fig. 2B). The N7 position in 7dzA is substituted with carbon and therefore lacks a hydrogen bond acceptor critical for Hoogsteen base-pair formation. Additionally, this C7 position carries a hydrogen atom that could disrupt Hoogsteen base-pairing geometry because of the steric clash with the N3 hydrogen in thymine. 2AP is similar to adenine, except that it lacks the N6 amino group, which participates in both Hoogsteen and Watson–Crick base pairing, and it harbors an amino group at the second position. Thus, 2AP still retains Watson–Crick hydrogen bond mediators at the N1 and N2 positions but lacks the N6 amino group used in Hoogsteen base pairing. In both cases, the efficiency of nucleotide incorporation should be reduced if Hoogsteen base pairing were involved.
Fig. 2.
Base analogs used. (A) Structure of A:T Watson–Crick and Hoogsteen base pairs. (B) Chemical structures of adenine analogs used in this study.
To examine the requirement of Watson–Crick base pairing for nucleotide incorporation opposite the 3′ T or the 5′ T of the TT dimer, we used the adenine analog 1meA. Because 1meA harbors a methyl group at the N1 position of adenine (Fig. 2B), that would disrupt the Watson–Crick hydrogen bonding with thymine.
Nucleotide Incorporation Opposite the 3′ T of the CPD by Dpo4 Occurs via an Abasic Site-Like Intermediate. Using base analogs, we first examined the mechanism of nucleotide incorporation opposite the 3′ T of the CPD by Dpo4 vs. Polη. For Dpo4, the efficiency of A incorporation opposite the 3′ T of a CPD was reduced by ≈1,000-fold compared with the efficiency of A incorporation opposite the undamaged 3′ T (Table 1). By contrast, Polη inserts an A opposite the 3′ T of the CPD almost as efficiently as opposite the undamaged 3′ T (Table 2). To determine whether Dpo4 utilizes Watson–Crick base pairing for incorporating an A opposite the 3′ T of the CPD, we assayed the efficiency of 1meA incorporation and also of 7dzA incorporation as a control, opposite this site. As shown in Table 1, opposite the undamaged 3′ T, Dpo4 incorporates 7dzA as efficiently as it incorporates an A, but 1meA is incorporated 250-fold less efficiently. As expected, these results are in keeping with Watson–Crick base pairing of the incoming A opposite the undamaged 3′ T. In striking contrast, opposite the 3′ T of the cis-syn TT dimer, all three nucleotides are inserted poorly by Dpo4. Whereas the efficiency of A or 7dzA incorporation opposite the 3′ T of the CPD drops ≈1,000-fold compared with that opposite the nondamaged T, the efficiency of 1meA incorporation opposite the 3′ T of CPD drops only ≈10-fold compared with that opposite the nondamaged 3′ T. In fact, opposite the 3′ T of the TT dimer, Dpo4 incorporates 1meA only ≈3-fold less well than an A or a 7dzA. Thus, the presence of the methyl group on 1meA does not impede its incorporation opposite the 3′ T of the CPD (Table 1).
Table 1. Efficiency of A, 7dzA, and 1meA incorporation opposite the 3′T of a cis-syn TT dimer by Dpo4.
| Template residue | Incoming dNTP | kcat, min-1 | Km, μM | kcat/Km | Efficiency relative to A |
|---|---|---|---|---|---|
| 3′T of ND | A | 22.8 ± 1.0 | 2.8 ± 0.3 | 8.1 | 1 |
| 7dzA | 24.3 ± 1.2 | 2.4 ± 0.3 | 10.1 | 1.2 | |
| 1meA | 8.8 ± 0.5 | 279 ± 43 | 3.2 × 10-2 | 4.0 × 10-3 (↓250×) | |
| 3′T of CPD | A | 2.7 ± 0.2 | 325 ± 64 | 8.3 × 10-3 | 1 |
| 7dzA | 4.1 ± 0.5 | 344 ± 140 | 1.2 × 10-2 | 1.4 | |
| 1meA | 0.73 ± 0.05 | 287 ± 48 | 2.5 × 10-3 | 0.3 (↓3×) | |
| AP site | A | 3.8 ± 0.7 | 205 ± 8 | 1.9 × 10-2 | 1 |
| 7dzA | 5.1 ± 0.2 | 297 ± 28 | 1.7 × 10-2 | 0.9 | |
| 1meA | 1.7 ± 0.2 | 223 ± 75 | 7.6 × 10-3 | 0.4 (↓2.5×) |
In the last column, the × in parentheses indicates the fold change in efficiency. ND, nondamaged; CPD, cis-syn TT dimer; AP, abasic.
