Abstract
Substitution of leucine for isoleucine at residue 50 (I50L) of human immunodeficiency virus (HIV) protease is the signature substitution for atazanavir (ATV) resistance. A unique phenotypic profile has been associated with viruses containing the I50L substitution, which produces ATV-specific resistance and increased susceptibility to most other approved HIV protease inhibitors (PIs). The basis for this unique phenotype has not been clearly elucidated. In this report, a direct effect of I50L on the susceptibility to the PI class is described. Cell-based protease assays using wild-type and PI-resistant proteases from laboratory and clinical isolates and in vitro antiviral assays were used to demonstrate a strong concordance between changes in PI susceptibility at the level of protease inhibition and changes in susceptibility observed at the level of virus infection. The results show that the induction of ATV resistance and increased susceptibility to other PIs by the I50L substitution is likely determined at the level of protease inhibition. Moreover, the I50L substitution functions to increase PI susceptibility even in the presence of other primary and secondary PI resistance substitutions. These findings may have implications regarding the optimal sequencing of PI therapies necessary to preserve PI treatment options of patients with ATV-resistant HIV infections.
Human immunodeficiency virus (HIV) protease inhibitors (PIs) are used in combination with other antiretroviral agents to treat HIV infections. Such combination therapies, known as highly active antiretroviral therapy (HAART), create enormous benefit for patients by suppressing HIV replication and delaying the progression of HIV disease (12, 13). Due to selective pressure exerted by the antiviral action of HAART, viruses that are resistant to inhibition by the antiviral agents can emerge over time (46). This decrease in susceptibility of circulating virus, or phenotypic resistance, leads to virologic rebound and treatment failure.
Phenotypic resistance to HIV PIs is typically associated with genotypic changes in the HIV protease and the substitution of key amino acid residues that alter PI susceptibility (19, 44). The protease is required during infection to carry out specific cleavages of the gag and gag-pol polyproteins (17), producing the mature structural proteins that make up viral capsids and the enzymes required for genome amplification (8, 18, 36). Key amino acid substitutions in PI-resistant proteases, known as primary substitutions, interfere with the binding of drug molecules within the protease active site (4, 7, 9, 14, 16, 21, 24, 32, 33, 40). These substitutions can also reduce the binding of natural substrates and, consequently, reduce the overall cleavage efficiency of the viral protease (7, 25, 39, 47, 50). This in turn may lead to impaired replication of the PI-resistant virus (7, 10, 27, 28, 50, 51). Additional secondary amino acid substitutions in the protease can occur which compensate for the deleterious effects of primary substitutions, thereby improving the replicative fitness of the drug-resistant virus (10, 28). Secondary substitutions do not impart resistance on their own, typically, but they may alter the resistance associated with primary substitutions or restore viability (5, 7, 42). Other compensatory substitutions can occur at the protease cleavage sites, allowing for improved processing by the drug-resistant protease (3, 9, 22, 25, 41, 51). The cumulative result of the different types of genotypic changes in PI-resistant HIV is an enhanced ability to replicate in the presence of drug concentrations that would normally suppress wild-type virus replication.
The development of phenotypic resistance can be monitored using in vitro tissue culture assays to measure antiviral susceptibility. These in vitro systems can also reveal important details about the mechanism by which phenotypic resistance emerges and about the potential consequences of resistance development that may ultimately help guide strategies for therapeutic intervention. In many instances the substitutions that cause resistance to one PI confer some level of cross-resistance to other PIs (2, 11, 15, 23, 29, 38, 45, 47). Less commonly, signature substitutions for specific PIs may arise that cause specific resistance to a given PI with little or no effect on the susceptibility to other PIs, such as the D30N substitution associated with nelfinavir resistance (34). The emergence of cross-resistance can confound the selection of drugs used to treat HIV infections and may result in fewer treatment options for the patient if PI therapies are not sequenced optimally. So understanding the consequences of resistance development for each PI is important with respect to potential treatment alternatives and successful outcomes for patients.
Atazanavir (ATV) is a potent, once-daily HIV PI approved for the treatment of HIV type 1 (HIV-1) infections. Evaluation of the in vitro drug susceptibilities of a large panel of ATV-naïve HIV clinical virus isolates revealed a distinct resistance profile relative to other PIs (6). Characterization of virus isolates from PI-naïve patients who failed ATV therapy led to the identification of a signature substitution of leucine for isoleucine at residue 50 (I50L) of the protease in 100% of ATV-resistant isolates (5). The same substitution emerged in ∼30% of PI-experienced patients who failed ATV therapy. Viruses containing the I50L substitution exhibited a unique phenotype of ATV-specific resistance coupled with significantly increased susceptibilities to other PIs. This occurred in clinical virus isolates with diverse genetic backgrounds, as well as in engineered laboratory strains designed to mimic ATV-resistant HIV (5). It was also found that the I50L substitution significantly impaired viral replication kinetics. The clinical significance of these observations remains to be elucidated, but the emergence of I50L-containing viruses during ATV therapy may, at a minimum, preserve susceptibility to other PIs and retain future PI treatment options. With the clear demonstration of increased susceptibilities to other PIs demonstrated at the virus level, the aim of the current study was to further understand the underlying mechanism(s) involved and determine if HIV-1 protease processivity played a role in this distinctive resistance profile.
MATERIALS AND METHODS
Expression vectors and provirus constructs.
To construct a gag-pol expression vector, a 3,460-bp region of HIV (NL4-3) DNA spanning the gag/gag-pol initiation codon to the end of the reverse transcriptase domain was amplified from a proviral clone using the primers 5′-ATCGCGGGATCCGCCACCATGGGTGCGAGAGCGTCGG-3′ and 5′-ATCCGGTTAACTTATAGTACTTTCCTGATTCCAGCAC-3′, which incorporated BamHI and EcoRV restriction sites (underlined), a consensus Kozak sequence, and a stop codon directly after the natural cleavage site between reverse transcriptase and integrase. Primers were obtained from Sigma Genosys (The Woodlands, TX). The BamHI-EcoRV fragment was cloned downstream of the T7 promoter in pcDNA3.1+neo (Invitrogen, Carlsbad, CA). HIV-1 NL4-3 proviral clones were obtained through the AIDS Research and Reference Reagent Program, National Institutes of Health. Recombinant HIV-1 strains with defined substitutions in the protease gene were generated by QuikChange site-directed mutagenesis as recommended by the manufacturer (Stratagene, La Jolla, CA). The chimeric NL4-3 100B proviral clone (5) was constructed by replacing the ApaI/SbfI fragment of the pNL4-3 clone with the corresponding fragment generated by reverse transcription-PCR from a patient virus isolate. The DNA sequence of all constructs was determined by the BMS Applied Genomics Sequencing Facility (Wallingford, CT).
Cells and viruses.
