Abstract
The oxazolidinones are a relatively new structural class of antibacterial agents that act by inhibiting bacterial protein synthesis. The oxazolidinones inhibit mitochondrial protein synthesis, as shown by [35S]methionine incorporation into intact rat heart mitochondria. Treatment of K562 human erythroleukemia cells with the oxazolidinone eperezolid resulted in a time- and concentration-dependent inhibition of cell proliferation. The cells remained viable, but an increase in doubling time was observed with eperezolid treatment. Inhibition was reversible, since washing and refeeding of cells in the absence of compound resulted in a resumption of growth. The growth-inhibitory effect of the oxazolidinones did not appear to be cell type specific, and inhibition of CHO and HEK cells also was demonstrated. Treatment of cells resulted in a decrease in mitochondrial cytochrome oxidase subunit I levels, consistent with an inhibition of mitochondrial protein synthesis. Eperezolid caused no growth inhibition of rho zero (ρ0) cells, which contain no mitochondrial DNA; however, the growth of the parent 143B cells was inhibited. These results provide a direct demonstration that the inhibitory effect of eperezolid in mammalian cells is the result of mitochondrial protein synthesis inhibition.
The oxazolidinone linezolid is a new antibacterial approved for marketing in 2000 that inhibits bacterial protein synthesis (5, 20, 41). It represents a new structural class of antibiotics, with activity against several gram-positive organisms, including several resistant strains. Linezolid has been shown to be effective in treating nosocomial pneumonia caused by methicillin-susceptible and -resistant Staphylococcus aureus or multidrug-resistant Streptococcus pneumonia and skin and soft tissue infections caused by methicillin-susceptible and -resistant Staphylococcus aureus, Streptococcus pyogenes, and Streptococcus agalactiae. It is also effective against community-acquired pneumonia caused by methicillin-susceptible S. aureus, multidrug-resistant S. pneumoniae, and vancomycin-resistant Enterococcus faecium infections (15, 31).
The oxazolidinones inhibit bacterial protein synthesis, although the exact details concerning the mechanism(s) of inhibition are still emerging. Early results demonstrated that the oxazolidinone eperezolid binds to 50S but not 30S ribosomal subunits. Furthermore, binding was inhibited by chloramphenicol and lincomycin (27). Cross-linking studies have been carried out to identify the sites of oxazolidinone binding. Using ribosomes from Escherichia coli, a number of nucleotide residues in domain V of 23S rRNA were identified, as well as residue A864 of 16S rRNA (28). Colca et al. carried out cross-linking experiments using intact Staphylococcus aureus and showed that tRNA, two ribosomal proteins, and nucleotide A2602 of 23S rRNA all were labeled by the cross-linker (10).
The results from mapping oxazolidinone resistance mutations agree with the cross-linking studies. Linezolid-resistant mutants of Halobacterium halobium were isolated and shown to contain single point mutations in the central loop of domain V of 23S rRNA (24). Likewise, Escherichia coli oxazolidinone-resistant mutants contained G2032A and G2447A mutations, which also are in domain V (4, 47). Introduction of the G2032A mutation back into linezolid-sensitive Escherichia coli conferred resistance to the oxazolidinone. Domain V of 23S rRNA is part of the peptidyl transferase center of the bacterial ribosome, and biochemical experiments demonstrate that the oxazolidinones affect steps in protein synthesis. Work by Eustice et al. showed that an early step in synthesis is inhibited at or before the initiation step (13). Linezolid inhibits the formation of the tRNA Met-mRNA-70S complex but not the synthesis of N-formylmethionyl-tRNA (fMet-tRNA) (2). Elongation factor G-dependent translocation of fMet-tRNA from the A site to the P site is inhibited, and the oxazolidinones also inhibit peptide synthesis, as shown by in vitro assays using fMet and puromycin (2, 4, 7, 34, 45). Taken together, all of the available data from mutation, resistance, and biochemical experiments are consistent with the hypothesis that the primary binding site of the oxazolidinones is located within the ribosomal peptidyl transferase center.
In contrast to the current knowledge of oxazolidinone action on bacteria, much less is understood about oxazolidinone effects on mammalian cells. Clinically, linezolid therapy results in minimal side effects for most patients. However, reversible myelosuppression, characterized by anemia and thrombocytopenia, has been infrequently observed in patients treated with linezolid for at least 21 days (25). The mechanism of this effect is not known, but it has been suggested that the myelosuppression may result from linezolid inhibition of mitochondrial protein synthesis. In fact, it has been shown that the oxazolidinones inhibit [35S]methionine incorporation into mitochondrial proteins (39).
In the present study we have examined the effect of an oxazolidinone, eperezolid, on the proliferation of a variety of mammalian cells. We show that treatment with eperezolid inhibits proliferation, and further, that the inhibition of proliferation results from eperezolid-mediated inhibition of mitochondrial protein synthesis.
MATERIALS AND METHODS
Cell culture.