Table 2. Efficiency of A, 7dzA, and 1meA incorporation opposite the 3'T of a cis-syn TT dimer by Polη.
| Template residue | Incoming dNTP | kcat, min-1 | Km, μM | kcat/Km | Efficiency relative to A |
|---|---|---|---|---|---|
| 3'T of ND | A | 4.9 ± 0.2 | 1.6 ± 0.3 | 3.1 | 1 |
| 7dzA | 4.2 ± 0.2 | 2.5 ± 0.4 | 1.7 | 0.5 (↓2×) | |
| 1meA | 2.3 ± 0.2 | 168 ± 38 | 1.4 × 10-2 | 4.5 × 10-3 (↓222×) | |
| 3'T of CPD | A | 3.8 ± 0.7 | 2.5 ± 0.9 | 1.5 | 1 |
| 7dzA | 2.8 ± 0.1 | 3.7 ± 0.6 | 0.8 | 0.5 (↓2×) | |
| 1meA | 1.4 ± 0.1 | 100 ± 25 | 1.4 × 10-2 | 9.3 × 10-3 (↓108×) |
In the last column, the × in parentheses indicates the fold change in efficiency. ND, nondamaged; CPD, cis-syn TT dimer.
The absence of a significant reduction in the efficiency of 1meA incorporation opposite the 3′ T of the CPD raised the possibility that Dpo4 does not directly insert the A opposite the 3′ T of the dimer via Watson–Crick base pairing, but rather it incorporates the A opposite an abasic site-like intermediate, as has been shown to be the case for T7 DNA Pol (31, 32), which also is highly inefficient at the incorporation of an A opposite the 3′ T of a TT dimer. To verify the validity of this idea, we compared the kinetics of A and 1meA incorporation by Dpo4 opposite the AP site vs. the 3′ T of the CPD. Our observations that Dpo4 incorporates these nucleotides opposite both these lesion sites with very similar efficiencies (Table 1) support the premise that A incorporation opposite the 3′ T of the CPD occurs via an abasic site-like intermediate and not via direct Watson–Crick base pairing.
Similar to Dpo4, Polη is inhibited at the incorporation of 1meA opposite the undamaged 3′ T (Table 2), consistent with the requirement of Watson–Crick base pairing for the replication of undamaged DNA by both these Pols. However, in contrast to Dpo4, for Polη, the efficiency of 1meA incorporation opposite the 3′ T of the CPD is reduced by >100-fold compared with the efficiency of A insertion, whereas 7dzA is incorporated almost as well as A (Table 2). In fact, overall, for Polη, the efficiencies of A, 7dzA, and 1meA incorporation opposite the 3′ T of the CPD and opposite the undamaged 3′ T are very similar. Also, compared with the efficient incorporation of an A opposite the 3′ T of the CPD, Polη is highly inefficient at incorporating an A opposite an abasic site (21, 33). All these observations support the conclusion that Polη incorporates an A opposite the 3′ T of a CPD using the intrinsic Watson–Crick base-pairing ability of the lesion.