BHK-21 cells were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, and antibiotics. A viral T7 RNA polymerase expression vector, MVA-T7 (modified vaccinia virus Ankara strain expressing T7 RNA polymerase, provided by Bernard Moss) (48), was propagated in BHK-21 cells and used in cell-based assays at a multiplicity of infection of 3.5 to 5 PFU per cell, prior to transfection with gag-pol expression vectors (see below). HEK 293T and MT-2 cells were obtained from the NIH AIDS Research and Reference Reagent Program. The 293T cells were propagated in DMEM supplemented with 10% heat-inactivated FBS, 10 mM HEPES buffer (pH 7.55), 2 mM l-glutamine, 100 units/ml penicillin G, 100 units/ml streptomycin, and 0.25 μg/ml amphotericin B. MT-2 cells were cultured in RPMI 1640 medium supplemented with 10% FBS, 2 mM l-glutamine, and 10 mM HEPES buffer. HIV-1 stocks were generated by transient transfection of proviral DNA into 293T cells using Lipofectamine reagent (Gibco BRL). The viruses were harvested 72 h posttransfection and amplified in MT-2 cells, and titers were determined in the same cells using a virus yield assay. For antiviral assays, MT-2 cells were infected with recombinant viruses at a multiplicity of infection of 0.005 50% tissue culture infective doses per cell. At 5 days 20 μl of culture medium was added to 40 μl of reaction cocktail {62.5 mM Tris HCl (pH 7.8), 100 mM KCl, 0.0625% NP-40, 2.5 mM dithiothreitol, 6.25 mM MgCl2, 1 mM EGTA, 6.25 μg/ml poly(rA), streptavidin SPA beads with bound biotinylated dT18, and 1 μCi [3H]TTP } and incubated at 37°C for 1.5 h. The reactions were performed in white opaque plates and quenched with 100 μl of EDTA per reaction. The SPA beads were allowed to settle for 1 h prior to being counted on a Trilux (Wallac) plate reader. Susceptibility of viruses to test compounds was determined by incubation in the presence of serial dilutions of the compound. The 50% effective concentration (EC50) was calculated by using the exponential form of the median effect equation where (Fa) = 1/[1 + (EC50/drug concentration)m].
Cell-based p24 processing assay.
BHK-21 cells seeded in 96-well culture plates at 2 × 104 to 3 × 104 cells/well were infected with MVA-T7 in 25 μl DMEM-2% FBS using a multiplicity of infection of 3.5 to 5 PFU per cell 1.5 h prior to transfection. Each well was transfected with 100 ng of a T7-gag-pol expression plasmid using Lipofectamine reagent and Plus reagent (Invitrogen) in a total volume of 62.5 μl, according to the manufacturer's instructions. The liposome-DNA mixture was left on cells for 5 to 6 h, after which culture supernatants were replaced with 70 μl DMEM plus 2% FBS and incubated for an additional 12 to 16 h. Inhibition studies were conducted with PIs serially diluted in the same medium containing 0.2% dimethyl sulfoxide. Cell monolayers were lysed in 12.5 μl/well Tris-buffered saline (TBS)-0.1% sodium dodecyl sulfate-1% NP-40-5 mM EDTA supplemented with 0.1 to 0.2 U/μl Benzonase nuclease (Novagen) and incubated for 5 to 10 min at 37°C prior to addition of 12.5 μl 2× Laemmli sample buffer. Samples were stored at −20°C in sealed PCR plates.
Western blots and protease 50% inhibitory concentration (IC50) determinations.
Dose-response measurements were performed by monitoring p24 accumulation in Western blots. Cell lysates were separated in 4 to 20% Tris-glycine gels (Invitrogen) and then transferred to immunoblot polyvinylidene difluoride membranes (Bio-Rad) in 1× Tris-glycine buffer (Bio-Rad)/20% methanol. Membranes were incubated overnight at 4°C with 5% Carnation nonfat dry milk in 1× TBS to block nonspecific binding. Blocked membranes were washed twice in 1× TBS-0.5% Tween (TBS-T) and then incubated with anti-p24 monoclonal antibody (MAb; Perkin-Elmer; NEA-9306) in TBS-T-1% bovine serum albumin for 1 h at room temperature. After four washes in 1× TBS-T, the blots were incubated with alkaline phosphatase-conjugated secondary antibodies (Bio-Rad) in TBS-T-1% bovine serum albumin for 1 h at room temperature. After four additional washes in 1× TBS-T, alkaline phosphatase was detected by placing the washed blots protein side down onto 0.2 ml enhanced chemifluorescent substrate (Amersham Pharmacia Biotech) and scanning them immediately using a Molecular Dynamics Storm 860 imager (Amersham Bioscience Corp., Piscataway, NJ). Western blot images were analyzed using ImageQuant 5.0 (Molecular Dynamics) to determine the level of p24/p25 as a fraction of total immunoreactive p24-related proteins.
Selection of lamivudine-resistant HIV using I50L-containing viruses.
MT-2 cells (5 × 105/ml) were incubated with the NL4-3 recombinant 100B (genotype I50L, L63P, K70R, A71V, and G73S virus) (5) at a multiplicity of infection of 0.1 for 90 min. Following virus adsorption, infected cells were washed free of the virus inoculum and resuspended in culture medium in the presence or absence (control) of lamivudine at a concentration two times the EC50. The lamivudine concentration was increased over sequential passages performed when cells exhibited significant cytopathic effects. Typically, 0.5 ml of infected culture supernatant was transferred to fresh MT-2 cells at a 1:5 (vol/vol) ratio every 4 to 6 days. The infected culture supernatant was collected and stored at −80°C for drug susceptibility testing and genome sequence analysis (reverse transcriptase and protease genes). Control cultures lacking lamivudine were maintained in parallel under conditions identical to those for the lamivudine-containing cultures.
RESULTS
gag processing assay.
Previous studies demonstrated that HIV-1 isolates with a protease containing an I50L substitution exhibited ATV-specific resistance, increased susceptibility to other PIs, and impaired viral replication (5). Enhanced susceptibility to nearly an entire class of antiretroviral agents is somewhat surprising, but since the equivalent increases are not observed when I50L-containing viruses are assayed for susceptibilities to other classes of antiretrovirals (unpublished data), this characteristic is not likely due to impaired growth properties of I50L-containing viruses.
To further investigate the role of the protease itself in PI susceptibility when an I50L substitution is present, a cell-based assay for HIV protease was developed. This assay monitored the proteolytic processing of p55 gag, the natural substrate of HIV protease, when T7 gag-pol expression plasmids were transfected into cells infected with a vaccinia virus (Ankara) derivative, MVA-T7, expressing T7 RNA polymerase (48). The transient cytosolic expression of both p55 gag and the gag-pol protease precursor inside cells resulted in specific cleavages of p55. These cleavages were monitored by assays with Western blots probed with a p24-specific MAb and quantified by using enhanced chemifluorescence detection.
Lysates of cells expressing the wild-type (NL4-3) gag-pol gene were used to reveal the time-dependent processing of intracellular p55 gag. After the initial accumulation of full-length p55 gag, Western blots indicated decreasing levels of p55 and increasing accumulation of p24 up to 24 h posttransfection (Fig. 1A). In addition to p24, the predicted cleavage intermediates p41 and p25 were also detected. The identification of the expected cleavage products indicated that p55 gag was cleaved at the appropriate primary (SP1/NC), secondary (MA/CA), and tertiary (CA/SP1) cleavage sites. Alteration of the catalytic aspartic acid residue at position 25 in the protease open reading frame to asparagine (D25N) resulted in the accumulation of intact p55 gag and of the gag-pol precursor, at much lower levels than gag, and eliminated the accumulation of all p24-related cleavage products (data not shown). These observations demonstrated a requirement for HIV protease activity and were consistent with the expression of both gag and gag-pol (the latter at reduced levels through ribosomal frameshifting), resulting in the enzymatic activation of HIV protease and the authentic processing of p55 gag.