The human osteosarcoma-derived cell line 143B (thymidine kinase deficient) and its mitochondrial DNA (mtDNA)-less derivative, 143B206 ρ0, were cultured in Dulbecco's modified Eagle's (DME) high glucose supplemented with 10% fetal bovine serum, 1 mM sodium pyruvate, and 50 μg/ml uridine as described by King and Attardi (23). The cells were a gift from G. Attardi and A. Chomyn (California Institute of Technology). Chinese hamster ovary (CHO) cells were grown in alpha minimal essential medium supplemented with 10% fetal bovine serum and 4 mM l-glutamine. Human embryonic kidney (HEK) cells were cultured in DME high-glucose medium supplemented with 10% fetal bovine serum, 4 mM l-glutamine, and 1 mM sodium pyruvate. Human erythroleukemia K562 cells were obtained from the American Type Culture Collection and grown in RPMI 1640 medium containing 25 mM HEPES, 1.5 mM Glutamax, and 10% fetal bovine serum. Cells were incubated at 37°C, 5% CO2 in the presence or absence of compound. Compound stocks were prepared in 100% dimethyl sulfoxide (DMSO), and the final DMSO concentration in the assay was 0.5%. Following incubation, quantitation of proliferation was carried out by cell counting using a Coulter counter.
K562 cell mitochondrial preparation.
K562 cells were incubated with compound or vehicle for 5 days, and cells were collected, washed once with phosphate-buffered saline (PBS), resuspended in 1 ml of TKM buffer (10 mM Tris HCl, pH 7.5, 10 mM KCl, and 0.15 mM NaCl), and homogenized with a hand-held homogenizer (Ultra-Turrax; IKA Labotechnik) at setting 3 for 15 s. An aliquot (300 μl) of TKM buffer containing 1 M sucrose was added, and the samples were centrifuged at 1,000 × g for 5 min. The supernatant fraction was respun at 5,000 × g for 20 min. The mitochondrial pellet was resuspended in TKM buffer containing 0.25 M sucrose in a 100-μl volume. Mitochondrial protein content was measured using the bicinchoninic acid protein assay kit (Pierce).
Western blot analysis.
Mitochondrial proteins (10 μg) were electrophoresed on 10% Criterion gels (Bio-Rad) and transferred to polyvinylidene difluoride membranes (Bio-Rad). After blocking overnight at 4°C with 1% bovine serum albumin (BSA) and 1% dry milk in Tris-buffered saline (Bio-Rad) containing 0.1% Tween 20, the membranes were incubated with 1 μg/ml cytochrome c oxidase subunit 1 (Cox-1) monoclonal antibody (Molecular Probes A-6403) for 3 h, followed by chemiluminescent detection (Amersham ECL plus) according to the manufacturer's instructions. Membranes were stripped by RE-blot Plus strong solution (Chemicon 2504) and incubated with 1 μg/ml Tom-20 polyclonal antibody (Gift from B. Wattenberg, University of Louisville), followed by chemiluminescent detection. The Cox-1 bands and Tom-20 bands in the X-ray films were scanned by a Personal Densitometer SL (Molecular Dynamics) and quantitated by Quantity One (Bio-Rad).
Rat heart mitochondria preparation.
The procedure for mitochondria preparation and the protein synthesis assay was adapted from that described by McKee et al. (29). All experiments involving animals were carried out in compliance with national legislation and subject to local ethical review. Briefly, male Sprague-Dawley rats (∼250 g) were anesthetized by inhalation of isoflurane and injected with heparin IV, and the heart was removed and rinsed in ice-cold MSE buffer (220 mM mannitol, 70 mM sucrose, 50 mM HEPES, 2 mM EGTA, pH 7.2) until free of blood. Hearts were minced in MSE buffer containing 10 mg/ml Nagarse (Sigma), washed once, and homogenized in MSE-Nagarse buffer using a Brinkman polytron. Protease inhibitor (Complete; Boehringer) then was added to the homogenate. The homogenate was centrifuged at 900 × g for 3 min, and the resulting supernatant was centrifuged at 10,000 × g for 10 min. The pellet was resuspended in 1.0 ml of ice-cold MSE buffer at a ratio of 20 ml/g ventricular tissue and centrifuged at a speed of 900 × g for 3 min at 4°C. The resulting supernatant was centrifuged at 10,000 × g for 10 min at 4°C. Pellets were resuspended in MSE buffer and protein was determined (Bio-Rad).
Mitochondrial protein synthesis (MPS) assay.
Mitochondrial protein synthesis was measured in a 50-μl total volume containing 90 mM KCl, 4 mM MgSO4, 2.5 mM KH2PO4, 50 mM HEPES, pH 7.2, 0.25 mM amino acids without methionine, 20 mM glutamate, 0.5 mM malate, 2 mM ADP, 1 mg/ml BSA (Sigma A3059), 0.1 mg/ml cycloheximide (Sigma D-335715), 1 μM cold methionine, [35S]methionine (40 μCi/ml), and mitochondrial protein (1 mg/ml) in the presence or absence of compound. The final concentration of DMSO was 1% in the assay. Incubation was carried out at 30°C for 90 min followed by addition of 200 μl 3% sulfosalicylic acid in 0.1 mM EDTA to precipitate proteins. Proteins were harvested onto filtermats (Wallac 1205--404) using a Tomtec harvester followed by counting in a Wallac BetaPlate scintillation counter. The 50% inhibitory concentrations (IC50) were determined by GraphPad Prism software, using a sigmodial dose response equation with a fixed bottom.