Nucleotide Incorporation Opposite the 5′ T of CPD by Dpo4 Occurs via Watson–Crick and Not via Hoogsteen Base Pairing. Next, we examined the incorporation of 7dzA and 2AP by Dpo4 and Polη opposite the 5′ T of the CPD. Although the CPD imparts a slight impediment to A insertion opposite the 5′ T by Dpo4 (≈4-fold lower than opposite nondamaged T; Table 3), to our surprise, 7dzA and 2AP were incorporated opposite the 5′ T of the CPD with efficiencies equal to, or only ≈3-fold lower than, respectively, for the incorporation of an A (Table 3). Polη incorporated an A opposite the undamaged 5′ T and the CPD counterpart with similar efficiencies, and it incorporated 7dzA and 2AP opposite the 5′ T of the CPD with efficiencies quite similar to those for the incorporation of an A (Table 3). Because for both Pols the efficiencies for the incorporation of 7dzA and 2AP vs. for the incorporation of an A opposite the 5′ T were very similar on the undamaged and CPD-containing templates, this indicated that in solution, Watson–Crick rather than Hoogsteen base pairing was used for the incorporation of dATP opposite the 5′ T of the CPD by Dpo4 and, as we expected, this was the case for Polη also.
Table 3. Efficiency of A, 7dzA, 2AP, and 1meA incorporation opposite the 5'T of a cis-syn TT dimer by Dpo4 and Polη.
| DNA Pol | Template residue | Incoming dNTP | kcat, min-1 | Km, μM | kcat/Km | Efficiency relative to A |
|---|---|---|---|---|---|---|
| Dpo4 | 5'T of ND | A | 13.8 ± 0.4 | 0.8 ± 0.1 | 17.3 | 1 |
| 7dzA | 15.9 ± 1.1 | 1.1 ± 0.3 | 14.5 | 0.8 | ||
| 2AP | 20.5 ± 0.9 | 6.0 ± 1.1 | 3.4 | 0.2 (↓5×) | ||
| 1meA | 8.7 ± 0.6 | 232 ± 34 | 3.8 × 10-2 | 2.2 × 10-3 (↓455×) | ||
| 5'T of CPD | A | 14.2 ± 0.9 | 3.2 ± 0.6 | 4.3 | 1 | |
| 7dzA | 12.2 ± 1.0 | 2.8 ± 0.7 | 4.4 | 1 | ||
| 2AP | 17.6 ± 2.5 | 14.4 ± 5.8 | 1.2 | 0.3 (↓3×) | ||
| 1meA | 3.8 ± 1.0 | 403 ± 208 | 9.4 × 10-3 | 2.2 × 10-3 (↓455×) | ||
| Polη | 5'T of ND | A | 4.8 ± 0.3 | 3.6 ± 0.7 | 1.3 | 1 |
| 7dzA | 4.2 ± 0.2 | 3.2 ± 0.8 | 1.3 | 1 | ||
| 2AP | 5.3 ± 0.4 | 1.2 ± 0.3 | 4 | 3.1 (↑3×) | ||
| 1meA | 2.1 ± 0.3 | 168 ± 40 | 1.3 × 10-2 | 1 × 10-2 (↓100×) | ||
| 5'T of CPD | A | 3.8 ± 0.5 | 2.6 ± 1.0 | 1.5 | 1 | |
| 7dzA | 2.8 ± 0.1 | 3.7 ± 0.6 | 0.8 | 0.5 (↓2×) | ||
| 2AP | 5.3 ± 0.5 | 1.3 ± 0.5 | 2.7 | 2.7 (↑3×) | ||
| 1meA | 1.4 ± 0.1 | 100 ± 22 | 1.4 × 10-2 | 9.0 × 10-3 (↓111×) |
In the last column, the × in parentheses indicates the fold change in efficiency. ND, nondamaged; CPD, cis-syn TT dimer.
To provide direct evidence for the involvement of Watson–Crick base pairing at the 5′ T of a CPD, we assayed the incorporation of 1meA by Dpo4 and also by Polη. As shown in Table 3, Dpo4 and Polη exhibit 455- and 100-fold reduction in efficiency, respectively, for 1meA insertion opposite the undamaged 5′ T residue, and both Pols exhibit a nearly identical reduction in the efficiencies of 1meA incorporation opposite the 5′ T of the CPD. Our observations thus support the conclusion that Dpo4 utilizes Watson–Crick base pairing for the incorporation of dATP opposite the 5′ T of a CPD and not Hoogsteen base pairing, and this conclusion holds for Polη as well.
