Abstract
Lateral gene transfer and recombination play important roles in the evolution of many parasitic bacteria. Here we investigate intragenic recombination in Wolbachia bacteria, considered among the most abundant intracellular bacteria on earth. We conduct a detailed analysis of the patterns of variation and recombination within the Wolbachia surface protein, utilizing an extensive set of published and new sequences from five main supergroups of Wolbachia. Analysis of nucleotide and amino acid sequence variations confirms four hypervariable regions (HVRs), separated by regions under strong conservation. Comparison of shared polymorphisms reveals a complex mosaic structure of the gene, characterized by a clear intragenic recombining of segments among several distinct strains, whose major recombination effect is shuffling of a relatively conserved set of amino acid motifs within each of the four HVRs. Exchanges occurred both within and between the arthropod supergroups. Analyses based on phylogenetic methods and a specific recombination detection program (MAXCHI) significantly support this complex partitioning of the gene, indicating a chimeric origin of wsp. Although wsp has been widely used to define macro- and microtaxonomy among Wolbachia strains, these results clearly show that it is not suitable for this purpose. The role of wsp in bacterium-host interactions is currently unknown, but results presented here indicate that exchanges of HVR motifs are favored by natural selection. Identifying host proteins that interact with wsp variants should help reveal how these widespread bacterial parasites affect and evolve in response to the cellular environments of their invertebrate hosts.
Surface proteins in pathogenic bacteria often function as antigens, and evidence indicates that their molecular evolution is driven by both positive selection and recombination (1, 28). Examples include the pilin genes from Neisseria species (17, 21), msp2 from Anaplasma marginale (12, 37), porB from Neisseria meningitidis (58), and ompL1 of the Leptospira genus (19). The examples above all involve pathogenic bacteria of vertebrates, and it is believed that recombination and rapid sequence evolution in their surface antigens are selectively advantageous by promoting avoidance of the vertebrate host immune response. Less well understood is the evolution of surface proteins of bacteria found strictly in invertebrates. Here we investigate the patterns of variation in the surface protein of Wolbachia, an intracellular bacterium found in arthropods and nematodes.
Wolbachia bacteria are among the most successful and intriguing intracellular bacteria in nature. It is estimated that 20 to 75% of insect species harbor Wolbachia bacteria (25, 62, 65), with infections also commonly found in terrestrial crustaceans, chelicerata, and filarial nematodes (3, 5, 14, 18, 49).
Transmission of Wolbachia bacteria within host populations is vertical (64); however, it is now well known that Wolbachia bacteria in arthropods can also shift host species, “jumping” to new unexplored cellular environments through mechanisms still unclear (horizontal transmission) (60, 63, 68).
As a parasite of arthropods, Wolbachia bacteria are best known to be manipulators of host reproduction. A major phenotypic effect of the symbiosis with Wolbachia bacteria is a distortion of the host sex ratio, through mechanisms enhancing the female proportion (the sex transmitting the bacterium), such as feminization of genetic males, parthenogenesis induction, and male killing (54, 64). Wolbachia bacteria are also able to induce cytoplasmic incompatibility between eggs from uninfected females and sperm from infected males, thus rapidly increasing the proportion of infected individuals in host populations, often to fixation (48). While in insects Wolbachia bacteria are primarily reproductive parasites, in filarial nematodes the symbiosis with Wolbachia bacteria appears to have evolved toward a mutualistic interaction (3, 4).
The genus Wolbachia (class Alphaproteobacteria, order Rickettsiales) is currently divided into six taxonomic supergroups (A to F) based primarily on 16S and ftsZ gene phylogenies. Phylogenies for these two genes are concordant at the supergroup level (31). A and B are the two main groups found in arthropods. C and D are found in filarial nematodes (3). Recently, two new supergroups, E and F, have been proposed. So far, supergroup E contains Wolbachia bacteria infecting springtails (class Collembola), a primitive insect group (15, 59), while supergroup F contains Wolbachia bacteria that infect termites and filarial species of the genus Mansonella (31, M. Casiraghi, S. R. Bordenstein, L. Baldo, N. Lo, T. Beninati, J. J. Wernegreen, J. H. Werren, and C. Bandi, unpublished data).
The vertical transmission of Wolbachia bacteria through the reproductive tissues of their hosts implied that these bacteria experience little recombination, as appears to be the case for other vertically inherited symbionts (e.g., Buchnera aphidicola) (56). However, the discordances between the phylogenies of some Wolbachia genes (27) and the discovery of recombination events within the Wolbachia surface protein (wsp) (47, 66) suggested that recombination may be more common in Wolbachia bacteria than some other endosymbiotic bacteria. Furthermore, the relatively frequent occurrence of multiple infections with different Wolbachia strains in the same hosts (23, 62, 63), the presence of phages and insertion elements within the Wolbachia genome (35, 68), and lateral transfer of phage among strains (7) are consistent with a recombinogenic genome.
The Wolbachia surface protein gene wsp encodes a major surface membrane protein showing sequence similarity to the major outer membrane proteins of closely related alphaproteobacteria (9). Among the Wolbachia genes for which a large sequence data set is currently available, wsp is the most variable, showing relatively high genetic divergence among strains. Analyses of the rates of synonymous and nonsynonymous substitutions along the gene sequences show discrete regions under strong positive selection in a background of overwhelming purifying selection (2, 28). Because of its variability, wsp has been used extensively in phylogenetic analyses and for microtaxonomic subdivision of the two major clades, A and B, into subgroups (68).
Localization of the protein at the interface between the two cellular environments and the presence of regions under strong positive selection suggest a key role of the protein in the arms race expected to occur between arthropod hosts and this intracellular parasite (2, 61). Furthermore, in nematode Wolbachia bacteria, wsp has been demonstrated to play an antigenic role in stimulating the immune response of the vertebrate animals that are infected by filarial worms (6, 10, 11).
Despite the fact that Wolbachia bacteria are not found in vertebrates, their outer surface membrane protein (Wsp) shows surprising analogies with antigenic proteins of pathogens: a heterogeneous pattern of variation characterized by hypervariable regions (HVRs) flanked by conserved regions (CRs) (2, 9), strong positive selection affecting the HVRs (2, 28), and evidence of recombination affecting its sequences (26, 47, 66). All this strongly suggests a potential role of this protein in host-Wolbachia interactions.
Previous evidence of recombination in wsp has come from three studies. Werren and Bartos (66) reported the first example of recombination within supergroup B, occurring between the two Wolbachia strains of a parasitoid wasp and the fly it parasitizes. More recently, Reuter and Keller (47) showed recombination to have occurred among five strains of Wolbachia bacteria belonging to supergroup A and infecting the same species of ant, Formica exsecta. Jiggins (26) provided an estimation of the rate of recombination in wsp and ftsZ, suggesting a high level of recombination in both genes of arthropod Wolbachia bacteria, but not for those found in nematodes.
The role of recombination in shaping the evolution of wsp has not yet been clarified. We therefore performed a molecular evolutionary study of wsp, with the following goals: (i) to clarify the pattern of DNA rearrangement occurring in wsp, (ii) to verify the extent of recombination by examining the large wsp sequence set now available in sequence databases, and (iii) to clarify the potential occurrence of horizontal DNA transfers and recombination among the different Wolbachia supergroups.
To pursue these goals, we conducted a detailed analysis of the pattern of variation and recombination within wsp, utilizing an extensive set of published and new sequences from five Wolbachia supergroups. The results reveal a complex chimeric structure of the gene, characterized at the protein level by shuffling of a set of amino acid motifs at each of four HVRs, which strongly supports the occurrence of multiple horizontal DNA transfers. Exchanges occur both within and between supergroups. Consistent with extensive recombination, striking discordances occur in phylogenetic trees of the different HVRs.
MATERIALS AND METHODS
Data set selection and alignment.
Initially, a set of 93 wsp sequences (available in GenBank or sequenced during this study) was examined for patterns of variation and recombination. Care was taken to select divergent amino acid sequences that represented the range of variability in the gene. The amino acid sequences were aligned based on ClustalX (57) and modified by eye in Bioedit (20). Analyses of the nucleotide sequence divergence were performed using DNAsp (50).
To have a data set more suitable for analysis and presentation, the 93 sequences were trimmed to 40 as follows. First we proceeded by randomly extracting only a single sequence per Wsp type, i.e., one sequence among strains sharing more than 95% amino acid sequence similarity (this threshold was arbitrarily selected). The final data set included strains from the five major supergroups, A, B, C, D, and F (Table 1). For the above sequences, we made a separate nucleotide sequence alignment. Because of the variable length of the gene and the great nucleotide sequence variability characterizing some of the regions (HVRs), we first aligned the CRs based on ClustalX and by eye. Then single HVRs were aligned by eye as follows. Within a single HVR, we grouped and aligned sequences sharing very similar amino acid motifs based on their homology and length, then we proceeded by aligning distinct groups of sequences among them, minimizing the insertion of gaps. Since HVRs show great nucleotide sequence variability, often requiring the insertion of long stretches of multiple gaps (especially at HVR3 and HVR4), we produced various alignments of each HVR for analysis. The final nucleotide sequence alignment had a length of 540 bp. The sequences were analyzed for recombination breakpoints and for phylogenetic discordances along the gene and between HVRs as described below.