RESULTS
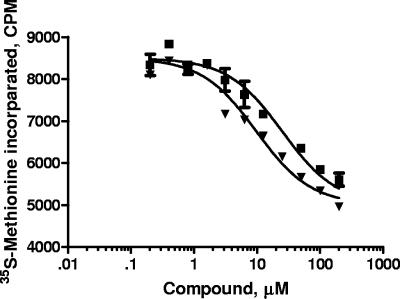
Eperezolid and linezolid (Fig. 1) are oxazolidinones that are active against gram-positive bacteria, and mechanistic studies have established that they inhibit bacterial protein synthesis (41). Their IC50 values for inhibiting [14C]leucine incorporation in the membrane-permeable E. coli strain UC6782 are approximately 1 μM, while their MICs at which 90% of the isolates tested are inhibited (MIC90) against gram-positive bacteria such as Staphylococcus aureus is 4 μg/ml (12 μM) (41). These compounds were tested in an in vitro mitochondrial protein synthesis assay. Isolated rat heart mitochondria were incubated with linezolid or eperezolid in the presence of [35S]methionine, and acid-precipitable radioactivity was measured following a 90-min incubation (Fig. 2). Both compounds inhibited synthesis, with IC50 values of 9.5 ± 1.5 μM (n = 2) and 16 ± 2 μM (n = 2) for eperezolid and linezolid, respectively. The value for linezolid is similar to that reported by Sciotti et al. (39).
FIG. 1.
Structures of oxazolidinones linezolid and eperezolid.
FIG. 2.
Linezolid and eperezolid inhibit rat heart mitochondrial protein synthesis. Mitochondrial protein synthesis was carried out in isolated rat heart mitochondria as described in Materials and Methods in the presence or absence of varying concentrations of linezolid ▪ or eperezolid ▾. Results shown are the average of duplicate determinations in one experiment; error bars show the range.
Given the inhibition of mitochondrial protein synthesis and the role of mitochondrial proteins in oxidative phosphorylation and energy metabolism, we investigated whether the compounds affect the growth of mammalian cells. Incubation of K562 human leukemia cells with varying concentrations of eperezolid resulted in a time- and concentration-dependent inhibition of cell number (Fig. 3). There was a 5% to 10% difference in cell number for the treated and control cultures at the 24-h time point. The maximal inhibitory effect on proliferation at each time point was achieved with a concentration of 100 μM. After 96 h of incubation with 100 μM compound, cell growth was significantly slowed. All the cells in the culture were viable throughout the time course, based on trypan blue exclusion (data not shown).
FIG. 3.
Eperezolid inhibits the growth of human K562 cells. K562 cells were incubated in the presence or absence of varying concentrations of eperezolid, and cell counts were monitored as described in Materials and Methods. A. Cells were treated with the following: ▪, vehicle (0.5% DMSO); ▴, 1.7 μM; ▾, 5 μM; ⧫, 15 μM; •, 45 μM; □, 135 μM; or Δ, 400 μM eperezolid. B. K562 cells were incubated with varying concentrations of eperezolid, and cell proliferation was monitored by cell counts after 120 h.
Fluorescence-activated cell sorter analysis demonstrated that the cells were not undergoing cell cycle arrest (data not shown). The percentage of cells in each phase of the cell cycle was approximately the same between control and treated cells. Rather, the major effect of the compound was to slow the growth by increasing the doubling time of the treated cultures. The time course data was used to plot the concentration dependency curve of growth inhibition (Fig. 3B). The IC50 for growth inhibition by eperezolid in K562 cells for a 5-day exposure was 12 μM.
The inhibition of proliferation by eperezolid was reversed upon withdrawal of compound from the growth media (Fig. 4). Cells were grown in either 15 or 30 μM compound for 72 h followed by removal of the medium, washing, and refeeding the cells with medium containing either vehicle (0.5% DMSO) or 15 or 30 μM eperezolid. Cells then were cultured for an additional 5 days. At the 5-day time point the cell numbers of cultures incubated in 15 μM compound and then washed was essentially the same as the untreated cultures. Reversibility also was observed for cells treated with 30 μM compound, although the cell number did not return completely to that of untreated cells at the 5-day time point.
FIG. 4.
Growth inhibition by eperezolid is reversible. K562 cells were grown in the presence of ▪, vehicle (0.5% DMSO); ▾, 15 μM eperezolid; or •, 30 μM eperezolid for 72 h. Cells were then washed in RPMI media and replated in media containing ▪, vehicle (0.5% DMSO); ▾, 15 μM eperezolid; ▴, vehicle (15 μM eperezolid recovery); •, 30 μM eperezolid; or ⧫, vehicle (30 μM eperezolid recovery) and cultured for an additional 5 days. Results shown are the average of triplicate determinations, and error bars show the standard errors of the means.
