Abstract
Dental caries is a major worldwide oral disease problem in children. Although caries are known to be influenced by dietary factors, the disease results from a bacterial infection; thus, caries susceptibility may be affected by host factors such as salivary antimicrobial peptides. This study aimed to determine a possible correlation between caries prevalence in children and salivary concentrations of the antimicrobial peptides human beta-defensin-3 (hBD-3), the cathelicidin LL37, and the alpha-defensins HNP1-3 (a mixture of HNP1, 2, 3). Oral examinations were performed on 149 middle school children, and unstimulated whole saliva was collected for immunoassays of the three peptides and for assay of caries-causing bacteria in saliva. The median salivary levels of hBD-3, LL37, and HNP1-3 were in the microgram/ml range but were highly variable in the population. While levels of LL37 and hBD-3 did not correlate with caries experience, the median HNP1-3 levels were significantly higher in children with no caries than in children with caries. Children with high caries levels did not have high levels of salivary Streptococcus mutans, and the HNP1-3 level was not correlated with salivary S. mutans. By immunohistochemistry we localized HNP1-3 in submandibular salivary duct cells. HNPs are also released by neutrophils into the gingival crevicular fluid. Both sources may account for their presence in saliva. Low salivary levels of HNP1-3 may represent a biological factor that contributes to caries susceptibility. This observation could lead to new ways to screen for caries susceptibility and to new means of assessing the risk for this common oral problem.
Dental caries is a common disease process that afflicts a large proportion of the world's population. Extensive research indicates that dental caries is the result of a bacterial infection (18) but also is influenced by host and dietary factors (11). Current research seeks to identify risk factors for caries as well as to identify natural oral defenses that may protect against or prevent caries development. Salivary defense systems play a significant role in maintaining the health of the oral cavity and preventing caries. These defenses include factors which inhibit or reverse demineralization of exposed tooth surfaces, such as simple mechanical rinsing, buffering action, and calcium phosphate binding proteins as well as antimicrobial activities including microorganism aggregation and clearance from the oral cavity, immune surveillance, and the secretion of antimicrobial peptides (AMPs) (38).
AMPs are natural antibiotics that provide a first line of defense against a wide spectrum of pathogens (8, 40, 41). These peptides may be particularly important in the oral cavity, where members of the microbial flora are present in high numbers at all times. The three main AMP families are defined by amino acid composition and three-dimensional structure: α-helical peptides without cysteine (the cathelicidins) (1), peptides with three disulfide bonds (the α- and β-defensins) (8, 9), and peptides with an unusually high proportion of specific amino acids, for example, the histatins (26).
Recent research suggests the importance of the defensins and the cathelicidins as antibacterial agents in the oral cavity (5), while histatins are primarily antifungal agents (26). The human β-defensins (hBDs) are widely expressed in oral tissues and in the gingival epithelium (6, 7, 42). hBD1 and -2 have also been detected in salivary glands and ducts and in saliva (3, 31). The α-defensins HNP1 to -3 are expressed in neutrophils and participate in nonoxidative microbial death (9) and have been identified in gingival crevicular fluid (21). The human cathelicidin peptide LL37 is found in neutrophils and inflamed epithelia as well as in saliva (23). Both the mRNA and protein for cathelicidin peptides have been localized to the salivary glands, specifically in acinar cells of the submandibular gland and palatine minor glands, as well as in lingual epithelium and palatal mucosa in mice (23) and submandibular duct cells in humans (39). This pattern of expression suggests that both the defensins and LL37 could have a role in protecting the tooth structure from caries as well as protecting oral mucosa.
The defensins and cathelicidins have broad antimicrobial activity against gram-negative and gram-positive bacteria and Candida albicans and are effective in vitro against oral microorganisms such as Streptococcus mutans, Porphyromonas gingivalis, and Actinobacillus actinomycetemcomitans (5, 13, 25, 35). The cathelicidins and α- and β-defensins act synergistically with other antimicrobials (19, 24). Thus, the coexpression in saliva of cathelicidins and defensins with peptides such as histatin, proline-rich proteins, and calprotectin may provide a natural antibiotic barrier (38). The purpose of this study was to determine a possible correlation between AMP levels in saliva and caries experience in children. We also tested for a known genetic polymorphism in the gene encoding hBD1 (14) and for salivary bacterial load of Streptococcus mutans, the major causative organism for caries (18). We show extensive variation in AMP levels between individuals. Our findings suggest that low salivary levels of HNP1-3 (a mixture of HNP1, 2, 3) may contribute to caries susceptibility and could be a new and useful measure of the risk for caries in children.
MATERIALS AND METHODS
Participants and oral examination.