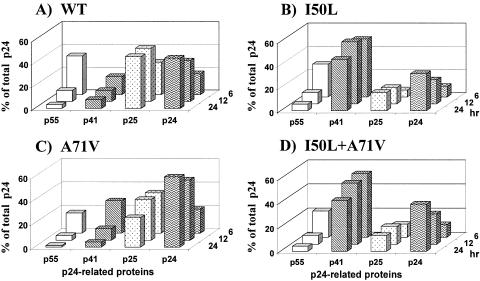
FIG. 1.
gag processing monitored by p24 Western blotting. The accumulation of p55 (open), p41 (hatched), p25 (stippled), and p24 (wavy) is plotted as a percentage of total p24-related protein. Cells were transfected with NL4-3 gag-pol expression vectors, and replicate samples were harvested after 6, 12, and 24 h. Western blots of total cell lysates were probed with anti-p24 MAb, followed by quantitation of individual protein bands, as described in Materials and Methods. (A) Wild-type (WT) gag-pol; (B) I50L gag-pol; (C) A71V gag-pol; (D) I50L+A71V gag-pol.
The I50L substitution was introduced into the gag-pol expression vector to determine its effect on protease cleavage. The A71V secondary substitution was also introduced, alone and in combination with I50L, because it is observed in conjunction with I50L in ∼50% of proteases from ATV-resistant HIV-1 isolates and has been reported to enhance the I50L phenotype (5). The individual I50L and A71V single mutants (Fig. 1B and C, respectively) and the I50L/A71V double mutant (Fig. 1D) each gave rise to the same cleavage products observed with wild-type protease. The I50L-containing mutants both cleaved the majority of p55 in the cells but produced a higher proportion of the cleavage intermediate p41, suggesting that the secondary cleavage step (MA/CA) may be impaired for these mutants. The I50L substitution resulted in a 50% reduction in the accumulation of total p24/p25 cleavage products in both the presence and absence of the A71V substitution (Fig. 1B and D).
PI susceptibility of I50L/A71V, I50L, and A71V proteases.
The addition of HIV PIs to the media of cell cultures expressing HIV gag-pol revealed dose-dependent inhibition of gag processing, indicated by the decreasing accumulation of p24/p25 and the increasing accumulation of unprocessed p55 gag with increasing drug concentrations. To derive IC50s for protease inhibition, the relative levels of the p24/25 proteins, expressed as the fraction of total p24-related proteins, were quantified and plotted as a function of drug concentration. Dose-response curves were generated with wild-type, I50L, A71V, and I50L/A71V proteases for a panel of marketed HIV PIs that included ATV, amprenavir (APV), indinavir (IDV), lopinavir (LPV), nelfinavir (NFV), ritonavir (RTV), and saquinavir (SQV). The wild-type protease was susceptible to inhibition by added PIs and exhibited IC50s in the range expected for each PI (Fig. 2).
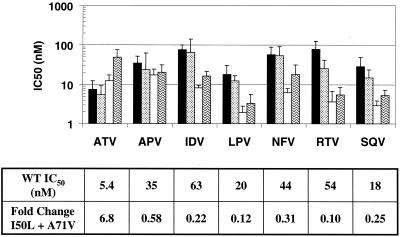
FIG. 2.
PI susceptibilities of I50L-containing proteases. IC50s were measured by Western blotting of total cell lysates for wild-type (WT) and ATV-resistant proteases after transfection of gag-pol expression vectors: wild-type NL4-3 (black), A71V alone (stippled), I50L alone (white), and I50L+A71V (wavy). The attached table shows IC50s for wild-type protease of each inhibitor along with the change in susceptibility for the I50L+A71V protease, relative to wild type (n = 10).
Introduction of the I50L substitution alone led to a twofold decrease in susceptibility to ATV and significant increases in the susceptibility to five of the six other marketed PIs. The A71V substitution alone had a modest and uniform effect on PI susceptibilities. The combination of I50L and A71V substitutions led to a further increase in resistance to ATV (to sevenfold) while maintaining the increased susceptibility to other PIs (Fig. 2). The susceptibilities of RTV and LPV were most dramatically affected by the I50L+A71V double substitution, with an average increase in susceptibility of 10-fold and 8-fold, respectively, relative to wild type. The susceptibilities to IDV and SQV were increased an average of four- to sixfold, while the increase in susceptibility to NFV was less dramatic (threefold). The susceptibility of protease to APV was increased only modestly, if at all, by the I50L+A71V substitutions, a result that is very similar to what has been reported with clinical virus isolates (5).
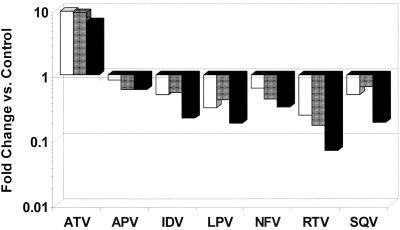
The reduced susceptibilities to ATV of I50L and I50L+A71V substituted proteases shown in Fig. 2 (twofold and sevenfold, respectively) were quite similar in magnitude to changes in antiviral susceptibility reported previously when these substitutions were introduced into the isogenic NL4-3 virus strain (5). Comparison of the relative PI susceptibilities of the I50L+A71V protease with the corresponding virus susceptibilities, both in the isogenic NL4-3 background and in genetically more diverse clinical isolates (also reported in reference 5), shows that changes in protease susceptibilities closely mirror the changes in antiviral susceptibilities (Fig. 3). These results suggest that phenotypic changes for HIV isolates with the I50L+A71V substitutions, including both ATV-specific resistance and broadly increased susceptibility to other PIs, are exhibited at the level of enzyme susceptibility to each PI.
FIG. 3.
Relative PI susceptibility comparison of I50L+A71V protease and viruses. Fold changes in PI susceptibility for clinical virus isolates (white) and HIV NL4-3 virus (shaded) with I50L+A71V substitutions (both from reference 5) plotted with changes for I50L+A71V protease (black) from Table 1.
I50L increases the susceptibility of engineered PI-resistant HIV proteases.
Combinations of primary PI resistance substitutions sometimes cause increased levels of PI resistance beyond that observed with either change alone (3, 30, 47). Given that the I50L resistance substitution conferred increased susceptibility to most PIs, it was of interest to determine whether this effect could be manifested in the presence of other primary resistance substitutions. For this, individual protease variants were generated with each of the prevalent primary PI substitutions D30N, M46L, G48V, V82A, I84V, and L90M commonly associated with PI resistance. Each substitution was introduced alone, in combination with A71V, and in combination with I50L+A71V. All of the mutant proteases gave rise to the same cleavage intermediates as wild-type protease and cleaved >85% of the available p55 by 24 h, with one exception. The D30N protease cleaved only 60% of p55 by 24 h, indicating that the D30N substitution impaired proteolytic processing (data not shown). This impairment led to a three- to fivefold reduction in the accumulation of p24/25, relative to wild-type protease, in the presence and absence of the A71V substitution. The remaining substitutions all reduced the accumulation of p24/25 by less than twofold, either alone or combined with A71V, with the M46L and L90M proteases producing wild-type levels of cleavage products (data not shown). As described earlier, the I50L substitution caused a twofold reduction in the accumulation of p24/25 in both the wild-type and A71V protease backgrounds. The I50L substitution also reduced the level of p24/25 accumulation by twofold when combined with A71V and each of the other primary resistance substitutions, clearly indicating that this facet of the I50L phenotype was manifested in the presence of other primary substitutions.
