Abstract
Two clinical Candida albicans isolates that exhibited high-level resistance to azoles and modest decreases in susceptibility to amphotericin B were cultured from unrelated patients. Both isolates harbored homozygous nonsense mutations in ERG3, which encodes an enzyme, sterol Δ5,6-desaturase, involved in ergosterol synthesis. Extraction and analysis of the sterols from both isolates confirmed the absence of sterol Δ5,6-desaturase activity. Although the loss of sterol Δ5,6-desaturase activity is known to confer resistance to azoles, this mechanism of resistance has rarely been seen in clinical isolates, suggesting that such mutants are at a competitive disadvantage. To test this hypothesis, the virulence of the erg3 mutants was assayed by using a mouse systemic infection model. The mutants were significantly less virulent than the wild-type comparator strains. However, the kidney fungal burdens in mice infected with the erg3 mutants were similar to those in mice infected with the wild-type strains. Similar results were obtained by using a laboratory-generated homozygous erg3 deletion mutant (D. Sanglard et al., Antimicrob. Agents Chemother. 47:2404-2412, 2003). Reintroduction of a wild-type ERG3 allele into the homozygous deletion mutant restored virulence, ergosterol synthesis, and susceptibility to azoles, confirming that these phenotypic changes were solely due to the inactivation of Erg3p.
Although Candida albicans is a commensal organism, in immunocompromised hosts it can cause a number of infections, many of which are fatal. There are several therapeutic options for the treatment of Candida infections; most currently available drugs target either the fungal cell wall or the membrane. For example, the azoles block ergosterol synthesis through inhibition of lanosterol 14α-demethylase (CYP51), the echinocandins disrupt cell wall synthesis through inhibition of β-glucan synthesis, and the polyenes bind to ergosterol and disrupt the fungal cell membrane (14).
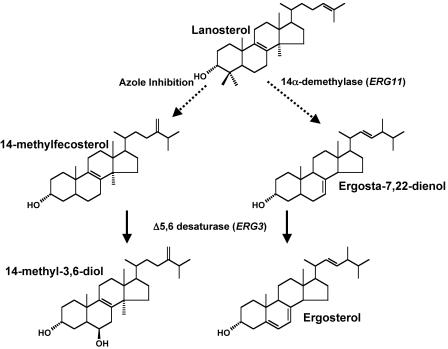
Resistance to antifungal drugs and, in particular, the azoles has been the subject of numerous studies (for a review, see reference 17). In Candida the most prevalent mechanisms of azole resistance appear to be decreased intracellular drug accumulation, which results from increased expression of efflux pump genes, and mutations in the drug target CYP51 (encoded by ERG11), which are proposed to impair drug binding. A third, less frequently observed mechanism involves inactivation of the sterol Δ5,6-desaturase (encoded by ERG3), a nonessential enzyme involved in the late stages of ergosterol synthesis (Fig. 1). Ordinarily, exposure of C. albicans to azoles results in the accumulation of 14α-methylfecosterol, which is subsequently converted by the sterol Δ5,6-desaturase to 14α-methylergosta-8,24 (28)dien-3β,6α-diol (14α-methyl-3,6-diol). Since 14α-methylfecosterol is capable of supporting fungal growth and 14α-methyl-3,6-diol is toxic to the cell, inactivation of the sterol Δ5,6-desaturase renders the cells resistant to azoles (6).
FIG. 1.
Schematic representation of a part of the ergosterol synthesis pathway highlighting the role of the Δ5,6-desaturase in conferring reduced susceptibility to azoles.
In this study we characterized two clinical C. albicans isolates that exhibited resistance to a wide range of azoles; both isolates were found to harbor homozygous nonsense mutations in ERG3. The loss of ERG3 appeared to attenuate virulence and the ability of the organism to undergo the yeast phase-mycelial phase transition, findings that may explain why this mechanism of resistance is rarely seen in clinical isolates.