The effect of eperezolid on cell growth of several other cell lines was investigated. Eperezolid inhibited proliferation of human HEK cells and Chinese hamster ovary (CHO) cells, as well as mouse BB88 cells (data not shown). Direct comparison of the effect of the oxazolidinone on the proliferation of different cell lines is complicated by the differences in culture conditions and doubling times; however, the cells were scored during the logarithmic phase of growth. The IC50 value for eperezolid varied somewhat between the various cell types and was 63 and 20 μM for CHO and HEK cells, respectively. In all cases, a progressive slowing of growth due to an increase in cell doubling time was observed. These experiments demonstrate that the antiproliferative effect of the oxazolidinones was not cell type specific.
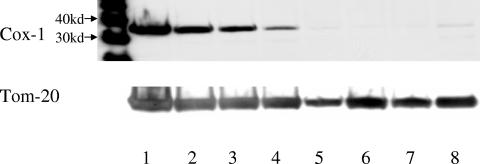
The oxazolidinones inhibit mitochondrial protein synthesis in an in vitro system and they inhibit cell proliferation, but the link, if any, between these two activities has not been investigated. Therefore, the level of mitochondrial proteins in K562 cells was examined to determine whether oxazolidinones inhibited mitochondrial protein synthesis in intact cells. K562 cells were incubated with varying concentrations of eperezolid for 5 days under the same conditions as in Fig. 3, and the cells were harvested and mitochondria were isolated. This mitochondrial fraction was run on Western blots and probed with a monoclonal antibody to subunit 1 of cytochrome oxidase, which is 1 of the 13 proteins encoded by the mitochondrial genome (Fig. 5). To normalize for mitochondrial protein loading, after exposure to the Cox-1 antibody the blots were stripped and reprobed with an antibody to Tom-20, an outer mitochondrial membrane protein encoded by the nucleus. Tom-20 protein levels were relatively constant across the gel. However, Cox-1 protein expression decreased with increasing concentrations of eperezolid, and 25 μM eperezolid resulted in a greater than 90% decrease in Cox-1 levels. As a control, cells also were treated with 100 μM chloramphenicol, a known mitochondrial protein synthesis inhibitor. Almost complete inhibition of Cox-1 levels was observed with this compound, which also inhibited the proliferation of the cells (data not shown). The decreased level of mitochondrial protein was not specific for Cox-1, since [35S]methionine labeling of mitochondria isolated from oxazolidinone-treated cells showed decreased incorporation of radioactivity into all 13 mitochondrial proteins (data not shown).
FIG. 5.
Eperezolid decreases mitochondrial Cox-1 protein levels. K562 cells were treated with vehicle (0.5% DMSO), 100 μM chloramphenicol, or varying concentrations of eperezolid for 5 days, and mitochondria were isolated. Aliquots of mitochondrial protein (10 μg) were electrophoresed on sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membranes, and probed with an antibody to Cox-1 (top gel). The blot was stripped and reprobed with an antibody to Tom-20 (bottom gel). Lane 1, vehicle; 2, 3.1 μM eperezolid; 3, 6.2 μM eperezolid; 4, 12 μM eperezolid; 5, 25 μM eperezolid; 6, 50 μM eperezolid; 7, 100 μM eperezolid; 8, 100 μM chloramphenicol. The 30-kDa and 40-kDa molecular mass markers are designated on the left.
Rho zero (ρ0) cells are derived from a human osteosarcoma parent cell line (143B), which was depleted of mtDNA by culturing and selection in the presence of ethidium bromide. These cells cannot carry out oxidative phosphorylation, require pyruvate and uridine for growth, and have glycolysis as their only source of ATP (23). Rho zero and parent 143B cells were treated with various concentrations of eperezolid, and the cell number was quantitated (Fig. 6). 143B cells had a doubling time of 17 ± 1.0 (n = 4) h, while that of the rho zero cells was longer, 24 ± 0.2 (n = 4) h. As expected, eperezolid resulted in a time- and concentration-dependent decrease in 143B cell numbers. At the 72-h time point, the IC50 value was ∼20 μM. In contrast, growth of rho zero cells was not inhibited by treatment with any concentration of the two mitochondrial protein synthesis inhibitors, eperezolid or chloramphenicol. As a control, to demonstrate that rho zero cells are not resistant to all types of growth arrest, cells were treated with etoposide, a topoisomerase II inhibitor, or with cycloheximide. As expected, both these compounds inhibited growth of rho zero cells, as well as the 143B cells.
FIG. 6.
Eperezolid inhibits cell proliferation in 143B cells but not in rho zero cells. Parent 143B (A) or rho zero (B) cells were incubated with □, vehicle (0.5% DMSO); ▪, 3 μM eperezolid; ▴, 10 μM eperezolid; ▾, 100 μM eperezolid; •, 1.7 μM etoposide; ⋄, 1.75 μM cycloheximide; or ○, 30 μM chloramphenicol, and cells were counted at 48, 72, 96, and 120 h.
DISCUSSION
The oxazolidinones inhibit bacterial protein synthesis, and results from resistance mutations and cross-linking experiments demonstrate that they interact with nucleotides in domain V of 23S rRNA, which contains the peptidyl transferase center. Details concerning the structure, mechanism(s), and activities of this region are emerging (17, 32). The crystal structures of the individual 50S and 30S subunits as well as the functional 70S complex have been solved (3, 8, 9). Results from cryoelectron microscopy (cryo-EM) studies have shown the conformational changes this region undergoes during peptide bond formation and translocation, including ratchet-like rotations of the 30S and 50S subunits (16). Proper alignment of the two tRNA substrates appears to be the driving force for peptide bond formation (42). Mutational and kinetic analysis demonstrated that nucleotides A2602, A2451, U2585, and C2063, which are located in the innermost layer of the active site, are critical for the catalysis of peptide release (34, 48).