TABLE 1.
List and features of the 40 Wolbachia strains analyzed in this study
| Host species | Host order | Wolbachia supergroupa | Strain identifying code | Accession no. |
|---|---|---|---|---|
| Drosophila melanogaster | Diptera | A | 1-DmelA | AE017259 |
| Callyrhytis glandium | Hymenoptera | A | 2-CglaA | AY095156 |
| Echinophthirius horridus | Phthiraptera | A | 3-EhorA | AY331986 |
| Bovicola bovis | Phthiraptera | A | 4-BbovA | AY331128 |
| Colpocephalum unciferum | Phthiraptera | A | 5-CuncA | AY330308 |
| Sitotroga cerealella | Lepidoptera | A | 6-ScerA | AY177735 |
| Pegoscapus herrei | Hymenoptera | A2 | 7-PherA2 | AF521151 |
| Formica exsecta (wFex5) | Hymenoptera | A | 8-FexsA | AY101200 |
| Blastophaga brownii | Hymenoptera | A | 9-BbroA | AF521165 |
| D.simulans (wRi) | Diptera | A | 10-DsimA | AF020070 |
| Perithemis tenera | Odonata | B | 11-PtenB | AF217725 |
| Tipula aino | Diptera | B | 12-TainB | AF481165 |
| Horridipamera nietneri | Hemiptera | B | 13-HnieB | AB109581 |
| Lutzomyia whitmani | Diptera | A | 14-LwhiB | AF237885 |
| Trichopria drosophilae | Hymenoptera | A | 15-TdroA | AF071910 |
| Pediculus humanus | Phthiraptera | ? | 16-Phum? | AY331114 |
| Chelymorpha alternans | Coleoptera | B | 17-CaltB | AY566421 |
| Elasmucha putoni | Heteroptera | B | 18-EputB | AB109614 |
| Acraea encedon | Lepidoptera | B | 19-AencB | AJ130716 |
| Paromius exiguus | Hemiptera | B | 20-PexiB | AB109580 |
| Blastophaga nipponica | Hymenoptera | B | 21-BnipB | AF521156 |
| Ostrinia scapulalis | Lepidoptera | B | 22-OscaB | AB077201 |
| Orseolia oryzae | Diptera | B | 23-OoryB | AF481164 |
| Drosophila innubila | Diptera | B | 24-DinnB | AY552552 |
| Pieris rapae | Lepidoptera | B | 25-PrapB | AB094372 |
| Protocalliphora sialia | Diptera | A1 | 26-PsiaB | AY188687 |
| Acraea encedon T | Lepidoptera | B | 27-AencTB | AJ271198 |
| Porcellio spinicornis | Isopoda | B | 28-PspiB | AJ276608 |
| Porcellionides pruinosus | Isopoda | B | 29-PpruB | AJ276605 |
| Armadillidium vulgare | Isopoda | B | 30-AvulB | AJ276598 |
| Pegoscapus gemellus | Hymenoptera | A | 31-PgemA | AF521152 |
| Dysdera erythrina | Araneae | A | 32-DeryA | AJ276615 |
| Ephestia cautella | Lepidoptera | A | 33-EcauA | AB024571 |
| Trinoton querquedulae | Phthiraptera | A | 34-TqueA | AY330316 |
| Dirofilaria immitis | Spirurida | C | 35-DimmC | AJ252062 |
| Onchocerca gibsoni | Spirurida | C | 36-OgibC | AJ252178 |
| Wuchereria bancrofti | Spirurida | D | 37-WbanD | AJ252180 |
| Brugia malayi | Spirurida | D | 38-BmalD | AJ252061 |
| Coptotermes lacteus | Isoptera | F | 39-ClacF | AJ833930 |
| Coptotermes acinaciformis | Isoptera | F | 40-CaciF | AJ833931 |
Supergroup identification for each strain is based on published literature. A question mark indicates that no identification of the supergroup is provided.
Sequencing of wsp gene.
Among the 40 wsp sequences selected for the main analysis, 37 were already available in GenBank. Sequences from Coptotermes lacteus and Coptotermes acinaciformis were obtained during this study. The two species were gifts from Michael Lenz (CSIRO Entomology) and John Holt (James Cook University), respectively, and were collected in Melbourne and Townsville. The guts of a single worker termite from each species were removed, and DNA was extracted from the remaining tissues as described previously (32). PCR was performed using the conditions described by Maekawa et al. (32), with primers WSPestF (5′-TTAGACTGCTAAAGTGGAATT) and WSPestR (5′-AAACCACTGGGATAAGAAGA).
Direct sequencing of the PCR product was performed using the BigDye v2.0 terminator sequencing kit and an ABI 3700 automated sequencer.
Analysis of recombination. (i) Detection of breakpoints.
To identify potential recombination breakpoints, we used the recombination detection program RDP2 (34), which implements different methods for detecting recombination. We primarily used the MAXCHI program (43, 52), which employs the following approach. For every possible sequence pair in the alignment, a window of set length with a partition in its center is moved along the sequences and a chi-square value is calculated, being an expression of the difference in the number of variable sites on either side of the central partition. A variable window size setting, with different proportions of variable sites (VI) per window was initially tested, providing basically similar results regardless of the VI proportions. To estimate the pattern of distribution of recombination events, the parameters were fixed as follows. Sequence triplets were scanned using a variable window size with a 0.3 fraction of VI and a highest acceptable P value of 0.001. For specific detection of breakpoints in the selected subsamples of sequences (see Fig. 3), we used the option “Manual MaxChi,” which permits the analysis to be performed selecting potential recombinant and parental sequences. The significance of chi-square peaks was more accurately determined by a permutation test (1,000 permutations). Peaks at which the observed chi-square values exceeded values in the 5% tail of the null distribution were considered significant.
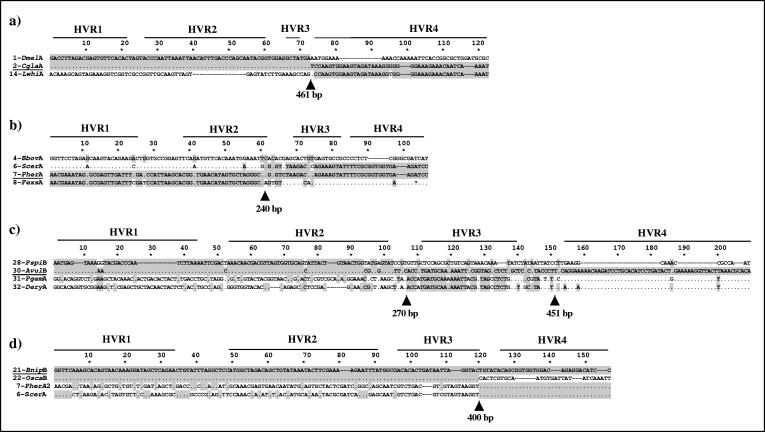
FIG. 3.
Examples of recombination breakpoints along wsp. For each alignment, only the polymorphic sites are shown. Also shown are the positions of the four HVRs. Polymorphisms shared with the underlined sequence are highlighted in gray. Arrows indicate significant breakpoints detected by MAXCHI and the approximate nucleotide position. In all cases, recombination breakpoints fall between HVRs.
(ii) Phylogenetic analyses.
Phylogenetic analyses were performed on different portions of wsp. The nucleotide sequence alignment was subdivided into four sectors of nearly equal lengths (about 135 bp each) starting from the first nucleotide. Partitioning was based on the observed pattern of nucleotide sequence divergence along the alignment and on the pattern of the ratios of dN/dS (the nonsynonymous/synonymous substitution rate ratio per codon site) reported for wsp by Baldo et al. (2), identifying three regions under positive selection in arthropod Wolbachia bacteria, separated by CRs (in that study, the last HVR was only partially included in the analyses). The four sectors were divided within the middle of each CR, allowing each segment to encompass a single HVR plus part of the flanking conserved domain. Since the vast majority of nucleotide sequence variability is at the HVRs, CRs are expected to have a minor effect on the phylogenetic reconstructions.
Phylogenetic analyses of each sector were conducted using Bayesian inference of phylogeny (BI) and maximum parsimony. For BI, the appropriate models of sequence evolution were estimated for each of the four gene partitions using the program Modeltest 3.06 (42). In each case, this was found to be GTR+I+G (GTR, general time-reversible model; I+G, invariable or gamma distributed rates of variation at sites).
The BI analyses were performed using MrBayes 3.0 (22). One hundred thousand trees were generated, with a sample frequency of 100. The first 500 trees were considered the burn-in and discarded. From the remaining 500 trees, 50% majority rule consensus trees were generated. Maximum-parsimony 50% majority rule bootstrap trees were estimated in PAUP* (55) (1,000 replicates, 10 random-addition replicates per bootstrap replicate). All characters were weighted equally, and gaps were treated as missing. Since HVR3 and -4 were difficult to align unambiguously, we performed phylogenetic analyses on various alignments of both regions.
Test for selective pressure at the HVRs.