MATERIALS AND METHODS
C. albicans strains.
C43, C655, C673, and C410 are clinical isolates from the Schering-Plough Research Institute (SPRI; Kenilworth, NJ) culture collection. DSY1751 (erg3Δ::hisG/erg3Δ::hisG-URA3-hisG erg11Δ::hisG/ERG11), a homozygous erg3 deletion mutant, and its progenitor, DSY1336 (erg3Δ::hisG-URA3-hisG/ERG3 erg11Δ::hisG/ERG11), were provided by Dominique Sanglard (18). ASC99 is a ura3 derivative of DSY1751 that was generated by plating DSY1751 on yeast extract-peptone-dextrose (YPD; QBiogene, Carlsbad, CA) agar supplemented with 0.01% (wt/vol) 5-fluoorotic acid and 10 mM uridine (Sigma Chemical Co., St. Louis, MO) to select for cells that had lost URA3 through genetic recombination between the hisG repeats. Strains were routinely cultured in either Sabouraud dextrose (QBiogene) or YPD broth. The identities of the clinical isolates were confirmed by using the Vitek identification system with the Yeast Biochemical Card (bioMérieux Vitek Inc., Hazelwood, MO).
Antifungal agents and susceptibility testing.
Posaconazole was prepared at SPRI as a micronized powder. Itraconazole and amphotericin B powders were obtained from Janssen Pharmaceutica Inc. (Beerse, Belgium) and Sigma Chemical Co., respectively. Voriconazole and fluconazole were obtained from Pfizer Inc. (New York, N.Y.). All drugs except voriconazole were dissolved in dimethyl sulfoxide; voriconazole was dissolved in water. Broth microdilution MIC testing in RPMI 1640 medium (BioWhittaker, Walkersville, MD) was performed as described in the CSLI (formerly the National Committee for Clinical Laboratory Standards) document M27-A2, Reference Method for Broth Dilution Susceptibility Testing of Yeasts (12). Agar-based MIC testing was performed by using Etest strips (AB Biodisk, Solna, Sweden) on RPMI 1640 agar plates supplemented with 2% glucose, as directed by the manufacturer.
DNA-typing techniques for strain identification.
Repetitive element PCR was performed by using a DiversiLab Candida kit, and the PCR was run on a Caliper 1000 analyzer, as described by the manufacturer (Bacterial BarCodes, Houston, TX). The resultant data were analyzed with DiversiLab System software.
Sterol identification.
Test strains were labeled with [14C]acetate, and nonsaponifiable lipids were extracted and resolved by high-pressure liquid chromatography as described previously (11). Sterol identity was confirmed by gas chromatography-mass spectroscopy (GC-MS).
DNA sequencing and quantitation of gene expression levels.
The ERG3- and ERG11-coding regions were amplified by PCR from purified chromosomal DNA as overlapping 500-bp fragments and were sequenced by GeneWiz, Inc. (North Brunswick, NJ). The sequences were compared to the following GenBank sequences: the ERG3 sequence with accession number AF069752 and the ERG11 sequence with accession number X13296. Gene expression levels were measured by a real-time quantitative multiplex PCR, as described previously (1).
Germ tube formation.
Cells were grown overnight in YPD at 30°C with shaking. Germ tube formation was induced by subculturing the cells into fresh YPD supplemented with 10% (vol/vol) serum (Serum Supreme; Biowhittaker) and incubating the cells at 37°C without shaking. Control cultures were grown at 30°C without serum or shaking.
Reversion of Δerg3 in C. albicans strain DSY1751.