In contrast to our understanding of the prokaryotic 70S ribosome, less is known about the structural details of the mitochondrial 55S and the mammalian 80S ribosomes. cryo-EM visualization of the 80S ribosome was first reported in 2004 with a resolution of 17 to 20 Å, and more recently, that structure has been refined to 9.5-Å resolution (18, 43). These structures do not provide details of the peptidyl transferase center of the 80S ribosome. Biochemical experiments comparing 70S, 55S, and 80S ribosomes in a peptide “fragment reaction” showed that antibiotics such as chloramphenicol and lincomycin inhibit bacterial protein synthesis with no significant effect on mammalian 80S protein synthesis (12). Although many aspects of protein translation are conserved between eukaryotes and prokaryotes, the specificity of these antibiotics is believed to result from key differences between these ribosomes.
In the case of the mitochondrial ribosome, results from cryoelectron microscopy (cryo-EM) along with extensive physical characterization of the mitoribosome have shown that there are a number of marked differences between bacterial and mitochondrial ribosomes (33, 40). The mitoribosome is larger in size and consists of 69% protein and 31% RNA compared to the 33% protein and 67% RNA of the bacterial ribosome. Of the approximately 80 ribosomal proteins of the mitoribosome, only half of them have bacterial homologs, and the homologous proteins are often larger in size than their bacterial counterparts. As a result, the mitoribosome contains several distinct topological differences. For example, the mRNA entry site is composed primarily of mitoribosome-specific proteins, which form a triangular gatelike structure that is not present in the bacterial ribosome. Also, the cryo-EM map of the polypeptide tunnel of the mitoribosome suggests that there are two possible pathways for proteins to exit (40).
Despite these differences, there are certain critical features of the ribosome that are conserved between bacteria and mitochondria (1). Mears et al. carried out comparative sequence analyses between rRNA of archaea, bacteria, eukaryotic nuclear, mitochondrial, and chloroplast ribosomes (30). The areas of greatest similarities map to key functional domains, including the mRNA binding site, the peptidyl transferase center, and the subunit interface, all of which contain the highest number of “universally conserved” nucleotides. An important aspect, however, is areas where there is the least similarity are primarily on the periphery of the rRNA three-dimensional structure in regions that may not be essential for protein translation. In addition, the polypeptide exit tunnel is considerably shortened in the mitochondrial ribosome compared to the bacterial ribosome.
Given the high degree of homology in the peptidyl transferase centers, it is not surprising that antibiotics that inhibit bacterial protein synthesis also inhibit mitochondrial protein synthesis. Denslow and O'Brien demonstrated that chloramphenicol inhibited E. coli 70S and bovine mitochondrial 55S peptidyl transferase activity with IC50 values of 40 and 100 μM, respectively. Apparent dissociation constants were calculated from Scatchard plots of the inhibition data. The results demonstrated that E. coli ribosomes have a somewhat higher affinity (100 μM) for chloramphenicol than mitochondrial ribosomes (29 μM) (11). The ability of antibiotics to inhibit mitochondrial protein synthesis has been used as evidence to support the idea that key structural and functional features are shared between bacteria and mitochondrial ribosomes (11, 21). The crystal structures of several different antibacterials with the bacterial 50S subunit have been reported and demonstrate that the compounds all interact at or near the sites of peptide bond formation. The crystal structure of chloramphenicol binding to Deinococcus radiodurans is different than that of its binding to Haloarcula marismortui, although it is bound in the peptidyl transferase center in both structures (19, 38). The ability of eperezolid to compete for chloramphenicol binding (27) as well as the resistance mapping indicates that the oxazolidinones bind in the peptidyl transferase region as well (47).
The oxazolidinones inhibited cell proliferation in a variety of cell types, suggesting there is no cell type specificity in their growth-inhibitory effect. The levels of the mitochondrial protein Cox-1 decreased concomitantly with oxazolidinone treatment. However, the most direct demonstration of the role of mitochondrial protein synthesis inhibition in the antiproliferative effect of the oxazolidinones was demonstrated in the rho zero cell experiments. Growth of the parent 143B cells was inhibited, but not that of the rho zero cells. The rho zero cells contain no mtDNA and synthesize no mitochondrial proteins, and in these cells the oxazolidinones had no effect on growth.
The role of mitochondrial protein synthesis inhibition in cell proliferation has been established in a number of studies, and the results with the oxazolidinones are consistent with those observations. Treatment with eperezolid resulted in a time-dependent lengthening of cell doubling time, consistent with a progressive loss of oxidative phosphorylation and an increasing reliance on glycolysis. A similar observation was reported by Jazayeri et al., who induced a rho zero phenotype in cells by inducible expression of a dominant-negative form of DNA polymerase-γ (22). This resulted in a depletion of mitochondrial DNA, loss of mitochondrial proteins and function, and an increase in cell doubling time. Storrie and Attardie treated HeLa cells with the mitochondrial protein synthesis inhibitor chloramphenicol and observed a slowing of the growth rate, with the cells ultimately entering a stationary phase. They concluded that this effect on cell growth was the result of decreased respiratory rate capacity in the cells, as the mitochondrial proteins were no longer being synthesized (44).