To investigate whether the HVRs show stabilizing or diversifying selection, we used an independent-contrast approach (13) to evaluate rates of synonymous versus nonsynonymous substitution (Ks and Ka) of phylogenetically independent sets of closely related sequences. Independent contrasts were selected based on the phylogenetic analysis. The advantages of this approach are that alignment issues are avoided due to similarity in sequences and relatively short-term trends in synonymous versus nonsynonymous divergence can be evaluated. Sets of phylogenetically independent contrasts were compared using a one-tailed Wilcoxon matched-pair signed-rank test (13).
RESULTS
Pattern of variation along wsp.
The pattern of nucleotide sequence divergence along wsp, based on 93 sequences, is shown in Fig. 1a. A heterogeneous distribution of variability is evident along the gene: four HVRs are seen as peaks, with similar maximum values of nucleotide sequence divergence of around 0.5, separated by regions under strong conservation (CRs). The corresponding amino acid pattern of variation mirrors this distribution: highly variable amino acid sequences at the HVRs interspersed by conserved protein sequences at CRs (Fig. 2). Based on this evidence, the major distinction among wsp protein sequences is in the specific amino acid sequences that each sequence has at the four HVRs.
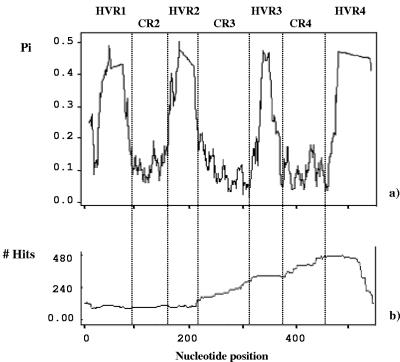
FIG. 1.
(a) Nucleotide divergence (Pi) of the alignment of 93 wsp sequences. Four peaks are identified, corresponding to the four HVRs. (b) Pattern of distribution of recombination events along the alignment of the 40 wsp nucleotide sequences, as detected by the MAXCHI program. The cumulative number of recombination events per site is given. Notably, all sites encompassing a single HVR experience similar numbers of events, suggesting that single HVRs are generally exchanged as unique tracts. HVR3 and -4 clearly appear to have undergone a proportionally higher number of recombination events than HVR1 and -2.
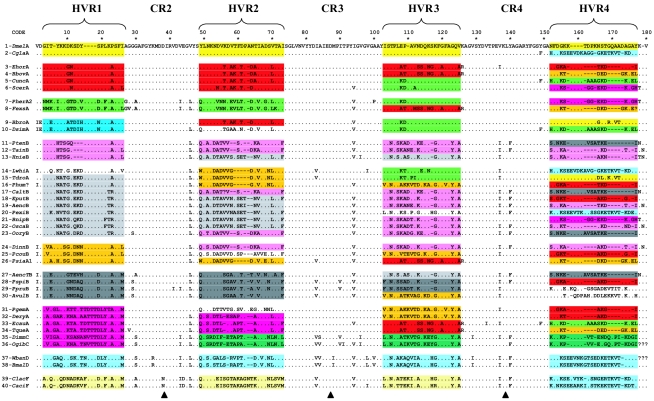
FIG. 2.
Amino acid alignment of 40 wsp sequences (180 amino acid in length). The reference sequence is 1-DmelA (from the host D. melanogaster). Amino acid motifs at the HVRs are color coded based on similarity of shared polymorphisms within each HVR. HVR1 is used as the initial reference region for sequence grouping. HVR motifs with uncertain groupings were left uncolored. Based on the pattern of colors along the gene, each of the first 34 arthropod sequences shows a unique combination of four HVR motifs and thus could be regarded as a distinct protein type. A clear shuffling of HVRs between sequences can be visualized. CR1 (about 145 bp) is not shown because the region was not completely sequenced for all of the strains. Arrowheads indicate the limits of the alignment sectors (HVR+) analyzed for phylogenetic reconstructions (see Fig. 4).
Despite the high level of divergence within each HVR at the nucleotide and amino acid sequence levels, a comparative analysis of shared amino acid sequence polymorphisms shows a limited set of well-distinguishable motifs within each HVR. This is shown in Fig. 2, where distinct motifs within single HVRs are indicated by different colors. Each motif within an HVR contains sequences very similar at both the amino acid and nucleotide sequence levels. For example, at HVR3, sequences 3, 4, 8, 26, 33, and 34 (motif in red) are nearly identical in nucleotide sequence (Pi = 0.025), as are sequences 5, 6, 7, 9, and 10 (motif in green, Pi = 0.011). Similarly, at HVR2, sequences 3, 4, 5, 6, and 9 (Pi = 0.016, motif in red) and sequences 13, 18, 19, 20, 21, and 22 (Pi = 0.023, motifs in light gray) show low levels of nucleotide sequence divergence within the motif. In contrast, comparison between motifs indicates high divergence. For the above example, Ka is 0.572 between the two motifs at HVR2 (red and light gray) but only 0.022 and 0.031 within each motif. Differences between motifs do not involve simple frameshifts: the sequences are divergent at the nucleotide sequence level. Their evolution often involves a combination of short insertions and deletions and nucleotide sequence changes. In fact, although alignments at the nucleotide sequence level within motifs were straightforward and reliable, the divergence between motifs in some cases made alignments difficult, particularly for HVR4. This merely reflects the pattern of considerable divergence between motifs, with relatively few transitional sequences. For this reason, we do not claim that the alignment between motifs at the HVRs (shown in Fig. 2) reflects actual nucleotide sequence homologies. The alignments are very useful, however, for showing conservation within and divergence between amino acid sequence motifs.
Recombination in wsp.
We have examined the wsp gene for recombination by three basic methods. First, we have evaluated the amino acid sequences for signatures of recombination between the HVRs by visual inspection. Examples of such recombination are readily apparent in Fig. 2, and a sample of these is described below. Second, we have analyzed the sequences for recombination occurrence and specific breakpoints using primarily the program MAXCHI. Third, we have used phylogenetic approaches to further support significant discordances at the nucleotide sequence level for the different HVRs of wsp and to show how relationships among sequences shift across the four HVRs.
Recombination between HVRs, which results in shuffling of motifs between sequences, is readily apparent by examination of Fig. 2. The patterns are easily visualized by the color coding of HVRs based on amino acid motifs. In HVR1, there are clear similarities among sequences 1 and 2, 3 to 6, 7 and 8, 9 and 10, 11 to 13, 14 to 23, 24 to 26, 27 to 30, 31 to 36, 37 and 38, and 39 and 40. However, these same groups of sequences can differ dramatically in their composition in other HVRs. For example, by HVR4, sequence 1 now groups with sequences 9 and 15 by amino acid motif, whereas sequence 2 now groups with 14 by amino acid motif. Basically, sequences showing almost identical amino acid motifs at some HVRs are dramatically divergent at different HVRs while grouping with different sets of sequences based upon shared motifs. Such a remarkable pattern of similarity or divergence among sequences along the gene strongly indicates a shuffling among a limited set of motifs. We note that all sequences shown in Fig. 2 are consistent with the above recombinant pattern. Below, we illustrate the recombinant pattern with three examples. These examples are also highlighted in the analysis of breakpoints at the nucleotide sequence level (Fig. 3) and phylogenetic analyses (Fig. 4).
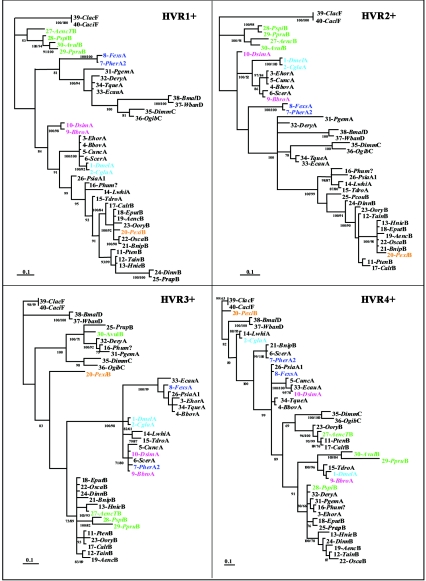
FIG. 4.
Phylogenetic trees of the four portions of wsp encompassing single HVR+s (135 bp each). The trees were generated by MrBayes (100,000 replicates, 50% majority rule) and are unrooted. Sequence identification corresponds to the description provided in Table 1. Colors highlight some of the examples discussed in the text (see Results) and show changes in phylogenetic association across HVR+s affecting all sequences shown. Sequences of supergroups A and B do not form separate groupings at any HVR. Posterior probability values higher than 70% are shown at the nodes. For nodes supported also by parsimonious analyses, the corresponding bootstrap value is shown under the posterior probabilities.
Example 1.
Sequences 1-DmelA (from the dipteran Drosophila melanogaster) and 2-CglaA (from the hymenopteran Callyrhytis glandium) share 100% amino acid identity at HVR1, -2, and -3, while at HVR4 the two sequences greatly diverge. At HVR4, sequence 1-DmelA is very similar to sequences 9-BbroA (from the hymenopteran Blastophaga brownii) and 15-TdroA (from the parasitic wasp Trichopria drosophilae) with 21/25 monomorphic sites (Ms) shared between the two. In contrast, sequence 2-CglaA is almost identical to sequence 14-LwhiA (the dipteran Lutzomyia whitmani, 24/25 Ms).
