Abstract
Crushed seeds of the Moringa oleifera tree have been used traditionally as natural flocculants to clarify drinking water. We previously showed that one of the seed peptides mediates both the sedimentation of suspended particles such as bacterial cells and a direct bactericidal activity, raising the possibility that the two activities might be related. In this study, the conformational modeling of the peptide was coupled to a functional analysis of synthetic derivatives. This indicated that partly overlapping structural determinants mediate the sedimentation and antibacterial activities. Sedimentation requires a positively charged, glutamine-rich portion of the peptide that aggregates bacterial cells. The bactericidal activity was localized to a sequence prone to form a helix-loop-helix structural motif. Amino acid substitution showed that the bactericidal activity requires hydrophobic proline residues within the protruding loop. Vital dye staining indicated that treatment with peptides containing this motif results in bacterial membrane damage. Assembly of multiple copies of this structural motif into a branched peptide enhanced antibacterial activity, since low concentrations effectively kill bacteria such as Pseudomonas aeruginosa and Streptococcus pyogenes without displaying a toxic effect on human red blood cells. This study thus identifies a synthetic peptide with potent antibacterial activity against specific human pathogens. It also suggests partly distinct molecular mechanisms for each activity. Sedimentation may result from coupled flocculation and coagulation effects, while the bactericidal activity would require bacterial membrane destabilization by a hydrophobic loop.
Diverse communities in the world traditionally use different natural agents from animal or vegetal sources as raw water additives to produce drinking water. Systematic studies have shown that among the different plant-derived materials tested, Moringa oleifera seeds seem to be one of the most effective primary water treatments (17, 28). Water-soluble proteins released from the crushed seed kernels function as natural flocculating agents, which have been proposed to bind and cross-link particles suspended in a colloidal structure, forming larger sedimenting particles (17, 18, 29). Microorganisms are generally attached to solid particles in raw water samples, and treatment employing Moringa seed powder can remove over 90% of the bacterial load (35). Unexpectedly, a study on the sedimentation effect of the seed-derived peptide termed Flo indicated also that it mediates bacterial disinfection, being able to kill antibiotic-resistant bacteria, including several human pathogens (62).
The cationic nature of the Flo polypeptide is characteristic of a wider group of cationic peptide antibiotics that are usually less than 10 kDa in size and display an overall net positive charge. Cationic antimicrobial peptides (AMPs) make part of the innate immunity response, which is the first line of defense against pathogens. Most of the AMPs are expressed constitutively by a wide range of organisms, such as protozoa, prokaryotes, plants, insects, and other higher organisms (4, 23, 59). Target cell specificities are variable among AMP family members, exhibiting bactericidal, fungicidal, and/or tumoricidal properties. Despite their functional variability, AMPs share at least two common structural features: (i) a net positive charge that facilitates the interaction with negatively charged microbial surfaces and (ii) the ability to assume amphiphilic structures, allowing their incorporation into cellular membranes. AMPs have been proposed to exert their antibacterial effects by a wide range of mechanisms, including leakage out of cellular content due to the formation of transmembrane channels, depolarization of bacterial membranes, scrambling of lipid distribution between the leaflets of the cell membrane bilayer, and/or damage of intracellular targets after peptide internalization (73).
A large proportion of the AMPs may display amphipathic α-helices acting as the bactericidal structure (71). The molecular basis of the specificity against microbial membranes has been proposed to rely on the double interaction of the hydrophobic and hydrophilic AMP surfaces with the lipid core and acidic phospholipids of the target bacterial membrane, respectively. Three models were proposed to explain how this interaction ultimately leads to membrane destabilization and cell death: amphipathic helices may mediate the formation of pores spanning the membrane bilayer either by interfacing regions of the lipid core region and aqueous environment within the pore (barrel stave model) (3, 11, 25) or by the formation of pores constituted by peptide and phospholipid heads from the bent membrane (aggregate channel model) (36, 46). Alternatively, a more general disorganization of the membrane hydrophobic and hydrophilic layers may occur, leading to their disruption (carpet model) (2, 24, 26, 52).
Because AMPs have been found to effectively kill bacterial pathogens that are otherwise resistant to many commonly used antibiotics, they have attracted considerable attention in recent years as novel and potentially efficacious therapeutics. AMP efficacy varies significantly in terms of their effective dose and specificity relative to the affected bacterial cells. Improvement has been achieved either by mutating the primary sequence, for instance to augment their amphiphilic character (9, 10, 14, 19, 63), or by multimerizing the active sequence (1, 33, 67).
Since the Flo polypeptide exhibits the unprecedented property of combining both water clarification and disinfection properties (62), we set up experiments to examine the molecular basis and structural determinants of each of the two activities by performing a structure-function analysis. We find that distinct Flo sequences, and hence mechanisms, mediate either activity. This study also indicates that Flo antibacterial activity may be associated with a motif composed of two amphipathic helices separated by a hydrophobic and kinked structure and that multimeric structures of this motif exhibit more efficient and specific antibacterial activities.
MATERIALS AND METHODS
Computational analysis and modeling of Flo structure.
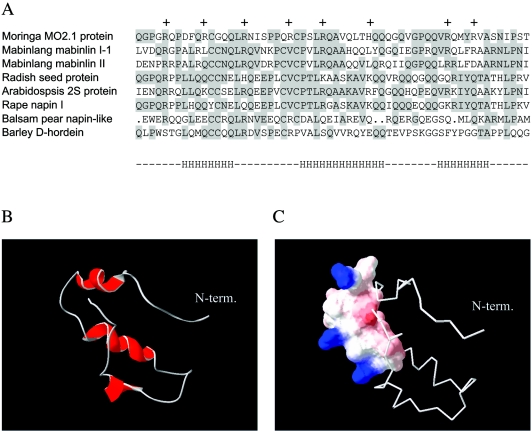
Multiple alignments of Flo (accession number P24303) and similar amino acid sequences were performed employing ClustalW, and secondary structure prediction analysis was performed by Jpred (http://www.ch.embnet.org/) using the following polypeptide sequences. sp P24303 MO2X_MOROL corresponds to the flocculent-active proteins MO2.1 and MO2.2 from Moringa oleifera. sp P80351 2SS1_CAPMA is the sweet protein mabinlin I-1, while sp P30233 2SS2_CAPMA is the precursor of sweet protein mabinlin II, both from Capparis masaikai (mabinlang). tr Q41169 is a fragment of a storage protein from Raphanus sativus (radish), while sp P15460 2SS4_ARATH is the precursor of 2S seed storage protein 4 from Arabidopsis thaliana. tr Q9S9E8 is the napin large chain from Brassica napus (rape), tr Q9S870 is a fragment of 7.9-kDa napin-like protein large chain from Momordica charantia (balsam pear), and tr Q40045 is a fragment of d-hordein from Hordeum vulgare (barley). MODELLER release 6.2 (58) was used to build all non-hydrogen atom models of the Flo polypeptide and derivatives based on the multiple alignment and using the napin B chain as a template (PDB code 1PNB). One hundred models were built for each target. Each model generated by MODELLER was energy minimized using the CHARMM program (7) using 30 steps of Steepest Descend followed by 30 steps of Adopted Basis Newton-Raphson and the CHARMM19 force field (42, 54), with a dielectric constant of 1 and a 20-Å cutoff. For each minimized structure, the ANOLEA energy of residues was determined (37). We used ANOLEA energy values averaged over a 5-residue sliding window. The heavy atoms RMSD of the hundred minimized structures were clustered using a hierarchical clustering method. The minimized structure belonging to the biggest cluster with the lowest ANOLEA energy was retained as the best candidate for each target. The model of Flo polypeptide in Fig. 1 was represented by employing the Deep View software (21, 22).
FIG. 1.
Multiple amino acid alignment of Flo-related proteins and structure modeling. (A) Multiple alignment of 2S seed albumin proteins from plants. Conserved amino acid residues are shaded. The first line corresponds to the flocculating protein MO2.1 from M. oleifera. The following partial sequences represent two members of the mabinlin family from C. masakai, 2S seed storage proteins from R. sativus and A. thaliana, napin protein family members from B. napus and M. charantia, and d-hordein from H. vulgare. Plus signs indicate positive charges in the Moringa protein, while the letter “H” at the bottom indicates the localization of α-helices as predicted from the sequence. The overall sequence identity of Flo and napin is 71%. (B) Three-dimensional representation of Flo polypeptide, as modeled from the known structure of the napin large chain. The N-terminal glutamine residue is indicated. (C) Hydrophobic and hydrophilic properties of the antibacterial Flo structure. The portion of Flo shown to mediate the antibacterial effect is shown using a space-filling model. Colored surfaces indicate polar areas of the molecule, with the positively charged side chains of arginines in blue, while uncharged surfaces are shown in white.