A linear PCR product that comprised the URA3 marker flanked on the 5′ side by the intact ERG3 gene and on the 3′ side by the sequences normally found downstream of ERG3 was generated as follows. Chromosomal DNA extracted from C. albicans C43 was used as the template for the PCRs. The open reading frames for ERG3 (5′-AGCATCCCTCTAATCTAAGAAATACTTTGT-3′ and 5′-CAGCTCTTTTTTTTGTTTCCGTTTATACCATCCAATCATTGTTCAACATATTCTCTA-3′) and URA3 (5′-GTTGACGATAGAGAATATGTTGAACAATGATTGGATGGTATAAACGGAAACA-3′ and 5′-AGTCAATGGTCCAAAACAAAGATGTACCAATCTAGAAGGACCACCTTTGATTG-3′), plus the region downstream of ERG3 (5′-CTATTTACAATCAAAGGTGGTCCTTCTAGATTGGTACATCTTTGTTTTGGACCA-3′ and 5′-TCTACTTCTAAACAAACTGAAATGGCTAACACT-3′), were amplified by PCR with the indicated primers. The primers were designed to provide 20 bp of overlapping homology between each fragment; subsequently, all three fragments were fused together in a single PCR by using flanking primers (5′-AGACTACGCGAGACCACACTTGCAT-3′ and 5′-AAGAAAGAAAGGTGTATTTAAAGTTCGATT-3′). The 3.97-kb product was used to transform ASC99 to prototrophy by using the Frozen-EZ Yeast Transformation II kit (Zymo Research, Orange, CA).
Mouse virulence models.
The virulence of the C. albicans strains was assayed by injecting 5 × 106 CFU into the tail vein of groups of 10 male CR-CF1 mice; survival was monitored for 10 days. For measurement of both survival and kidney fungal burdens, groups of 20 mice were inoculated as described above and then randomly assorted into two groups; one group was monitored for survival. Half the mice from the second group were euthanized after 24 h, and the kidney fungal burdens were determined by plating serial dilutions of homogenized tissue. Samples from the remaining five mice were cultured as they died; the survivors were euthanized on day 10 and samples were cultured. Differences in survival were analyzed for statistical significance by using the log-rank test.
RESULTS
Source of clinical C. albicans isolates.
Fourteen days after receiving hematopoietic stem cell transplantation, a patient was diagnosed with candidemia and was given fluconazole. C. albicans strains C673 and C410 were isolated 15 and 17 days posttransplantation, respectively. After 13 days of fluconazole therapy there was no response and the patient was switched to liposomal amphotericin B; seven days later the patient was culture negative. However, the patient died 4 days later, 38 days after receiving the transplant. Strain C655 was isolated from a second patient at a different hospital; no further details are available.
Susceptibility profiles of the clinical C. albicans isolates.
Strains C410 and C655 exhibited significant reductions in susceptibility to azoles and a modest reduction in susceptibility to amphotericin B (Table 1). Strains C673 and C43 (strain C43 is a clinical isolate from the SPRI culture collection and was included for comparative purposes) were susceptible to all drugs tested. DNA fingerprinting confirmed that the isolates from the patient who had received a hematopoietic stem cell transplant (C410 and C673) were closely related to each other but distinct from C43 and C655 (data not shown).
TABLE 1.
Antifungal susceptibilities of C. albicans isolates
| Strains | ERG3 genotype | MIC (μg/ml)
|
||||
|---|---|---|---|---|---|---|
| Posaconazole | Itraconazole | Fluconazole | Voriconazole | Amphotericin B | ||
| C43 | ERG3/ERG3 | <0.03 | 0.06 | 0.25 | 0.03 | 0.5 |
| C673 | ERG3/ERG3 | 0.016 | 0.03 | 0.25 | 0.016 | 0.5 |
| C410 | erg3/erg3 | >8 | >16 | >256 | >16 | 1 |
| C655 | erg3/erg3 | >8 | >8 | >256 | >16 | 2 |
| DSY1366a | ERG3/erg3 | 0.003 | 0.006 | 0.75 | 0.012 | NTb |
| DSY1751a | erg3/erg3 | >32 | >32 | >256 | >32 | NT |
| ASC100a | ERG3/erg3 | 0.002 | 0.003 | 0.5 | 0.008 | NT |
MIC values were obtained by using Etest strips.
NT, not tested.
Azole-resistant clinical C. albicans isolates harbor homozygous nonsense mutations in ERG3.
