Abstract
Translocation of proteins across the mitochondrial inner membrane is an essential process requiring an import motor having mitochondrial Hsp70 (mtHsp70) at its core. The J protein partner of mtHsp70, Pam18, is an integral part of this motor, serving to stimulate the ATPase activity of mtHsp70. Pam16, an essential protein having an inactive J domain that is unable to stimulate mtHsp70's ATPase activity, forms a heterodimer with Pam18, but its function is unknown. We set out to test the importance of three properties of Pam16: (i) a stable interaction between Pam16 and Pam18, (ii) the inability of Pam16's degenerate J domain to stimulate Ssc1's ATPase domain, and (iii) the innately lower stimulatory activity of the Pam16:Pam18 heterodimer, compared to Pam18 alone. Neither substantial reduction in the ability of Pam18 to stimulate Ssc1's ATPase activity, nor the presence of an active J domain in Pam16, had deleterious effects on cell growth, indicating the lack of importance of two of these biochemical properties. However, a stable interaction between Pam16's degenerate J domain and Pam18's J domain was found to be critical for function. Alterations that destabilized the Pam16:Pam18 heterodimer had deleterious effects on cell growth and mitochondrial protein import; intragenic suppressors that restored robust growth also restored heterodimer stability. Our results support the idea that Pam16's J-like domain strongly interacts with Pam18's J domain, leading to a productive interaction of Pam18 with mtHsp70 at the import channel.
Keywords: mitochondria, translocation, Hsp40, Pam18, heterodimer
Mitochondrial function depends on the efficient import of hundreds of proteins that are synthesized on cytosolic ribosomes and then translocated through proteinaceous channels into the matrix (1–3). Typically, N-terminal targeting sequences are driven across the inner membrane through the presequence translocase (Tim23 complex) by the membrane potential (4–6). Translocation of the remainder of the protein through the Tim23 channel requires the action of the import motor that is associated with the channel at the inner membrane (6–9). Five critical components of this import motor, all of which are highly conserved among eukaryotes, have been identified (3, 6).
MtHsp70 (Ssc1 in yeast), the core of the import motor, interacts with translocating polypeptides in an ATP-dependent process to drive import (7–9). Like any typical Hsp70, Ssc1 functions in the import process with two cochaperones that facilitate its reaction cycle. Pam18 (Tim14) serves as its J protein partner, stimulating ATP hydrolysis and thus, stabilizing the interaction with translocating polypeptide (10–12). Pam18, like all J proteins, contains a J domain with a conserved histidine, proline, aspartic acid (HPD) tripeptide that is essential for function (10–12). Mge1 is a nucleotide release factor that facilitates release of ADP and rebinding of ATP, thus destabilizing the polypeptide interaction (8, 13, 14). Tim44, the fourth essential motor component, is a peripheral membrane protein that serves to tether Ssc1 to the Tim23 translocase until it binds to a translocating polypeptide (7–9, 15, 16). Interaction of Ssc1 with a translocating polypeptide triggers release of Ssc1 from the channel, hence, completing one cycle of the translocation process.
Pam16 (Tim16), the most recently discovered essential component, displays some similarities with Pam18 (17, 18). Both are tightly associated with the inner membrane, presumably via their hydrophobic N-terminal regions. In addition, the C-terminal region of Pam16 has significant sequence similarity to Pam18's C-terminal J domain. However, Pam16 lacks the critical HPD motif, having DKE in its place, and does not stimulate Ssc1's ATPase activity (19). Pam16 and Pam18 form a complex that has a reduced ability to stimulate Ssc1's ATPase activity compared to Pam18 alone. This inhibition has been suggested to play an important role in protein translocation (19, 20).
To better understand Pam16's function, we asked whether this innately lower stimulatory activity of the Pam16:Pam18 heterodimer and Pam16's inability to stimulate Ssc1's ATPase activity were critically important in vivo. We found that neither substantial reduction in the activity of Pam18 to stimulate Ssc1's ATPase activity nor the presence of an active J domain in Pam16 had deleterious effects on cell growth. However, a stable interaction between Pam16's degenerate J domain and Pam18's J domain was found to be critical for function.
Materials and Methods
Plasmids and Genetic Analysis. A PAM16:PAM18 chimera was constructed by introducing an AvrII site at the codons for amino acids 106–107 of PAM18 and between amino acids 52 and 53 of PAM16 through site-directed mutagenesis using the QuikChange protocol (Stratagene). This mutation causes a K107R change in Pam18. The N terminus of Pam18 was epitope tagged with three tandem copies of the hemagglutinin (HA) peptide by introducing a NotI site after PAM18's initiation codon and inserting a NotI fragment encoding the HA tag from pGTEPI (21).
To obtain temperature-sensitive (Ts) mutants, a library of random mutations in pRS315-PAM18 (10) was created through PCR amplification of the coding region of PAM18. Δpam18 carrying pRS316-PAM18 was transformed with the library and incubated at 30°C on leucine omission plates. Transformants were patched onto 5-fluoroorotic acid plates to select for candidates that could grow in the absence of the wild-type copy of PAM18 and then replicated to leucine omission plates and incubated at 37°C.