Preparation of synthetic polypeptides.
Polypeptides derived from the Flo amino acid sequence were synthesized on an Applied Biosystems synthesizer 433A or an Abimed AMS 422 multiple peptide synthesizer. Amino acid derivatives were obtained from Novabiochem (Läufelfingen, Switzerland), and other reagents were from Fluka and Sigma Chemie (Buchs, Switzerland). The full-length Flo polypeptide, as well as the P5, P6, and branched derivatives P4aMAP and P4dMAP, was synthesized at the Protein and Peptide Chemistry Facility of the University of Lausanne. The synthesis was performed using a fivefold excess of 9-fluorenylmethoxy carbonyl (Fmoc) amino acid derivatives dicyclohexylcarbodiimide and N-hydroxybenzotriazole as activating agents, and a 60-min coupling time on a p-alkoxybenzylalcohol resin (Wang, Bachem, Bubendorf, Switzerland). Side-chain-protecting groups included the following: the triphenylmethyl group for Asn, Gln, Cys, and His; the pentamethyl-chroman-sulfonyl group for Arg; the t-butyloxycarbonyl group for Lys and Trp; and the t-butyl group for Asp, Glu, Ser, Thr, and Tyr. The peptides were deprotected and cleaved from the resin by treatment with 2.5% H2O-5% triethylsilane in trifluoroacetic acid (TFA) for 2 h at room temperature. After removal of the resin by filtration, the peptides were precipitated with tert-butyl methyl ether and centrifuged and the pellets were resuspended in 1% acetic acid-water and lyophilized. The lyophilized material was then subjected to analytical high-performance liquid chromatography (HPLC) and to matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) or electrospray ionization-TOF analyses. External calibration was performed to allow accurate mass measurement.
MAPs (multiple-antigen peptides) were prepared as an asymmetric four-branch structure as described previously (65), except that an extra Fmoc-Lys (Boc) was added as the C-terminal amino acid on the resin followed by an Fmoc-Ala followed by Fmoc-Lys (Fmoc) to serve as the first branching point. In short, MAPs were synthesized by stepwise solid-phase synthesis in a single operation starting with the C-terminus core matrix using deprotected Fmoc-Lys (Fmoc) to create the branching points (65, 68). After deprotection of Fmoc by piperidine, a spacer comprising G-G was added, followed by Fmoc-Lys (Fmoc), which served as the second branching point of the MAP. After deprotection of Fmoc, the peptide sequences were sequentially elongated to the lysine core matrix on the resin to form the desired four-branch structure. The amino acid compositions of the branched peptides were determined by amino acid analysis (8) and were in agreement with expectations.
Purification and analysis of synthetic polypeptides.
Crude MAPs were reconstituted in 1% acetic acid-water, and low-molecular-weight contaminants were removed by gel filtration on Sephadex G-25. The materials eluted in the void volume were lyophilized, reconstituted in 1 ml water, and subjected to reverse-phase (RP) HPLC on a Vydac column (250 by 22 mm, 10 to 15 μm; Bucher Biotec, Basel, Switzerland), Xterra Prep MS C18 (5 μm, 10 mm by 100 mm; Waters Corporation, Milford, Mass.), or monolithic Chromolith Performance RP-C18 column (4.6 by 100 mm; Merck, Darmstadt, Germany). The Chromolith column was eluted at a flow rate of 9 ml/min by a linear gradient of 0.1% TFA-acetonitrile on 0.1% TFA-H2O, rising within 60 min from 10% to 100%. The optical density of the eluate was monitored at 220 or 280 nm. Fractions were collected and analyzed by mass spectrometry. Fractions containing the peptide of the expected molecular weight were pooled, lyophilized, and reconstituted in water. The purified material was then subjected to mass spectrometry and analytical HPLC in a 250- by 4-mm, 5-μm C18 nucleosil column (Agilent Technologies AG, Geneva, Switzerland). The column was eluted at 1 ml/min by a linear gradient of 0.1%-acetonitrile on 0.1% TFA-H2O, rising within 30 min from 0 to 60%, and detection was performed at 220 and 280 nm using a Waters 991 photodiode array detector. The concentration of peptides was estimated from the absorbance of peptide bonds at 214 nm and/or by amino acid analysis performed using the DABS-Cl (dimethylamino azobenzenesulfonyl chloride) method as described in reference 8. The purity of the peptides was estimated to be at least 80%.
Mass spectrometry and amino acid analysis.
Mass spectrometry analyses were performed using a PerSeptive Biosystems MALDI-TOF Voyager DE-RP mass spectrometer (Framingham, MA) operated in the delayed extraction and linear mode or an LCT instrument (Micromass, Manchester, United Kingdom) combining an electrospray source and a TOF analyzer. α-Cyano-4-hydroxycinnamic acid was purchased from Sigma Chemical Co. (St. Louis, Mo.). All other chemicals were of the highest purity and were used without further purification.
Water clarification assays.
Water clarification activity was evaluated using a 1-mg/ml suspension of 3.5- to 7-μm-diameter glass particles (Spheriglass 5000; Potters-Ballotini, Barnsley, United Kingdom) in 2 ml of 50 mM phosphate buffer, pH 7.0, to mimic turbid water (62). Stirring was continuous at 800 rpm, and optical density (OD) was recorded at 600 nm in a spectrophotometer (LabVIEW software; National Instruments Corporation). After 5 min of continuous stirring, the compound to be tested was added to a final concentration of 10 μg/ml, unless otherwise noted, and stirring was continued for 15 min. The sedimentation efficiency was estimated from a linear regression performed on time points corresponding to 4 min before the addition of the polypeptide solution (basal sedimentation) and 4 min after its addition (coagulation-mediated sedimentation).
Bacterial strains and antibacterial activity assays.
Escherichia coli ER2566 (New England Biolabs, Hitchin, Hertfordshire, United Kingdom), Salmonella enterica serovar Typhimurium (72), Pseudomonas aeruginosa PAO1 ATCC 15692, methicillin-resistant Staphylococcus aureus P8 (12), Streptococcus pyogenes ATCC 700294, and Enterococcus faecalis (clinical isolate) were grown at 37°C in either brain heart infusion (Oxoid Ltd., Hampshire, England), in Luria Bertani broth (LB; Difco Laboratories, Detroit, MI), on LB agar (Difco), or on Columbia agar (Oxoid) supplemented with 3% blood.
Antibacterial activity was assayed as described previously (62). In short, exponentially growing bacteria (OD620 = 0.5) were washed and resuspended in the same volume of 10 mM phosphate buffer, pH 7.0. Synthetic peptides were added to a final concentration of 300 μM, unless indicated otherwise in the figure legend, and incubated with bacteria for 2 h. Cell viability was assessed by plating treated or nontreated bacteria on nonselective agar plates. Unless otherwise noted, the average of the survival ratio and standard error of at least three independent experiments are depicted on the figures.
Confocal laser microscopy.
E. coli cells from 100 μl of an exponential-phase culture were incubated as described above with 300 μM of Flo derivatives for 4 h at 37°C. Control and treated bacteria were exposed to the Live/Dead BacLight bacterial viability stains according to the manufacturer's instructions (Molecular Probes, Eugene, Oreg.). One hundred microliters of the undiluted bacterial suspension was mixed with 0.3 μl of a mixture of equal parts of SYTO 9 and propidium iodide and incubated in the dark for 20 min. Two microliters of the stained suspension was deposited on a glass slide and covered with a coverslip. Image acquisition was performed with a confocal scanning laser microscope (model TCS SL; Leica Lasertechnik GmbH, Heidelberg, Germany). Confocal illumination was provided by Ar and HeNe lasers (488-nm and 543-nm laser excitation, respectively). The SYTO 9 signal was collected in the range 503 to 523 nm, and the propidium iodide signal was collected between 577 and 631 nm. The sensitivities of the photomultipliers and the laser intensity were adjusted and thereafter kept constant throughout the experiments. Randomly selected regions of each sample were imaged using a ×20 oil immersion objective. The area of each section was transformed into a digital image of 512 by 512 pixels. A similar incubation protocol was used for the analysis of bacterial cell aggregation by phase-contrast microscopy, except that a 10-fold-larger amount of bacteria was used.