Example 2.
Sequence 7-PherA2 (from the fig wasp Pegoscapus herrei) is almost identical to sequence 8-FexsA (from the ant Formica exsecta) at HVR1 and HVR2 (respectively, 21/22 and 23/24 Ms), while at HVR3 the two sequences diverge. Sequence 7-PherA2 becomes almost identical to sequences 5-CuncA (from the louse Colpocephalum unciferum), 6-ScerA (from the lepidopteran Sitotroga cerealella), 9-BbroA, and 10-DsimA (from the fruit fly Drosophila simulans) (17/18 Ms), and sequence 8-FexsA converges to sequences 3-EhorA (from the louse Echinophthirius horridus), 4-BbovA (from the louse Bovicola bovis), 26-PsiaA1 (from the dipteran Protocalliphora sialia), 33-EcauA (from the lepidopteran Ephestia cautella), and 34-TqueA (from the louse Trinoton querquedulae) (17/18 Ms). Then, at HVR4, sequence 7-PherA2 diverges from all the previous sequences and becomes identical to sequences 6-ScerA and 21-BnipB (from the hymenopteran Blastophaga nipponica), while sequence 8-FexsA remains very close only to sequences 4-BbovA 26-PsiaA1, and 34-TqueA (24/25 Ms).
Example 3.
Sequence 30-AvulB is from a Wolbachia bacterium present in the isopod Armadillidium vulgare and induces feminization in this host. Phylogenetically, this Wolbachia bacterium is embedded within a clade of bacteria otherwise found in insects and therefore probably represents a major host shift from insects to isopods (8). At HVR1 and -2, this sequence groups with sequences 29-PpruB (from the isopod Porcellionides pruinosus, 22/22 and 21/24 Ms, respectively, at HVR1and HVR2), 28-PspiB (from the isopod Porcellio spinicornis, 21/22 and 22/24 Ms), and 27-AencTB (from the lepidopteran Acraea encedon, 17/22 and 20/24 Ms). At HVR3, it diverges dramatically from the previous grouping and, interestingly, groups strongly with some sequences from supergroup A, 31-PgemA (from the hymenopteran Pegoscapus gemellus, 13/18 Ms) and 32-DeryA (from the spider Dysdera erythrina, 14/18 Ms), and with 16-Phum? (from the louse Pediculus humanus, 13/18 Ms). It then diverges from these in HVR4, where it does not show strong similarities to any other HVR4 in the data set (but it shows high similarity with other wsp sequences from isopod hosts not in the data shown; e.g., sequences with accession no. AJ276599, AJ276600, and AJ276606).
(i) Recombination breakpoints.
The pattern of recombination in wsp appears highly complex. Analyses performed with the MAXCHI program did not identify one partitioning of wsp describing all the recombinant patterns, since breakpoint locations are quite different among sequences and recombination can involve segments of different lengths.
For this reason, we first characterized the general pattern of recombination within the gene. We then analyzed some of the same examples previously reported to show the recombination events at the nucleotide sequence level. Figure 1b presents an analysis using MAXCHI, which calculates the number of recombination blocks among the 40 sequences for each position along the gene. The number of detected hits can differ considerably, depending on the parameter settings (e.g., the window size), ranging from 480 to 1,600 for variable window sizes ranging from 0.1 to 0.5 of variable sites (VI). However, despite minor changes in parameter values, the pattern of distribution of hits along the gene is quite consistent. Data indicate that potential recombination hits have involved all sites along the gene. The number of hits per site increases around position 215 of the alignment (in CR3, after HVR2), levels off in HVR3, increases again around position 380 (before HVR4), and levels off in HVR4. The fact that all sites within a single HVR experience nearly identical numbers of events indicates that single HVRs are generally exchanged as whole tracts. We cannot exclude the possibility that in some cases recombination could have occurred within HVRs, involving small sequence tracts within these regions, but there is no significant evidence for this from our analyses of breakpoints. Regions with an increasing number of hits (CR3 and CR4) involve sites that have undergone an unequal number of recombination events, thus reflecting the occurrence of recombination breakpoints. This suggests that recombination events are more likely to occur between HVR2 and -3 and between HVR3 and -4 than between HVR1 and -2. This observation is confirmed also by visual inspection of the protein alignment (Fig. 2) and phylogenetic analyses (Fig. 4) showing most of the incongruences in relationships occurring between the regions cited above.
Recombinant segments can involve either single HVRs or longer segments, encompassing two or more HVRs at a time (in all cases, CRs can also be recombined). Some sequences show a single recombinant breakpoint, while some show several breakpoints leading to partitions of the gene in multiple recombinant blocks. Consequently, recombinant sequences can be characterized by an x segment mosaic (from two to four segments).
Figure 3 shows four cases of recombination breakpoints at the nucleotide sequence level. For 1-DmelA, 2-CglaA, and 14-LwhiA, a single significant breakpoint is present in the region around position 461 of the nucleotide sequence alignment, corresponding to the 5′ end of HVR4 (Fig. 3a). The breakpoint divides the alignment into two portions: the first portion encompasses HVR1, -2, and -3 (CRs included), while the second involves only HVR4. Before the breakpoint, sequence 2-CglaA is identical to 1-DmelA, sharing all 73 polymorphisms with it, while in the second portion it clearly becomes identical to sequence 14-LwhiA at 48/49 polymorphic sites. Statistically significant strings of associated polymorphisms strongly indicate recombination (53): the probability of getting a string of 73 matching polymorphisms followed by a string of 48 is remarkably low (P < 10−12 based on the permutation probability).
Figure 3b shows a more complex pattern, involving four sequences and a single recombinant breakpoint around nucleotide position 240, corresponding to the 3′ end of HVR2. Based on the shared polymorphisms, the breakpoint divides sequences into two portions, clearly grouping sequences 4-BbovA with 6-ScerA and sequences 7-PherA with sequences 8-BbroA before the breakpoint and with sequences 4-BbovA with 8-BbroA and sequences 6-ScerA with 7-PherA after the breakpoint. This represents either independent recombinant events with a common breakpoint (e.g., recombination hotspot) or a reciprocal exchange event.
Figure 3c shows the recombinant pattern involving sequence 30-AvulB. It shows similarity at the nucleotide sequence level to sequence 28-PspiB before position 270, with a breakpoint occurring within CR3, and to sequences 31-PgemA and 32-DeryA after. A second breakpoint is localized around position 451, with 28-PspiB having high similarity to 31-PgemA and 32-DeryA, while 30-AvulB dramatically diverges from all the sequences. This suggests recombination occurring between A and B sequences.
Similar to the previous example, Fig. 3d shows recombination involving A and B sequences. Sequence 21-BnipB is identical to sequence 22-OscaB before position 400 (in CR4), sharing all of the 119 polymorphisms with it. After that, it becomes identical to sequences 7-PherA2 and 6-ScerA, sharing all of the remaining 36 polymorphisms with them.
The origin or direction of these recombination events cannot be reliably inferred because the DNA exchange history in wsp appears too reticulated to be resolved. A preliminary attempt to resolve relationships among some strains by examining additional genes has so far been unsuccessful. Other genes also provided ambiguous inferences about strain relationships, probably reflecting the widespread recombination in Wolbachia genomes (L. Baldo, J. H. Werren, and S. R. Bordenstein, unpublished data).
(ii) Phylogenetic analyses.
As indicated by the patterns of amino acid similarity or divergence along the sequences described above, phylogenetic relationships predicted for the four HVRs are in conflict. To provide further statistical significance for this pattern and to show how relationships shift across HVRs in the whole data set, we compared the nucleotide sequence phylogenies of the four regions of wsp. The wsp gene was divided into four sectors; the breakpoints for these sectors are indicated in Fig. 2 (each sector is indicated as HVR+ to point out that it contains both the HVR and a portion of the flanking CRs).
Shown in Fig. 4 are the four consensus Bayesian trees describing the phylogeny of the four sectors of wsp. The tree topologies from bootstrap analysis were generally congruent with those estimated from Bayesian analysis, and associated bootstrap values were similar. The use of different alignments of HVR3+ and HVR4+ resulted in consistent phylogenies for the main clades: for this reason, only one phylogeny of these regions, based on the alignment showed in Fig. 2, is reported.
The goal of this analysis was not to define the precise phylogenetic relationships between motifs, as these will be influenced by alignment issues (i.e., small insertions or deletions that complicate alignment between motifs). Rather, the analysis is used to show phylogenetic relationships within motifs (where alignment is not an issue) and the shuffling of these relationships between HVRs.
As shown in Fig. 4, the evolutionary relatedness of the 40 sequences varies considerably across the four regions. The four trees greatly differ both in their topologies and in the branch lengths found between pairs of sequences, revealing striking phylogenetic incongruences.