Suppressors of the Pam18L150W Ts phenotype were selected by generating a mutagenized library of pRS315-pam18L150W as described above. The haploid, Δpam18 carrying pRS316-pam18L150W, was transformed with the library, and the resulting transformants were replica plated onto 5-fluoroorotic acid plates and incubated at 37°C. Suppressors of the Ts phenotype caused by the Pam16:Pam18 fusion protein in a Δpam16 strain was performed essentially as described above, except that the template for mutagenesis was the AvrII–XhoI fragment encoding the Pam18 J domain. In all cases, plasmid DNA was recovered from Ts transformants and used to transform the parent strain to verify the phenotype. All in vivo experiments were carried out in the W303 genetic background in derivatives of PJ53 (22).
Protein Purification. Six histidine codons were introduced at the 5′ end of genes encoding Pam18 and Pam16:Pam18 and at the 3′ end of Pam16. All were cloned into the plasmid pET3a. Overexpression was carried out in Escherichia coli strain C41 (23) by allowing growth at 20°C or 28°C to an A600 of 0.6, followed by induction using 0.5 mM IPTG for 4 or 6 h. Protein was purified by standard affinity chromatography using Ni-NTA agarose. The Ssc1His (24) was purified from yeast as described. Antibodies were affinity purified and crosslinked to protein A-Sepharose beads as described (8).
Coimmunoprecipitation (co-IP). Purified protein (7 μg) was incubated with Pam18- or Pam16-specific antibody beads in co-IP buffer (20 mM Mops-KOH, pH 7.4/250 mM sucrose/80 mM KCl/0.2% Triton X-100/1 mM PMSF) at 4°C for 1 h. The Pam16- and Pam18-bound beads were washed three times in co-IP buffer and blocked with 0.1% BSA at 23°C for 20 min. Approximately 90% of Pam18 or Pam16 was immunoprecipitated. For analysis of complex formation, 7 μg of either Pam18 or Pam16 was added, and the mixture incubated in a final volume of 200 μl at 23°C for 30 min. The beads were washed four times and precipitated material analyzed by SDS/PAGE, followed by Coomassie staining.
For mitochondria, 1 mg was preincubated at 30°C or 37°C for 15 min to induce the mutant phenotype and lysed on ice by addition of 1% Triton X-100 in 20 mM Mops-KOH, pH 7.4/250 mM sucrose/80 mM KCl/5 mM EDTA/1 mM PMSF. Lysates were centrifuged for 10 min at 20,800 × g at 4°C. Beads (15 μl bed volume) crosslinked to specific antibodies were incubated with the supernatants for 1 h at 4°C, and the samples were processed as described above. In the case of HA-Pam18, the lysates were incubated with HA-specific antibody for 2 h before the co-IP by using protein A-Sepharose.
Size Exclusion Chromatography. Superdex 200HR-10/30 was run at 0.5 ml/min in Sizing buffer (25 mM Hepes-KOH, pH 7.4/100 mM KCl/5% glycerol/0.2% Triton X-100) at 4°C. Fractions (300 μl) were collected between 7 and 30 ml; aliquots from alternate fractions were subjected to SDS/PAGE and immunoblot analysis using Pam16- and Pam18-specific antibodies.
Miscellaneous. Affinity-purification of antibodies (8), ATPase assays (8), co-IP of Ssc1-Tim44 complex (9), mitochondria purification (24), and in vitro import assays (25, 26) were carried out as described. For import assays, mitochondria isolated from cells grown at 30°C were preincubated at 37°C for 10 min to induce the mutant phenotype before the addition of precursor protein at 30°C. Immunoblot analysis was carried out by using the ECL system (Amersham Pharmacia) according to the manufacturer's instructions.
Results
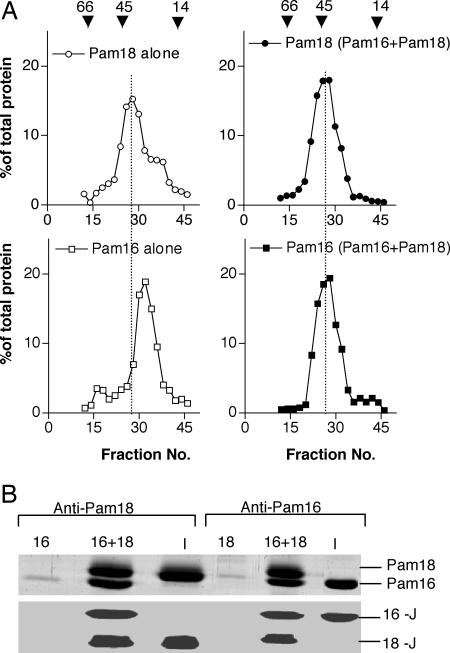
Pam16 and Pam18 Can Form Homodimers. In mitochondria depleted of Pam16, Pam18 can be cross-linked to itself (17), suggesting that Pam18 might be able to form homodimers. We assessed the quaternary structure of purified Pam18 by using size-exclusion chromatography. Pam18, a 18-kDa protein, eluted in fractions 24–30, corresponding to an estimated molecular mass of 43–45 kDa (Fig. 1A Upper Left). Under similar conditions, Pam16, a 16-kDa protein, eluted primarily in fractions 28–34, corresponding to an estimated molecular mass of 39–40 kDa (Fig. 1 A Lower Left). These estimated molecular masses are consistent with formation of homodimers by both Pam16 and Pam18.
Fig. 1.