Hemolytic activity assay.
One hundred microliters of fresh human blood (109 erythrocytes/ml) was mixed with 5 U of heparin (Sigma, St. Louis, Mo.) and 900 μl of 0.9% NaCl (Amino AG, Neuehof, Switzerland). Human erythrocytes were purified from plasma and buffy coat by centrifugation and diluted 1% (vol/vol) in 0.9% NaCl followed by incubation with synthetic peptide at the indicated concentrations for 1 h at 37°C. Hemolysis was determined from the supernatant absorbance at 540 nm (OD540) after sedimentation of the intact cells in a microcentrifuge. Red blood cell lysis was normalized to the absorbance of supernatant value obtained from human erythrocytes similarly incubated with 0.1% Triton X-100, which was scored as 100% hemolysis.
RESULTS
The Flo polypeptide, either expressed as a recombinant peptide or synthesized chemically, was shown to possess both a bactericidal activity and water clarification properties. For instance, it was shown to decrease the viability of gram-negative or gram-positive bacterial cells and to mediate the aggregation of negatively charged particles in suspension, such as bacterial cells, clay, or silicate microspheres (6, 62). This finding raised the possibility that both activities might have related mechanisms and/or structural determinants. To address this possibility, we set out to perform a structure-function analysis of the polypeptide in order to localize the polypeptide sequence responsible for each activity. As a first step toward the synthesis of Flo variants, its secondary and tertiary conformations were modeled according to the known solution structure of related proteins.
Computational modeling of Flo secondary and tertiary structures.
Amino acid sequence analysis and multiple alignments indicated a high degree of similarity between Flo and several polypeptides belonging to the family of 2S albumin seed storage proteins from various plant species (Fig. 1A). These proteins are synthesized as a precursor polypeptide, which is cleaved to form the A and B chains of the mature protein. For instance, the napins are initially synthesized as a precursor protein, which is proteolytically cleaved to generate a smaller A chain of 4,000 Da and a larger B chain of 9,000 Da held together by disulfide bonds (13, 16). Flo is homologous to a part of the large chain of both napin and mabinlin, implying that it corresponds to the B chain of the Moringa 2S seed protein.
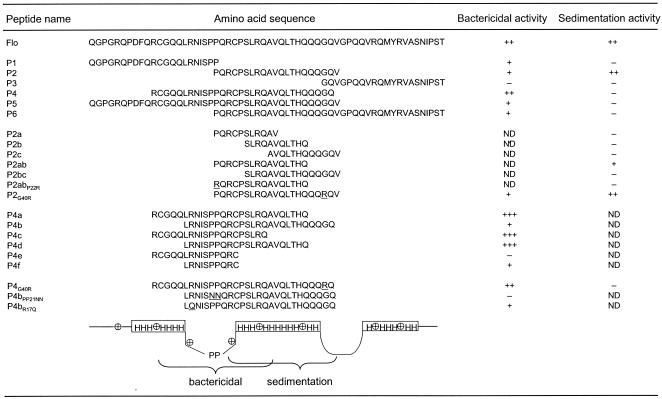
Nuclear magnetic resonance analysis of the A and B subunits of the napins indicated that they contain several α-helices, namely three in the B subunit (47, 56). When analyzing the homologous Flo sequence with several structure prediction programs (20, 55), a consensus model of Flo secondary structure arose, showing a high probability of α-helices in the three regions that form helical structures in the corresponding parts of napin B (Fig. 1A, bottom line). A molecular model for Flo tertiary structure was built, taking these observations and the structure of napin B into account (Fig. 1B). From this model, three short derivatives of Flo were designed so as to contain amino acid sequences corresponding to each of the three putative α-helices (polypeptides P1, P2, and P3, Table 1). These peptides were chemically synthesized and purified by HPLC, and the purity and identity of these and subsequent peptides were confirmed by analytical HPLC and mass spectrometry analysis.
TABLE 1.
Summary of the structure and antibacterial and water clarification effects of the Flo polypeptide and derivativesa
The name, sequence, and activity summary of all linear peptides are shown along with the parental Flo sequence. The underlined amino acid residues on the primary sequence indicate punctual sequence changes. Antibacterial activities are summarized with a minus sign, indicating greater than 10% bacterial survival, or with +, ++, or +++ representing 1 to 10%, 0.01 to 1%, or less than 0.01% survival, respectively, in the antibacterial assay. Flocculation activities depicted by a minus sign indicate lack of detectable activity, while ++ indicates an activity corresponding to 60 to 100% of that observed for the full-length Flo. + represents lower but detectable activity. ND, not determined. At the bottom, a schematic representation of Flo structural and functional features displays the position of the positive charges and the regions predicted to form α-helical structures (H), loops, and the two prolines within a loop.
Analysis of the flocculation activity determinants.
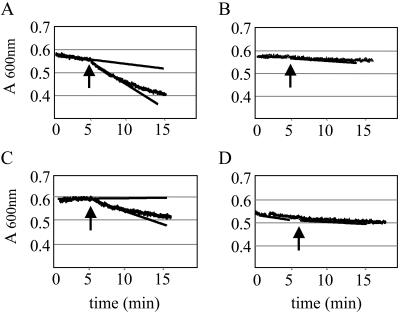
Previous results indicated that the recombinant and synthetic forms of Flo polypeptide are equally active in sedimenting negatively charged microparticles suspended in water, mimicking those found in natural turbid water (6, 62, 69). In this semiquantitative assay, flocculation is recorded as the decrease of the absorbance of the suspension, as resulting from the sedimentation of the flocculated glass microparticles. Efficient flocculation was observed in the presence of 20 μg/ml of chemically synthesized Flo and P2 (Fig. 2A and C), which exhibited comparable activities at this subsaturating concentration (62). In titration experiments, detectable activity was lost between 0.1 and 0.05 μg/ml of either Flo or P2 (data not shown). In contrast, no detectable activity was observed using P1 or P3, even at the high concentration of 50 μg/ml (Fig. 2B and D; see Table 1 for summary). Peptides containing either the N- or C-terminal half of P2 (P2a, P2c) or a peptide of similar length covering the central portion of P2 (P2b) did not show detectable flocculation activity. This suggests that the P2 sequence mediates the flocculation effect of the full-length Flo polypeptide.
FIG. 2.
Effect of Flo, P1, P2, and P3 on water clarification. Sedimentation assays employing glass particle suspension were performed in spectrophotometer cells as described in Materials and Methods, and optical density measurements were taken at 600 nm at 1-s intervals. Individual data points are displayed by the broad curves. After 5 min of stirring, Flo (A), peptide P1 (B), peptide P2 (C), and peptide P3 (D), respectively, were added to a final concentration of 20 μg/ml (A and C) or 50 μg/ml (B and D), as indicated by the arrow. The slopes of the sedimentation curves, before and after addition of the compound to be tested, were estimated by linear regression calculations and are shown as straight lines.
The flocculation activity of Flo may conceivably involve the interaction of the amino acid side chains with the suspended particles through various types of interactions (e.g., hydrophobic, polar, or electrostatic interactions). For instance, Flo positively charged amino acids might associate with, and neutralize, the negative charges present on the surface of the glass microparticles. Consistently, addition of Flo to the microparticles neutralized the zeta potential value of the later from −48 mV to values close to zero (data not shown), indicating that conditions mediating the maximal sedimentation rate of the suspension correspondingly result in nearly complete charge neutralization.
Removal of sequences encompassing one or both of the positively charged arginines from P2 abolished flocculation (P2bc and P2c peptides, Table 1). Deletion of a polyglutamine stretch in P2ab also decreased but did not fully abolish flocculation. Further deletion into a sequence bearing the partial positive charge of histidine and additional glutamines abrogated activity fully in P2a. Addition of a positive charge to the polyglutamine-deleted P2ab peptide, yielding P2ab-P22R, did not restore flocculation, indicating that the additional positive charge cannot compensate for the removal of the glutamine-rich stretch. Similarly, replacement of the glycine of P2 with an arginine in P2-G40R to increase the net positive charge did not increase the flocculation activity either. Taken together, these observations are consistent with a model for a sedimentation mechanism involving both the positive charges and the polyglutamine-containing stretch of P2. Consistently, the P1 and P3 peptides that bear two or three positive charges with a similar distribution to that found for P2, but that lack a polar polyglutamine stretch, do not mediate flocculation. In addition to these features of the polypeptide, other structural constraints may also influence the flocculation activity, as suggested by the lack of activity of the P5 and P6 peptides.