Discussion
Based upon the ternary crystal structure of Dpo4 complexed with a cis-syn TT dimer, it has been proposed that Dpo4 incorporates an A opposite the 3′ T of a CPD via Watson–Crick base pairing, whereas the A opposite the 5′ T of the CPD adopts a syn conformation and is incorporated via Hoogsteen base pairing (24). This mechanism of CPD bypass advanced for Dpo4, in particular the conclusion that the incorporation of an A opposite the 5′ T of CPD occurs via Hoogsteen pairing, has been difficult to rationalize, especially in view of the fact that a CPD does not significantly alter the ability of the two Ts to engage in Watson–Crick base pairing with the incoming As. Furthermore, based upon observations with the crystal structure of Dpo4, the suggestion has been advanced that Polη utilizes the same mechanisms for replicating through the two residues of a CPD as Dpo4. Here, we have examined the mechanism of CPD bypass by Dpo4 in biochemical experiments by using nucleotide analogs to determine whether the mechanism of CPD bypass inferred from the crystal structure does in fact operate in solution under normal physiological conditions.
A CPD presents a strong block to synthesis by Dpo4, and this is because Dpo4 is strongly inhibited at incorporating an A opposite the 3′ T of the CPD. From steady-state kinetic analyses, we have determined that the efficiency of A incorporation opposite the 3′ T of the CPD is reduced by ≈1,000-fold relative to the insertion of an A opposite the undamaged T. Insertion of an A opposite the 5′ T by Dpo4, however, is not significantly impeded. In its highly inefficient ability of nucleotide incorporation opposite the 3′ T of the CPD, Dpo4 resembles the various replicative/repair Pols, which also are strongly inhibited at the 3′ T. For example, the efficiency of A insertion opposite the 3′ T of a CPD by T7 Pol is ≈3,000-fold lower than the efficiency of inserting an A opposite an undamaged T (31). Polη, by contrast, incorporates an A opposite the 3′ T of the CPD with the same efficiency as opposite the undamaged T.
The 3′ T of a CPD presents a strong block to synthesis by most DNA Pols, because the cyclobutane linkage of the two Ts in the CPD prevents the flipping of the 5′ T out of the active site. Biochemical and structural studies with the T7 Pol have shown that this Pol inserts an A opposite the 3′ T of a CPD with the photoproduct lying outside the active site, whereas insertion opposite the 5′ T occurs with the CPD in the active site (31, 32, 34). Thus, the A incorporation opposite the 3′ T of a CPD is not template-directed but rather occurs via an abasic site-like intermediate.
Our biochemical studies indicate that the mechanism of CPD bypass adopted by Dpo4 resembles that used by T7, in which nucleotide incorporation opposite the 3′ T of the CPD involves an abasic site-like intermediate. In this regard, our observations with 1meA are particularly instructive. 1meA incorporation opposite the undamaged 3′ T is reduced by 250-fold, consistent with the requirement of Watson–Crick base pairing for nucleotide incorporation opposite the undamaged template residue. However, even though the efficiency of A incorporation opposite the 3′ T of the CPD is reduced by 1,000-fold, compared with that opposite the undamaged T, the incorporation of 1meA opposite the 3′ T of the CPD occurs with almost the same efficiency as opposite the nondamaged T. If A incorporation opposite the 3′ T of the CPD had required Watson–Crick base pairing, then 1meA incorporation opposite the 3′ T of the CPD would have declined by the same magnitude (≈250-fold) as occurs opposite the undamaged T. The absence of any significant reduction in the efficiency of 1meA incorporation opposite the 3′ T of the CPD beyond what is observed for A incorporation supports the inference that A incorporation opposite the 3′ T of the CPD by Dpo4 does not entail Watson–Crick base pairing. Additionally, the observation that Dpo4 incorporates an A and 1meA opposite the 3′ T of the CPD and opposite the abasic site with very similar efficiencies supports the conclusion that A incorporation opposite the 3′ T of the CPD occurs opposite an abasic site-like intermediate, rather than directly opposite the 3′ T with the CPD remaining in the active site.