In vitro analysis of Pam16–Pam18 complex formation. (A) Size exclusion chromatography. Three separate reactions were carried out: (i) 50 μg of Pam18 alone, (ii) 50 μg of Pam16 alone, and (iii) 50 μg of Pam18 and Pam16 incubated in size exclusion buffer for 30 min at 23°C. Reaction mixtures were subjected to size exclusion chromatography. Alternate fractions were analyzed by SDS/PAGE followed by immunoblot analysis using Pam16- and Pam18-specific antibodies. Signals were quantified by using densitometry and plotted as percent of total Pam16 or Pam18 protein. A dashed line is drawn to indicate the peak fraction of Pam16. Note the shift of position of Pam16 in presence of Pam18. Positions of migration of molecular standards are indicated by arrowheads: BSA, 66 kDa; ovalbumin, 45 kDa; lysozyme, 14.3 kDa. (B) Co-IP. Prebound 2 μM of either Pam16 (16) and/or Pam18 (18), 4 μM of J-like domain, amino acids 48–148 of Pam16 (16-J) and/or amino acids 101–168 of Pam18 (18-J) were incubated for 30 min, subjected to immunoprecipitation using indicated antibodies, and analyzed by SDS/PAGE followed by staining with Coomassie blue. A sample containing the input (I) served as a control for co-IP efficiency.
Previous analyses indicated that Pam16 and Pam18 could form a heterodimer (18, 19). Consistent with these findings, we found that upon mixing equimolar amounts of full-length Pam16 and Pam18, >75% of Pam16 was coimmunoprecipitated with antibodies specific for Pam18 (Fig. 1B). Conversely, >75% of Pam18 could be coimmunoprecipitated with antibodies specific for Pam16. Thus, Pam16 and Pam18 efficiently formed heteromeric complexes. To assess the size of these heterodimeric complexes, mixtures of Pam18 and Pam16 were separated by gel filtration. The elution of Pam16 and Pam18 was coincident (Fig. 1 A Right) at a position corresponding to a molecular size of 43–45 kDa. This estimated molecular mass is consistent with that of a Pam18:Pam16 heterodimer. Based on these co-IP and chromatographic results, we conclude that, although Pam16 and Pam18 can form homodimers, heterodimers form preferentially. These results are consistent with Pam16 and Pam18 having a higher affinity for each other than for themselves.
To determine whether the J and J-like domains of the two proteins are sufficient for heterodimer formation, we mixed fragments containing amino acids 48–149 of Pam16 and amino acids 101–168 of Pam18 together (Fig. 2A) and performed immunoprecipitation with Pam18- and Pam16-specific antibodies. As with full-length protein, these smaller fragments containing the J domains coimmunoprecipitated (Fig. 1B). Pam16 contains a 30-aa C-terminal segment beyond the J-like domain. A fragment containing only the J-like domain, amino acids 48–119, could also be coprecipitated with the Pam18 J domain (data not shown). Thus, the J domains are sufficient for heterodimer formation.
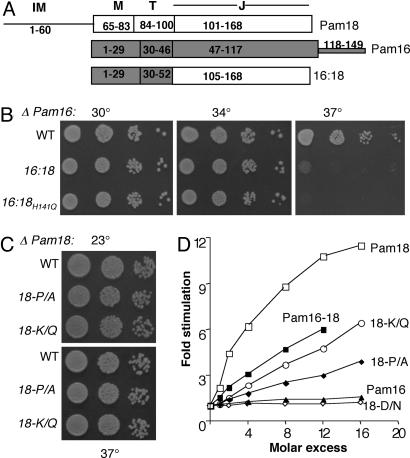
Fig. 2.
Analysis of Pam16:Pam18 chimera and Pam18 J domain mutants. (A) Predicted domains of Pam16, Pam18, and Pam16:Pam18 (16:18) based on sequence alignment. The amino acids corresponding to predicted regions are highlighted. Regions: intermembrane space region (IM), membrane association region (M), predicted mitochondria targeting region (T), and J domain (J). (B and C) Growth phenotype. Tenfold serial dilutions of Δpam16 or Δpam18 cells expressing the indicated genes were spotted onto minimal media and incubated at the indicated temperatures for 2 days. (B) Δpam16 cells expressing Pam16 (WT), Pam16:Pam18 (16:18), or Pam16:Pam18H141Q (16:18H141Q). (C) Δpam18 cells expressing pam18 (WT), Pam18P142A (18-P/A), and Pam18K144Q (18-K/Q). (D) Stimulation of Ssc1 ATPase activity. Ssc1:[α-32P]ATP complex (0.25 μM) was incubated at 23°C in the presence of indicated concentrations of Pam18 or Pam18 mutants: P142A (P/A), K144Q (K/Q), D143N (D/N), and/or Pam16. The rate of ADP formation was measured and plotted as the fold stimulation over the basal rate determined in the absence of any additional component (set at one) versus the ratio of the Pam protein relative to Ssc1 concentration (molar excess).