One hundred forty-nine children participated in the study. A brief health history survey was completed by parents of the subjects. Oral examinations were performed by trained calibrated clinicians using standardized procedures. The study was conducted with the permission of school officials, and informed consent of subjects and parents was obtained through an educational session and written bilingual consent in accordance with a protocol approved by the University of Washington Institutional Review Board. Examiners were instructed to rank subjects separately for active caries and for filled surfaces as follows: 0, no decayed or filled surfaces; 1, mild (one or two affected surfaces); 2, moderate (three to six affected surfaces); 3, severe (more than six affected surfaces). The final caries experience score was determined as the sum of the scores for active decay and filled surfaces. Oral examinations and sample collection were conducted over a 2-day period primarily in the morning.
Salivary analysis.
Unstimulated saliva was collected (3 to 5 ml), the detergent Nonidet P-40 (Sigma, St. Louis, MO) was added to a final concentration of 0.1%, and the sample was frozen for later analysis. Saliva was thawed and cleared by centrifugation twice at 15,000 rpm for 10 min. Total protein concentration was evaluated in the supernatant (cleared unfractionated saliva) by bicinchoninic acid assay (Pierce Inc., Rockford, IL). Cleared unfractionated saliva was also used for the HNP1-3 enzyme-linked immunosorbent assay according to the manufacturer's instructions (HyCult Biotechnology, Uden, The Netherlands). Aliquots (200 μl) of supernatant were acid extracted by the addition of an equal volume of 1 M HCl/1% trifluoroacetic acid overnight with mixing in the cold (23). The sample was centrifuged, and the supernatant was concentrated by vacuum evaporation and resuspended in distilled water equal to the starting sample volume. Acid-extracted saliva was used for immunoassay of LL37 and hBD3. LL37 was assayed by slot blot using an assay kit (Phoenix Pharmaceuticals Inc., Belmont, CA). hBD3 was assayed by slot blot using polyclonal antibody to hBD3 (Orbigen Inc., San Diego, CA). The peptide standard for hBD3 was from Peptides International, Inc. (Louisville, KY). Statistical analysis for association of AMP levels with caries score was done using the Kruskal-Wallis nonparametric test based on rank and designed for nonnormally distributed data.
Salivary bacterial analysis.
The levels of S. mutans and closely related streptococcal species in whole saliva were determined using DNA-DNA hybridization with radiolabeled rRNA probes (29). Briefly, 50 μl of cleared undiluted saliva and 100 μl of saliva serially diluted 10-fold (10−1 to 10-5) were applied to GeneScreenPlus membranes (Perkin-Elmer Life Sciences, Boston, MA) using a slot blot apparatus under vacuum. The membranes were prepared as previously described to precipitate the bacterial DNA in place and then hybridized with 32P-labeled oligonucleotide probes including SM010, SM002, and SSP001 for detection of bacterial ribosome RNA sequences encoded in DNA. The probes are specific for mutans streptococci (S. mutans and Streptococcus sobrinus), but not Streptococcus mitis, Streptococcus sanguis, or Streptococcus pneumoniae. Positive controls were included with each experiment. The number of the greatest dilution at which positive hybridization was detected reflects the salivary load of bacteria (mainly S. mutans) associated with caries. Data were analyzed using Pearson's chi-square test.
Genetic analysis.
Buccal swabs were collected and genomic DNA extracted using the Epicenter (Madison, WI) kit. The genomic DNA was purified with the QIAGEN (Valencia, CA) QIA Quick kit and used for single-nucleotide polymorphism (SNP) analysis. DEFB1 SNP analysis was performed by the TaqMan technique as previously described (14) and completed for 144 samples.
Immunohistochemistry.
Formalin-fixed sections were evaluated for expression of LL37 and HNP1-3 using the ABC technique (Vector Laboratories, Burlingame, CA). Briefly, sections were deparaffinized, rehydrated, and treated with antigen-unmasking solution (Vector Laboratories). Endogenous peroxide was blocked using 1% hydrogen peroxide/Tris-buffered saline for 30 min. Sections were blocked with appropriate sera and incubated with the primary antibody overnight before visualization with ABC reagents using 3,3′-diaminobenzidine as the substrate. Methyl green counterstain (KPL, Gaithersburg, MD) was used to visualize tissue morphology. The antibodies used were polyclonal rabbit anti-LL37 (Phoenix Pharmaceuticals, Inc., Belmont, CA) and monoclonal antibody clone D21 anti-HNP1-3 (Cell Sciences, Canton, MA). Histological sections of minor salivary glands were obtained from the Division of Oral Pathology, School of Dentistry, University of Washington, in accordance with Institutional Review Board procedures (29). Commercially available histological sections of human submandibular glands were from Spring Biosciences (Fremont, CA).
RESULTS
Demographics and caries experience.