The very inefficient incorporation of dATP opposite the 3′ T of the CPD by Dpo4 implies that, despite the fact that both residues of the CPD occupy the active site in the crystal structure (24), this Pol is impeded in performing catalysis, presumably because of the steric constraints that arise when both the Ts of the dimer are in the active site. Consequently, the Pol flips the CPD out of its active site and then inserts an A opposite the resultant abasic site-like intermediate (Fig. 3A). Hence, for Pols that are considerably inhibited at the 3′ T of the CPD, the adoption of an abasic-like form could provide for a general mechanism for mediating a very inefficient synthesis through this site.
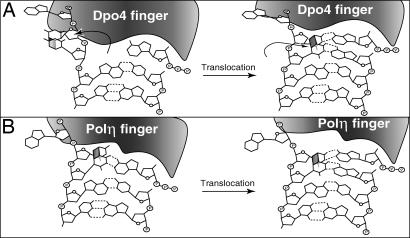
Fig. 3.
Mechanism of TT dimer bypass by Dpo4 and Polη.(A) Although in the crystal structure, Dpo4 is seen to hold a CPD in its active site, this form does not permit nucleotide insertion opposite the 3′ T of the CPD. Only by flipping the CPD out of the active site, which generates an AP site-like intermediate, does Dpo4 carry out the nucleotide insertion reaction. After this, the Pol translocates, and the CPD enters the active site. Dpo4 then incorporates an A opposite the 5′ T of the CPD by using Watson–Crick and not Hoogsteen base pairing, as was inferred from the crystal structure. (B) Polη can accommodate and maintain the two residues of the CPD in its active site. The incorporation of an A opposite the 3′ T is directed by the formation of Watson–Crick base pairing. Subsequently, the Pol translocates, placing the 5′ T of the CPD in the active site. Watson–Crick base pairing is used to direct the incorporation of an A opposite the 5′ T of the dimer.
dATP incorporation opposite the 5′ T of the TT dimer by Dpo4 occurs with the CPD in the active site of the enzyme and that utilizes the Watson–Crick and not the Hoogsteen base pairing (Fig. 3A). This conclusion is supported by our observation that 7dzA is incorporated opposite the 5′ T of the CPD as efficiently as an A, and 2AP is incorporated only slightly less efficiently. Because the incorporation of these nucleotide analogs, which would impair Hoogsteen base pairing, occurs as efficiently as the incorporation of an A, Hoogsteen base pairing cannot be the means adopted by Dpo4 for incorporating an A opposite the 5′ T of the CPD. Furthermore, our observation that the efficiency of 1meA incorporation opposite the 5′ T of the CPD is reduced to the same extent (>450-fold) as opposite the undamaged 5′ T provides compelling evidence for the requirement of Watson–Crick base pairing at the 5′ T of the CPD.
Polη carries out proficient replication through a cis-syn TT dimer, because it can accommodate both the nucleotides of the CPD in its active site (22), and also because it can carry out efficient dATP incorporation opposite both the Ts when the dimer is in the active site (Fig. 3B). This implies the absence of any steric constraints in the Polη active site to hinder efficient nucleotide incorporation from occurring. Furthermore, our observations that, opposite both the 3′ T and 5′ T of the CPD, 1meA is incorporated very poorly, whereas 7dzA is incorporated very efficiently, indicate that opposite both these lesion sites, Polη incorporates an A via Watson–Crick base pairing. The mechanism of nucleotide incorporation inferred previously from kinetic and structural studies (19–22) and further supported from the biochemical studies reported here provides for an efficient means of CPD bypass, and Polη is the only Pol known to have such an ability.
Because of its proficient ability to replicate through CPDs, Polη makes a major contribution to DNA synthesis through these lesions; consequently, inactivation of Polη in humans causes the variant form of xeroderma pigmentosum. However, because Dpo4 is very inefficient at incorporating nucleotides opposite the 3′ site, we expect it to contribute to CPD bypass in a far less significant way.