The Absence of a Functional J Domain in Pam16 Is Not Functionally Important. Pam16 is not capable of stimulating Ssc1's ATPase activity and thus, is not a functional J protein partner of Ssc1 (19). However, whether the absence of this activity is an important aspect of Pam16's function is not known. To address this question, a Pam16 molecule containing an active J domain capable of stimulating Ssc1's ATPase activity was needed. Simply replacing DKE with HPD was not sufficient to convert Pam16's J-like domain to an active J domain (ref. 19 and data not shown). Therefore, we replaced Pam16's J-like domain with Pam18's functional J domain, forming a chimeric protein, Pam16:Pam18, composed of the N-terminal 52 aa of Pam16 and the J domain of Pam18 (amino acid s106–168; Fig. 2 A). We purified Pam16:Pam18 to determine whether it could, in fact, stimulate the ATPase activity of Ssc1. The fusion was 70% as active as Pam18 when added to preformed radiolabeled ATP-Ssc1 complexes. When present at a 2.5-fold excess over Ssc1, Pam18 and the fusion protein stimulated the ATPase activity of Ssc1 4- and 2.8-fold, respectively.
We then tested whether the fusion could substitute for Pam16 in vivo. Δpam16 cells expressing Pam16:Pam18 (Δpam16 [Pam16:Pam18]) at normal levels were viable, growing as well as wild-type cells at 30 and 34°C (Fig. 2B). Thus, presence of the J-like domain of Pam16 is not uniquely required for viability. However, Δpam16 [Pam16:Pam18] did not grow at 37°C. This Ts growth could indicate that the presence of an active J domain in Pam16 had deleterious consequences. To test whether this stimulatory activity of the chimeric protein affected growth in vivo, we mutated the DNA encoding the Pam18 HPD tripeptide such that a glutamine would replace the histidine, an alteration we had previously shown to obviate the ability of Pam18 to stimulate the ATPase activity of Ssc1 and rescue the inviability of Δpam18 cells (10). However, the H/Q alteration in the chimeric protein did not improve growth of the Δpam16 strain (Fig. 2B). We conclude that the inability of the degenerate J domain of Pam16 to act as a J domain is unimportant to Pam16's essential in vivo function.
Pam18 with Significantly Reduced Stimulatory Activity Is Functional in Vivo. The J domain of Pam18 is less active in stimulating Ssc1's ATPase activity when in complex with a fragment of Pam16 (19). In agreement with these results, we found that full-length Pam16:Pam18 heterodimer was 50–60% as active as Pam18 alone in stimulating Ssc1's ATPase activity over a range of protein concentrations (Fig. 2D). To begin to assess whether this inhibition by Pam16 of Pam18's stimulatory capacity is an important part of Pam16's function, as suggested (19), we wanted to determine whether a 50–60% reduction in the stimulatory ability of Pam18 had consequencess in vivo. Therefore, we constructed several PAM18 mutants encoding single amino acid alterations in or near the crucial HPD tripeptide: P142A, D143N, and K144Q. Pam18D143N, like the previously analyzed Pam18H141Q, was neither able to rescue the inviability of Δpam18 cells nor measurably stimulate the ATPase activity of Ssc1 (Fig. 2D). However, pam18P142A and pam18K144Q cells grew as well as wild-type cells under a variety of conditions (Fig. 2C and data not shown). Protein import into mitochondria of these mutants was also unaffected (Fig. 5, which is published as supporting information on the PNAS web site). On the other hand, the ability of Pam18P142A and Pam18K144Q to stimulate Ssc1's ATPase activity was reduced ≈70 and 60%, respectively (Fig. 2D). As expected, formation of heterodimers between the mutant Pam18 proteins and Pam16 resulted in further reduction in ATPase stimulation (Fig. 6, which is published as supporting information on the PNAS web site). Together, these results indicate that a decrease in the ability of Pam18 to stimulate Ssc1's ATPase activity is well tolerated. We conclude that the extent of the decrease in ATPase stimulation caused by the interaction of Pam18 with Pam16 is not sufficient to have significant effects on mitochondrial protein import or cell growth.
Suppression of the Ts Phenotype of Δpam16 [Pam16:Pam18]. We next tested the hypothesis that heterodimer formation between Pam16 and Pam18 is critical in vivo for transport of proteins into mitochondria. To approach this question, we used the Pam16:Pam18 chimera containing the Pam18 J domain for our initial analysis. This chimera was chosen because Δpam16 [Pam16:Pam18] cells are Ts for growth, indicating that the Pam18 J domain does not completely substitute for the Pam16 J-like domain. Although J domains of Pam16 and Pam18 are sufficient for dimer formation, the Pam18 homodimer is less stable than the Pam16:Pam18 heterodimer. We reasoned that the Ts growth could be due to the less stable interaction between the chimera and Pam18 that presumably heterodimerize by interaction between the two Pam18 J domains.
As a first step, we analyzed the chimera more thoroughly. Making use of an in organellar translocation assay, we asked whether protein import into the matrix of the mitochondria of Δpam16 [Pam16:Pam18] cells was defective. Saturating amounts of purified Cytb2(47)-DHFR, a fusion protein in which the matrix targeting sequence of cytochrome b2 is fused to dihydrofolate reductase (DHFR), was added to Δpam16 [Pam16:Pam18] mitochondria that had been preincubated at 37°C to induce a mutant phenotype. Thirty minutes after addition of saturating amounts of precursor protein, only 20% as much DHFR was imported into Δpam16 [Pam16:Pam18] mitochondria compared to wild-type mitochondria (Fig. 3A).