Eighty-eight females and 61 males participated in the study. All children were between 11 and 15 years of age. Most of the population was Hispanic with some Native Americans and Caucasians. Overall, the children were healthy, with 92% having no history of major illness or disease. The most commonly reported medication was for asthma. One subject reported current use of an antibiotic. Oral examination showed that 80% of the children had permanent dentition, 20% had mixed dentition, 6% had missing teeth, and 11% had loose teeth. Sixty-five percent of the population reported having regular dental care. Gingivitis was noted in only a small number of subjects (less than five); one subject had a stainless steel crown and two subjects had orthodontic appliances. Fifty-three subjects (36%) had no decay; 37 (24%), 39 (27%), and 20 (13%) had caries scores of 1, 2, and 3 or greater, respectively.
Salivary analysis.
The median protein concentration of unstimulated saliva samples (n = 144) was 1,485 μg/ml (range from 421 to 7,052 μg/ml). The salivary protein concentration showed no correlation with age, gender, or caries score. This value agrees with previously reported total protein concentration for this age group (2). AMP concentrations were in the μg/ml range. AMP levels were also normalized to the protein concentration in whole saliva for each sample. Results are summarized in Table 1. HNP1-3, hBD3, and LL37 all showed extensive variation in concentration in our population, even when normalized to total salivary protein levels. Median values for HNP1-3, hBD3, and LL37 were 0.61, 0.31, and 3.07 μg/ml, respectively.
TABLE 1.
Salivary antimicrobial peptide levels
| Peptide | Sample no. | Concn (μg/ml)
|
Concn relative to total salivary protein (μg/mg salivary protein)
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Median | 25%-75% | Minimum | Maximum | Median | 25%-75% | Minimum | Maximum | ||
| HNP1-3 | 143 | 0.61 | 0.39-1.09 | 0.06 | 10.50 | 0.43 | 0.25-0.74 | 0.04 | 6.96 |
| LL37 | 127 | 3.07 | 1.72-4.83 | 0.12 | 12.0 | 2.29 | 1.05-3.15 | 0.07 | 25.33 |
| hBD3 | 133 | 0.31 | 0.10-0.89 | 0 | 6.21 | 0.23 | 0.06-0.68 | 0 | 4.36 |
| Total protein | 144 | 1,485 | 1,101-1,905 | 421 | 7,052 | ||||
Relationship of salivary AMPs and caries experience.
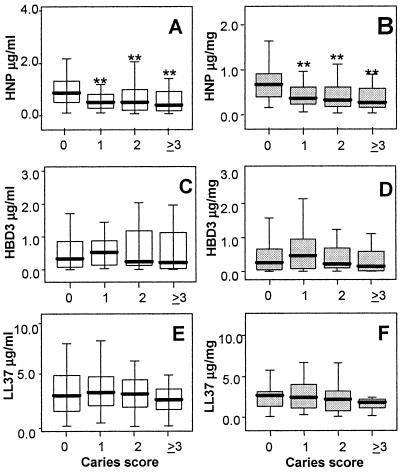
In order to evaluate the relationship of AMP expression and caries experience in the population, we used the Kruskal-Wallis nonparametric test based on rank. We found a significant difference in the level of HNP1-3 among different caries groups (P = 0.004). Differences were observed for both the median level of salivary HNP1-3 concentration (μg/ml) and salivary HNP1-3 relative to salivary protein (μg/mg). The median HNP1-3 concentration was 0.89 μg/ml, with an (interquartile range of 0.24 to 0.9) for the caries-free group (n = 51) and 0.5 μg/ml (interquartile range, 0.24 to 0.9) for all subjects with evidence of caries (n = 92). The HNP1-3 value relative to total salivary protein was 0.67 μg/mg protein (0.38 to 0.93) in the caries-free group and 0.33 μg/mg protein (0.19 to 0.59) in the combined caries group (P = 0.004) (Fig. 1A and B). Similar analysis for LL37 is shown in Fig. 1E and F. The results showed the same trend with higher levels of LL37 in the no caries group than in those with caries, but results were not statistically significant. hBD3 concentration in saliva and the level of hBD3 relative to protein showed no significant difference among the population or between the different caries groups (Fig. 1C and D).
FIG. 1.
AMP levels in saliva as a function of caries score. (A, C, and E) HNP1-3 (A), hBD3 (B), and LL37 (C) concentrations in saliva, expressed as μg/ml; (B, D, and F) HNP1-3, hBD3, and LL37 levels relative to salivary protein in μg/mg protein. The caries-free group showed significantly higher HNP1-3 concentration (A) than each of the groups with caries.(**, P < 0.01) and significantly higher concentration than the combined caries groups (P = 0.004). Also, the HNP1-3 level relative to salivary protein (B) in the caries-free group is significantly higher than each of the caries groups (**, P < 0.01) and the combined caries groups (P < 0.004). For LL37, even though no significant difference was found among the groups with or without caries, the LL37 level relative to salivary protein (F) shows a trend of decreasing level in higher caries score groups. There is no evidence of association of hBD3 with caries (C and D). Box plots show the median and 25 to 75% range, with error bars indicating 5% and 95% intervals.