Previously, we used real-time quantitative PCR to measure the levels of expression of genes encoding efflux pumps (CDR1, CDR2, and MDR1), ERG11, and two control genes (ACT1 and PMA1) in a collection of 14 fluconazole-susceptible C. albicans isolates (1). For each gene, the average expression levels and the variations within the population were determined. The azole-resistant strains (C410 and C655) were analyzed in the same fashion. The expression levels of CDR1, CDR2, MDR1, and ERG11 in both strains were comparable to those found in the azole-susceptible isolates (data not shown). In the absence of changes in the levels of expression of efflux pumps, we screened for mutations in ERG11 and ERG3. There were no missense mutations in ERG11 in strains C410 and C673. Strain C655 harbored a homozygous mutation in ERG11 that resulted in the substitution of glutamate for aspartate at residue 116. However, this mutation has been found in several azole-susceptible isolates and is not thought to play a role in conferring azole resistance (16). There were no mutations in ERG3 in azole-susceptible strains C673 and C43. In contrast, strains C410 and C655 harbored homozygous nonsense mutations in ERG3: in C410, codon 266 was mutated from leucine to an opal stop codon; in isolate C655, codon 228 was mutated from tryptophan to an opal stop codon.
Sterol analysis confirms the absence of sterol Δ5,6-desaturase activity in the erg3 mutants.
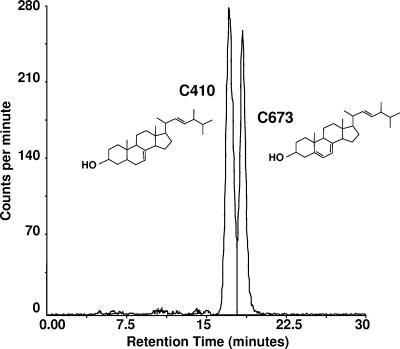
The predominant sterol extracted from 24-h cultures of azole-susceptible strains C43 and C673 was ergosterol (data not shown). The major sterol extracted from the erg3 mutants (C410 and C655) eluted approximately 1 min earlier than ergosterol. This was confirmed by coinjecting the sterols isolated from C673 and C410; there was a clear separation of the two sterols (Fig. 2). GC-MS analysis confirmed that the sterol extracted from both erg3 mutants was ergosta-7,22-dien-3-ol, an ergosterol precursor that retains a saturated bond between C-5 and C-6. Consistent with the loss of Δ5,6-desaturase activity, exposure of strain C655 to sub-MICs of posaconazole resulted in the accumulation of 14-methylfecosterol, as well as smaller amounts of eburicol, obtusifoliol, and lanosterol (data not shown).
FIG. 2.
High-pressure liquid chromatography chromatogram of the sterols extracted from C. albicans strains C673 (wild type) and C410 (homozygous erg3 mutant) run on the same column; the major peaks were identified by GC-MS as ergosterol and ergosta-7,22-dien-3-ol, respectively.
C. albicans erg3 mutants exhibit reduced virulence.
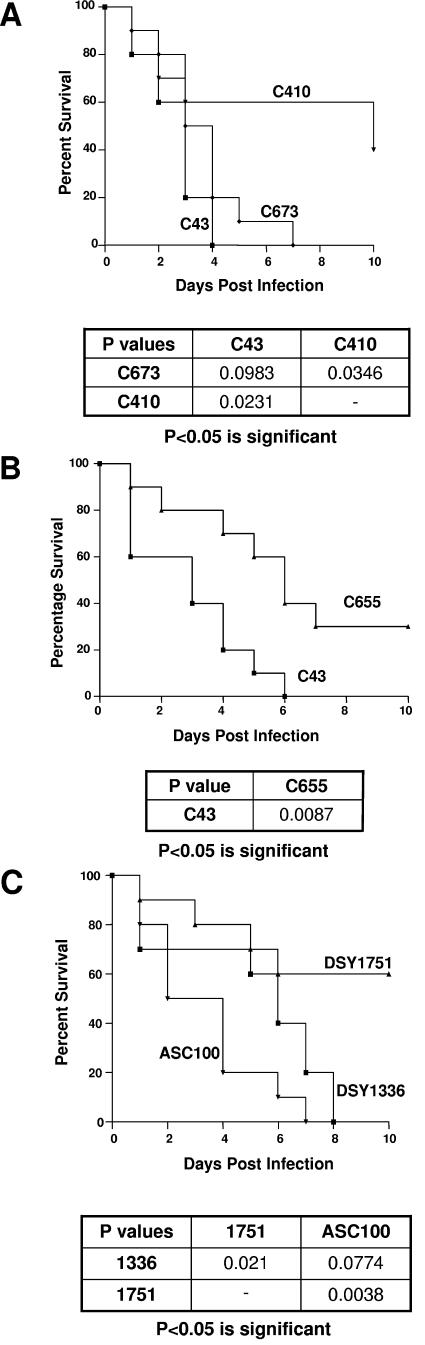
The virulence of strains C43, C410, C673, and C655 was assayed in a mouse systemic infection model using survival as an end point. At comparable inoculation levels, azole-susceptible wild-type strains C43 and C673 were equally virulent; in two independent tests, all mice succumbed by days 6 to 10 (Fig. 3A). In contrast, mice infected with the erg3 mutants (C410 and C655) survived longer than those infected with the wild-type strains (Fig. 3A and B).