Localization of the antibacterial activity determinants.
To determine if a similar portion of Flo may also mediate its known antibacterial activity, Flo-derived peptides were assayed for their effects on the viability of E. coli enteric bacteria, a common marker of raw water contamination. Exponentially growing bacterial cultures were incubated with the various peptides in a nutrient-poor buffer to mimic natural raw waters (62), and viability was scored as the ability to form colonies after plating on a solid growth medium.
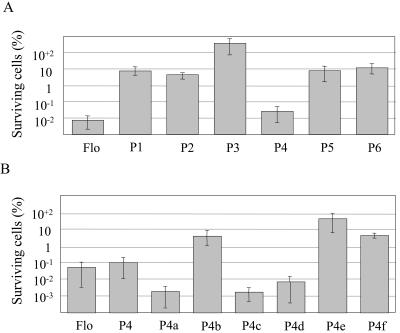
Viability was decreased by 4 orders of magnitude after incubation with full-length Flo peptide, as shown previously (62). Incubation of E. coli cells with 300 μM of P1 and P2 yielded a 10- to 15-fold decrease in viability, whereas P3 was inactive at this or higher concentrations (Fig. 3A) (data not shown). Because P1 and P2 are moderately active as compared to Flo, a new polypeptide was synthesized to comprise the full-length P2 and the C-terminal half of P1, yielding P4. The activity exerted by P4 was found to be comparable to that of Flo (Fig. 3; see Table 1 for summary). Further extensions of P4 sequence towards the C- or the N-terminal extremities decreased activity, suggesting that P4 bears all of the antibacterial determinants (P5 and P6, Fig. 3A and Table 1).
FIG. 3.
Effect of Flo-derived polypeptides on cell viability. Exponentially growing E. coli cells were incubated as described in Materials and Methods with the indicated polypeptides at a final concentration of 300 μM. Bacterial suspensions were plated on nonselective solid medium and incubated overnight at 37°C. Cell viability is expressed as CFU, and values are normalized to those obtained for cells supplemented with buffer only, which were assigned a value of 100%. Initial bacterial loads ranged from 1 × 109 to 5 × 109 CFU/ml. Panel A represents treatments after using Flo-derived polypeptides. Panel B displays the activity of P4 derivatives, with full-length Flo as a standard.
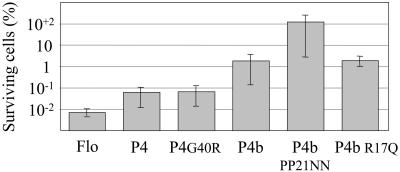
These results suggested that the amino acid sequence responsible for the antibacterial effect may span a sequence overlapping the P1 and P2 peptides, which contains two successive helix-breaking prolines bracketed by positively charged arginines. This possibility is supported by the observation that C-terminal deletions within the second α-helix of P4 do not decrease antibacterial activity (P4a and P4c, Fig. 3B and Table 1). Similarly, deletion of the first α-helix from the N terminus did not affect the antibacterial activity significantly, when comparing P4a with P4d, and it had only a moderate effect when comparing P4 and P4b. However, the complete deletion of helix 2 abrogated activity, when comparing P4c and P4e. Deletions from both ends, up to the amino acid preceding the arginines bracketing the prolines, yielded low activity too (P4f, Fig. 3B). P4b exhibited a weaker effect compared to P4d, and a similar observation was made when comparing P4 with P4a, indicating that the addition of the glutamine-rich stretch interferes with the antimicrobial activity. Taken together, these results localize the minimal antibacterial domain to the proline-containing structure separating the putative helices 1 and 2, while full activity further requires flanking amphipathic helix-forming sequences.
The two prolines at positions 21 and 22 introduce a kink into the peptide, which may expose these hydrophobic residues to the solvent (Fig. 1). This kink structure is bracketed by hydrophilic side chains, the positively charged guanido groups of arginines R17, R24, and R29. Thus, the antibacterial activity might be mediated by the interaction of positively charged amino acids with the negatively charged surface of the bacterial membrane, with the kink pointing into the hydrophobic membrane layers, as a first step toward bacterial membrane disruption. This possibility was addressed by designing peptides where the two prolines at positions 21 and 22 were replaced by asparagines in P4bPP21NN, as they occupy approximately the same volume but are more hydrophilic. This was found to fully abrogate the antibacterial activity (Fig. 4 and Table 1). Alternatively, arginine 17 was replaced by an uncharged glutamine in P4b R17Q, so as to decrease the net positive charge while conserving a high hydrophilicity and approximately the same volume, or a positive charge was added to P4 by replacing G20 with an arginine (P4G20R). The two latter changes did not affect the activity significantly, suggesting that multiple amino acids mediate the proposed interaction of Flo with the bacterial membrane.
FIG. 4.
Effect of mutated polypeptides on cell viability. The E. coli antibacterial assay was performed as described in the legend to Fig. 3, using peptides at a concentration of 300 μM.
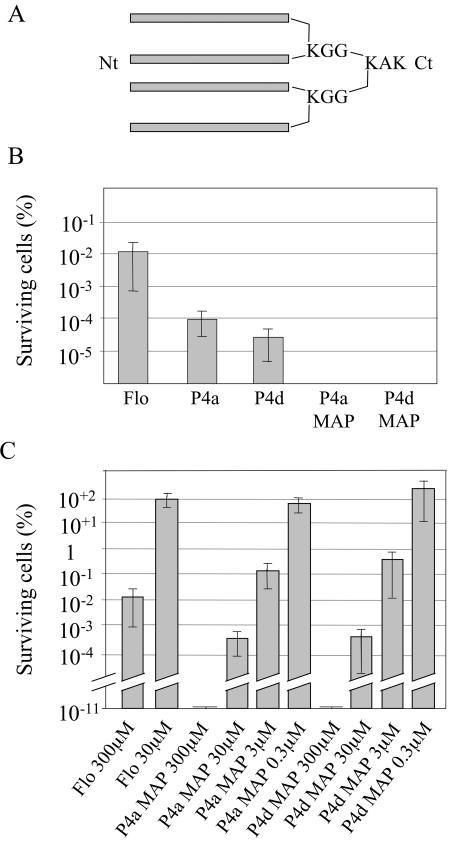
After localizing the sequence responsible for the antibacterial activity, we attempted to identify derivatives of this domain that would mediate higher activity and/or function at reduced concentration. Branched peptides known as MAPs have been used as more efficient antigens than their linear counterparts in immunization studies, which may indicate increased interaction with antibodies (64, 66). MAPs containing four copies of the P4a and P4d peptides were thus synthesized in parallel to their linear counterparts, and E. coli cultures were challenged as before. Branched peptides exhibited improved antibacterial effect as compared to their linear counterparts, and they reduced viable cell counts by more than 10 orders of magnitude (Fig. 5B and C). Moreover, titration experiments indicated that concentrations as low as 3 μM of the branched polypeptides were effective, yielding similar antibacterial activity to 300 μM of Flo (Fig. 5C). This corresponds to titers of 39 and 32 μg/ml for P4a MAP and P4d MAP, respectively.
FIG. 5.
Antibacterial activity of multimerized peptides. (A) Schematic representation of the branched MAPs employed in this study. Each bar represents a monomer of either P4a or P4d peptide. The N terminus and C terminus are depicted by Nt and Ct, respectively. (B) Antibacterial activities of P4a and P4d and corresponding MAP multimers. The antibacterial assay was performed as in Fig. 3, using the indicated peptides at a concentration of 300 μM. (C) Titration of P4a and P4d and corresponding MAP multimers, performed as in the previous panel.