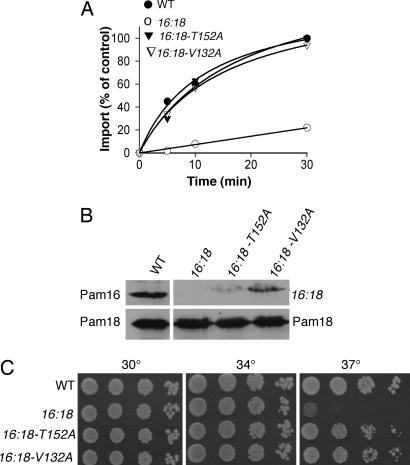
Fig. 3.
Analysis of intragenic suppressors of PAM16:PAM18 phenotype. (A) In vitro import. Cytb2(47)-DHFR was added to 50 μg of preincubated indicated mitochondria at 37°C for 15 min. Reaction mixtures were separated by SDS/PAGE and imported i-Cytb2(47)-DHFR was quantified after immunoblotting using anti-DHFR antibodies. Import in WT mitochondria after 30 min was set to 100%. (B and C) The following genes were used: WT, pam16:pam18 (16:18), pam16:pam18-T152A (16:18-T152A), and pam16:pam18-V132A (16:18-V132A). (B) In organellar analysis of Pam18–Pam16 complex. Mitochondria from cells expressing Pam18 with an HA-tag were lysed and subjected to co-IP using HA-specific antibodies. Immunoblotting was carried out by using Pam16-specific (Upper) or Pam18-specific (Lower) antibodies. (C) Growth phenotypes. Tenfold serial dilutions of Δpam16 cells expressing indicated genes were spotted onto minimal medium and incubated at the indicated temperatures for 2 days.
Next, we tested the stability of the Pam18:chimera complex in vivo. To carry out this test, we first generated a HA-tagged Pam18 to allow us to distinguish between Pam18 and the Pam16:Pam18 chimera. Extracts of detergent solubilized mitochondria from cells expressing this tagged Pam18, which supports growth like wild-type Pam18 in vivo (data not shown), were subjected to immunoprecipitation using HA-specific antibodies. As expected from previous results (17, 18), Pam16 was coimmunoprecipitated with the tagged Pam18. However, in mitochondrial lysates from cells expressing the Pam16:Pam18 chimera in place of wild-type Pam16, no coimmunoprecipitation was observed (Fig. 3B, data not shown).
This apparent decrease in stability of the chimera and the defect in protein translocation is consistent with the idea that the import and growth defects of cells expressing the fusion protein are caused by the instability of the heterodimer. To test this idea, we carried out an intragenic suppressor analysis of Δpam16 [Pam16:Pam18], selecting for growth at 37°C after mutagenesis of the DNA encoding the J domain of the chimera. Two suppressors encoding single amino acid alterations within the J domain were isolated: T152A and V132A (Fig. 4B). Both partially suppressed the growth defect at 37°C caused by the chimera (Fig. 3C). Mitochondria isolated from the suppressor strains imported Cytb2(47)-DHFR nearly as efficiently as wild-type mitochondria, indicating that the amino acid alterations in the J domain suppressed both growth and translocation defects imparted by the chimeric protein (Fig. 3A). To assess the interaction between Pam18 and the chimera containing the suppressor amino acid substitution, co-IP assays were carried out. Chimeric suppressor protein was coimmunoprecipitated with Pam18, although not as efficiently as wild-type Pam16 (Fig. 3B), indicating that the suppressor mutations caused a partial restoration of a stable interaction between Pam18 and the chimera.
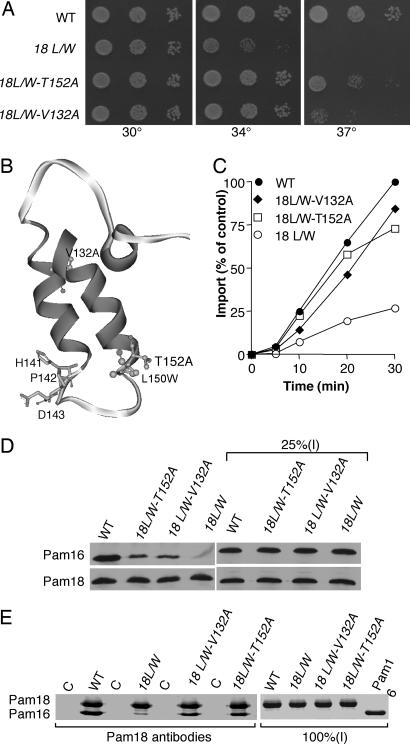
Fig. 4.
Analysis of pam18(L150W) and its intragenic suppressors. pam18(L150W), 18L/W; pam18(L150W-T152A), 18L/W-T152A; pam18(L150W-V132A), 18L/W-V132A. (A) Growth phenotypes. Tenfold serial dilutions of Δpam18 expressing indicated PAM18 genes were spotted onto rich media and incubated at indicated temperatures for 2 days. (B) Model of Pam18 J domain. Amino acid sequence corresponding to the J domain of Pam18 (108–164) was modeled by using coordinates of HDJ1 (34) and drawn by using Micron Separations Web Lab VIEWERPRO software. Relevant amino acids are highlighted. (C) In vitro import. Import reactions were carried out as in Fig. 3B. (D) In organellar analysis of Pam18–Pam16 complex formation. A total of 25% of lysate was loaded as a control (25% I). (E) In vitro co-IP analysis of complex formation. Proteins (2μM) were incubated for 30 min at 23°C: Pam16 (16), Pam18 (18), Pam18 L150W (18L/W), Pam18L150W-V132A (18L/W-V132A), and Pam18L150W-T152A (18L/W-T152A). Co-IPs were carried out by using Pam18-specific antibodies. Samples were analyzed by SDS/PAGE and stained with Coomassie blue. As a control (C), Pam18 and mutants were omitted. All (100%) of the input was loaded.