Each of the single (primary substitution), double (primary substitution + A71V), and triple (primary substitution + A71V + I50L) mutant proteases were subsequently tested for inhibition using the panel of marketed PIs. The effects of adding A71V to primary resistance substitutions were generally small in magnitude (<2.5-fold) and showed no consistent trends, increasing susceptibility slightly in some cases and decreasing them slightly in others (data not shown). For the double mutants, there were nine instances in which PI susceptibility was reduced by 2.5-fold or greater: D30N with NFV, M46L with ATV and IDV, G48V with SQV, I84V with APV, ATV and IDV, and L90M with SQV and IDV (Fig. 4, white bars), and these reductions corresponded generally to the effects on antiviral susceptibility reported previously for each of the primary substitutions (1, 44). The V82A substitution with A71V had only modest effects on all PIs tested, reducing susceptibility of no PI by more than 2.5-fold. These results for the other primary substitutions were in contrast to the effect of the I50L substitution, described above (Fig. 2).
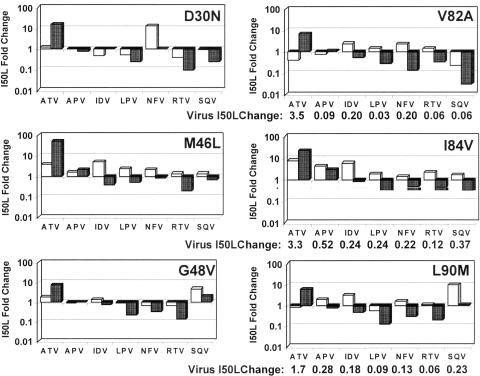
FIG. 4.
PI susceptibilities of proteases containing primary resistance substitutions with and without I50L. PI susceptibilities (IC50 of mutant/IC50 of wild type) were measured using Western blotting of total cell lysates after transfection of gag-pol expression vectors (n = 3) and plotted for primary resistance substitutions + A71V (double mutants, white bars) and for primary resistance substitutions + I50L/A71V (triple mutants, shaded bars). Parallel dose-response experiments using infectious virus mutants with V82A, I84V, and L90M substitutions (+A71V) with and without I50L were conducted. The I50L fold change for each virus (EC50 of triple mutant/EC50 of double mutant) was calculated. These values are shown on the line below the plots for the corresponding proteases (virus I50L change).
Addition of an I50L substitution to each double mutant protease had two major effects on PI susceptibility. First, as expected, it reduced susceptibility to ATV in all six PI-resistant protease backgrounds (Fig. 4, shaded bars). Second, the I50L substitution dramatically increased the susceptibility of PI-resistant proteases to most of the other PIs. Susceptibility to APV again appeared to be the exception, as APV inhibition was only modestly impacted by the presence of an I50L substitution, if at all, and trended neither higher or lower. Excluding ATV and APV, the addition of an I50L substitution led to increased protease susceptibility in 29 out of 30 cases (five drugs versus six proteases) involving IDV, LPV, NFV, RTV, and SQV. While the sizes of these increases were not all of significant magnitude (i.e., >2.5-fold), they did show a clear and broad trend toward increased PI susceptibility associated with the I50L substitution.
The greatest increases in susceptibility were again observed with LPV and RTV, with average increases of fourfold and fivefold, respectively, across all protease backgrounds. Increases in susceptibility to NFV and SQV were also observed consistently, though increases for the M46L and G48V backgrounds were of smaller magnitude (2- to 2.5-fold). Significant increases in susceptibility to IDV were observed in four of the PI-resistant protease backgrounds, but not in the presence of G48V or D30N. Remarkably, in six of the seven instances where primary substitutions conferred ≥2.5-fold resistance to one or more PIs (excluding ATV), the addition of I50L reversed the presence of PI resistance, increasing susceptibility to the level of wild-type protease with an average increase of sixfold. The lone exception was APV resistance associated with an I84V substitution, for which susceptibility was only marginally increased (Fig. 4).
To examine the effects of the I50L substitution on virus susceptibility in the presence of other primary resistance substitutions, three pairs of double and triple mutant PI-resistant proteases (V82A, I84V, and L90M) were transferred to the cloned NL4-3 provirus, as described in Materials and Methods. Dose-response studies with these mutant viruses showed that the primary substitutions caused significant reductions in PI susceptibility, relative to wild-type NL4-3 virus, and that, as in the protease assay, addition of the I50L substitution to each mutant backbone produced a broad trend toward increased susceptibility to other PIs. In all of 15 virus susceptibility comparisons (three viruses with five PIs: IDV, LPV, NFV, RTV, and SQV), addition of the I50L substitution increased PI susceptibility of the virus (Fig. 4, virus I50L change). Similar to the observations made using the protease assay, antiviral resistance to PIs caused by other primary resistance substitutions was eliminated by the addition of the I50L substitution in eight of eight cases. Not surprisingly, introduction of an I50L substitution also coincided with decreased susceptibility to ATV in all three mutant viruses.
I50L increases susceptibility of proteases from PI-resistant clinical isolates.
The results above describe the impact of I50L in the context of an engineered laboratory strain containing only primary PI resistance substitutions and A71V. The protease analysis was extended to determine whether these results are truly predictive of what would be expected of I50L variants from patients failing treatment with ATV-containing drug combinations. In the clinical setting, while the I50L substitution is characteristic of ATV resistance in treatment-naïve patients, it has also emerged in PI-experienced subjects whose virus isolates have other primary and secondary resistance substitutions prior to treatment with ATV. To determine whether these diverse backbones could influence expression of the I50L phenotype, the protease regions from three paired baseline and on-treatment patient samples from ATV clinical trials were cloned into the gag-pol expression vector. Individual clones were then identified with matching genotypes except for the absence and presence of I50L in the baseline and on-treatment samples, respectively. Due to prior exposure to PIs at study entry, each of these isolates had multiple backbone substitutions known to affect resistance to PIs, including one or two primary PI resistance substitutions (e.g., D30N, M46I, V82A, and L90M) in addition to 8 to 11 secondary substitutions (Table 1).
TABLE 1.
Impact of I50L substitution on protease susceptibility from patient isolates before and after treatment with ATVa
| Patient protease backbone | I50L Change
|
||||||
|---|---|---|---|---|---|---|---|
| ATV | APV | IDV | LPV | NFV | RTV | SQV | |
| Q7E, L10I, K14R, D30N, L38I, M46I, Q61E, L63P, I72V, V77I, N88D, I93L | 7.4 | 0.65 | 1.2 | 0.26 | 0.24 | 0.15 | 0.41 |
| T12D, K14R, V32I, K43T, M46I, I47V, I62V, L63P, V82A, I93L | 4.3 | 0.50 | 0.14 | 0.83 | 0.66 | 0.33 | 0.24 |
| L10I, T12P, K14R, K20I, Q61E, I62V, L63P, I64L, A71V, T74P, L90M, I93L | 16 | 1.4 | 0.27 | 0.36 | 0.69 | 0.23 | 0.33 |
I50L change (IC50 on treatment [with I50L]/IC50 baseline [no I50L]) was calculated after measuring protease susceptibility by Western blot assays of total cell lysates after transfection of gag-pol expression vectors (n = 3). Primary resistance substitutions are underlined.