We have shown here a marked effect of the oxazolidinone eperezolid on cell growth. The lack of effect of eperezolid on the proliferation of rho zero cells demonstrates that the effect of the compound is due to mitochondrial protein synthesis inhibition. Treatment of wild-type cells with eperezolid in effect causes them to approach a rho zero phenotype, as seen by an increase in the cell doubling time and a loss of mitochondrial proteins. Cultured cells, as used here, have been shown to derive much of their energy from glycolysis when grown in high-glucose medium (36). However, elimination of the contribution of oxidative phosphorylation by mitochondrial protein inhibition by eperezolid or by chloramphenicol or dominant-negative DNA polymerase (as outlined above) nonetheless impacts cell metabolism as demonstrated by the increase in doubling time.
Deficits in respiratory chain function resulting from mtDNA mutations are known to affect a variety of different tissues. The degree to which a particular cell type or tissue is affected reflects the specific “threshold capacity” (35). If a given tissue has a high threshold capacity, then a greater loss in mitochondrial proteins can be sustained without a loss in oxidative phosphorylation function. This has been shown for cultured cell lines as well as for different tissues (35, 46). For example, the threshold value for complex IV is higher (around 86%) in the liver, kidney, and brain compared to the value in muscle and heart (around 67%) (37). In the case of the oxazolidinones, erythroid precursor cells may be particularly sensitive to mitochondrial protein synthesis inhibition, resulting in the anemia observed in some patients with long-term treatment. In fact, the reversible anemia observed with the mitochondrial protein synthesis inhibitor chloramphenicol or with human immunodeficiency virus reverse transcriptase inhibitors (which inhibit mtDNA synthesis and thus decrease mitochondrial protein levels) also has been attributed to a loss in mitochondrial function in hematopoietic cells (6, 14, 26, 49).
Acknowledgments
The contributions of Gregory N. Cosma, Michael A. Fisher, and David P. Blakeman to the mitochondrial protein synthesis assay are gratefully acknowledged.
REFERENCES
- 1.Anderson, S., M. H. de Bruijn, A. R. Coulson, I. C. Eperon, F. Sanger, and I. G. Young. 1982. Complete sequence of bovine mitochondrial DNA. Conserved features of the mammalian mitochondrial genome. J. Mol. Biol. 156:683-717. [DOI] [PubMed] [Google Scholar]
- 2.Aoki, H., L. Ke, S. M. Poppe, T. J. Poel, E. A. Weaver, R. C. Gadwood, R. C. Thomas, D. L. Shinabarger, and M. C. Ganoza. 2002. Oxazolidinone antibiotics target the P site on Escherichia coli ribosomes. Antimicrob. Agents Chemother. 46:1080-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ban, N., P. Nissen, J. Hansen, P. B. Moore, and T. A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289:905-920. [DOI] [PubMed] [Google Scholar]
- 4.Bobkova, E. V., Y. P. Yan, D. B. Jordan, M. G. Kurilla, and D. L. Pompliano. 2003. Catalytic properties of mutant 23 S ribosomes resistant to oxazolidinones. J. Biol. Chem. 278:9802-9807. [DOI] [PubMed] [Google Scholar]
- 5.Bozdogan, B., and P. C. Appelbaum. 2004. Oxazolidinones: activity, mode of action, and mechanism of resistance. Int. J. Antimicrob. Agents 23:113-119. [DOI] [PubMed] [Google Scholar]
- 6.Brinkman, K., H. J. ter Hofstede, D. M. Burger, J. A. Smeitink, and P. P. Koopmans. 1998. Adverse effects of reverse transcriptase inhibitors: mitochondrial toxicity as common pathway. AIDS 12:1735-1744. [DOI] [PubMed] [Google Scholar]
- 7.Burghardt, H., K. L. Schimz, and M. Muller. 1998. On the target of a novel class of antibiotics, oxazolidinones, active against multidrug-resistant gram-positive bacteria. FEBS Lett. 425:40-44. [DOI] [PubMed] [Google Scholar]
- 8.Cate, J. H., M. M. Yusupov, G. Z. Yusupova, T. N. Earnest, and H. F. Noller. 1999. X-ray crystal structures of 70S ribosome functional complexes. Science 285:2095-2104. [DOI] [PubMed] [Google Scholar]
- 9.Clemons, W. M., Jr., J. L. May, B. T. Wimberly, J. P. McCutcheon, M. S. Capel, and V. Ramakrishnan. 1999. Structure of a bacterial 30S ribosomal subunit at 5.5 A resolution. Nature 400:833-840. [DOI] [PubMed] [Google Scholar]