Amino Acid Substitutions in J Domain of Pam18 Destabilize the Pam16:Pam18 Interaction. The data presented above supports the idea that the interaction between Pam18 and Pam16 is important for function of the translocation machinery. However, to obtain independent confirmation of this idea, we screened for Ts mutations in PAM18. One PAM18 mutant was isolated that grew like wild-type at the optimum growth temperature of 30°C, but poorly at 37°C (Fig. 4A). The growth defect was caused by a single amino acid change in the J domain, L150W, at a site predicted to be at the beginning of helix III, near the opposite end of the loop from the HPD tripeptide (Fig. 4B). To test for defects in protein import in pam18(L150W) mitochondria, henceforth called pam18L/W, in vitro import experiments were conducted. Import of Cytb2(47)-DHFR was substantially reduced in pam18L/W mitochondria compared to wild-type (Fig. 4C).
To test for biochemical defects that could be responsible for the reduced import efficiency, we purified the mutant protein and tested its ability to stimulate the ATPase activity of Ssc1. Pam18L150W was >75% as active as wt Pam18. For example, at a 2.5-fold excess of Pam18 over Ssc1, Pam18L150W and wild-type Pam18 stimulated Ssc1 3.1- and 4-fold, respectively. This level of activity is significantly above that required for both normal growth and rates of protein import, as indicated by the results of the experiments reported above.
We also assessed the Pam18:Pam16 complex in mitochondrial extracts. Co-IP of Pam16 with Pam18L150W was significantly less than co-IP of wild-type Pam18. Interactions between other components of the import motor, Ssc1 and Tim44, and Ssc1 and Mge1, were also tested; no defects in these interactions were found in mutant mitochondria (Fig. 7, which is published as supporting information on the PNAS web site). To more directly analyze heterodimer formation, purified proteins were used. Under the conditions tested, 7-fold less Pam18L150W was found in complex with Pam16 than wild-type Pam18 (Fig. 4E). In sum, both in organellar and in vitro experiments indicate decreased stability of the Pam16:Pam18L150W heterodimer compared to wild-type heterodimer.
Intragenic Suppressors of PAM18 Ts Mutations Compensate for the Import Defect. We selected for intragenic suppressors of the growth defect of pam18L/W. Two suppressors, both of which grew like wild-type at 34°C, but slowly at 37°C, were identified: pam18L/W-V132A and pam18L/W-T152A (Fig. 4A). These amino acid alterations are the same as those able to suppress the temperature sensitivity of Δpam16 cells expressing the Pam16:Pam18 chimera. V132 and T152 are predicted to lie within helix II and helix III of the J domain, respectively (Fig. 4B).
To determine whether the suppressor mutations compensated for the import defect of pam18L/W, we carried out in organellar import assays using saturating amounts of Cytb2(47)-DHFR. Import was substantially improved in both suppressors compared to pam18L/W (Fig. 4C). To test whether the defect in heterodimer formation was also suppressed, we performed co-IP analysis using lysates of pam18L/W-T152A and pam18L/W-V132A mitochondria. More Pam16 was coimmunoprecipitated by Pam18-specific antibodies from lysates of suppressors than from lysates of pam18L/W; however, less Pam16 was coimmunoprecipitated than from wild-type lysates (Fig. 4D), consistent with the partial suppression of the growth defect. In addition, the purified suppressor Pam18 proteins were more efficiently coimmunoprecipitated with Pam16, than was Pam18L150W (Fig. 4E). In sum, the suppressor mutations that partially correct both the growth and translocation defect of pam18L/W cells also partially restore stability of the Pam16:Pam18 heterodimer.
Discussion
The idea that the ability of J proteins to stimulate the ATPase activity of their partner Hsp70s is critically important is both well accepted and substantiated. However, the results presented here demonstrate that, at least in some cases, decreased stimulatory capacity can be tolerated with little if any effects on cell growth. A Pam18 mutant protein having only 30% of wild-type activity supported vigorous growth under all conditions tested. Whether such reduction in activity of other J proteins would be as well tolerated is not clear, because, to our knowledge, no studies correlating stimulatory ability of J proteins and in vivo function have previously been reported. Such robustness may be restricted to cases such as Pam18 and Ssc1 in which both components are tethered to their site of action, resulting in extremely high local concentrations. Consistent with this idea, the ability of mutant Pam18 to enhance Ssc1's ATPase activity approached that of wild-type at higher concentrations, at which point the stimulatory capacity of WT began to plateau.