From our biochemical studies, we conclude that the mechanisms inferred for TT dimer bypass from the crystal structure of Dpo4 (24) are not those adopted by this Pol in solution, and therefore they are not a valid reflection of the Dpo4 action mechanism. Thus, Watson–Crick base pairing is not used for nucleotide incorporation opposite the 3′ T of the CPD, as was indicated from the crystal structure; instead, incorporation opposite the 3′ site occurs opposite an abasic site-like intermediate. And, opposite the 5′ T of the CPD, instead of Hoogsteen base pairing inferred from the crystal structure, we find that Watson–Crick base pairing is important.
How could one account for the structures seen in the Dpo4 crystals but not supported by our biochemical data? Although crystal structures are crucial for revealing the structural features of proteins and other macromolecules, from which action mechanisms could be inferred, they yield no direct information about the efficiency of the action mechanism. Also, because crystallization can, at times, trap intermediates that are not in the reaction pathway, the functional implications of the crystal structure, particularly when it suggests a highly unusual action mechanism, need to be confirmed by independent means. Biochemical studies, because they measure the efficiencies of reaction mechanisms, are important for validating the functional significance of the structure adopted by the crystal form. Because our biochemical studies provide no verification for the mechanisms inferred from the crystal structure, we surmise that the base pairing seen in Dpo4 structures is imposed by the crystal form and therefore is of little biological relevance.
Acknowledgments
We are grateful to Richard P. Hodge and the Synthetic Organic Chemistry Core Laboratory, supported by National Institute of Environmental Health Sciences Center Grant ES006676, for generating the 1meA. This work was supported by National Institutes of Health Grants CA094006 and ES012411.
Author contributions: R.E.J., L.P., and S.P. designed research; R.E.J. and L.P. performed research; R.E.J. and S.P. analyzed data; and R.E.J., L.P., and S.P. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CPD, cyclobutane pyrimidine dimer; Pol, polymerase; 7dzA, 7-deaza-2′-deoxyadenosine-5′-triphosphate; 2AP, 2-aminopurine-2′-deoxyribose-5′-triphosphate; 1meA, N1-methyl-2′deoxyadenosine-5′-triphosphate.
References
- 1.Ciarrocchi, G. & Pedrini, A. M. (1982) J. Mol. Biol. 155, 177–183. [DOI] [PubMed] [Google Scholar]
- 2.Husain, I., Griffith, J. & Sancar, A. (1988) Proc. Natl. Acad. Sci. USA 85, 2558–2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kemmink, J., Boelens, R., Koning, T., van der Marel, G. A., van Boom, J. H. & Kaptein, R. (1987) Nucleic Acids Res. 15, 4645–4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park, H., Zhang, K., Ren, Y., Nadji, S., Sinha, N., Taylor, J.-S. & Kang, C. (2002) Proc. Natl. Acad. Sci. USA 99, 15965–15970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doublie, S., Tabor, S., Long, A. M., Richardson, C. C. & Ellenberger, T. (1998) Nature 391, 251–258. [DOI] [PubMed] [Google Scholar]
- 6.Kiefer, J. R., Mao, C., Braman, J. C. & Beese, L. S. (1998) Nature 391, 304–307. [DOI] [PubMed] [Google Scholar]
- 7.Li, Y., Korolev, S. & Waksman, G. (1998) EMBO J. 17, 7514–7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kool, E. T. (2002) Annu. Rev. Biochem. 71, 191–219. [DOI] [PubMed] [Google Scholar]
- 9.Prakash, S., Johnson, R. E. & Prakash, L. (2005) Ann. Rev. Biochem. 74, 317–353. [DOI] [PubMed] [Google Scholar]