Additional analyses showed that HNP1-3 concentration was positively correlated with total salivary protein (Spearman's rank correlation [r] = 0.239; P < 0.001). HNP1-3 concentration was also correlated with LL37 (r = 0.506, P < 0.001). As suggested above, HNP1-3 concentration was negatively correlated with caries score (r = −0.281), and the correlation is significant at the 0.001 level (P = 0.001). No correlation was found between salivary hBD3 level and any other variables.
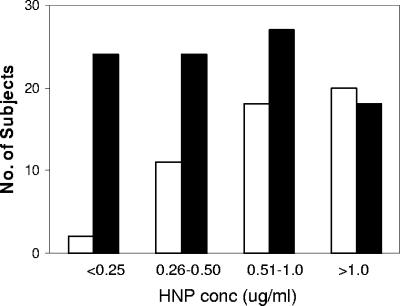
To further examine the relationship of HNP1-3 with caries, the HNP concentration range was evaluated in subjects with no caries compared to those with caries (Fig. 2). An increasing proportion of subjects had no caries as the HNP concentration increased; 90% of the subjects with HNP1-3 levels of ≤0.25 μg/ml (n = 26) had caries, but only 47% of the subjects with HNP1-3 levels greater than 1.0 μg/ml (n = 38) had caries.
FIG. 2.
HNP1-3 values and caries in the population. The number of subjects with no caries (open bars) compared to those with caries (filled bars) with HNP1-3 concentrations (μg/ml saliva) in the ranges indicated. Note that an increasing proportion of subjects had no caries as the HNP concentration increased.
Immunohistochemical localization of AMPs.
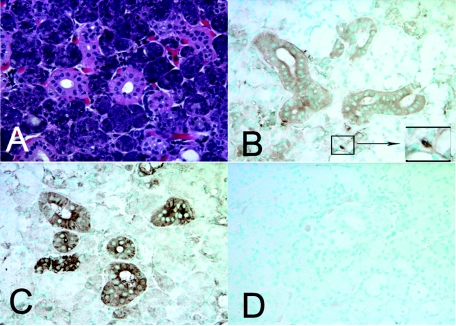
The presence of HNP1-3 in saliva, especially in some individuals with levels more than twice the median concentration, and the overall greater level in caries-free children, were unexpected. There was no evidence that those with high HNP1-3 levels had loose teeth, which might lead to the presence of increased blood and neutrophils in their saliva, which would be expected to contribute to elevated HNP1-3. To clarify the possible source of AMPs in saliva, immunohistochemistry was performed on previously biopsied minor salivary gland samples from adult subjects (unrelated to the subjects in the caries study) and in commercially available histological sections of human submandibular glands (Spring Biosciences, Fremont, CA). Positive immunohistochemical reaction for both HNP1-3 and LL37 was seen in duct cells in submandibular glands (Fig. 3B and C) and in minor salivary glands (not shown). As expected, neutrophils in these biopsy samples stained positively for ΗΝP1-3 and LL37. No staining for HNP1-3 was detected in the mucosal epithelium present in the same biopsy samples, while, in contrast, a strong reaction for LL37 was found in the mucosal epithelium (not shown).
FIG. 3.
AMP expression in submandibular salivary gland. Immunohistochemistry for HNP1-3 and LL37 shows positive reactions in duct cells of submandibular glands. (A) Hematoxylin and eosin stain. (B) HNP1-3 immunostaining with monoclonal antibody D21 was positive in salivary duct cells and neutrophils (inset). (C) LL37 immunostaining was also positive in duct cells. (D) Negative control without primary antibody. An isotype-specific control for the HNP1-3 monoclonal antibody was also negative (not shown). Original magnification, ×40.
Salivary S. mutans.
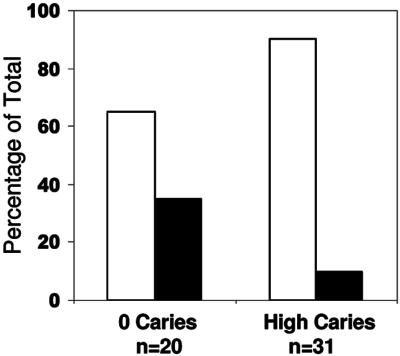
We next asked if salivary AMP peptide levels were associated with the presence of S. mutans in whole saliva. Fifty-one samples were assayed for the level of salivary mutans streptococci, 31 from the caries group (with caries score ≥2) and 20 from the caries-free group, using DNA-DNA hybridization with radiolabeled DNA probes for rRNA sequences (29). All samples gave a positive hybridization signal with the initial saliva sample (dilution, 1:2). Subjects in the caries group showed detectable salivary bacterial DNA at dilutions of 1:2, 1:10, and 1:100 in 90% of the subjects. In contrast, the caries-free group showed detectable bacterial DNA over a wide range of dilutions (1:2 to 1:100,000), with 65% only at the lower dilution, 1:2 to 1:100, levels (Fig. 4). Thus, the caries group generally had lower levels of salivary mutans streptococci than the caries-free group. The salivary bacterial level did not correlate with presence of active decay or total decay. Salivary concentrations of ΗΝP1-3, hBD3, and LL37 were analyzed as possible explanations for the variation in bacterial levels in saliva, but no correlation was detected with the AMP levels.