FIG.3.
Impact of nonsense mutations in ERG3 on the virulence of C. albicans. Azole-resistant clinical isolates C410 and C655 (both isolates harbored homozygous nonsense mutations in ERG3) and azole-susceptible isolates C43 and C673 were tested in a murine systemic infection model. For all strains, 5 × 106 CFU was injected into the tail vein. Statistical differences in virulence, as measured by survival, are shown under each Kaplan-Meier plot (A and B). A laboratory-generated homozygous erg3 deletion mutant (DSY1751) along with its heterozygous progenitor (DSY1366) and an engineered revertant (ASC100) were similarly tested (C).
The virulence assay was repeated using a matched pair of laboratory strains; either one or both copies of ERG3 were deleted from DSY1336 and DSY1751, respectively (note both strains have a deletion of one copy of ERG11). Consistent with previous results, the homozygous erg3 deletion mutant, but not the heterozygous progenitor, exhibited reduced susceptibility to azoles (note that these strains gave inconsistent MICs when they were tested by the broth microdilution methodology; consequently, the values reported in Table 1 were obtained with Etest strips). Similarly, in the mouse model, DSY1751 was significantly less virulent than DSY1336 (Fig. 3C). To confirm that both the virulence and resistance phenotypes were a direct result of the loss of Δ5,6-desaturase activity a wild-type ERG3 gene was reintroduced into a ura3 derivative of DSY1751, ASC99 (see the Materials and Methods section for a description of ASC99). The resultant strain, ASC100, was susceptible to azoles (Table 1), synthesized ergosterol (data not shown), and was significantly more virulent than the homozygous erg3 mutant (Fig. 3C).
Reduced virulence is not accompanied by a reduction in kidney fungal burdens.
To determine if the observed reduction in virulence reflected an inability of the erg3 mutants to exit the bloodstream and enter tissues, we examined the kidney fungal burdens 24 h after infection. There were no significant differences (P ≥ 0.05) in the number of cells recovered from the kidneys of mice infected with either wild-type or the mutant strains (Table 2). The fungal burdens in mice infected with an erg3 mutant were also measured at day 10 to determine if cell numbers changed over time. At day 10 there appeared to be a 10-fold increase in the number of colonies recovered; however, the increase was not statistically significant (P = 0.051).
TABLE 2.
Kidney fungal burdens in mice 24 h after infection with C. albicans isolates
| Strain | ERG3 genotype | Average fungal burden (log10 CFU) | Variance | Comparison of fungal burdens (P values) |