To test the null hypothesis that the phylogenies for each portion (HVR) of the gene were not significantly different, we compared sets of the most probable trees given by MrBayes for each HVR+. MrBayes generated by MCMC search a set of about 500 trees for each HVR+, sorted by a posterior probability (P) of <1 and with a cumulative P equal to 100 (expressed as a percentage). The probability that two HVRs share the same underlying tree was then determined by multiplying their joint probabilities for all trees within the set. By this analysis, no two regions shared any common trees, so the proportion of shared trees for any of the four regions with any other was zero. This indicates that the different sectors have very different phylogenetic histories. To specifically infer significant conflicts in subphylogenies inferred by the different sectors, we estimated the posterior cumulative probability at each region that two given sequences form a cluster (the P value for a given clade at one sector is the sum of the posterior probabilities given by all the trees in a set inferring that clade). Comparison of the P values given by each HVR provides a simple way to support incongruences of relationships. For instance, a discordance among phylogenies is highly significant when the P values for a given cluster in two different HVRs are, respectively, equal to 100 and 0.
Comparison of the four tree topologies shows significant shuffling of the HVRs occurring within a single supergroup, as well as between supergroups (see both Fig. 2 and 4 for comparison). Some of the examples previously mentioned in terms of amino acid motifs are highlighted in the phylogenetic analysis (Fig. 4). As can be seen, they show highly supported clusters at some HVRs and divergence at others. The two examples below show recombination between supergroups A and B.
Example 1.
Within supergroup A, sequence 7-PherA2 strongly clusters with sequence 8-FexsA at HVR1+ and HVR2+ (P = 100), while at HVR3+ the two sequences diverge. Sequence 7-PherA2 now clusters with sequences 5-CuncA, 6-ScerA, 9-BbroA, and 10-DsimA (P = 71), and sequence 8-FexsA forms a strong monophyletic group with sequences 3-EhorA, 4-BbovA, 26-PsiaA1, 33-EcauA, and 34-TqueA (P = 100). Then, at HVR4+, sequence 7-PherA2 again diverges from the previous cluster while strongly grouping with sequence 6-ScerA and, interestingly, with a sequence from supergroup B, 21-BnipB (P = 99). Sequence 8-FexsA remains phylogenetically close to sequence 26-PsiaA1.
Example 2.
HVR1+ and -2+ show a consistent grouping of sequences 30-AvulB, 29-PpruB, and 28-PspiB (in both cases, P = 100). But at HVR3+, the cluster is no longer supported; 30-AvulB now forms a monophyletic group with supergroup A sequences 31-PgemA and 32-DeryA and with 16-Phum? and 25-PcouB (P = 100). Again, at HVR4+, 30-AvulB radically diverges and significantly groups with sequence 29-PpruB (P = 100), even if the two are separated by a relatively high level of genetic divergence (as indicated by the branch length leading to 30-AvulB).
Noticeable also is the shift in phylogenetic relationships across the HVRs of nematode sequences from supergroups C and D with respect to each other and relative to arthropod Wolbachia sequences. Specifically, the two supergroups are phylogenetically close at HVR1+ and -2+ but significantly diverge at HVR3+ and -4+, where each of the two supergroups significantly clusters with different arthropod sequences. This discordance can be seen also in the alignment in Fig. 2.
Another interesting example of amino acid shuffling within wsp appears to have occurred among several supergroups, including recently proposed supergroup F (containing Wolbachia bacteria that infect termites). The wsp sequences from termites have not been previously described. The two sequences analyzed, 39-ClacF (from Coptotermes lacteus) and 40-CaciF (from Coptotermes acinaciformis), strongly cluster across all four HVRs while considerably varying in their relatedness with respect to the other sequences. They form a phylogenetically distant clade at HVR1+ and -2+, but branch lengths decrease at HVR3+, accompanied by a relatively close association with the two sequences from nematode Wolbachia bacteria of supergroup D, 37-WbanD (from Wuchereria bancrofti) and 38-BmalD (from Brugia malayi). At HVR4+, sequence 20-PexiB (from the hemipteran Paromius exiguus), which clustered with sequences 18-EputB and 19-AencB at HVR1+ (P = 100) and -2 (P = 100), is now close to those of termites (P = 80), which together are still relatively close to sequences from supergroup D in nematodes (P = 80). The P value that sequence 20-PexiB clusters with sequences 18-EputB and 19-AencB at HVR3+ and HVR4+ is, in both cases, equal to 0. The conflict in relationships inferred by the diverse HVRs among termites and members of supergroups D and B suggests a common origin of HVR4 for the three supergroups, possibly due to intragenic recombination. Indeed, visual examination of the HVR4 protein sequence (Fig. 2) suggests a common motif shared among the two group D nematode Wolbachia sequences and insect sequences 39-ClacF, 40-CaciF, 2-CglaA, 14-LwhiA, and 20-PexiB. The above relationships are also supported by parsimony analyses of HVR4+, which groups these sequences in a single cluster with a bootstrap value of 98% (although the same cluster is not supported by a posterior probability of >0.5 in a Bayesian analysis of HVR4+).
As underlined by comparison of branch lengths among trees, several of the above sequences that shift position from one cluster to another across the HVRs (within supergroups A and B and between them) still show high nucleotide sequence identities (>95%) within both clusters. This is unlikely to be due to divergent evolution of sequences along the gene. Instead, it strongly indicates recombination among a set of wsp sequence portions. The remarkable nucleotide sequence conservation among motifs shared by distinct sequences also suggests either relatively recent horizontal DNA transfers or pressure for nucleotide sequence conservation acting on the recombined segments at synonymous sites subsequent to intragenic exchange.
Diversifying selection at the HVRs.
We further investigated whether the HVRs are experiencing stabilizing or diversifying selection at the amino acid level by comparing rates of synonymous versus nonsynonymous substitutions (Ks and Ka). Previous studies have used the program PAML and detected evidence of diversifying selection in wsp, particularly within HVR1 to -3, whereas HVR4 was only partially included or excluded from the analyses due to alignment issues (2, 28). However, these analyses did not take into consideration recombination within the wsp gene. Here we augment those earlier studies by using an independent-contrast approach (13). We compare independent sets of closely related sequences at each HVR (based on the phylogenetic analysis) for rates of Ks and Ka (Table 2). This approach avoids problems of recombination and alignment by analyzing only closely related sequences and evaluates them for evidence of diversifying versus stabilizing selection. Although sample sizes for independent contrasts are relatively small within each HVR, a significantly higher Ka was found in HVR1 (Wilcoxon W = 21, mean Ka − Ks = 0.0256 to 0.005, P < 0.05, n = 6) and HVR4 (W = 21, mean Ka − Ks = 0.022 to 0.000, P < 0.05, n = 6), and the same trend was found in the other two HVRs, though differences were not significant. Pooling across all HVRs, we analyzed a total of 31 sets of independent contrasts, finding a strong pattern of elevated rates of amino acid substitution. Indeed, among the 31 sets, 19 showed Ka > Ks, 11 showed Ka = Ks, and only one set showed Ka < Ks (Wilcoxon W = 198, mean Ka − Ks = 0.0181 to 0.002, P < 0.02, n = 20). This is further evidence that the HVRs of wsp are subject to strong diversifying selection. All the analyzed sets of paired contrast sequences at single HVRs differ only by single nucleotide polymorphisms, and there were no insertions or deletions (indels). This suggests that the primary engine for early variation at HVR motifs are single nucleotide substitutions.
TABLE 2.
Average Ks and Ka among closely related sequences at each HVR motif
| Groupa | Sequencesb | Ksc | Kac |
|---|---|---|---|
| HVR1 | |||
| a | 1;2 | 0 | 0 |
| b | 3;4;5;6 | 0 | 0.0233 |
| c | 7;8 | 0 | 0.0221 |
| d | 12;13 | 0 | 0 |
| e | 18;19;20;21;22;23 | 0 | 0.0573 |
| f | 24;25 | 0 | 0.0234 |
| g | 28;29;30 | 0 | 0.0268 |
| h | 32;33;34 | 0.0386 | 0.0519 |
| HVR2 | |||
| a | 1;2 | 0 | 0 |
| b | 3;4;5 | 0 | 0 |
| c | 6;9 | 0 | 0.0374 |
| d | 7;8 | 0 | 0.0183 |
| e | 11;17 | 0 | 0 |
| f | 13;18;19;21;22 | 0 | 0.0314 |
| g | 28;29 | 0 | 0.0186 |
| h | 33;34 | 0 | 0 |
| i | 39;40 | 0 | 0 |
| HVR3 | |||
| a | 1;2 | 0 | 0 |
| b | 3,4;34 | 0 | 0 |
| c* | 5;6;7;9;10 | 0.0316 | 0.0077 |
| d | 11;17;23 | 0 | 0.0131 |
| e | 18;22,24 | 0 | 0.0131 |
| f | 39;40 | 0 | 0.0381 |
| HVR4 | |||
| a | 2;14 | 0 | 0.0176 |
| b | 3;16;31 | 0 | 0.0500 |
| c | 4;8;26;34 | 0 | 0.0227 |
| d | 6;7;21 | 0 | 0 |
| e | 10;33 | 0 | 0 |
| f | 11;17;23;27 | 0 | 0.0220 |
| g | 12;19 | 0 | 0.0300 |
| h | 18;25 | 0 | 0.0362 |
Groups were extrapolated from phylogenies of HVR+s in Fig. 4. Only very closely related groups of sequences, i.e., showing Ks and Ka of <0.06, were considered. All groups but one (indicated with an asterisk), show Ks ≤ Ka.