FIG. 4.
Salivary mutans streptococcus (MS) levels in no-caries and high-caries (caries score ≥ 2) children by DNA-DNA hybridization with radiolabeled rRNA probes. White bars indicate the percentages of the group with positive MS detectible in the range of 1:2 to 1:100 dilution. Black bars indicate percentages with MS detectible in the range of 1:1,000 to 1:10,000. In the high-caries group, a significantly higher percentage of subjects had detectible MS only at low salivary dilutions. Thus, 90% of children with high caries (compared to 65% of children with no caries) had low salivary MS bacterial levels. Pearson's chi-square test yielded a P of 0.001.
DEFB1 SNP analysis.
Finally, we examined a genetic SNP in the DEFB1 gene, encoding hBD1. hBD1 is a constitutively expressed β-defensin, although the expression of hBD1 mRNA is known to vary between individuals (17). The level of salivary hBD1 peptide was not determined due to the lack of sensitive immunoassay. However, two SNPs lie within the 5′ untranslated region of the mRNA that could alter the amount of peptide expressed. We previously showed that the DEFB1 SNP (−44 C→G) is associated with oral Candida levels (14), and this SNP is also associated with resistance to human immunodeficiency virus (HIV) infection in infants of HIV-positive mothers (4). We assayed the SNPs at positions −20 (A→G) and −44 (C→G) by a TaqMan assay. The frequencies of the −20 and −44 SNPs were 61% and 28%, in agreement with previous work (15). There was no significant association of either hBD1 SNP with caries experience. In addition, for samples in which the −20/−44 haplotype could be determined unambiguously (n = 108), there was no association of any haplotype with caries experience.
DISCUSSION
There are several new findings of this study. First, HNP1-3, LL37, and hBD3 are all detectable in saliva but show extensive variation in concentration between subjects. The concentration of AMPs in unstimulated saliva of children has not been previously reported, although healthy adults had a mean value of 0.8 μg/ml HNP1-3 (22). Second, salivary HNP1-3 is significantly greater in caries-free children than in those with caries. Third, HNP1-3 is detectable in salivary duct cells, suggesting that salivary glands express α-defensins as well as β-defensins and LL37. Finally, two other potential risk indicators, the hBD1 SNP, previously shown to be correlated with oral Candida load, and the salivary mutans streptococci level, do not correlate with caries experience. Further, salivary mutans streptococcus level is not correlated with the concentration of the AMPs tested.
Mutans streptococci are considered to be the main etiological agent for caries, and salivary levels have been used as an indictor of caries risk (27, 33, 37), although use of salivary mutans streptococci by itself as a risk factor for caries is controversial due to the multifactorial nature of the disease process (10, 16). Our results showing the tendency to lower levels of salivary mutans streptococci in children with caries may be a reflection of increased adherence of bacteria to the tooth surface or may be due to the presence of strains that are more adherent and/or more cariogenic (30). Many salivary protein components, such as proline-rich glycoprotein, mucins, immunoglobulins, agglutinin, lactoferrin, cystatins, and lysozyme, are thought to have a role in defense in the oral cavity (38). Numerous studies have investigated the correlation between these salivary proteins and glycoproteins and caries experience, but no studies have shown reliable association between a single salivary component and caries experience (16, 36).
The importance of antimicrobial peptides in innate immunity and oral health is now widely recognized. For instance, deficiencies in LL37 and in α-defensins are related to the occurrence of early-onset periodontal disease in the morbus Kostmann syndrome (28). The AMP levels found in saliva in this study are in the range of effective antimicrobial function, especially considering the low salt concentration in saliva and the synergistic action of the peptides (19, 24). Thus, the correlation of a salivary cationic AMP with caries experience suggests the possible protective effect of HNP1-3. Conversely, low levels of HNP1-3 may result in increased susceptibility to caries.