|---|---|---|---|---|
| C43 | ERG3/ERG3 | 4.9 | 0.23 | C43 vs C673 (0.28) |
| C673 | ERG3/ERG3 | 5.2 | 0.17 | C43 vs C410 (0.05) |
| C410 | erg3/erg3 | 5.8 | 0.57 | C410 vs C673 (0.16) |
| DSY1336 | ERG3/erg3 | 4.4 | 0.08 | DSY1336 vs DSY1751 (0.16) |
| DSY1751 | erg3/erg3 | 4.0 | 0.37 |
C. albicans erg3 mutants are impaired in their ability to filament.
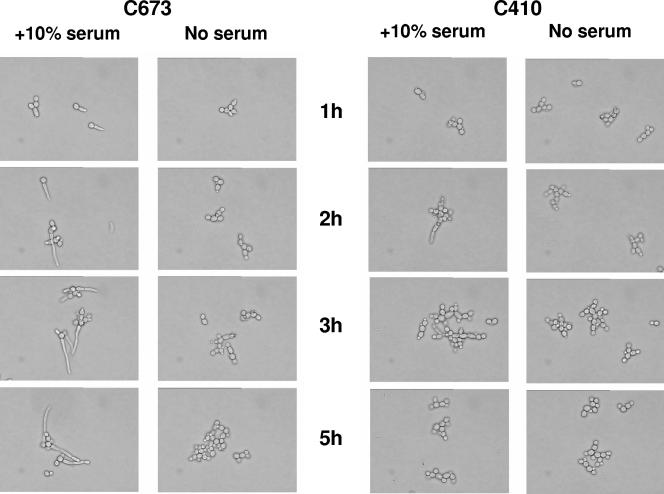
The ability of C. albicans to transition from the yeast form to a filamentous form is essential for virulence (19). To determine if the erg3 mutants retained the ability to form filaments they were exposed to serum at 37°C. Wild-type strains C43 and C673 readily formed filaments within 2 to 3 h of being exposed to serum; over the same time period, the erg3 mutants C655 and C410 did not elaborate hyphae (Fig. 4).
FIG. 4.
Impact of nonsense mutations in ERG3 on the ability of C. albicans to transition to the hyphal form. Strains C673 (wild type) and C410 (homozygous erg3 mutant) were subcultured into medium supplemented with serum and incubated at 37°C. At the indicated time points, they were examined microscopically for filament formation.
DISCUSSION
Therapeutic intervention inevitably selects for those microorganisms with mutations that confer reduced susceptibility to the challenge drug. In the case of azoles, reduced susceptibility has primarily been associated with either alterations in the drug target site or increased efflux resulting from the overexpression of the genes encoding efflux pumps. In rare instances, isolates with nonsense mutations in ERG3 have been recovered (5, 6, 13). In an effort to combat resistance, as well as to improve the spectrum of activity, a new group of triazoles (e.g., posaconazole, itraconazole, voriconazole, and ravuconazole) has been developed. Despite the increased potencies of these new drugs, C. albicans isolates with reduced susceptibility to the newer azoles have been described. In the case of voriconazole and itraconazole, at least two substitutions in CYP51 were required to confer resistance (D. Sanglard, F. Ischer, and J. Bille, Abstr. 42nd Intersci. Conf. Antimicrob. Agents Chemother., abstr. M-221, 2002). For posaconazole, multiple (up to five) substitutions in CYP51 were associated with significant reductions in susceptibility (1, 7). Given the rarity with which isolates with multiple mutations might be expected to occur, it is curious that such isolates appear to be recovered both from patients (3, 4, 8, 15, 16, 21) and from in vitro experiments (2) more frequently than isolates with homozygous nonsense mutations in ERG3 (6, 13). One explanation for this apparent anomaly is that erg3 mutants are at a competitive disadvantage, possibly due to a significant reduction in either growth rate or virulence. To address this issue we analyzed two unrelated clinical C. albicans isolates with homozygous nonsense mutations in ERG3. Growth rate measurements did not reveal any significant differences between wild-type strains and erg3 mutants (data not shown). However, in a mouse model both erg3 mutants were significantly less virulent than the wild-type comparator strains. Similar findings were obtained by using matched laboratory-generated deletion mutants; in this case, the homozygous erg3 deletion mutant was significantly less virulent than its heterozygous progenitor. Reintroduction of a wild-type ERG3 gene into the homozygous erg3 deletion mutant restored both virulence and sensitivity to azoles, confirming that these phenotypic changes were due solely to the loss of sterol Δ5,6-desaturase activity.