Numbers refer to sequence codes in Table 1.
Estimation is based on the formula of Nei and Gojobori (39a).
DISCUSSION
Recombination in the wsp gene has implications for (i) potential functional and evolutionary interactions of this surface protein with the host cytoplasm and (ii) the widespread use of wsp for determining phylogenetic relationships among Wolbachia bacteria.
In recent years, new insights into mechanisms shaping prokaryotic genomes and their molecular evolution have highlighted a fluidity among microbial genomes associated with lateral gene transfer and recombination events (30, 44). Consistent with these new findings, we have found evidence of extensive recombination in the Wolbachia surface protein encoded by wsp, which results in shuffling of HVR motifs. We can assume that these recombinant proteins have been selectively favored, and previous analyses of synonymous and nonsynonymous substitutions across wsp support this view (2, 28).
Recombination provides an effective means for introducing variability in bacterial genomes. More specifically, homologous intragenic and intergenic recombinations have important implications for gene evolution. While intergenic recombination only transfers a gene type to a different background genome (giving rise to a new “genome type”), intragenic recombination may create new variants of the gene and promote the evolution of novel phenotypes through rearrangement of sequence combinations (46). If the two recombinant segments are highly divergent in portions of their sequences, intragenic recombination can be responsible for a dramatic change in the protein sequence. For instance, homologous recombination involving target amino acid motifs, which does not disrupt the correct functioning of the protein, could represent a powerful engine for protein innovation, providing otherwise “clonal” bacteria with a tool for counteracting the effects of slow accumulation of mutations, thereby escaping Muller's ratchet (38, 39). The importance of recombinational events depends, however, on the selective advantages introduced into the novel product.
The complex pattern of recombination detected by the present study in wsp suggests a long history of recombination for the gene, which has led to a marked mosaicism of its representative sequences. Analyses of the four HVRs of wsp in the selected data set revealed a strict conflicting pattern of similarities and differences among sequences, leading to a clear partitioning of the alignment into segments with incongruent phylogenetic relationships. High posterior probabilities and bootstrap values support the shuffling of amino acid motifs within the four HVRs through horizontal DNA transfer events. The presence of a high number of polymorphisms and indels in the HVRs and the limited set of HVR motifs make it highly unlikely that the observed mosaic pattern has been simply shaped by random substitutions, via homoplasic events. Since two paralogs of wsp (wspB and wspC) have been recently annotated in the wMel genome, we also evaluated the hypothesis that these genes could represent a source of sequence fragments for recombination in wsp. However, no similarity was found between any wsp HVRs and the two paralog gene sequences. Furthermore, a BLAST search of the HVRs of the wsp sequence from wMel against the whole wMel genome but wsp resulted in no significant hits. These results appear to exclude a potential role of intragenomic recombination (with either other surface proteins or different portions of the genome) in shaping wsp.
Recombination in wsp involves both CRs and HVRs. However, recombination affecting CRs appears to have a minor effect on the amino acid structure of the gene, being masked by the high protein conservation between recombining segments in these regions. The gene rearrangement affects to a greater extent the protein sequence of HVRs.
Previous studies using the program PAML indicate that the HVRs have been subject to strong positive selection, with ratios of dN/dS of ≫1 (2, 28). Recombination may not necessarily invalidate these previous results since the phenomenon would basically work by recombining a preexisting variability. However, using a different approach in this study, we were able to show elevated levels of amino acid substitutions relative to synonymous substitutions in closely related HVR sequences, thus avoiding the problems of recombination and alignment. Furthermore, this approach allowed us to demonstrate positive selection in the whole of HVR4, which was only partially inferred in previous analyses due to problems of aligning more distantly related sequences. It is worth noting that similar sequences within a motif of an HVR are typically found in different insect species, often in different orders of insects. Therefore, it is unclear whether selection for amino acid changes is a result of adaptation to new host environments, of antagonistic coevolution between the host and wsp HVRs, or of some combination of these two processes. Overall, both recombination and diversifying selection appear to be responsible for the extensive divergence among wsp sequences.
As an outer membrane protein-encoding gene, wsp shows sequence similarity with genes coding for the major surface proteins of Rickettsiales bacteria (9). A BLAST search in the Conserved Domain Database (available at the National Center for Biotechnology Information) identified significant domain homology between the wsp product and proteins that mediate various pathogen-host cell interactions from several pathogenic proteobacteria (e.g., Neisseria species). The genetic similarity of wsp with these genes is restricted to motifs at CRs, while the HVRs of wsp do not show any significant homology with any sequence in GenBank (data not shown). This could suggest a basic structural role for the conserved motifs and distinct functional roles for the HVRs among the different surface proteins. The function of wsp is unknown, and its three-dimensional molecular structure, as well as its precise location in the outer membrane, has yet to be characterized. However, wsp was found to be the most abundant protein expressed by infected Drosophila eggs (9), suggesting a potentially strong influence of the protein and its surface domains in host-bacterium interactions (27, 66).
Intragenic recombination has been frequently found to affect the outer membrane protein-encoding genes of several parasitic bacteria, i.e., the intimin genes of Escherichia coli (36), ospC and ospD of Borrelia burgdorferi (24, 33, 45), and recently ompL1 of the genus Leptospira (19). Similar to the recombinant pattern found for wsp, in leptospiral ompL1 recombination involves four variable regions encoding surface-exposed loops whose variants may represent specific adaptations to host environmental constraints. Regarding wsp, it will be interesting to determine what host proteins bind to the HVR domains and whether these proteins have been subject to divergent selection in infected host species.
The wsp gene has been widely used to identify phylogenetic associations among Wolbachia strains (14, 25, 41, 49, 51, 68). In addition, a nucleotide sequence divergence of 2.5% among wsp sequences has been used to infer subgroup affiliation and to identify novel lineages (25, 29, 49, 68). Because of the high level of intragenic recombination within wsp, use of this gene for phylogenetic reconstructions could be misleading. For instance, Kikuchi and Fukatsu (29) proposed a new subgroup of Wolbachia bacteria using the wsp sequence from the heteropteran Elasmucha putoni (18-EputB). However, as can be seen in our analysis, the wsp gene from this strain contains no unique elements but rather portions of different existing wsp sequences characterized by high nucleotide sequence similarities. An example of partitioned similarity in this sequence is its similarity with sequence 19-AencB at HVR1 and -2, 24-DinnB at HVR3, and 25-PcouB at HVR4 (Fig. 2 and 4). Therefore, although the combination of HVR motifs appears to be novel (thus leading to its appearance as a new subgroup), its HVR motifs are not. The bacterium may well be divergent, but its phylogenetic status cannot be inferred simply based on wsp. The above example suggests that caution should be taken to avoid false subgroup affiliation of strains and confusion between novel “Wolbachia lineages” and new “wsp recombinant genotypes.” Phylogenetic relationships among Wolbachia strains can be potentially clarified through comparative analyses of different portions of the genomes (such as analysis of the flanking regions of wsp and different genes).
Our results strongly suggest that recombination has occurred not only within supergroups but also between them. Nucleotide identities of motifs shared by sequences from supergroups A and B are in some cases very high, as in Sitotroga cerealella, Pegoscapus herrei, and Blastophaga nipponica, sharing 100% nucleotide sequence identity at HVR4. Such a high homology in an HVR strongly suggests recent horizontal DNA transfer between supergroups A and B. An interesting transfer of motifs at HVR4 may also have occurred between sequences from the heteropteran species Paromius exiguus and those from termites (supergroup F).
As a result, the shuffling among HVRs leads to a strong alteration of the phylogenetic signal, which not only affects the relative relatedness of the strains within a supergroup but can also weaken the major taxonomic organization of the genus into supergroups.
wsp has likely undergone both intragenic and intergenic recombination (lateral gene transfer). It is well known that Wolbachia strains experience frequent horizontal transmission, even if in some cases demonstrations are based on incongruences between wsp and host phylogenies, which leads to some circularity. However, horizontal movement of Wolbachia bacteria has also been detected using much more conserved genes such as ftsZ (63) and 16S (40, 63). It is clear that horizontal transmission of Wolbachia strains cannot be inferred solely on the basis of the wsp gene, since lateral gene transfer, rather than bacterial transfer, could be responsible. All three of these phenomena (intragenic and intergenic recombination and horizontal transmission) have the major effect of producing artifactual phylogenies. For this reason, wsp phylogeny alone should no longer be considered to represent bacterial strain phylogeny, to invoke discrepancies between Wolbachia and host systematics (51, 68), or as a reference phylogeny to support potential lateral gene transfer in comparative gene phylogenies (7). For instance, the reported incompatibility between wsp and ftsZ phylogenies (27) could instead be explained in light of intragenic recombination in wsp rather than lateral transfer of the ftsZ gene.