In the transient expression assay, the normal profile of gag cleavage intermediates and cleavage products was observed for each clinical isolate protease and, as with the engineered mutants described above, the on-treatment isolates with I50L substitutions typically produced two- to threefold-less p24/p25 than the baseline isolates (data not shown). The results of dose-response titrations demonstrated reduced susceptibility to ATV and a broad trend toward increased susceptibility to other PIs upon the emergence of I50L in each of the three multiply substituted proteases (Table 1). Increases in protease susceptibility of various magnitudes were observed in 14 out of 15 cases for five PIs (IDV, LPV, NFV, RTV, and SQV). As observed with the laboratory strain proteases, no significant changes were observed for APV as a result of adding the I50L substitution.
Phenotypic analysis of clinical isolates with and without I50L.
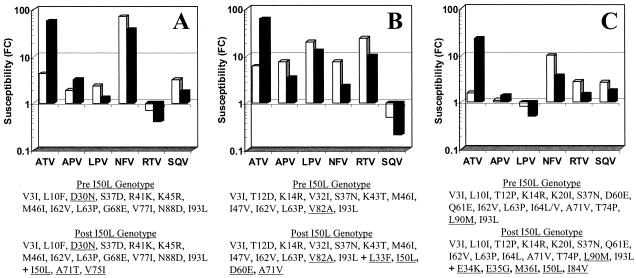
To better understand the potential clinical significance of this finding, a survey was conducted of clinical isolates evaluated during the course of ATV clinical trials. Three such subjects who had either a D30N, V82A, or L90M substitution and subsequently developed an I50L substitution on ATV therapy are profiled in Fig. 5. Apart from their primary substitution, all three had 11 to 14 additional secondary and polymorphic changes throughout the protease prior to emergence of an I50L substitution. Phenotypic profiles showed a very familiar picture, with increased resistance levels to ATV and increased susceptibility to LPV, NFV, RTV, and SQV upon emergence of an I50L substitution (Fig. 5). Susceptibility to APV increased modestly in two of the cases, again in line with the in vitro results discussed above in which the I50L substitution had a minimal effect on APV susceptibility. IDV susceptibility was not determined in these studies.
FIG. 5.
Susceptibilities of clinical isolates at baseline and after emergence of I50L substitutions while on treatment with ATV. Susceptibilities (FC) of clinical isolates at baseline (no I50L) (white bars) or after emergence of I50L substitution (black bars) are measured relative to a wild-type reference strain (from ViroLogic). Protease genotypes are shown for both baseline and on-treatment samples with primary resistance substitutions underlined.
Stability of I50L substitution.
While the clinical significance of these findings remains to be determined in clinical studies focused on PI sequencing, an in vitro study was conducted to determine the stability of the I50L substitution in the absence of ATV. A recombinant NL4-3 proviral clone containing the protease gene from an ATV-resistant clinical isolate originating in a study with the substitutions I50L, L63P, K70R, A71V, and G73S (NL4-3 100B [5]) was passaged in cell culture in the absence of any inhibitors. Despite the growth impairment exhibited by this recombinant virus, there was no evidence of reversion to isoleucine at residue 50 over 11 passages covering 54 days (unpublished data). To ensure that sufficient viral replication was taking place during these passages to select for changes, a second study was conducted in which the same I50L-containing virus was passaged in the presence of increasing concentrations of lamivudine. Results showed that, following 11 passages encompassing 54 days in culture, the I50L substitution was retained while an M184I lamivudine resistance substitution emerged by passage 7 (36 days). These results suggest that viruses containing an I50L substitution do not readily eliminate this substitution in the absence of ATV, despite the consequence of growth impairment. While I50L viruses are unlikely to compete in a mixed population with viruses that are not growth impaired, the I50L substitution may have clinical utility in patients with sustained antiretroviral suppression.
DISCUSSION
The emergence of HIV protease resistance substitutions during antiviral therapy with PIs is often associated with reduced PI susceptibility and poses a significant problem for the treatment of HIV-1-infected patients. Resistance-associated substitutions can arise during treatment with all of the PIs currently available and generally confer reduced susceptibility to multiple PIs, not just the one being used at the time of treatment failure. Less commonly, a signature substitution can arise that results in specific resistance to only a single PI without affecting susceptibility to other PIs, e.g., D30N with NFV (34). ATV is similar to NFV in selecting for a unique signature I50L substitution in the HIV protease but differs in that this is an exclusive resistance pathway for ATV in PI-naïve subjects. This substitution not only is ATV specific but does not result in cross-resistance to any other PI and consistently increases the antiviral susceptibility to most other PIs currently used for treating HIV infections (5). This finding suggests a potential strategy for preserving and perhaps even enhancing the treatment options using HIV PIs for patients failing ATV therapy.
We found that the unique increases in PI susceptibility resulting from the presence of an I50L substitution are unlikely to be due solely to viral growth impairment, since other classes of antiretroviral agents fail to show the uniform increases observed with PIs. To explore the underlying mechanism further, a cell-based HIV protease assay in a system devoid of viral replication was developed in order to characterize how properties of I50L-containing protease might relate to the virus phenotypes. Introduction of the I50L substitution led to a twofold reduction in protease activity regardless of the protease backbone employed, both wild type and PI resistant. Reduced proteolytic cleavage efficiency has been associated with other primary substitutions (35, 43, 50), and reduced replicative capacity due to resistance substitutions is a common feature of PI-resistant HIV (28, 37, 50). It was reported previously that the I50L substitution in HIV protease also led to impaired replication kinetics of I50L-containing viruses (5). It appears likely now that the impaired replication of I50L-containing viruses is a direct result of the reduced proteolytic activity of I50L-containing proteases.
Protease susceptibility assays clearly demonstrated that the I50L substitution confers reduced susceptibility of the protease to ATV and increased susceptibility to IDV, LPV, NFV, RTV, and SQV. Only modest effects on APV susceptibility were observed in protease assays, though a similar trend toward increased susceptibility was observed. The changes in PI susceptibility for the I50L+A71V protease closely mirrored the changes in antiviral susceptibility reported for HIV strains containing the I50L+A71V substitutions (5). The striking similarity between profiles for the protease assayed in isolation and that of replicating viruses strongly suggests that the increased virus susceptibility to other PIs is likely determined at the level of protease susceptibility.
The results of assays with PI-resistant proteases in both engineered laboratory strains and clinical isolates strongly indicated that the I50L substitution can also counteract the reduced susceptibility caused by other primary substitutions known to confer PI resistance. Five of the six clinically relevant primary resistance substitutions tested conferred significant resistance to one or more PIs in the cell-based protease assay, exhibiting resistance levels of 3- to 13-fold (Fig. 4). Addition of an I50L substitution to each mutant background counteracted this PI resistance, increasing protease susceptibility by an average of sixfold and achieving wild-type levels of inhibition. The lone exception was APV resistance due to the I84V substitution, which was not counteracted by the I50L substitution. If the increased susceptibility to PIs is related directly to changes in PI binding caused by the I50L substitution, then perhaps it is possible that leucine at this position interferes with and precludes enhanced binding of APV.