Within the oral cavity, LL37, and β-defensins are expressed in oral epithelial tissues including salivary glands (3, 5, 7, 23, 39). In addition, we show expression of HNP1-3 by immunohistochemistry in submandibular salivary duct cells. The presence of both HNP1-3 and LL37 in submandibular glands, which are the major source of unstimulated saliva, suggests that these cells may be a source of AMPs in saliva. An additional source for both of these peptides in saliva is the neutrophils that migrate into the oral cavity via gingival crevicular fluid. In healthy individuals it has been estimated that 30,000 neutrophils per minute enter the oral cavity via this route through the junctional epithelium surrounding the teeth (32). Our results suggest that both neutrophils and duct cells are a source of HNP1-3 and that α-defensins are part of the extensive antimicrobial armamentarium of saliva.
Salivary AMP concentrations showed large variation between individuals, with a significantly higher level of salivary α-defensins HNP1 to -3 in children with no caries. The salivary levels of HNP1 to -3 antimicrobial peptides may represent a genetically determined factor that contributes to caries susceptibility. The large variation in the concentration of α-defensins in saliva could be due to previously demonstrated polymorphisms in sequence and copy number in the genes encoding these peptides (12, 20), as well as to variations in neutrophil efflux via the gingival fluid.
The prediction of caries risk has been of long-standing interest and is very important for development of new preventative strategies for caries. This is especially significant for young children and for children with special health care needs. Saliva is an easily available fluid which can be collected noninvasively and used to measure and monitor the risk for caries (34). The assay for HNP1-3 is easy to perform and can be done in less than 0.5 ml crude whole saliva. Based on our study design, we cannot yet determine if the level of salivary α-defensin is predictive of future caries, but we have shown that children with caries have a significantly lower median value of α-defensins based on both the volume of saliva and relative to the concentration of total salivary protein. Thus, low salivary levels of α-defensins (HNP1 to -3) could be a new and useful measure of the risk for caries in children. Future studies could lead to development of means to enhance endogenous oral peptide expression, utilization of these peptides as therapeutics, and to a simple test for clinical evaluation of caries risk.
Acknowledgments
We acknowledge excellent technical assistance of Mai Bai, Albany Molecular Research, Inc., for the TaqMan assays; the help of Oscar Suarez, Dorja Wojtkowski, Andrea Pinnick, Helen Hoekman, and Jesus Jimenez with sample collection; and Nancy Chino, whose bilingual skills and dedication were important to the success of the project. We also thank Leonor de Maldonado, the school principal, and her staff for their cooperation with the study and the Yakima Valley Farmworker's clinic for use of mobile dental units. We acknowledge the UW Oral Health Collaborative and the Tooth Fairy Academy for their interest in education and dental health in rural Washington State.
This work was supported by grants from the National Institute of Dental and Craniofacial Research including a pilot project from the Comprehensive Center for Oral Health Research (P60 DE13061) and project support from the Northwest/Alaska Center for Research to Reduce Oral Health Disparities (U54 DE14254) and by an American Association of Dental Research student research fellowship (to M.T.T.). We thank Penelope Leggott, Paul Robertson, Murray Robinovitch, and Whasun O. Chung for critical reading of the manuscript and Lloyd Mancl for assistance with statistical analysis.
REFERENCES
- 1.Bals, R., and J. M. Wilson. 2003. Cathelicidins—a family of multifunctional antimicrobial peptides. Cell. Mol. Life Sci. 60:711-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ben-Aryeh, H., M. Fisher, R. Szargel, and D. Laufer. 1990. Composition of whole unstimulated saliva of healthy children: changes with age. Arch. Oral Biol. 35:929-931. [DOI] [PubMed] [Google Scholar]
- 3.Bonass, W. A., A. S. High, P. J. Owen, and D. A. Devine. 1999. Expression of beta-defensin genes by human salivary glands. Oral Microbiol. Immunol. 14:371-374. [DOI] [PubMed] [Google Scholar]
- 4.Braida, L., M. Boniotto, A. Pontillo, P. A. Tovo, A. Amoroso, and S. Crovella. 2004. A single-nucleotide polymorphism in the human beta-defensin 1 gene is associated with HIV-1 infection in Italian children. AIDS 18:1598-1600. [DOI] [PubMed] [Google Scholar]
- 5.Dale, B. A., and L. P. Fredericks. 2004. Antimicrobial peptides in the oral environment: expression and function in health and disease, p. 223-251. In R. L. Gallo (ed.), Antimicrobial peptides in human health and disease. Horizon Bioscience, Wymondham, United Kingdom.