The results presented here support the idea that Pam18 and Pam16 interact via their J and J-like domains. To our knowledge, the idea of two proteins dimerizing via their J domains is unprecedented. Several J proteins are homodimers, but interact via sequences distinct from their J domains. For example, Sis1 and Ydj1 of the yeast cytosol have been shown to homodimerize via C-terminal coiled-coils located at their C termini, far from the N-terminal J domain (27, 28). Although E. coli DnaJ is dimeric (29), extensive analysis indicates that its J domain is monomeric (30, 31). However, interaction between Pam16 and Pam18 through their J-like domains is supported by both biochemical and genetic analyses. First, fragments of Pam16 and Pam18 containing only their J domains are competent to form a complex. Second, a single amino acid alteration within Pam18's J domain destabilizes the heterodimer, a defect that can be suppressed by a second change in the J domain. In addition, both Pam18 and Pam16 form homodimers in vitro, consistent with increased cross linking of Pam18 to itself upon depletion of Pam16 and vice versa (17).
Our data also support the idea that this interaction between Pam16 and Pam18 is critical in vivo. In both the case of the Pam16:Pam18 chimera and the Pam18 Ts mutant, Pam18L150W, the stability of the heterodimer was reduced compared to the wild-type Pam16:Pam18 interaction. Intragenic suppressors of the growth defect not only corrected the translocation defect, but also partially restored stability of the dimer. Although it is conceivable that the suppressor mutations revert some unknown biochemical property of the mutant protein that is crucial for function, our data are most consistent with the idea that a stable interaction between Pam16 and Pam18 is critical for mitochondrial protein import and thus cell growth.
Combining the data reported here with earlier results, a picture emerges of a heterodimer of Pam16 and Pam18 formed by two distantly related proteins interacting through their most highly related sequences, their J-like domains. We think it likely that PAM16 and PAM18, which are present in higher eukaryotes, evolved from a single progenitor PAM gene, which encoded a protein that formed a homodimer that functioned in the import process. However, upon gene duplication, two specialized proteins coevolved in a manner consistent with the subfunctionalization model of protein evolution (32, 33). One of them, the ortholog of Pam18, maintained an active J domain. The other, the ortholog of Pam16, lost its functionality as a J protein, but its degenerate J domain evolved to stably interact with Pam18's J domain.
Although the data presented here provides evidence for the functional importance of the Pam16 J-like domain, the function of Pam16's N terminus, and thus, the heterodimer as a whole, remains to be clarified. Although Pam18 itself is a transmembrane protein (10–12), we think that the most likely function of the N terminus of Pam16 is to precisely position Pam18 in juxtaposition to Ssc1 at the translocon to maximize ATPase stimulation. The N terminus of Pam16, the region required for its membrane association, is essential (data not shown). The N terminus of Pam16 may also serve a regulatory function during protein import. The Pam16:18 heterodimer could toggle between having an active, Pam18, and inactive, Pam16, J domain juxtaposed to Ssc1 depending on whether a translocating polypeptide is in the import channel and the ATPase activity of Ssc1 needs to be stimulated. Other types of regulatory scenarios envisioned are much less likely in light of the data presented in this report. For example, any model based on the innately lower stimulatory activity of the Pam18:Pam16 heterodimer compared to Pam18 alone or the absence of an active J domain in Pam16 is unlikely because an active J domain supports Pam16 function as well as an inactive one and robust growth, even at 37°C, is supported by a mutant Pam18 J domain that is only 30% as active as wild-type Pam18; thus, Pam16's binding to Pam18, which decreases activity ≈50%, would not be sufficient for significant regulation of activity.
In summary, our data are most consistent with the idea that Pam16's degenerate J domain serves the critical function of forming a stable interaction with Pam18 and to position Pam18 precisely at the channel for optimal activity. It is quite possible that such interactions and possible conformational changes caused by the presence of translocating polypeptides could serve to alter the positioning of Pam18's J domain at the translocon, thus dramatically affecting its ability to stimulate Ssc1. Critical testing of such models awaits further reconstitution of the import machinery.
Supplementary Material
Acknowledgments
We thank Jaroslaw Marszalek for helpful comments on the manuscript. This work was supported by National Institutes of Health Grant GM278709 (to E.A.C.) and American Heart Association Fellowship 0420049Z (to P.R.D.).
Author contributions: P.R.D., B.S., and E.A.C. designed research; P.R.D., B.S., and W.W. performed research; P.R.D., B.S., and E.A.C. analyzed data; and P.R.D. and E.A.C. wrote the paper.
Abbreviations: HA, hemagglutinin; Ts, temperature sensitive; co-IP, coimmunoprecipitation.