Previously, it was found that C. albicans strains that were unable to transition from the yeast to the hyphal form exhibited reduced virulence in mice (19). Interestingly, despite their lack of virulence, the strains were able to leave the blood vessels and establish infections in organs. Both the clinical and the laboratory-generated erg3 mutants described above exhibited a similar phenotype: they were impaired in their ability to form hyphae in the presence of serum in vitro, and they exhibited reduced virulence in mice. Yet, there were no significant differences in the kidney fungal burdens in mice infected with wild-type and mutant strains. The molecular basis for these observations remains to be determined. Previously, azoles (including posaconazole; data not shown) were shown to inhibit hyphal formation (4). A subsequent study suggested that by blocking ergosterol synthesis, the azoles inhibited the formation of ergosterol-rich “rafts” that are associated with the leading edge of the developing hyphae (9). It is conceivable that the loss of sterol Δ5,6-desaturase activity and, as a consequence, the replacement of ergosterol by ergosta-7,22-dien-3-ol also affect raft formation. However, it should be noted that inhibition of hyphal formation was not caused by the accumulation of ergosta-7,22-dien-3-ol per se. Exposure of the erg3 mutants to azoles, which, as described above, results in the synthesis of 14-methylfecosterol rather than ergosta-7,22-dien-3-ol, did not restore the ability of the mutants to grow as hyphae (data not shown). Alternatively, the change in sterol composition may negatively affect one (or more) of the membrane-localized components of the signal transduction pathway(s) that regulate the change in cell morphology. Prior work highlighted the sensitivity of membrane-localized proteins to changes in membrane composition (10); for example, treatment of C. albicans with azoles resulted in uncoordinated activation of chitin synthase (20). The recent construction of C. albicans strains that are able to grow as filaments in the absence of serum (19), presumably bypassing the signal transduction pathway(s), may allow testing of this hypothesis.
In summary, we have shown that although nonsense mutations in ERG3 confer high-level resistance to azoles and a moderate decrease in susceptibility to AMB, they also attenuate virulence in C. albicans. The loss of virulence most likely appears to be a consequence of the erg3 mutant's inability to form hyphae and may go some way to explaining why such mutants are rarely recovered from patients.
Acknowledgments
We thank Dominique Sanglard (University Hospital Lausanne, Lausanne, Switzerland) for supplying C. albicans strains DSY1336 and DSY1751, Andriani Patera (SPRI) for performing survival curve analysis, and Scott Walker (SPRI) for critical reading of the manuscript.
REFERENCES
- 1.Chau, A. S., C. A. Mendrick, F. J. Sabatelli, Loebenberg, and D. P. M. McNicholas. 2004. Application of real-time quantitative PCR to the molecular analysis of Candida albicans strains exhibiting reduced susceptibility to azoles. Antimicrob. Agents Chemother. 48:2124-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cowen, L. E., D. Sanglard, D. Calabrese, C. Sirjusingh, J. B. Anderson, and L. M. Kohn. 2000. Evolution of drug resistance in experimental populations of Candida albicans. J. Bacteriol. 182:1515-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Favre, B., M. Didmon, and N. S. Ryder. 1999. Multiple amino acid substitutions in lanosterol 14alpha-demethylase contribute to azole resistance in Candida albicans. Microbiology 145:2715-2725. [DOI] [PubMed] [Google Scholar]
- 4.Ha, K. C., and T. C. White. 1999. Effects of azole antifungal drugs on the transition from yeast cells to hyphae in susceptible and resistant isolates of the pathogenic yeast Candida albicans. Antimicrob. Agents Chemother. 43:763-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kakeya, H., Y. Miyazaki, H. Miyazaki, K. Nyswaner, B. Grimberg, and J. E. Bennett. 2000. Genetic analysis of azole resistance in the Darlington strain of Candida albicans. Antimicrob. Agents Chemother. 44:2985-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelly, S. L., D. C. Lamb, D. E. Kelly, N. J. Manning, J. Loeffler, H. Hebart, U. Schumacher, and H. Einsele. 1997. Resistance to fluconazole and cross-resistance to amphotericin B in Candida albicans from AIDS patients caused by defective sterol delta5,6-desaturation. FEBS Lett. 400:80-82. [DOI] [PubMed] [Google Scholar]