- 6.Dale, B. A., J. R. Kimball, S. Krisanaprakornkit, F. Roberts, M. Robinovitch, R. O'Neal, E. V. Valore, T. Ganz, G. M. Anderson, and A. Weinberg. 2001. Localized antimicrobial peptide expression in human gingiva. J. Periodontal Res. 36:285-294. [DOI] [PubMed] [Google Scholar]
- 7.Dunsche, A., Y. Acil, H. Dommisch, R. Siebert, J. M. Schroder, and S. Jepsen. 2002. The novel human beta-defensin-3 is widely expressed in oral tissues. Eur. J. Oral Sci. 110:121-124. [DOI] [PubMed] [Google Scholar]
- 8.Ganz, T. 2003. Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 3:710-720. [DOI] [PubMed] [Google Scholar]
- 9.Ganz, T., M. E. Selsted, D. Szklarek, S. S. Harwig, K. Daher, D. F. Bainton, and R. I. Lehrer. 1985. Defensins. Natural peptide antibiotics of human neutrophils. J. Clin. Investig. 76:1427-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansel Petersson, G., S. Twetman, and D. Bratthall. 2002. Evaluation of a computer program for caries risk assessment in schoolchildren. Caries Res. 36:327-340. [DOI] [PubMed] [Google Scholar]
- 11.Hicks, J., F. Garcia-Godoy, and C. Flaitz. 2003. Biological factors in dental caries: role of saliva and dental plaque in the dynamic process of demineralization and remineralization (part 1). J. Clin. Pediatr. Dent. 28:47-52. [DOI] [PubMed] [Google Scholar]
- 12.Hollox, E. J., J. A. Armour, and J. C. Barber. 2003. Extensive normal copy number variation of a beta-defensin antimicrobial-gene cluster. Am. J. Hum. Genet. 73:591-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joly, S., C. Maze, P. B. McCray, Jr., and J. M. Guthmiller. 2004. Human β-defensins 2 and 3 demonstrate strain-selective activity against oral microorganisms. J. Clin. Microbiol. 42:1024-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jurevic, R. J., M. Bai, R. B. Chadwick, T. C. White, and B. A. Dale. 2003. Single-nucleotide polymorphisms (SNPs) in human β-defensin 1: high-throughput SNP assays and association with Candida carriage in type I diabetics and nondiabetic controls. J. Clin. Microbiol. 41:90-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jurevic, R. J., P. Chrisman, L. Mancl, R. Livingston, and B. A. Dale. 2002. Single-nucleotide polymorphisms and haplotype analysis in beta-defensin genes in different ethnic populations. Genet. Test. 6:261-269. [DOI] [PubMed] [Google Scholar]
- 16.Kirstila, V., P. Hakkinen, H. Jentsch, P. Vilja, and J. Tenovuo. 1998. Longitudinal analysis of the association of human salivary antimicrobial agents with caries increment and cariogenic microorganisms: a two-year cohort study. J. Dent. Res. 77:73-80. [DOI] [PubMed] [Google Scholar]
- 17.Krisanaprakornkit, S., A. Weinberg, C. N. Perez, and B. A. Dale. 1998. Expression of the peptide antibiotic human beta-defensin 1 in cultured gingival epithelial cells and gingival tissue. Infect. Immun. 66:4222-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loesche, W. J. 1986. Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 50:353-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maisetta, G., G. Batoni, S. Esin, F. Luperini, M. Pardini, D. Bottai, W. Florio, M. R. Giuca, M. Gabriele, and M. Campa. 2003. Activity of human β-defensin 3 alone or combined with other antimicrobial agents against oral bacteria. Antimicrob. Agents Chemother. 47:3349-3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mars, W. M., P. Patmasiriwat, T. Maity, V. Huff, M. M. Weil, and G. F. Saunders. 1995. Inheritance of unequal numbers of the genes encoding the human neutrophil defensins HP-1 and HP-3. J. Biol. Chem. 270:30371-30376. [DOI] [PubMed] [Google Scholar]
- 21.McKay, M. S., E. Olson, M. A. Hesla, A. Panyutich, T. Ganz, S. Perkins, and E. F. Rossomando. 1999. Immunomagnetic recovery of human neutrophil defensins from the human gingival crevice. Oral Microbiol. Immunol. 14:190-193. [DOI] [PubMed] [Google Scholar]
- 22.Mizukawa, N., K. Sugiyama, T. Ueno, K. Mishima, S. Takagi, and T. Sugahara. 1999. Levels of human defensin-1, an antimicrobial peptide, in saliva of patients with oral inflammation. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 87:539-543. [DOI] [PubMed] [Google Scholar]
- 23.Murakami, M., T. Ohtake, R. A. Dorschner, and R. L. Gallo. 2002. Cathelicidin antimicrobial peptides are expressed in salivary glands and saliva. J. Dent. Res. 81:845-850. [DOI] [PubMed] [Google Scholar]