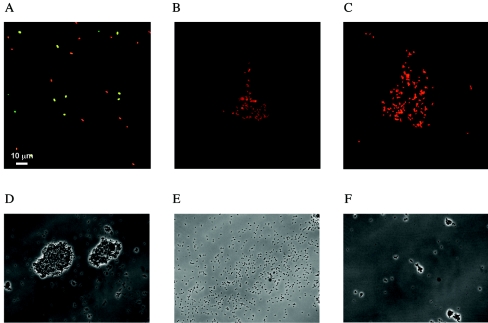
Concomitantly to its effect on cell viability, Flo has been shown to aggregate bacteria at concentrations of 300 μM or higher (6, 62), and similar observations were made here with the MAPs. A previous analysis had indicated that the observed loss of colony formation potential of Flo-treated bacteria was due to a true bactericidal effect, but did not merely result from cellular aggregation. To further assess whether the observed decrease in CFU reflects true antimicrobial effects, treated or untreated cells were observed by confocal microscopy after vital staining using fluorescent dyes probing membrane permeability, followed by several washes to decrease cell aggregation. Most of the bacteria incubated in phosphate buffer alone were viable, as indicated by the green fluorescence (Fig. 6A). On the contrary, essentially all bacteria were red after treatment with P4a MAP, indicating a massive decrease in viability both on free cells and on cells that remained aggregated (Fig. 6C). Identical results were obtained in presence of P4d MAP (data not shown). A partial effect was observed with P4, with cells staining with both the green and red dyes, which is consistent with the lower bactericidal activity of this peptide as compared to the MAPs in the plating assay (Fig. 6B and Table 1). These results thus imply that the Flo-derived peptides strongly decrease cell viability by permeabilizing or disrupting the bacterial membrane.
FIG. 6.
Effect of Flo derivatives on the integrity of E. coli cell membrane. Exponentially growing E. coli cells were incubated briefly with buffer (A) or with 300 μM of P4 (B) or of MAP P4a (C), followed by staining for membrane integrity using the BacLight SYTO 9 and propidium iodide dye assay. Viable (green) and nonviable (red) E. coli cell fluorescence was visualized by confocal microscopy. Alternatively, E. coli cells were incubated with 300 μM of P2 (D), P2ab (E), or P4 (F), washed with phosphate buffer, and visualized by phase-contrast microscopy.
Large aggregates of E. coli cells were clearly observed after incubation with the P2 peptide (Fig. 6D), in agreement with its high activity in the flocculation assay. A similar incubation with P2ab, which displays reduced flocculation activity, yielded reduced bacterial aggregation, with occasional small flocs being dispersed within a population of isolated cells (Fig. 6E) (data not shown). When a similar assay was performed using P4, which displays high bactericidal activity and low flocculation effect, few intact cells remained, either within flocs or as isolated cells (Fig. 6F), suggesting that a massive amount of cell lysis had occurred. Overall, these results are consistent with those obtained with the flocculation assay, and they further document the bacterial disruption activities of Flo derivatives.
To further assess the antibacterial activity of Flo, P4a MAP, and P4d MAP, several gram-negative and gram-positive bacteria were selected from strains that are potential human pathogens. As for E. coli, the antibacterial activity of the MAP polypeptides was much stronger than that noted for Flo (Table 2). Both P4a MAP and P4d MAP were highly effective against P. aeruginosa and S. pyogenes. E. faecalis was less sensitive to the MAPs, whereas S. enterica was only slightly sensitive to P4a MAP. In each case, P4a MAP was somewhat more effective than P4d MAP. S. aureus appeared to be resistant. Finally, Flo presented no antifungal activity against Candida albicans and Cladosporum cucumerinum (data not shown).
TABLE 2.
Summary of antibacterial effect of Flo, P4a MAP, and P4d MAP on pathogenic bacteria
| Organism | Viability (log CFU/ml)a
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Nontreatedb | Flo (300 μM) | P4a MAP
|
P4d MAP
|
|||||
| 300 μM | 30 μM | 3 μM | 300 μM | 30 μM | 3 μM | |||
| P. aeruginosa | 7.16 ± 0.12 | 5.53 ± 0.83 | — | 2.51 ± 1.68 | 6.79 ± 0.06 | — | 4.04 ± 1.10 | 7.01 ± 0.24 |
| S. enterica | 7.91 ± 0.24 | 7.60 ± 0.18 | 5.02 ± 0.22 | 7.29 ± 0.17 | ND | 6.15 ± 0.12 | 7.73 ± 0.73 | ND |
| E. faecalis | 7.98 ± 0.09 | 6.50 ± 0.23 | 3.47 ± 0.43 | 5.69 ± 0.12 | 7.55 ± 0.14 | 3.68 ± 0.39 | 5.07 ± 1.25 | 7.52 ± 0.17 |
| S. aureus | 8.51 ± 0.52 | 7.57 ± 0.11 | 7.61 ± 0.01 | ND | ND | 7.21 ± 0.18 | ND | ND |
| S. pyogenes | 7.10 ± 0.31 | 6.95 ± 0.44 | — | 2.13 ± 0.49 | 6.71 ± 0.35 | — | 4.88 ± 1.47 | 7.02 ± 0.99 |
Viability was determined after 2 h of exposure to Flo, MAP P4a, or MAP P4d. ND, not determined since a 10-times higher concentration showed little or no antibacterial effect. —, total loss of viability.
Viable count after 2 h in phosphate buffer.
Hemolytic activity.
To assess possible membrane disruption effects of the MAPs on eukaryotic cells, the hemolytic activities of the linear and branched peptides were assayed on red blood cells isolated from human blood samples. As can be seen from Table 3, little or no hemolytic activity could be noted at the effective concentrations of 30 or 3 μM. Some hemolytic activity could be noted at the highest concentration (300 μM), either with the parental Flo or with the branched or linear derivatives. Thus, the bactericidal effect can be observed at concentrations that are 1 to 2 orders of magnitude lower than those that elicit disruptive effects on mammalian cells.
TABLE 3.
Summary of antibacterial and hemolytic activities of Flo and derivativesa
| Peptide | Conc (μM) | Surviving cells (%) | Hemolytic activity (%) |
|---|---|---|---|
| Flo | 300 | 10−2-10−1 | 10 |
| 30 | 100 | ND | |
| P4a | 300 | 10−4-10−3 | 41 |
| P4d | 300 | 10−5-10−3 | 34 |
| P4a MAP | 300 | <10−11 | 22 |
| 30 | 10−4-10−3 | 0 | |
| 3 | 10−1 | 0 | |
| 0.3 | 10-100 | 1 | |
| P4d MAP | 300 | <10−11 | 19 |
| 30 | 10−4-10−3 | 6 | |
| 3 | 10−1-1 | 0 | |
| 0.3 | 100 | 1 |
The antibacterial effect was determined on E. coli and depicts the percentage of surviving cells after treatment. Nontreated cultures (100%) correspond to an average of 1012 bacteria per assay. Hemolytic activity was determined by recording the absorbance of red blood cell supernatant after incubation with the indicated peptide and centrifugation. One hundred percent hemolysis corresponds to the absorbance value of human erythrocytes treated with 0.1% Triton X-100.
DISCUSSION
Moringa oleifera seed extracts have been used as a traditional water clarification agent that sediments colloidal particles and suspended bacteria (6, 40, 44, 45, 62). The Flo Moringa seed peptide was recently found to mediate flocculation and to decrease bacterial viability from nutrient-poor contaminated natural waters as well as from rich medium-based minimal bactericidal concentration or MIC assays (6, 62). In the present structure-function study, we wished to analyze the structural basis and possible relationship between the water clarification and disinfection activities of the Flo peptide.
The determinants of Flo flocculation activity were found within the putative central α-helical portion. This study suggests two types of determinants for the sedimentation effect, the most important being a positively charged amino acid sequence, another being a glutamine-rich stretch required for maximal activity. This finding is consistent with a model of the sedimentation of particulate material involving a two-step sequential process (38). The stability of colloidal suspensions of bacteria or clay depends on the repulsive forces between the negative electrostatic charges of the particles and/or their counter-ion layer. Destabilization of colloids thus involves the adsorption of a positively charged compound to the surface of the particle, reducing these repulsive forces. If subsequent collisions lead to stable interactions of the particles, as mediated by attractive forces such as van der Waals forces, coagulation and sedimentation may occur. In addition to charge neutralization by coagulants, flocculating agents act by cross-linking the particles at a distance, forming loose aggregates that will also increase the sedimentation rate. As a consequence, effective water clarification often requires at least two different agents: i.e., cationic metals such as iron, aluminum, or magnesium for coagulation and synthetic or natural polymers for flocculation (28, 30, 31).