The impact of recombination on Wolbachia genomes is still to be clarified. As recently reported, the complete genome of wMel encodes the necessary apparatus for recombination, including RecA, and shows unusual features likely to be derived from frequent intragenomic recombination and lateral DNA transfers (67). The results of our study indicate a greater impact of recombination in the Wolbachia genome than previously appreciated. However, we do not know how unique wsp is with regard to intragenic recombination. The high number of distinct recombinant sequences detected for wsp and the complex patterns of recombination affecting some of them suggest that contact of DNA among different strains has occurred quite frequently within the Wolbachia genus. Mechanisms leading to contact and exchange of DNA among distant strains are still poorly understood, although multiple infections in the same hosts provide one obvious avenue (7, 66).
The implications of widespread recombination are clearly of great interest. Over the long term, the phenomenon disrupts bacterial clonal history and obfuscates the understanding of microbial evolution based on sequence comparisons (16). However, it also provides a potential motor for evolutionary change and the acquisition of new mechanisms among bacteria. The patterns of recombination in Wolbachia genomes could clarify important aspects of the evolution of this host-symbiont system, such as the evolution of similar phenotypes (e.g., host feminization, parthenogenesis induction, or male killing) among distantly related strains and the long persistence of these widespread parasitic bacteria in their invertebrate hosts.
Acknowledgments
We thank Cheryl Hayashi for suggestions for improving the manuscript. Claudio Bandi is thanked for discussions and for providing the opportunity of L.B. to visit the Werren laboratory, where this study was initiated. John Holt, Michael Lenz, John Jaenike, and Kelly Dyer are thanked for providing insects or DNA for this study.
The U.S. National Science Foundation (EF0328363), the Italian Ministry for Universities and Research (MIVR), and the Australian Research Council are kindly thanked for providing funds for this research.
REFERENCES
- 1.Andrews, T. D., and T. Gojobori. 2004. Strong positive selection and recombination drive the antigenic variation of the PilE protein of the human pathogen Neisseria meningitidis. Genetics 166:25-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldo, L., J. D. Bartos, J. H. Werren, C. Bazzocchi, M. Casiraghi, and S. Panelli. 2002. Different rates of nucleotide substitutions in Wolbachia endosymbionts of arthropods and nematodes: arms race or host shifts? Parasitologia 44:179-187. [PubMed] [Google Scholar]
- 3.Bandi, C., C. G. Anderson, C. Genchi, and M. L. Blaxter. 1998. Phylogeny of Wolbachia in filarial nematodes. Proc. R. Soc. Lond. B Biol. Sci. 265:2407-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bandi, C., T. J. C. Anderson, C. Genchi, and M. Blaxter. 2001. The Wolbachia endosymbionts of filarial nematodes, p. 25-43. In M. W. Kennedy and W. Harnett (ed.), Parasitic nematodes. CAB International, Wallingford, Oxon, United Kingdom.
- 5.Bandi, C., A. J. Trees, and N. W. Brattig. 2001. Wolbachia in filarial nematodes: evolutionary aspects and implications for the pathogenesis and treatment of filarial diseases. Vet. Parasitol. 98:215-238. [DOI] [PubMed] [Google Scholar]
- 6.Bazzocchi, C., F. Ceciliani, J. W. McCall, I. Ricci, C. Genchi, and C. Bandi. 2000. Antigenic role of the endosymbionts of filarial nematodes: IgG response against the Wolbachia surface protein in cats infected with Dirofilaria immitis. Proc. R. Soc. Lond. B Biol. Sci. 267:2511-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bordenstein, S. R., and J. J. Wernegreen. 2004. Bacteriophage flux in endosymbionts (Wolbachia): infection frequency, lateral transfer, and recombination rates. Mol. Biol. Evol. 21:1981-1991. [DOI] [PubMed] [Google Scholar]
- 8.Bouchon, D., T. Rigaud, and P. Juchault. 1998. Evidence for widespread Wolbachia infection in isopod crustaceans: molecular identification and host feminization. Proc. R. Soc. Lond. B Biol. Sci. 265:1081-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braig, H. R., W. Zhou, S. Dobson, and S. L. O'Neill. 1998. Cloning and characterization of a gene encoding the major surface protein of the bacterial endosymbiont Wolbachia. J. Bacteriol. 180:2373-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brattig, N. W., U. Rathjens, M. Ernst, F. Geisinger, A. Renz, and F. W. Tischendorf. 2000. Lipopolysaccharide-like molecules derived from Wolbachia endobacteria of the filaria Onchocerca volvulus are candidate mediators in the sequence of inflammatory and antiinflammatory responses of human monocytes. Microbes Infect. 2:1147-1157. [DOI] [PubMed] [Google Scholar]
- 11.Brattig, N. W., C. Bazzocchi, C. J. Kirschning, N. Reiling, D. W. Büttner, F. Ceciliani, F. Geisinger, H. Hochrein, M. Ernst, H. Wagner, C. Bandi, and A. Hoerauf. 2004. The major surface protein of Wolbachia endosymbionts in filarial nematodes elicits immune responses through TLR2 and TLR4. J. Immunol. 173:437-445. [DOI] [PubMed] [Google Scholar]
- 12.Brayton, K. A., G. H. Palmer, A. Lundgren, J. Yi, and A. F. Barbet. 2002. Antigenic variation of Anaplasma marginale msp2 occurs by combinatorial gene conversion. Mol. Microbiol. 43:1151-1159. [DOI] [PubMed] [Google Scholar]
- 13.Burt, A. 1989. Comparative methods using phylogenetically independent contrasts. Oxf. Surv. Evol. Biol. 6:33-53. [Google Scholar]
- 14.Cordaux, R., A. Michel-Salzat, and D. Bouchon. 2001. Wolbachia infection in crustaceans: novel hosts and potential routes for horizontal transmission. J. Evol. Biol. 14:237-243. [Google Scholar]
- 15.Czarnetzki, A. B., and C. C. Tebbe. 2004. Detection and phylogenetic analysis of Wolbachia in Collembola. Environ. Microbiol. 6:35-44. [DOI] [PubMed] [Google Scholar]
- 16.Doolittle, W. F. 2004. If the tree of life fell, would we recognize the sound? p. 119-133. In J. Sapp (ed.), Microbial evolution: concepts and controversies. Oxford University Press, New York, N.Y.
- 17.Gibbs, C. P., B. Y. Reimann, E. Schultz, A. Kaufmann, R. Haas, and T. F. Meyer. 1989. Reassortment of pilin genes in Neisseria gonorrhoeae occurs by two distinct mechanisms. Nature 338:651-652. [DOI] [PubMed] [Google Scholar]
- 18.Gotoh, T., H. Noda, and X. Y. Hong. 2003. Wolbachia distribution and cytoplasmic incompatibility based on a survey of 42 spider mite species (Acari: Tetranychidae) in Japan. Heredity 91:208-216. [DOI] [PubMed] [Google Scholar]
- 19.Haake, D. A., M. A. Suchard, M. M. Kelley, M. Dundoo, D. P. Alt, and R. L. Zuerner. 2004. Molecular evolution and mosaicism of leptospiral outer membrane proteins involves horizontal DNA transfer. J. Bacteriol. 186:2818-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 21.Howell-Adams, B., and H. S. Seifert. 2000. Molecular models accounting for the gene conversion reactions mediating gonococcal pilin antigenic variation. Mol. Microbiol. 37:1146-1158. [DOI] [PubMed] [Google Scholar]
- 22.Huelsenbeck, J. P., and F. Ronquist. 2001. MrBayes: bayesian inference of phylogeny. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 23.Jamnongluk, W., P. Kittayapong, V. Baimai, and S. L. O'Neill. 2002. Wolbachia infections of tephritid fruit flies: molecular evidence for five distinct strains in a single host species. Curr. Microbiol. 45:255-260. [DOI] [PubMed] [Google Scholar]
- 24.Jauris-Heipke, S., G. Liegl, V. Preac-Mursic, D. Rossler, E. Schwab, E. Soutschek, G. Will, and B. Wilske. 1995. Molecular analysis of genes encoding outer surface protein C (OspC) of Borrelia burgdorferi sensu lato: relationship to ospA genotype and evidence of lateral gene exchange of ospC. J. Clin. Microbiol. 33:1860-1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeyaprakash, A., and M. A. Hoy. 2000. Long PCR improves Wolbachia DNA amplification: wsp sequences found in 76% of sixty-three arthropod species. Insect Mol. Biol. 9:393-405. [DOI] [PubMed] [Google Scholar]
- 26.Jiggins, F. M. 2002. The rate of recombination in Wolbachia bacteria. Mol. Biol. Evol. 19:1640-1643. [DOI] [PubMed] [Google Scholar]