Antiviral assay results for three viruses bearing engineered protease substitutions (Table 1) and three chimeric viruses bearing proteases from clinical isolates paralleled the results of protease assays. In each case, the presence of the I50L substitution was clearly associated with reduced susceptibility to ATV and increased virus susceptibility to other PIs. The magnitudes of susceptibility increases corresponded quite well between the protease assay and the antiviral assay for engineered mutants in the NL4-3 laboratory strain but were somewhat more varied for the genetically more complex clinical isolates. Still, as in the protease assays, there was no evidence of cross-resistance to any other PI for any of the virus mutants evaluated.
There is no precedent for the unique phenotype of broadly increased susceptibility to others PIs associated with the I50L substitution. From the results reported here, it appears very unlikely that either reduced protease activity or reduced replicative capacity per se can account for this unique phenotypic profile. As determined here, four of the six other primary protease substitutions examined had reduced protease activity, yet none of these displayed any tendency toward increased susceptibility to PIs. The D30N substitution that confers NFV resistance provides a valuable point of comparison in this regard, as it was by far the most significantly impaired of all the mutant proteases examined. A direct comparison based on our results indicates that the I50L protease probably has slightly greater proteolytic activity than the D30N protease. If reduced protease activity were to enhance the susceptibility of HIV protease to any PI, then the D30N protease might exhibit significant increases in susceptibility to other PIs. The results clearly showed no such trend for increased susceptibility of the D30N protease. Similarly, an indirect effect of impaired virus replication on HIV susceptibility to other inhibitors is not likely to explain the unique phenotype of I50L-containing viruses. If it did, many previously described HIV isolates with impaired replication kinetics might display a similar phenotypic profile, yet the I50L phenotypic profile is novel. Also, HIV containing the D30N substitution exhibits significantly impaired viral replication kinetics (28, 37), yet phenotypic studies have shown that D30N confers NFV resistance without affecting susceptibility to other PIs (5, 26, 31, 34). Taken together, these observations suggest that neither reduced protease activity nor impaired replication kinetics alone is sufficient to confer enhanced PI susceptibility on HIV.
The phenotype of I50L-containing proteases is quite unique and distinct from the effects of other primary PI resistance substitutions, including the I50V (valine) primary substitution associated with APV resistance (23, 45). The I50V substitution reduces enzymatic activity of the protease, but unlike the I50L protease, I50V contributes to reduced susceptibility to both RTV and LPV and is associated with increased susceptibility to SQV and IDV in vitro (33, 35). While viruses containing the I50V substitution also display impaired replication kinetics (22), they typically display cross-resistance to other PIs (though not to ATV) with no trend toward increased susceptibility to other PIs (20, 23, 33, 35, 45). The frequency and magnitude of the observed I50L changes are distinct from those observed for I50V for APV. Viruses containing I50V or I84V substitutions displayed the greatest reductions in APV susceptibility, and four distinct genetic pathways to APV resistance have been described among clinical isolates from patients treated with APV: I50V, I54L/M, I84V, and V32I+I47V. Apart from the I84V pathway, the other three APV resistance pathways occur with near-equal frequency, and the most frequent substitution observed in the presence of I50V was M46I/L. In contrast, the I50L substitution mediates the increased susceptibility to other PIs and has a direct impact on changes in virus susceptibility, presumably through a unique mechanism that is dependent on this specific substitution.
Conceivably, the I50L substitution may exploit a unique structural feature of ATV to cause reduced binding to the protease and, in turn, reduced susceptibility of the virus to ATV. This change might then result in either enhanced accessibility of the enzyme active site to other inhibitors or increased binding of inhibitors directly. The location of I50L in the flap region of the enzyme structure positions it to have such an effect through potential alterations of the binding pocket. Seemingly small modifications, such as a change from the normal Ile to Leu or Val can result in ATV or APV resistance, and in the case of Leu, appears to significantly enhance susceptibilities to the other PIs. Recent studies on the structural and molecular basis for the I50L phenotype have indicated that PI binding affinities to I50L-containing proteases directly parallel the I50L phenotypic pattern observed in resistant viruses (49).
While future studies will be needed to determine the true clinical utility of these findings, the appropriate sequencing of PI therapies may be important to exploit this property of the I50L substitution and to optimize PI treatments for HIV-infected patients.
Acknowledgments
We thank Burt Rose for providing DNA for NL4-3 I50L and I50L+A71V mutants.
REFERENCES
- 1.Boden, D., and M. Markowitz. 1998. Resistance to human immunodeficiency virus type 1 protease inhibitors. Antimicrob. Agents Chemother. 42:2775-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caride, E., K. Hertogs, B. Larder, P. Dehertogh, R. Brindeiro, E. Machado, C. A. De Sa, W. A. Eyer-Silva, F. S. Sion, L. F. Passioni, J. A. Menezes, A. R. Calazans, and A. Tanuri. 2001. Genotypic and phenotypic evidence of different drug-resistance mutation patterns between B and non-B subtype isolates of human immunodeficiency virus type 1 found in Brazilian patients failing HAART. Virus Genes 23:193-202. [DOI] [PubMed] [Google Scholar]
- 3.Carrillo, A., K. Stewart, H. Sham, D. Norbeck, W. Kohlbrenner, J. Leonard, D. Kempf, and A. Molla. 1998. In vitro selection and characterization of human immunodeficiency virus type 1 variants with increased resistance to ABT-378, a novel protease inhibitor. J. Virol. 72:7532-7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, Z., Y. Li, H. Schock, D. Hall, E. Chen, and L. Kuo. 1995. Three-dimensional structure of a mutant HIV-1 protease displaying cross-resistance to all protease inhibitors in clinical trials. J. Biol. Chem. 270:21433-21436. [DOI] [PubMed] [Google Scholar]
- 5.Colonno, R., R. Rose, C. Mclaren, A. Thiry, N. Parkin, and J. Friborg. 2004. Identification of I50L as the signature atazanavir (ATV)-resistance mutation in treatment-naive HIV-1-infected patients receiving ATV-containing regimens. J. Infect. Dis. 189:1802-1810. [DOI] [PubMed] [Google Scholar]