Consistently, sedimentation of suspended particles by Flo or its shorter P2 derivative likely involves the neutralization of negative charges in a coagulation mechanism, as indicated by the neutralization of the microparticles' zeta potential. However, a simple charge neutralization mechanism cannot fully account for the sedimentation effect of P2. For instance, sedimentation does not occur with other positively charged portions of Flo having similar charge distribution and isoelectric points, but it requires the glutamine-rich sequence found in P2. Polyglutamine stretches are well known to mediate promiscuous protein-protein interactions, as exemplified by the interactions of numerous transcription-regulating proteins or the formation of insoluble protein aggregates by polyglutamine proteins (32, 49, 60, 61). These results thus point to a mechanism whereby the positively charged part of the peptide would associate with—and neutralize—negative charges on the surface of the particles in a coagulation effect, while the glutamine-rich stretch would mediate interaction of particle-bound peptides, effectively cross-linking particles and resulting in floc formation. The double mode of action of this peptide may be the underlying mechanism of its potent water clarification activity.
Interaction of Flo with the bacterial membrane may conceivably also mediate its antibacterial activity. Nevertheless, we find that the flocculation determinants of Flo can be separated from its antibacterial activity. For instance, the P4 derivative displays potent antibacterial activity, but it is inactive as a flocculation agent. Conversely, P2 and derivatives have high flocculation activity, but they are poor antibacterial agents. Therefore, we conclude that the two activities require distinct but overlapping structures and that the antibacterial activity cannot simply result from a cell aggregation mechanism but requires additional activities.
Flo is homologous to other plants' 2S seed proteins, such as the mabinlins, napins, and d-hordein, that may have the dual function of metabolic storage and defense against microbial attacks (5, 43, 70). The antibacterial effect of such antimicrobial peptides has often been ascribed to peptides that are poorly structured in aqueous solution, but which fold in an amphipathic α-helical structure upon association with the bacterial membrane (39, 71). Consistent with this possibility, the Flo peptide is poorly structured in aqueous solution, according to circular dichroism and nuclear magnetic resonance studies, but it may gain some helical structure upon incubation with synthetic membranes (M.S., unpublished results). Association of multiple amphipathic helices at the interface of the hydrophobic and hydrophilic layers of the membrane would then lead to membrane destabilization and/or to the formation of pores disrupting the transmembrane potential (71). However, further studies will be required to determine the active structure of Flo derivatives.
Unexpectedly, the minimal antibacterial portion of Flo lies within the proline-containing loop that separates two positively charged sequences prone to form α-helices. Structural analysis indicates that it may consist of a hydrophobic patch containing the two prolines bracketed by the positive charges of arginines pointing outward from the folded polypeptide (Fig. 1C). This finding suggests a mechanism whereby interaction of the positively charged side chains with the negatively charged bacterial membrane would ideally place the hydrophobic patch for internalization into the hydrophobic core of bacterial membrane. Indeed, replacing the two hydrophobic prolines with polar asparagines is the most deleterious amino acid substitution, as it totally abrogates antibacterial activity. This would then lead to membrane disruption as proposed for the amphipathic α-helical antimicrobial peptides. Consistently, we find that the MAP derivatives formed from this portion of Flo efficiently permeabilize E. coli membrane, as indicated by the vital dye assay.
Taken together, these observations point to the following mechanistic model. The positively charged portion of Flo involved in both flocculation and antibacterial activities, as found in the P2ab peptide, would interact with the negatively charged bacterial membrane. This portion of Flo would then lead to the flocculation of bacteria when acting in conjunction with the polyglutamine stretch in P2, while it would cooperate with the hydrophobic loop structure to destabilize the bacterial membrane in P4a. The presence of both the polyglutamine and hydrophobic loop as in P4 decreases both the antibacterial and the water clarification effects found with shorter derivatives. This indicates that if the initial step of the interaction of the peptide with the bacterial membrane may be common to both pathways, the peptide conformation or interactions required for further steps toward flocculation or membrane disruption should differ.
The proposed hydrophobic loop of Flo is reminiscent of similar structures found in other antibacterial peptides, such as the type III′ β-turn connecting both strands of the β-sheet on the MDG1 mussel defensin (27, 57). A 9-amino-acid peptide corresponding to this loop exhibited bacteriostatic activity after being cyclized by a non-naturally occurring disulfide bridge. Other antimicrobial loop structures stabilized by disulfide bridges include the C-terminal loop of the hemipteran insect thanatin (15, 34) or the disulfide-constrained loop on the β-hairpin polyphemusin from horseshoe crab (41, 53). In all of these polypeptides, loops are stabilized by the formation of natural or artificially made disulfide bridges that lock hydrophobic and positively charged amino acids in a sharp hairpin structure. In the case of Flo, the cysteines required to form these disulfide bridges are not present. Therefore, the loop may be stabilized by the kink produced by prolines at positions 21, 22, and 26 (Fig. 1C), which is reminiscent of the proline hinge of buforin II that was also proposed to play a key role in membrane penetration (48). Further studies will be required to determine if these proline-containing loop structures represent a new class of antibacterial structure.
The tetramer MAP employed in this study enhanced the interaction of the polypeptides with the target, diminishing the bacterial survival fraction by several orders of magnitude compared to linear polypeptides. This may result from the increase in effective molarity of the monomeric units and the increase of avidity resulting in a decrease of entropy of self-assembly into the bacterial membrane, mimicking the high-order assembly of antimicrobial peptides during their membranolytic action (67). Viability tests proved that MAPs have a strong antibacterial potential against gram-negative and gram-positive bacteria, and they are particularly active on the opportunistic pathogen P. aeruginosa. S. aureus is the only organism tested that appeared to be completely resistant to the MAPs. This resistance may be explained by S. aureus' known mechanism of protection against cationic peptides (50, 51). The finding of an antibacterial activity of MAP derivatives in the micromolar range, its specificity toward some human pathogens developing multidrug resistance to common antibiotics, and the lack of hemolytic activity at effective concentrations thus place Flo derivatives among the attractive potential antibacterial agents, for instance for topical applications.
Acknowledgments
We thank A. McNair for helpful comments on the manuscript and D. Brassard, Y. Poirier, Gisella Stocker, Ian Marison, and C. Doerries for support and helpful comments during the project.
This work was supported by a grant from the Swiss Commission for Technology and Innovation, the Etat de Vaud, and Optima Environnement SA.