- 10.Colca, J. R., W. G. McDonald, D. J. Waldon, L. M. Thomasco, R. C. Gadwood, E. T. Lund, G. S. Cavey, W. R. Mathews, L. D. Adams, E. T. Cecil, J. D. Pearson, J. H. Bock, J. E. Mott, D. L. Shinabarger, L. Xiong, and A. S. Mankin. 2003. Cross-linking in the living cell locates the site of action of oxazolidinone antibiotics. J. Biol. Chem. 278:21972-21979. [DOI] [PubMed] [Google Scholar]
- 11.Denslow, N. D., and T. W. O'Brien. 1978. Antibiotic susceptibility of the peptidyl transferase locus of bovine mitochondrial ribosomes. Eur. J. Biochem. 91:441-448. [DOI] [PubMed] [Google Scholar]
- 12.Denslow, N. D., and T. W. O'Brien. 1974. Susceptibility of 55S mitochondrial ribosomes to antibiotics inhibitory to prokaryotic ribosomes, lincomycin, chloramphenicol and PA114A. Biochem. Biophys. Res. Commun. 57:9-16. [DOI] [PubMed] [Google Scholar]
- 13.Eustice, D. C., P. A. Feldman, I. Zajac, and A. M. Slee. 1988. Mechanism of action of DuP721: inhibition of an early event during initiation of protein synthesis. Antimicrob. Agents Chemother. 32:1218-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Firkin, F. C. 1972. Mitochondrial lesions in reversible erythropoietic depression due to chloramphenicol. J. Clin. Investig. 51:2085-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ford, C. W., J. C. Hamel, D. M. Wilson, J. K. Moerman, D. Stapert, R. J. Yancey, Jr., D. K. Hutchinson, M. R. Barbachyn, and S. J. Brickner. 1996. In vivo activities of U-100592 and U-100766, novel oxazolidinone antimicrobial agents, against experimental bacterial infections. Antimicrob. Agents Chemother. 40:1508-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frank, J., and R. K. Agrawal. 2000. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature 406:318-322. [DOI] [PubMed] [Google Scholar]
- 17.Gregory, S. T., and A. E. Dahlberg. 2004. Peptide bond formation is all about proximity. Nature Structural Mol. Biol. 11:586-587. [DOI] [PubMed] [Google Scholar]
- 18.Halic, M., T. Becker, J. Frank, C. M. T. Spahn, and R. Beckmann. 2005. Localization and dynamic behavior of ribosomal protein L30e. Nat. Struct. Mol. Biol. 12:467-468. [DOI] [PubMed] [Google Scholar]
- 19.Hansen, J. L., P. B. Moore, and T. A. Steitz. 2003. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J. Mol. Biol. 330:1061-1075. [DOI] [PubMed] [Google Scholar]
- 20.Hutchinson, D. K. 2003. Oxazolidinone antibacterial agents: a critical review. Curr. Top. Med. Chem. 3:1021-1042. [DOI] [PubMed] [Google Scholar]
- 21.Ibrahim, N. G., J. P. Burke, and D. S. Beattie. 1974. The sensitivity of rat liver and yeast mitochondrial ribosomes to inhibitors of protein synthesis. J. Biol. Chem. 249:6806-6811. [PubMed] [Google Scholar]
- 22.Jazayeri, M., A. Andreyev, Y. Will, M. Ward, C. M. Anderson, and W. Clevenger. 2003. Inducible expression of a dominant negative DNA polymerase-gamma depletes mitochondrial DNA and produces a rho0 phenotype. J. Biol. Chem. 278:9823-9830. [DOI] [PubMed] [Google Scholar]
- 23.King, M. P., and G. Attardi. 1989. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science 246:500-503. [DOI] [PubMed] [Google Scholar]
- 24.Kloss, P., L. Xiong, D. L. Shinabarger, and A. S. Mankin. 1999. Resistance mutations in 23 S rRNA identify the site of action of the protein synthesis inhibitor linezolid in the ribosomal peptidyl transferase center. J. Mol. Biol. 294:93-101. [DOI] [PubMed] [Google Scholar]
- 25.Kuter, D. J., and G. S. Tillotson. 2001. Hematologic effects of antimicrobials: focus on the oxazolidinone linezolid. Pharmacotherapy 21:1010-1013. [DOI] [PubMed] [Google Scholar]
- 26.Lewis, L. D., S. Amin, C. I. Civin, and P. S. Lietman. 2004. Ex vivo zidovudine (AZT) treatment of CD34+ bone marrow progenitors causes decreased steady state mitochondrial DNA (mtDNA) and increased lactate production. Hum. Exp. Toxicol. 23:173-185. [DOI] [PubMed] [Google Scholar]
- 27.Lin, A. H., R. W. Murray, T. J. Vidmar, and K. R. Marotti. 1997. The oxazolidinone eperezolid binds to the 50S ribosomal subunit and competes with binding of chloramphenicol and lincomycin. Antimicrob. Agents Chemother. 41:2127-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matassova, N. B., M. V. Rodnina, R. Endermann, H. P. Kroll, U. Pleiss, H. Wild, and W. Wintermeyer. 1999. Ribosomal RNA is the target for oxazolidinones, a novel class of translational inhibitors. RNA 5:939-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKee, E. E., B. L. Grier, G. S. Thompson, and J. D. McCourt. 1990. Isolation and incubation conditions to study heart mitochondrial protein synthesis. Am. J. Physiol. 258:E492-E502. [DOI] [PubMed] [Google Scholar]