References
- 1.Neupert, W. & Brunner, M. (2002) Nat. Rev. Mol. Cell. Biol. 8, 555–565. [DOI] [PubMed] [Google Scholar]
- 2.Wiedemann, N., Frazier, A. E. & Pfanner, N. (2004) J. Biol. Chem. 279, 14473–14476. [DOI] [PubMed] [Google Scholar]
- 3.Koehler, C. M. (2004) Annu. Rev. Cell. Dev. Biol. 20, 309–335. [DOI] [PubMed] [Google Scholar]
- 4.Jensen, R. E. & Johnson, A. E. (2001) Nat. Struct. Biol. 8, 1008–1010. [DOI] [PubMed] [Google Scholar]
- 5.Huang, S., Ratliff, K. S. & Matouschek, A. (2002) Nat. Struct. Biol. 9, 301–307. [DOI] [PubMed] [Google Scholar]
- 6.Rehling, P., Brandner, K. & Pfanner, N. (2004) Nat. Rev. Mol. Cell. Biol. 5, 519–530. [DOI] [PubMed] [Google Scholar]
- 7.Schneider, H.-C., Berthold, J., Bauer, M. F., Dietmeier, K., Guiard, B., Brunner, M. & Neupert, W. (1994) Nature 371, 768–774. [DOI] [PubMed] [Google Scholar]
- 8.Liu, Q., D'Silva, P., Walter, W., Marszalek, J. & Craig, E. A. (2003) Science 300, 139–141. [DOI] [PubMed] [Google Scholar]
- 9.D'Silva, P., Liu, Q., Walter, W. & Craig, E. A. (2004) Nat. Struct. Mol. Biol. 11, 1084–1091. [DOI] [PubMed] [Google Scholar]
- 10.D'Silva, P. D., Schilke, B., Walter, W., Andrew, A. & Craig, E. A. (2003) Proc. Natl. Acad. Sci. USA 100, 13839–13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Truscott, K. N., Voos, W., Frazier, A. E., Lind, M., Li, Y., Geissler, A., Dudek, J., Muller, H., Sickmann, A., Meyer, H. E., et al. (2003) J. Cell Biol. 163, 707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mokranjac, D., Sichting, M., Neupert, W. & Hell, K. (2003) EMBO J. 22, 4945–4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneider, H. C., Westermann, B., Neupert, W. & Brunner, M. (1996) EMBO J. 15, 5796–5803. [PMC free article] [PubMed] [Google Scholar]
- 14.Miao, B., Davis, J. E. & Craig, E. A. (1997) J. Mol. Biol. 265, 541–552. [DOI] [PubMed] [Google Scholar]
- 15.Maarse, A. C., Blom, J., Grivell, L. A. & Meijer, M. (1992) EMBO J. 11, 3619–3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horst, M., Jeno, P., Kronidou, N. G., Bollinger, L., Oppliger, W., Scherer, P., Manning-Krieg, U., Jascur, T. & Schatz, G. (1993) EMBO J. 12, 3035–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kozany, C., Mokranjac, D., Sichting, M., Neupert, W. & Hell, K. (2004) Nat. Struct. Mol. Biol. 11, 234–241. [DOI] [PubMed] [Google Scholar]
- 18.Frazier, A., Dudek, J., Guiard, B., Voos, W., Li, Y., Lind, M., Meisinger, C., Geissler, A., Sickmann, A., Meyer, H., et al. (2004) Nat. Struct. Mol. Biol. 11, 226–233. [DOI] [PubMed] [Google Scholar]
- 19.Li, Y., Dudek, J., Guiard, B., Pfanner, N., Rehling, P. & Voos, W. (2004) J. Biol. Chem. 279, 38047–38054. [DOI] [PubMed] [Google Scholar]
- 20.Chacinska, A., Lind, M., Frazier, A. E., Dudek, J., Meisinger, C., Geissler, A., Sickmann, A., Meyer, H. E., Truscott, K. N., Guiard, B., et al. (2005) Cell 120, 817–829. [DOI] [PubMed] [Google Scholar]
- 21.Roof, D. M., Meluh, P. B. & Rose, M. D. (1992) J. Cell Biol. 118, 95–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.James, P., Pfund, C. & Craig, E. (1997) Science 275, 387–389. [DOI] [PubMed] [Google Scholar]
- 23.Miroux, B. & Walker, J. E. (1996) J. Mol. Biol. 260, 289–298. [DOI] [PubMed] [Google Scholar]
- 24.Liu, Q., Krzewska, J., Liberek, K. & Craig, E. A. (2001) J. Biol. Chem. 276, 6112–6118. [DOI] [PubMed] [Google Scholar]
- 25.Voisine, C., Craig, E. A., Zufall, N., von Ahsen, O., Pfanner, N. & Voos, W. (1999) Cell 97, 565–574. [DOI] [PubMed] [Google Scholar]
- 26.Lim, J. H., Martin, F., Guiard, B., Pfanner, N. & Voos, W. (2001) EMBO J. 20, 941–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sha, B., Lee, S. & Cyr, D. M. (2000) Structure Fold. Des. 8, 799–807. [DOI] [PubMed] [Google Scholar]
- 28.Wu, Y., Li, J., Jin, Z., Fu, Z. & Sha, B. (2005) J. Mol. Biol. 346, 1005–1011. [DOI] [PubMed] [Google Scholar]
- 29.Zylicz, M., Yamamoto, T., McKittrick, N., Sell, S. & Georgopoulos, C. (1985) J. Biol. Chem. 260, 7591–7598. [PubMed] [Google Scholar]
- 30.Szyperski, T., Pellecchia, M., Wall, D., Georgopoulos, C. & Wuthrich, K. (1994) Proc. Natl. Acad. Sci. USA 91, 11343–11347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pellecchia, M., Szyperski, T., Wall, D., Georgopoulos, C. & Wuthrich, K. (1996) J. Mol. Biol. 260, 236–250. [DOI] [PubMed] [Google Scholar]
- 32.Force, A., Lynch, M., Pickett, F. B., Amores, A., Yan, Y. L. & Postlethwait, J. (1999) Genetics 151, 1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prince, V. E. & Pickett, F. B. (2002) Nat. Rev. Genet. 3, 827–837. [DOI] [PubMed] [Google Scholar]
- 34.Peitsch, M. C. (1996) Biochem. Soc. Trans. 24, 274–279. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.