REFERENCES
- 1.Azuma, M., T. Kojima, I. Yokoyama, H. Tajiri, K. Yoshikawa, S. Saga, and C. A. Del Caprio. 1999. Antibacterial activity of multiple antigen peptides homologous to a loop region in human lactoferrin. J. Pept. Res. 54:237-241. [DOI] [PubMed] [Google Scholar]
- 2.Bechinger, B. 1999. The structure, dynamics and orientation of antimicrobial peptides in membranes by multidimensional solid-state NMR spectroscopy. Biochim. Biophys. Acta 1462:157-183. [DOI] [PubMed] [Google Scholar]
- 3.Boheim, G. 1974. Statistical analysis of alamethicin channels in black lipid membranes. J. Membr. Biol. 19:277-303. [DOI] [PubMed] [Google Scholar]
- 4.Boman, H. G. 1991. Antibacterial peptides: key components needed in immunity. Cell 65:205-207. [DOI] [PubMed] [Google Scholar]
- 5.Broekaert, W. F., B. P. A. Cammue, M. F. C. D. Bolle, K. Thevissen, G. W. D. Samblanx, and R. W. Osborn. 1997. Antimicrobial peptides from plants. Crit. Rev. Plant Sci. 16:297-323. [Google Scholar]
- 6.Broin, M., C. Santaella, S. Cuine, K. Koukou, G. Peltier, and T. Joet. 2002. Flocculent activity of a recombinant protein from Moringa oleifera Lam. seeds. Appl. Microbiol. Biotechnol. 60:114-119. [DOI] [PubMed] [Google Scholar]
- 7.Brooks, B. R., R. E. Bruccoleri, B. D. Olafson, D. J. States, S. Swaminathan, and M. Karplus. 1983. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comp. Chem. 4:187-217. [Google Scholar]
- 8.Chang, J. Y., R. Knecht, and D. G. Braun. 1981. Amino acid analysis at the picomole level. Application to the C-terminal sequence analysis of polypeptides. Biochem. J. 199:547-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dathe, M., H. Nikolenko, J. Meyer, M. Beyermann, and M. Bienert. 2001. Optimization of the antimicrobial activity of magainin peptides by modification of charge. FEBS Lett. 501:146-150. [DOI] [PubMed] [Google Scholar]
- 10.Dathe, M., M. Schumann, T. Wieprecht, A. Winkler, M. Beyermann, E. Krause, K. Matsuzaki, O. Murase, and M. Bienert. 1996. Peptide helicity and membrane surface charge modulate the balance of electrostatic and hydrophobic interactions with lipid bilayers and biological membranes. Biochemistry 35:12612-12622. [DOI] [PubMed] [Google Scholar]
- 11.Ehrenstein, G., and H. Q. Lecar. 1977. Electrically gated ionic channels in lipid bilayers. Rev. Biophys. 10:1-34. [DOI] [PubMed] [Google Scholar]
- 12.Entenza, J. M., Y. A. Que, J. Vouillamoz, M. P. Glauser, and P. Moreillon. 2001. Efficacies of moxifloxacin, ciprofloxacin, and vancomycin against experimental endocarditis due to methicillin-resistant Staphylococcus aureus expressing various degrees of ciprofloxacin resistance. Antimicrob. Agents Chemother. 45:3076-3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ericson, M. L., J. Rodin, M. Lenman, K. Glimelius, L. G. Josefsson, and L. Rask. 1986. Structure of the rape seed 1.7S storage protein, napin, and its precursor. J. Biol. Chem. 261:14576-14581. [PubMed] [Google Scholar]
- 14.Farnaud, S., C. Spiller, L. C. Moriarty, A. Patel, V. Gant, E. W. Odell, and R. W. Evans. 2004. Interactions of lactoferricin-derived peptides with LPS and antimicrobial activity. FEMS Microbiol. Lett. 233:193-199. [DOI] [PubMed] [Google Scholar]
- 15.Fehlbaum, P., P. Bulet, S. Chernysh, J. P. Biand, J. P. Roussel, L. Letellier, C. Hertu, and J. A. Hoffman. 1996. Structure-activity analysis of thanatin, a 21-residue inducible insect defense peptide with sequence homology to frog skin antimicrobial peptide. Proc. Natl. Acad. Sci. USA 93:1221-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferl, R., A. E. Simon, K. M. Tengbarge, and M. L. Crouch. 1983. cDNA clones for Brassica napus seed storage proteins: evidence from nucleotide sequence analysis that both subunits of napin are cleaved from a precursor polypeptide. J. Mol. Appl. Genet. 2:273-283. [PubMed] [Google Scholar]
- 17.Fladerer, F., H. Krist, G. Rohowkki, and W. Schneider. 1995. A comparative study of methods of disinfecting drinking water in developing countries. Deutsche Gesellschaft für Technische Zusammenarbeit (GTZ) GmbH, Eschborn, Germany.
- 18.Folkard, G., J. Sutherland, and R. Shaw. 1999. Water clarification using Moringa oleifera seed coagulant. Water Lines 17:15-17. [Google Scholar]
- 19.Friedrich, C. L., D. Moyles, T. J. Beveridge, and R. E. Hancock. 2000. Antibacterial action of structurally diverse cationic peptides on gram-positive bacteria. Antimicrob. Agents Chemother. 44:2086-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gasteiger, E., A. Gattiker, C. Hoogland, I. Ivanyi, R. D. Appel, and A. Bairoch. 2003. ExPASy—the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31:3784-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guex, N., A. Diemand, and M. C. Peitsch. 1999. Protein modelling for all. Trends Biochem. Sci. 24:364-367. [DOI] [PubMed] [Google Scholar]
- 22.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 23.Hancock, R. E. W., T. J. Falla, and M. Brown. 1995. Cationic bactericidal peptides. Adv. Microb. Physiol. 37:135-175. [DOI] [PubMed] [Google Scholar]
- 24.He, K., S. J. Ludtke, W. T. Heller, and H. W. Huang. 1996. Mechanism of alamethicin insertion into lipid bilayers. Biophys. J. 71:2669-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He, K., S. J. Ludtke, H. W. Huang, and D. L. Worcester. 1995. Antimicrobial peptide pores in membranes detected by neutron in-plane scattering. Biochemistry 34:15614-15618. [DOI] [PubMed] [Google Scholar]
- 26.Heller, W. T., A. J. Waring, R. I. Lehrer, T. A. Harroun, T. M. Weiss, L. Yang, and H. W. Huang. 2000. Membrane thinning effect of the beta-sheet antimicrobial protegrin. Biochemistry 39:139-145. [DOI] [PubMed] [Google Scholar]
- 27.Hubert, F., T. Noel, and P. Roch. 1996. A member of the arthropod defensin family from edible Mediterranean mussels (Mytilus galloprovincialis). Eur. J. Biochem. 240:302-306. [DOI] [PubMed] [Google Scholar]
- 28.Jahn, S. A. A. 2001. Drinking water from Chinese rivers: challenges of clarification. J. Water Supply Res. Technol. 50:15-27. [Google Scholar]
- 29.Jahn, S. A. A. 1986. Proper use of African natural coagulants for rural water supplies—research in the Sudan and a guide for new projects. Deutsche Gesellschaft für Technische Zusammenarbeit (GTZ) GmbH, Eschborn, Germany.
- 30.Kemmer, F. N. 1988. Coagulation and flocculation. The NALCO water handbook, 2nd ed. McGraw-Hill Book Company, New York, N.Y.
- 31.Kerth, A., A. Erbe, M. Dathe, and A. Blume. 2004. Infrared reflection absorption spectroscopy of amphipathic model peptides at the air/water interface. Biophys. J. 86:3750-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim, S., E. Nollen, K. Kitagawa, V. Bindokas, and R. Morimoto. 2002. Polyglutamine protein aggregates are dynamic. Nat. Cell Biol. 4:826-831. [DOI] [PubMed] [Google Scholar]
- 33.Lee, J. H., M. S. Kim, J. H. Cho, and S. C. Kim. 2002. Enhanced expression of tandem multimers of the antimicrobial peptide buforin II in Escherichia coli by the DEAD-box protein and trxB mutant. Appl. Microbiol. Biotechnol. 58:790-796. [DOI] [PubMed] [Google Scholar]
- 34.Lee, M. K., L. Cha, S. H. Lee, and K. Hahm. 2002. Role of the amino acid residues within the disulfide loop of thanatin, a potent antibiotic peptide. J. Biochem. Mol. Biol. 35:291-296. [DOI] [PubMed] [Google Scholar]
- 35.Madsen, M., J. Schlundt, and E. F. Omer. 1987. Effect of water coagulation by seeds of Moringa oleifera on bacterial concentrations. J. Trop. Med. Hyg. 90:101-109. [PubMed] [Google Scholar]
- 36.Matsuzaki, K., O. Murase, N. Fujii, and K. Miyajima. 1996. An antimicrobial peptide, magainin 2, induced rapid flip-flop of phospholipids coupled with pore formation and peptide translocation. Biochemistry 35:11361-11368. [DOI] [PubMed] [Google Scholar]
- 37.Melo, F., and E. Feytmans. 1998. Assessing protein structures with a non-local atomic interaction energy. J. Mol. Biol. 277:1141-1152. [DOI] [PubMed] [Google Scholar]
- 38.Montgomery, J. M. 1985. Water treatment principles and design, p. 116-134. Wiley Intersciences Publications, New York, N.Y.