- 6.Colonno, R. J., A. Thiry, K. Limoli, and N. Parkin. 2003. Activities of atazanavir (BMS-232632) against a large panel of human immunodeficiency virus type 1 clinical isolates resistant to one or more approved protease inhibitors. Antimicrob. Agents Chemother. 47:1324-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Croteau, G., L. Doyon, D. Thibeault, G. McKercher, L. Pilote, and D. Lamarre. 1997. Impaired fitness of human immunodeficiency virus type 1 variants with high-level resistance to protease inhibitors. J. Virol. 71:1089-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darke, P., R. Nutt, S. Brady, V. Garsky, T. Ciccarone, C. Leu, P. Lumma, R. Freidinger, D. Veber, and I. Sigal. 1988. HIV-1 protease specificity of peptide cleavage is sufficient for processing of gag and pol polyproteins. Biochem. Biophys. Res. Commun. 156:297-303. [DOI] [PubMed] [Google Scholar]
- 9.Dauber, D. S., R. Ziermann, N. Parkin, D. J. Maly, S. Mahrus, J. L. Harris, J. A. Ellman, C. Petropoulos, and C. S. Craik. 2002. Altered substrate specificity of drug-resistant human immunodeficiency virus type 1 protease. J. Virol. 76:1359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doyon, L., G. Croteau, D. Thibeault, F. Poulin, L. Pilote, and D. Lamarre. 1996. Second locus involved in human immunodeficiency virus type 1 resistance to protease inhibitors. J. Virol. 70:3763-3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gong, Y.-F., B. S. Robinson, R. E. Rose, C. Deminie, T. P. Spicer, D. Stock, R. J. Colonno, and P.-F. Lin. 2000. In vitro resistance profile of the human immunodeficiency virus type 1 protease inhibitor BMS-232632. Antimicrob. Agents Chemother. 44:2319-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hammer, S., K. Squires, M. Hughes, J. Grimes, L. Demeter, J. Currier, J. Eron, J. Feinberg, H. Balfour, L. Deyton, J. Chodakewitz, M. Fischl, et al. 1997. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. N. Engl. J. Med. 337:725-733. [DOI] [PubMed] [Google Scholar]
- 13.Hirsch, M., R. Steigbigel, S. Staszewski, J. Mellors, E. Scerpella, B. Hirschel, J. Lange, K. Squires, S. Rawlins, A. Meibohm, and R. Leavitt. 1999. A randomized, controlled trial of indinavir, zidovudine, and lamivudine in adults with advanced human immunodeficiency virus type 1 infection and prior antiretroviral therapy. J. Infect. Dis. 180:659-665. [DOI] [PubMed] [Google Scholar]
- 14.Kaplan, A., S. Michael, R. Wehbie, M. Knigge, D. Paul, L. Everitt, D. Kempf, D. Norbeck, J. Erickson, and R. Swanstrom. 1994. Selection of multiple human immunodeficiency virus type 1 variants that encode viral proteases with decreased sensitivity to an inhibitor of the viral protease. Proc. Natl. Acad. Sci. USA 91:5597-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kempf, D. J., J. D. Isaacson, M. S. King, S. C. Brun, Y. Xu, K. Real, B. M. Bernstein, A. J. Japour, E. Sun, and R. A. Rode. 2001. Identification of genotypic changes in human immunodeficiency virus protease that correlate with reduced susceptibility to the protease inhibitor lopinavir among viral isolates from protease inhibitor-experienced patients. J. Virol. 75:7462-7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klabe, R. M., L. T. Bacheler, P. J. Ala, S. Erickson-Viitanen, and J. L. Meek. 1998. Resistance to HIV protease inhibitors: a comparison of enzyme inhibition and antiviral potency. Biochemistry 37:8735-8742. [DOI] [PubMed] [Google Scholar]
- 17.Kohl, N., E. Emini, W. Schleif, L. Davis, J. Heimbach, R. Dixon, E. Scolnick, and I. Sigal. 1988. Active human immunodeficiency virus protease is required for viral infectivity. Proc. Natl. Acad. Sci. USA 85:4686-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kramer, R. A., M. D. Schaber, A. M. Skalka, K. Ganguly, F. Wong-Staal, and E. P. Reddy. 1986. HTLV-III gag protein is processed in yeast cells by the virus pol-protease. Science 231:1580-1584. [DOI] [PubMed] [Google Scholar]
- 19.Kuritzkes, D. 2002. Resistance to protease inhibitors. J. HIV Ther. 7:87-91. [PubMed] [Google Scholar]
- 20.Lam, E., and N. T. Parkin. 2003. Amprenavir resistance imparted by the I50V mutation in HIV-1 protease can be suppressed by the N88S mutation. Clin. Infect. Dis. 37:1273-1274. [DOI] [PubMed] [Google Scholar]
- 21.Lin, Y., X. Lin, L. Hong, S. Foundling, R. Heinrikson, S. Thaisrivongs, W. Leelamanit, D. Raterman, M. Shah, B. Dunn, and J. Tang. 1995. Effect of point mutations on the kinetics and the inhibition of human immunodeficiency virus type I protease: relationship to drug resistance. Biochemistry 34:1143-1152. [DOI] [PubMed] [Google Scholar]
- 22.Maguire, M., R. Guinea, P. Griffin, S. Macmanus, R. C. Elston, J. Wolfram, N. Richards, M. H. Hanlon, D. J. T. Porter, T. Wrin, N. Parkin, M. Tisdale, E. Furfine, C. Petropoulos, B. W. Snowden, and J.-P. Kleim. 2002. Changes in human immunodeficiency virus type 1 Gag at positions L449 and P453 are linked to I50V protease mutants in vivo and cause reduction of sensitivity to amprenavir and improved viral fitness in vitro. J. Virol. 76:7398-7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maguire, M., D. Shortino, A. Klein, W. Harris, V. Manohitharajah, M. Tisdale, R. Elston, J. Yeo, S. Randall, F. Xu, H. Parker, J. May, and W. Snowden. 2002. Emergence of resistance to protease inhibitor amprenavir in human immunodeficiency virus type 1-infected patients: selection of four alternative viral protease genotypes and influence of viral susceptibility to coadministered reverse transcriptase nucleoside inhibitors. Antimicrob. Agents Chemother. 46:731-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahalingam, B., J. M. Louis, C. C. Reed, J. M. Adomat, J. Krouse, Y.-F. Wang, R. W. Harrison, and I. T. Weber. 1999. Structural and kinetic analysis of drug resistant mutants of HIV-1 protease. Eur. J. Biochem. 263:238-245. [DOI] [PubMed] [Google Scholar]
- 25.Mammano, F., C. Petit, and F. Clavel. 1998. Resistance-associated loss of viral fitness in human immunodeficiency virus type 1: phenotypic analysis of protease and gag coevolution in protease inhibitor-treated patients. J. Virol. 72:7632-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Markowitz, M., M. Conant, A. Hurley, R. Schluger, M. Duran, J. Peterkin, S. Chapman, A. Patick, A. Hendricks, G. J. Yuen, W. Hoskins, N. Clendeninn, and D. D. Ho. 1998. A preliminary evaluation of nelfinavir mesylate, an inhibitor of human immunodeficiency virus (HIV)-1 protease, to treat HIV infection. J. Infect. Dis. 177:1533-1540. [DOI] [PubMed] [Google Scholar]
- 27.Markowitz, M., H. Mo, D. Kempf, D. Norbeck, T. N. Bhat, J. Erickson, and D. Ho. 1995. Selection and analysis of human immunodeficiency virus type 1 variants with increased resistance to ABT-538, a novel protease inhibitor. J. Virol. 69:701-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martinez-Picado, J., A. V. Savara, L. Sutton, and R. T. D'Aquila. 1999. Replicative fitness of protease inhibitor-resistant mutants of human immunodeficiency virus type 1. J. Virol. 73:3744-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mo, H., L. Lu, T. Dekhtyar, K. D. Stewart, E. Sun, D. J. Kempf, and A. Molla. 2003. Characterization of resistant HIV variants generated by in vitro passage with lopinavir/ritonavir. Antivir. Res. 59:173-180. [DOI] [PubMed] [Google Scholar]