- 7.Li, X., N. Brown, A. S. Chau, J. L. López-Ribot, C. A. Mendrick, D. Loebenberg, R. S. Hare, B. J. DiDomenico, and P. M. McNicholas. 2004. Changes in susceptibility to posaconazole (POS) in clinical isolates of Candida albicans. J. Antimicrob. Chemother. 53:74-80. [DOI] [PubMed] [Google Scholar]
- 8.Marichal, P., L. Koymans, S. Willemsens, D. Bellens, P. Verhasselt, W. Luyten, M. Borgers, F. C. Ramaekers, F. C. Odds, and H. V. Bossche. 1999b. Contribution of mutations in the cytochrome P450 14α-demethylase (Erg11p, Cyp51p) to azole resistance in Candida albicans. Microbiology 145:2701-2713. [DOI] [PubMed] [Google Scholar]
- 9.Martin, S. W., and J. B. Konopka. 2004. Lipid raft polarization contributes to hyphal growth in Candida albicans. Eukaryot. Cell 3:675-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miwa, T., Y. Takagi, M. Shinozaki, C. W. Yun, W. A. Schell, J. R. Perfect, H. Kumagai, and H. Tamaki. 2004. Gpr1, a putative G-protein-coupled receptor, regulates morphogenesis and hypha formation in the pathogenic fungus Candida albicans. Eukaryot. Cell 3:919-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munayyer, H., A. S. Chau, P. A. Mann, T. Yarosh-Tomaine, L. Heimark, K. J. Shaw, R. S. Hare, J. Greene, R. E. Palermo, and P. M. McNicholas. 2004. Posaconazole is a potent inhibitor of sterol 14α-demethylation in yeasts and molds. Antimicrob. Agents Chemother. 48:3690-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.National Committee for Clinical Laboratory Standards. 2002. Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard. Document M27-A2. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 13.Nolte, F. S., T. Parkinson, D. J. Falconer, S. Dix, J. Williams, C. Gilmore, R. Geller, and J. R. Wingard. 1997. Isolation and characterization of fluconazole- and amphotericin B-resistant Candida albicans from blood of two patients with leukemia. Antimicrob. Agents Chemother. 41:196-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Odds, F. C., A. J. Brown, and N. A. Gow. 2003. Antifungal agents: mechanisms of action. Trends Microbiol. 11:272-279. [DOI] [PubMed] [Google Scholar]
- 15.Perea, S., J. L. Lopez-Ribot, W. R. Kirkpatrick, R. K. McAtee, R. A. Santillan, M. Martinez, D. Calabrese, D. Sanglard, and T. F. Patterson. 2001. Prevalence of molecular mechanisms of resistance to azole antifungal agents in Candida albicans strains displaying high-level fluconazole resistance isolated from human immunodeficiency virus-infected patients. Antimicrob. Agents Chemother. 45:2676-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanglard, D., F. Ischer, L. Koymans, and J. Bille. 1998. Amino acid substitutions in the cytochrome P-450 lanosterol 14α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob. Agents Chemother. 42:241-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanglard, D., and F. C. Odds. 2002. Resistance of Candida species to antifungal agents: molecular mechanism and clinical consequences. Lancet Infect. Dis. 2:73-85. [DOI] [PubMed] [Google Scholar]
- 18.Sanglard, D., F. Ischer, T. Parkinson, D. Falconer, and J. Bille. 2003. Candida albicans mutations in the ergosterol biosynthetic pathway and resistance to several antifungal agents. Antimicrob. Agents Chemother. 47:2404-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saville, S. P., A. L. Lazzell, C. Monteagudo, and J. L. Lopez-Ribot. 2004. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot. Cell 2:1053-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vanden Bossche, H. 1985. Biochemical targets for antifungal azole derivatives: hypothesis on the mode of action. Curr. Top. Med. Mycol. 1:313-351. [DOI] [PubMed] [Google Scholar]
- 21.White, T. C. S. Holleman, F. Dy, F. D. Mirels, and D. A. Stevens. 2002. Resistance mechanisms in clinical isolates of Candida albicans. Antimicrob. Agents Chemother. 46:1704-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]