- 24.Nagaoka, I., S. Hirota, S. Yomogida, A. Ohwada, and M. Hirata. 2000. Synergistic actions of antibacterial neutrophil defensins and cathelicidins. Inflamm. Res. 49:73-79. [DOI] [PubMed] [Google Scholar]
- 25.Nishimura, E., A. Eto, M. Kato, S. Hashizume, S. Imai, T. Nisizawa, and N. Hanada. 2004. Oral streptococci exhibit diverse susceptibility to human beta-defensin-2: antimicrobial effects of hBD-2 on oral streptococci. Curr. Microbiol. 48:85-87. [DOI] [PubMed] [Google Scholar]
- 26.Oppenheim, F. G., T. Xu, F. M. McMillian, S. M. Levitz, R. D. Diamond, G. D. Offner, and R. F. Troxler. 1988. Histatins, a novel family of histidine-rich proteins in human parotid secretion. Isolation, characterization, primary structure, and fungistatic effects on Candida albicans. J. Biol. Chem. 263:7472-7477. [PubMed] [Google Scholar]
- 27.Powell, L. V. 1998. Caries prediction: a review of the literature. Community Dent. Oral Epidemiol. 26:361-371. [DOI] [PubMed] [Google Scholar]
- 28.Putsep, K., G. Carlsson, H. G. Boman, and M. Andersson. 2002. Deficiency of antibacterial peptides in patients with morbus Kostmann: an observation study. Lancet 360:1144-1149. [DOI] [PubMed] [Google Scholar]
- 29.Roberts, M. C., C. A. Riedy, S. E. Coldwell, S. Nagahama, K. Judge, M. Lam, T. Kaakko, J. L. Castillo, and P. Milgrom. 2002. How xylitol-containing products affect cariogenic bacteria. J. Am. Dent. Assoc. 133:435-441. [DOI] [PubMed] [Google Scholar]
- 30.Rudney, J. D., and R. K. Staikov. 2002. Simultaneous measurement of the viability, aggregation, and live and dead adherence of Streptococcus crista, Streptococcus mutans and Actinobacillus actinomycetemcomitans in human saliva in relation to indices of caries, dental plaque and periodontal disease. Arch. Oral Biol. 47:347-359. [DOI] [PubMed] [Google Scholar]
- 31.Sahasrabudhe, K. S., J. R. Kimball, T. H. Morton, A. Weinberg, and B. A. Dale. 2000. Expression of the antimicrobial peptide, human beta-defensin 1, in duct cells of minor salivary glands and detection in saliva. J. Dent. Res. 79:1669-1674. [DOI] [PubMed] [Google Scholar]
- 32.Schroeder, H. E. 1986. The periodontium. Springer-Verlag, Berlin, Germany.
- 33.Splieth, C., and O. Bernhardt. 1999. Prediction of caries development for molar fissures with semiquantitative mutans streptococci test. Eur. J. Oral Sci. 107:164-169. [DOI] [PubMed] [Google Scholar]
- 34.Streckfus, C. F., and L. R. Bigler. 2002. Saliva as a diagnostic fluid. Oral Dis. 8:69-76. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka, D., K. T. Miyasaki, and R. I. Lehrer. 2000. Sensitivity of Actinobacillus actinomycetemcomitans and Capnocytophaga spp. to the bactericidal action of LL-37: a cathelicidin found in human leukocytes and epithelium. Oral Microbiol. Immunol. 15:226-231. [DOI] [PubMed] [Google Scholar]
- 36.Tenovuo, J., H. Jentsch, T. Soukka, and L. Karhuvaara. 1992. Antimicrobial factors of saliva in relation to dental caries and salivary levels of mutans streptococci. J. Biol. Buccale 20:85-90. [PubMed] [Google Scholar]
- 37.Thibodeau, E. A., and D. M. O'Sullivan. 1999. Salivary mutans streptococci and caries development in the primary and mixed dentitions of children. Community Dent. Oral Epidemiol. 27:406-412. [DOI] [PubMed] [Google Scholar]
- 38.Van Nieuw Amerongen, A., J. G. Bolscher, and E. C. Veerman. 2004. Salivary proteins: protective and diagnostic value in cariology? Caries Res. 38:247-253. [DOI] [PubMed] [Google Scholar]
- 39.Woo, J. S., J. Y. Jeong, Y. J. Hwang, S. W. Chae, S. J. Hwang, and H. M. Lee. 2003. Expression of cathelicidin in human salivary glands. Arch. Otolaryngol. Head Neck Surg. 129:211-214. [DOI] [PubMed] [Google Scholar]
- 40.Yang, D., A. Biragyn, D. M. Hoover, J. Lubkowski, and J. J. Oppenheim. 2004. Multiple roles of antimicrobial defensins, cathelicidins, and eosinophil-derived neurotoxin in host defense. Annu. Rev. Immunol. 22:181-215. [DOI] [PubMed] [Google Scholar]
- 41.Zasloff, M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389-395. [DOI] [PubMed] [Google Scholar]
- 42.Zhao, C., I. Wang, and R. I. Lehrer. 1996. Widespread expression of beta-defensin hBD-1 in human secretory glands and epithelial cells. FEBS Lett. 396:319-322. [DOI] [PubMed] [Google Scholar]