- 30.Mears, J. A., J. J. Cannone, S. M. Stagg, R. R. Gutell, R. K. Agrawal, and S. C. Harvey. 2002. Modeling a minimal ribosome based on comparative sequence analysis. J. Mol. Biol. 321:215-234. [DOI] [PubMed] [Google Scholar]
- 31.Moellering, R. C. 2003. Linezolid: the first oxazolidinone antimicrobial. Ann. Intern. Med. 138:135-142. [DOI] [PubMed] [Google Scholar]
- 32.Nissen, P., J. Hansen, N. Ban, P. B. Moore, and T. A. Steitz. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289:920-930. [DOI] [PubMed] [Google Scholar]
- 33.O'Brien, T. W. 2003. Properties of human mitochondrial ribosomes. IUBMB Life 55:505-513. [DOI] [PubMed] [Google Scholar]
- 34.Polacek, N., S. Swaney, D. Shinabarger, and A. S. Mankin. 2002. SPARK-a novel method to monitor ribosomal peptidyl transferase activity. Biochemistry 41:11602-11610. [DOI] [PubMed] [Google Scholar]
- 35.Rossignol, R., B. Faustin, C. Rocher, M. Malgat, J. P. Mazat, and T. Letellier. 2003. Mitochondrial threshold effects. Biochem. J. 370:751-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rossignol, R., R. Gilkerson, R. Aggeler, K. Yamagata, S. J. Remington, and R. A. Capaldi. 2004. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res. 64:985-993. [DOI] [PubMed] [Google Scholar]
- 37.Rossignol, R., T. Letellier, M. Malgat, C. Rocher, and J. P. Mazat. 2000. Tissue variation in the control of oxidative phosphorylation: implication for mitochondrial diseases. Biochem. J. 347:45-53. [PMC free article] [PubMed] [Google Scholar]
- 38.Schlunzen, F., R. Zarivach, J. Harms, A. Bashan, A. Tocilj, R. Albrecht, A. Yonath, and F. Franceschi. 2001. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413:814-821. [DOI] [PubMed] [Google Scholar]
- 39.Sciotti, R. J., M. Pliushchev, P. E. Wiedeman, D. Balli, R. Flamm, A. M. Nilius, K. Marsh, D. Stolarik, R. Jolly, R. Ulrich, and S. W. Djuric. 2002. The synthesis and biological evaluation of a novel series of antimicrobials of the oxazolidinone class. Bioorg. Med. Chem. Lett. 12:2121-2123. [DOI] [PubMed] [Google Scholar]
- 40.Sharma, M. R., E. C. Koc, P. P. Datta, T. M. Booth, L. L. Spremulli, and R. K. Agrawal. 2003. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell 115:97-108. [DOI] [PubMed] [Google Scholar]
- 41.Shinabarger, D. L., K. R. Marotti, R. W. Murray, A. H. Lin, E. P. Melchior, S. M. Swaney, D. S. Dunyak, W. F. Demyan, and J. M. Buysse. 1997. Mechanism of action of oxazolidinones: effects of linezolid and eperezolid on translation reactions. Antimicrob. Agents Chemother. 41:2132-2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sievers, A., M. Beringer, M. V. Rodnina, and R. Wolfenden. 2004. The ribosome as an entropy trap. Proc. Natl. Acad. Sci. USA 101:7897-7901. (Erratum, 101: 12397-12398.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spahn, C. M., E. Jan, A. Mulder, R. A. Grassucci, P. Sarnow, and J. Frank. 2004. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: the IRES functions as an RNA-based translation factor. Cell 118:465-475. [DOI] [PubMed] [Google Scholar]
- 44.Storrie, B., and G. Attardi. 1972. Expression of the mitochondrial genome in HeLa cells. 13. Effect of selective inhibition of cytoplasmic or mitochondrial protein synthesis on mitochondrial nucleic acid synthesis. J. Mol. Biol. 71:177-199. [DOI] [PubMed] [Google Scholar]
- 45.Swaney, S. M., H. Aoki, M. C. Ganoza, and D. L. Shinabarger. 1998. The oxazolidinone linezolid inhibits initiation of protein synthesis in bacteria. Antimicrob. Agents Chemother. 42:3251-3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Villani, G., and G. Attardi. 2000. In vivo control of respiration by cytochrome c oxidase in human cells. Free Radic. Biol. Med. 29:202-210. [DOI] [PubMed] [Google Scholar]
- 47.Xiong, L., P. Kloss, S. Douthwaite, N. M. Andersen, S. Swaney, D. L. Shinabarger, and A. S. Mankin. 2000. Oxazolidinone resistance mutations in 23S rRNA of Escherichia coli reveal the central region of domain V as the primary site of drug action. J. Bacteriol. 182:5325-5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Youngman, E. M., J. L. Brunelle, A. B. Kochaniak, and R. Green. 2004. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell 117:589-599. [DOI] [PubMed] [Google Scholar]
- 49.Yunis, A. A. 1988. Chloramphenicol: relation of structure to activity and toxicity. Annu. Rev. Pharmacol. Toxicol. 28:83-100. [DOI] [PubMed] [Google Scholar]