- 30.Molla, A., M. Korneyeva, Q. Gao, S. Vasavanonda, P. Schipper, H. Mo, M. Markowitz, T. Chernyavskiy, P. Niu, N. Lyons, A. Hsu, G. Granneman, D. Ho, C. Boucher, J. Leonard, D. Norbeck, and D. Kempf. 1996. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat. Med. 2:760-766. [DOI] [PubMed] [Google Scholar]
- 31.Nunez, M., C. de Mendoza, L. Valer, E. Casas, S. Lopez-Calvo, A. Castro, B. Roson, D. Podzamczer, A. Rubio, J. Berenguer, and V. Soriano. 2002. Resistance mutations in HIV-infected patients experiencing early failure with nelfinavir-containing triple combinations. Med. Sci. Monit. 8:620-623. [PubMed] [Google Scholar]
- 32.Otto, M. J., S. Garber, D. L. Winslow, C. D. Reid, P. Aldrich, P. K. Jadhav, C. E. Patterson, C. N. Hodge, and Y. S. Cheng. 1993. In vitro isolation and identification of human immunodeficiency virus (HIV) variants with reduced sensitivity to C-2 symmetrical inhibitors of HIV type 1 protease. Proc. Natl. Acad. Sci. USA 90:7543-7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Partaledis, J. A., K. Yamaguchi, M. Tisdale, E. E. Blair, C. Falcione, B. Maschera, R. E. Myers, S. Pazhanisamy, O. Futer, A. B. Cullinan, C. M. Stuver, R. A. Byrn, and D. J. Livingston. 1995. In vitro selection and characterization of human immunodeficiency virus type 1 (HIV-1) isolates with reduced sensitivity to hydroxyethylamino sulfonamide inhibitors of HIV-1 aspartyl protease. J. Virol. 69:5228-5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patick, A. K., M. Duran, Y. Cao, D. Shugarts, M. R. Keller, E. Mazabel, M. Knowles, S. Chapman, D. R. Kuritzkes, and M. Markowitz. 1998. Genotypic and phenotypic characterization of human immunodeficiency virus type 1 variants isolated from patients treated with the protease inhibitor nelfinavir. Antimicrob. Agents Chemother. 42:2637-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pazhanisamy, S., C. M. Stuver, A. B. Cullinan, N. Margolin, B. G. Rao, and D. J. Livingston. 1996. Kinetic characterization of human immunodeficiency virus type-1 protease-resistant variants. J. Biol. Chem. 271:17979-17985. [DOI] [PubMed] [Google Scholar]
- 36.Peng, C., B. K. Ho, T. W. Chang, and N. T. Chang. 1989. Role of human immunodeficiency virus type 1 specific protease in core protein maturation and viral infectivity. J. Virol. 63:2550-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perrin, V., and F. Mammano. 2003. Parameters driving the selection of nelfinavir-resistant human immunodeficiency virus type 1 variants. J. Virol. 77:10172-10175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prado, J. G., T. Wrin, J. Beauchaine, L. Ruiz, C. Petropoulos, S. D. Frost, B. Clotet, R. T. D'Aquila, and J. Martinez-Picado. 2002. Amprenavir-resistant HIV-1 exhibits lopinavir cross-resistance and reduced replication capacity. AIDS 16:1009-1017. [DOI] [PubMed] [Google Scholar]
- 39.Rose, R., Y.-F. Gong, J. Greytok, C. Bechtold, B. Terry, B. Robinson, M. Alam, R. Colonno, and P.-F. Lin. 1996. Human immunodeficiency virus type 1 viral background plays a major role in development of resistance to protease inhibitors. Proc. Natl. Acad. Sci. USA 93:1648-1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sardana, V., A. Schlabach, P. Graham, B. Bush, J. Condra, J. Culberson, L. Gotlib, D. Graham, N. Kohl, R. LaFemina, et al. 1994. Human immunodeficiency virus type 1 protease inhibitors: evaluation of resistance engendered by amino acid substitutions in the enzyme's substrate binding site. Biochemistry 33:2004-2010. [DOI] [PubMed] [Google Scholar]
- 41.Schock, H. B., V. M. Garsky, and L. C. Kuo. 1996. Mutational anatomy of an HIV-1 protease variant conferring cross-resistance to protease inhibitors in clinical trials. J. Biol. Chem. 271:31957-31963. [DOI] [PubMed] [Google Scholar]
- 42.Servais, J., J.-M. Plesseria, C. Lambert, E. Fontaine, I. Robert, V. Arendt, T. Staub, F. Schneide, R. Hemme, and J.-C. Schmit. 2002. Genotypic correlates of resistance to HIV-1 protease inhibitors on longitudinal data: the role of secondary mutations. Antivir. Ther. 6:239-248. [PubMed] [Google Scholar]
- 43.Sigiura, W., Z. Matsuda, Y. Yokomaku, K. Hertogs, B. Larder, T. Oishi, A. Okano, T. Shiino, M. Tatsumi, M. Matsuda, H. Abumi, N. Takata, S. Shirahata, K. Yamada, H. Yoshikua, and Y. Nagai. 2002. Interference between D30N and L90M in selection and development of protease inhibitor-resistant human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 46:708-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soriano, V., and C. de Mendoza. 2002. Genetic mechanisms of resistance to protease inhibitors and entry inhibitors. HIV Clin. Trials 3:249-257. [DOI] [PubMed] [Google Scholar]
- 45.Tisdale, M., R. Myer, B. Maschera, N. Parry, N. Oliver, and E. Blair. 1995. Cross-resistance analysis of human immunodeficiency virus type 1 variants individually selected for resistance to five different protease inhibitors. Antimicrob. Agents Chemother. 39:1704-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wainberg, M., and G. Friedland. 1998. Public health implications of antiretroviral therapy and HIV drug resistance. JAMA 279:1977-1983. [DOI] [PubMed] [Google Scholar]
- 47.Watkins, T., R. W., D. Irlbeck, and R. Swanstrom. 2003. Selection of high-level resistance to human immunodeficiency virus type 1 protease inhibitors. Antimicrob. Agents Chemother. 47:759-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wyatt, L., B. Moss, and S. Rozenblatt. 1995. Replication-deficient vaccinia virus encoding bacteriophage T7 RNA polymerase for transient gene expression in mammalian cells. Virology 210:202-205. [DOI] [PubMed] [Google Scholar]
- 49.Yanchunas, J., Jr., D. R. Langley, L. Tao, R. E. Rose, J. Friborg, R. J. Colonno, and M. L. Doyle. 2005. Molecular basis for increased susceptibility of isolates with atazanavir resistance-conferring substitution I50L to other protease inhibitors. Antimicrob. Agents Chemother. 49:3825-3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zennou, V., F. Mammano, S. Paulous, D. Mathez, and F. Clavel. 1998. Loss of viral fitness associated with multiple Gag and Gag-Pol processing defects in human immunodeficiency virus type 1 variants selected for resistance to protease inhibitors in vivo. J. Virol. 72:3300-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang, Y. M., H. Imamichi, T. Imamichi, H. C. Lane, J. Falloon, M. B. Vasudevachari, and N. P. Salzman. 1997. Drug resistance during indinavir therapy is caused by mutations in the protease gene and its Gag substrate cleavage site. J. Virol. 71:6662-6670. [DOI] [PMC free article] [PubMed] [Google Scholar]