- 39.Moreman, L., S. Bosteels, W. Noppe, J. Willems, E. Clynen, L. Schoofs, K. Thevissen, J. Tytgat, J. V. Eldere, J. van der Walt, and F. Verdonk. 2002. Antibacterial and antifungal properties of alpha-helical, cationic peptides in the venom of scorpions from southern Africa. Eur. J. Biochem. 269:4799-4810. [DOI] [PubMed] [Google Scholar]
- 40.Muyibi, S., and L. Evison. 1995. Moringa oleifera seeds for softening hardwater. Water Res. 29:1099-1105. [Google Scholar]
- 41.Nakamura, T., H. Furunaka, T. Miyata, F. Tokunaga, T. Muta, S. Iwanaga, M. Niwa, T. Takao, and Y. Shimonishi. 1988. Tachyplesin, a class of antimicrobial peptide from the hemocytes of the horseshoe crab (Tachypleus tridentatus). Isolation and chemical structure. J. Biol. Chem. 263:16709-16713. [PubMed] [Google Scholar]
- 42.Neria, E., S. Fischer, and M. Karplus. 1996. Simulation of activation free energies in molecular systems. J. Chem. Phys. 105:1902-1921. [Google Scholar]
- 43.Ngai, P., and T. Ng. 2004. A napin-like polypeptide with translation-inhibitory, trypsin-inhibitory, antiproliferative and antibacterial activities from kale seeds. J. Pept. Res. 64:202-208. [DOI] [PubMed] [Google Scholar]
- 44.Okuda, T., A. U. Baes, W. Nishijima, and M. Okada. 2001. Coagulation mechanism of salt solution-extracted active component in Moringa oleifera seeds. Water Res. 35:830-834. [DOI] [PubMed] [Google Scholar]
- 45.Okuda, T., A. U. Baes, W. Nishijima, and M. Okada. 2000. Isolation and characterization of coagulant extracted from Moringa oleifera seed salt solution. Water Res. 35:405-410. [DOI] [PubMed] [Google Scholar]
- 46.Oren, Z., and Y. Shai. 1998. Mode of action of linear amphipathic alpha-helical antimicrobial peptides. Biopolymers 47:451-463. [DOI] [PubMed] [Google Scholar]
- 47.Pantoja-Uceda, D., O. Palomares, M. Bruix, M. Villalba, R. Rodriguez, M. Rico, and J. Santoro. 2004. Solution structure and stability against digestion of rproBnIb, a recombinant 2S albumin from rapeseed: relationship to its allergenic properties. Biochemistry 43:16036-16045. [DOI] [PubMed] [Google Scholar]
- 48.Park, C. B., K. S. Yi, K. Matsuzaki, M. S. Kim, and S. C. Kim. 2000. Structure-activity analysis of buforin II, a histone H2A-derived antimicrobial peptide: the proline hinge is responsible for the cell-penetrating ability of buforin II. Proc. Natl. Acad. Sci. USA 97:8245-8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perutz, M. F., T. Johnson, M. Suzuki, and J. T. Finch. 1994. Glutamine repeats as polar zippers: their possible role in inherited neurodegenerative diseases. Proc. Natl. Acad. Sci. USA 91:5355-5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peschel, A., and L. V. Collins. 2001. Staphylococcal resistance to antimicrobial peptides of mammalian and bacterial origin. Peptides 22:1651-1659. [DOI] [PubMed] [Google Scholar]
- 51.Peschel, A., R. W. Jack, M. Otto, L. V. Collins, P. Staubitz, G. Nicholson, H. Kalbacher, W. F. Nieuwenhuizen, G. Jung, A. Tarkowski, K. P. van Kessel, and J. A. van Strijp. 2001. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with L-lysine. J. Exp. Med. 193:1067-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pouny, Y., D. Rapaport, A. Mor, P. Nicolas, and Y. Shai. 1992. Interaction of antimicrobial dermaseptin and its fluorescently labeled analogues with phospholipid membranes. Biochemistry 31:12416-12423. [DOI] [PubMed] [Google Scholar]
- 53.Powers, J. P., A. Rozek, and R. E. Hancock. 2004. Structure-activity relationships for the beta-hairpin cationic antimicrobial peptide polyphemusin I. Biochim. Biophys. Acta 1698:239-250. [DOI] [PubMed] [Google Scholar]
- 54.Reiher, W. E., III. 1985. Theoretical studies of hydrogen bonding. Harvard University, Cambridge, Mass.
- 55.Rice, P., I. Longden, and A. Bleasby. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276-277. [DOI] [PubMed] [Google Scholar]
- 56.Rico, M., M. Bruix, C. Gonzalez, R. I. Monsalve, and R. Rodrıguez. 1996. 1H NMR assignment and global fold of napin BnIb, a representative 2S albumin seed protein. Biochemistry 35:15672-15682. [DOI] [PubMed] [Google Scholar]
- 57.Romestand, B., F. Molina, V. Richard, P. Roch, and C. Granier. 2003. Key role of the loop connecting the two beta strands of mussel defensin in its antimicrobial activity. Eur. J. Biochem. 270:2805-2813. [DOI] [PubMed] [Google Scholar]
- 58.Sali, A., and T. L. Blundell. 1993. Comparative protein modeling by satisfaction of spacial restraints. J. Mol. Biol. 234:779-815. [DOI] [PubMed] [Google Scholar]
- 59.Schonwetter, B. S., E. D. Stolzenberg, and M. A. Zasloff. 1995. Epithelial antibiotics induced at sites of inflammation. Science 267:1645-1648. [DOI] [PubMed] [Google Scholar]
- 60.Stott, K., J. M. Blackburn, P. J. Butler, and M. Perutz. 1995. Incorporation of glutamine repeats makes protein oligomerize: implications for neurodegenerative diseases. Proc. Natl. Acad. Sci. USA 92:6509-6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Su, W., S. Jackson, R. Tjian, and H. Echols. 1991. DNA looping between sites for transcriptional activation: self-association of DNA-bound Sp1. Genes Dev. 5:820-826. [DOI] [PubMed] [Google Scholar]
- 62.Suarez, M., J. M. Entenza, C. Doerries, E. Meyer, L. Bourquin, J. Sutherland, I. Marison, P. Moreillon, and N. Mermod. 2003. Expression of a plant-derived peptide harboring water-cleaning and antimicrobial activities. Biotechnol. Bioeng. 81:13-20. [DOI] [PubMed] [Google Scholar]
- 63.Tachi, T., R. F. Epand, R. M. Epand, and K. Matsuzaki. 2002. Position-dependent hydrophobicity of the antimicrobial magainin peptide affects the mode of peptide-lipid interactions and selective toxicity. Biochemistry 41:10723-10731. [DOI] [PubMed] [Google Scholar]
- 64.Tam, J. P. 1989. High-density multiple antigen-peptide system for preparation of antipeptide antibodies. Methods Enzymol. 168:7-15. [DOI] [PubMed] [Google Scholar]
- 65.Tam, J. P. 1988. Synthetic peptide vaccine design: synthesis and properties of a high-density multiple antigenic peptide system. Proc. Natl. Acad. Sci. USA 85:5409-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tam, J. P., and Y. A. Lu. 1989. Vaccine engineering: enhancement of immunogenicity of synthetic peptide vaccines related to hepatitis in chemically defined models consisting of T- and B-cell epitopes. Proc. Natl. Acad. Sci. USA 86:9084-9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tam, J. P., L. Yi-An, and Y. Jin-Long. 2002. Antimicrobial dendrimeric peptides. Eur. J. Biochem. 269:923-932. [DOI] [PubMed] [Google Scholar]
- 68.Tam, J. P., and F. Zavala. 1989. Multiple antigen peptide. A novel approach to increase detection sensitivity of synthetic peptides in solid-phase immunoassays. J. Immunol. Methods 124:53-61. [DOI] [PubMed] [Google Scholar]
- 69.Tauscher, B. 1994. Water treatment by flocculants compounds of higher plants. Plant Res. Dev. 40:56-70. [Google Scholar]
- 70.Terras, F. R. G., H. M. E. Schoofs, K. Thevissen, R. W. Osborn, J. Vanderleyden, B. P. A. Cammue, and W. F. Broekaert. 1993. Synergistic enhancement of the antifungal activity of wheat and barley thionins by radish and oilseed rape 2S albumins and by barley trypsin inhibitors. Plant Physiol. 103:1311-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tossi, A., L. Sandri, and A. Giangaspero. 2000. Amphipathic, alpha-helical antimicrobial peptides. Biopolymers 55:4-30. [DOI] [PubMed] [Google Scholar]
- 72.Turner, A. K., M. A. Lovell, S. D. Hulme, L. Zhang-Barber, and P. A. Barrow. 1998. Identification of Salmonella typhimurium genes required for colonization of the chicken alimentary tract and for virulence in newly hatched chicks. Infect. Immun. 66:2099-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang, L., A. Rozek, and R. E. W. Hancock. 2001. Interactions of cationic antimicrobial peptides with model membranes. J. Biol. Chem. 276:35714-35722. [DOI] [PubMed] [Google Scholar]