- 27.Jiggins, F. M., J. H. von Der Schulenburg, G. D. Hurst, and M. E. Majerus. 2001. Recombination confounds interpretations of Wolbachia evolution. Proc. R. Soc. Lond. B Biol. Sci. 268:1423-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiggins, F. M., G. D. Hurst, and Z. Yang. 2002. Host-symbiont conflicts: positive selection on an outer membrane protein of parasitic but not mutualistic Rickettsiaceae. Mol. Biol. Evol. 19:1341-1349. [DOI] [PubMed] [Google Scholar]
- 29.Kikuchi, Y., and T. Fukatsu. 2003. Diversity of Wolbachia endosymbionts in heteropteran bugs. Appl. Environ. Microbiol. 69:6082-6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lawrence, J. G., and H. Hendrickson. 2003. Lateral gene transfer: when will adolescence end? Mol. Microbiol. 50:739-749. [DOI] [PubMed] [Google Scholar]
- 31.Lo, N., M. Casiraghi, E. Salati, C. Bazzocchi, and C. Bandi. 2002. How many Wolbachia supergroups exist? Mol. Biol. Evol. 19:341-346. [DOI] [PubMed] [Google Scholar]
- 32.Maekawa, K., N. Lo, O. Kitade, T. Miura, and T. Matsumoto. 1999. Molecular phylogeny and geographic distribution of wood-feeding cockroaches in East Asian islands. Mol. Phylogenet. Evol. 13:360-376. [DOI] [PubMed] [Google Scholar]
- 33.Marconi, R. T., D. S. Samuels, R. K. Landry, and C. F. Garon. 1994. Analysis of the distribution and molecular heterogeneity of the ospD gene among the Lyme disease spirochetes: evidence for lateral gene exchange. J. Bacteriol. 176:4572-4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin, D., C. Williamson, and D. Posada. 2005. RDP2: recombination detection and analysis from sequence alignments. Bioinformatics 21:260-262. [DOI] [PubMed] [Google Scholar]
- 35.Masui, S., S. Kamoda, T. Sasaki, and H. Ishikawa. 2000. Distribution and evolution of bacteriophage WO in Wolbachia, the endosymbiont causing sexual alterations in arthropods. J. Mol. Evol. 51:491-497. [DOI] [PubMed] [Google Scholar]
- 36.McGraw, E. A., J. Li, R. K. Selander, and T. S. Whittam. 1999. Molecular evolution and mosaic structure of alpha, beta, and gamma intimins of pathogenic Escherichia coli. Mol. Biol. Evol. 16:12-22. [DOI] [PubMed] [Google Scholar]
- 37.Meeus, P. F., K. A. Brayton, G. H. Palmer, and A. F. Barbet. 2003. Conservation of a gene conversion mechanism in two distantly related paralogues of Anaplasma marginale. Mol. Microbiol. 47:633-643. [DOI] [PubMed] [Google Scholar]
- 38.Moran, N. 1996. Accelerated evolution and Muller's ratchet in endosymbiotic bacteria. Proc. Natl. Acad. Sci. USA 93:2873-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Müller, H. J. 1964. The relation of recombination to mutational advance. Mutat. Res. 1:2-9. [DOI] [PubMed] [Google Scholar]
- 39a.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 40.O'Neill, S. L., R. Giordano, A. M. Colbert, T. L. Karr, and H. M. Robertson. 1992. 16S rRNA phylogenetic analysis of the bacterial endosymbionts associated with cytoplasmic incompatibility in insects. Proc. Natl. Acad. Sci. USA 89:2699-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pintureau, B., S. Chaudier, F. Lassabliere, H. Charles, and S. Grenier. 2000. Addition of wsp sequences to the Wolbachia phylogenetic tree and stability of the classification. J. Mol. Evol. 51:374-377. [DOI] [PubMed] [Google Scholar]
- 42.Posada, D., and K. A. Crandall. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 43.Posada, D., and K. A. Crandall. 2001. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc. Natl. Acad. Sci. USA 98:13757-13762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Posada, D., K. A. Crandall, and E. C. Holmes. 2002. Recombination in evolutionary genomics. Annu. Rev. Genet. 36:75-97. [DOI] [PubMed] [Google Scholar]
- 45.Qiu, W. G., S. E. Schutzer, J. F. Bruno, O. Attie, Y. Xu, J. J. Dunn, C. M. Fraser, S. R. Casjens, and B. J. Luft. 2004. Genetic exchange and plasmid transfers in Borrelia burgdorferi sensu stricto revealed by three-way genome comparisons and multilocus sequence typing. Proc. Natl. Acad. Sci. USA 101:14150-14155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rajalingam, R., P. Parham, and L. Abi-Rached. 2004. Domain shuffling has been the main mechanism forming new hominoid killer cell Ig-like receptors. J. Immunol. 172:356-369. [DOI] [PubMed] [Google Scholar]
- 47.Reuter, M., and L. Keller. 2003. High levels of multiple Wolbachia infection and recombination in the ant Formica exsecta. Mol. Biol. Evol. 20:748-753. [DOI] [PubMed] [Google Scholar]
- 48.Rousset, F., and M. Raymond. 1991. Cytoplasmic incompatibility in insects: why sterilize females? Trends Ecol. Evol. 6:54-57. [DOI] [PubMed] [Google Scholar]
- 49.Rowley, S. M., R. J. Raven, and E. A. McGraw. 2004. Wolbachia pipientis in Australian spiders. Curr. Microbiol. 49:208-214. [DOI] [PubMed] [Google Scholar]
- 50.Rozas, J., and R. Rozas. 1999. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174-175. [DOI] [PubMed] [Google Scholar]
- 51.Shoemaker, D. D., C. A. Machado, D. Molbo, J. H. Werren, D. M. Windsor, and E. A. Herre. 2002. The distribution of Wolbachia in fig wasps: correlations with host phylogeny, ecology and population structure. Proc. R. Soc. Lond. B. Biol. Sci. 269:2257-2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith, J. M. 1992. Analyzing the mosaic structure of genes. J. Mol. Evol. 34:126-129. [DOI] [PubMed] [Google Scholar]
- 53.Smith, J. M. 1999. The detection and measurement of recombination from sequence data. Genetics 153:1021-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stouthamer, R., J. A. J. Breeuwer, and G. D. Hurst. 1999. Wolbachia pipientis: microbial manipulator of arthropod reproduction. Annu. Rev. Microbiol. 53:71-102. [DOI] [PubMed] [Google Scholar]
- 55.Swofford, D. L. 2000. PAUP*: phylogenetic analysis using parsimony (*and other methods). Sinauer Associates, Sunderland, Mass.
- 56.Tamas, I., L. Klasson, B. Canback, A. K. Naslund, A. S. Eriksson, J. J. Wernegreen, J. P. Sandstrom, N. A. Moran, and S. G. Andersson. 2002. 50 million years of genomic stasis in endosymbiotic bacteria. Science 296:2376-2379. [DOI] [PubMed] [Google Scholar]
- 57.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Urwin, R., E. C. Holmes, A. J. Fox, J. P. Derrick, and M. C. J. Maiden. 2002. Phylogenetic evidence for frequent positive selection and recombination in the meningococcal surface antigen PorB. Mol. Biol. Evol. 19:1686-1694. [DOI] [PubMed] [Google Scholar]
- 59.Vandekerckhove, T. M. T., S. Watteyne, S. Willems, J. G. Swings, J. Mertens, and M. Gillis. 1999. Phylogenetic analysis of the 16S rDNA of the cytoplasmic bacterium Wolbachia from the novel host Folsomia candida (Hexapoda, Collembola) and its implications for the Wolbachia taxonomy. FEMS Microbiol. Lett. 180:279-286. [DOI] [PubMed] [Google Scholar]
- 60.van Meer, M. M. M., J. Witteveldt, and R. Stouthamer. 1999. Phylogeny of the arthropod endosymbiont Wolbachia based on wsp gene. Insect Mol. Biol. 8:399-408. [DOI] [PubMed] [Google Scholar]
- 61.van Valen, L. 1973. A new evolutionary law. Evol. Theory 1:1-30. [Google Scholar]
- 62.Werren, J. H., D. Windsor, and L. R. Guo. 1995. Distribution of Wolbachia among neotropical arthropods. Proc. R. Soc. Lond. B Biol. Sci. 262:197-204. [Google Scholar]
- 63.Werren, J. H., W. Zhang, and L. R. Guo. 1995. Evolution and phylogeny of Wolbachia: reproductive parasites of arthropods. Proc. R. Soc. Lond. B Biol. Sci. 261:55-63. [DOI] [PubMed] [Google Scholar]
- 64.Werren, J. H. 1997. Biology of Wolbachia. Annu. Rev. Entomol. 42:587-609. [DOI] [PubMed] [Google Scholar]
- 65.Werren, J. H., and D. M. Windsor. 2000. Wolbachia infection frequencies in insects: evidence of a global equilibrium? Proc. R. Soc. Lond. B Biol. Sci. 267:1277-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Werren, J. H., and J. D. Bartos. 2001. Recombination in Wolbachia. Curr. Biol. 11:431-435. [DOI] [PubMed] [Google Scholar]
- 67.Wu, M., L. V. Sun, J. Vamathevan, M. Riegler, R. Deboy, J. C. Brownlie, E. A. McGraw, W. Martin, C. Esser, N. Ahmadinejad, C. Wiegand, R. Madupu, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, A. S. Durkin, J. F. Kolonay, W. C. Nelson, Y. Mohamoud, P. Lee, K. Berry, M. B. Young, T. Utterback, J. Weidman, W. C. Nierman, I. T. Paulsen, K. E. Nelson, H. Tettelin, S. L. O'Neill, and J. A. Eisen. 2004. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2:E69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou, W., F. Rousset, and S. L. O'Neill. 1998. Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc. R. Soc. Lond. B Biol. Sci. 265:509-515. [DOI] [PMC free article] [PubMed] [Google Scholar]