- 10.Johnson, R. E., Prakash, S. & Prakash, L. (1999) Science 283, 1001–1004. [DOI] [PubMed] [Google Scholar]
- 11.Masutani, C., Kusumoto, R., Yamada, A., Dohmae, N., Yokoi, M., Yuasa, M., Araki, M., Iwai, S., Takio, K. & Hanaoka, F. (1999) Nature 399, 700–704. [DOI] [PubMed] [Google Scholar]
- 12.Johnson, R. E., Prakash, S. & Prakash, L. (1999) J. Biol. Chem. 274, 15975–15977. [DOI] [PubMed] [Google Scholar]
- 13.McDonald, J. P., Levine, A. S. & Woodgate, R. (1997) Genetics 147, 1557–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang, Y.-C., Maher, V. M., Mitchell, D. L. & McCormick, J. J. (1993) Mol. Cell. Biol. 13, 4276–4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waters, H. L., Seetharam, S., Seidman, M. M. & Kraemer, K. H. (1993) J. Invest. Dermatol. 101, 744–748. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, R. E., Kondratick, C. M., Prakash, S. & Prakash, L. (1999) Science 285, 263–265. [DOI] [PubMed] [Google Scholar]
- 17.Stary, A., Kannouche, P., Lehmann, A. R. & Sarasin, A. (2003) J. Biol. Chem. 278, 18767–18775. [DOI] [PubMed] [Google Scholar]
- 18.Yu, S.-L., Johnson, R. E., Prakash, S. & Prakash, L. (2001) Mol. Cell. Biol. 21, 185–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, R. E., Washington, M. T., Prakash, S. & Prakash, L. (2000) J. Biol. Chem. 275, 7447–7450. [DOI] [PubMed] [Google Scholar]
- 20.Washington, M. T., Johnson, R. E., Prakash, S. & Prakash, L. (2000) Proc. Natl. Acad. Sci. USA 97, 3094–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Washington, M. T., Prakash, L. & Prakash, S. (2003) Proc. Natl. Acad. Sci. USA 100, 12093–12098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trincao, J., Johnson, R. E., Escalante, C. R., Prakash, S., Prakash, L. & Aggarwal, A. K. (2001) Mol. Cell 8, 417–426. [DOI] [PubMed] [Google Scholar]
- 23.Boudsocq, F., Iwai, S., Hanaoka, F. & Woodgate, R. (2001) Nucleic Acids Res. 29, 4607–4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ling, H., Boudscq, F., Plosky, B. S., Woodgate, R. & Yang, W. (2003) Nature 424, 1083–1087. [DOI] [PubMed] [Google Scholar]
- 25.McCulloch, S. D., Koroska, R. J., Masutani, C., Iwai, S., Hanaoka, F. & Kunkel, T. A. (2004) Nature 428, 97–100. [DOI] [PubMed] [Google Scholar]
- 26.Wolfle, W. T., Washington, M. T., Prakash, L. & Prakash, S. (2003) Genes Dev. 17, 2191–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haracska, L., Johnson, R. E., Unk, I., Phillips, B., Hurwitz, J., Prakash, L. & Prakash, S. (2001) Mol. Cell. Biol. 21, 7199–7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trincao, J., Johnson, R. E., Wolfle, W. T., Escalante, C. R., Prakash, S., Prakash, L. & Aggarwal, A. K. (2004) Nat. Struct. Mol. Biol. 11, 457–462. [DOI] [PubMed] [Google Scholar]
- 29.Koivisto, P., Duncan, T., Lindahl, T. & Sedgwick, B. (2003) J. Biol. Chem. 278, 44348–44354. [DOI] [PubMed] [Google Scholar]
- 30.Creighton, S., Bloom, L. B. & Goodman, M. F. (1995) Methods Enzymol. 262, 232–256. [DOI] [PubMed] [Google Scholar]
- 31.Li, Y., Dutta, S., Doublie, S., Bdour, H. M., Taylor, J.-S. & Ellenberger, T. (2004) Nat. Struct. Mol. Biol. 11, 784–790. [DOI] [PubMed] [Google Scholar]
- 32.Sun, L., Wang, M., Kool, E. T. & Taylor, J.-S. (2000) Biochemistry 39, 14603–14610. [DOI] [PubMed] [Google Scholar]
- 33.Haracska, L., Washington, M. T., Prakash, S. & Prakash, L. (2001) J. Biol. Chem. 276, 6861–6866. [DOI] [PubMed] [Google Scholar]
- 34.Smith, C. A., Baeten, J. & Taylor, J.-S. (1998) J. Biol. Chem. 273, 21933–21940. [DOI] [PubMed] [Google Scholar]