Abstract
The opportunistic human pathogen Pseudomonas aeruginosa causes persistent airway infections in patients with cystic fibrosis (CF). To establish these chronic infections, P. aeruginosa must grow and proliferate within the highly viscous sputum in the lungs of CF patients. In this study, we used Affymetrix GeneChip microarrays to investigate the physiology of P. aeruginosa grown using CF sputum as the sole source of carbon and energy. Our results indicate that CF sputum readily supports high-density P. aeruginosa growth. Furthermore, multiple signals, which reduce swimming motility and prematurely activate the Pseudomonas quinolone signal cell-to-cell signaling cascade in P. aeruginosa, are present in CF sputum. P. aeruginosa factors critical for lysis of the common CF lung inhabitant Staphylococcus aureus were also induced in CF sputum and increased the competitiveness of P. aeruginosa during polymicrobial growth in CF sputum.
Pseudomonas aeruginosa is a ubiquitous gram-negative bacterium isolated readily from water and soil environments. P. aeruginosa also causes human infections, particularly in patients with compromised systemic immunity or impaired mucosal defenses. These infections can be devastating, because P. aeruginosa is inherently resistant to many antibiotics and it produces an array of virulence factors, including proteases, lipases, exotoxins, and a number of secondary metabolites (45). In some settings, P. aeruginosa causes persistent infections, provoking chronic inflammation that steadily destroys host tissues (21, 26, 37).
One of the most notorious chronic infections caused by P. aeruginosa occurs in the lungs of patients with cystic fibrosis (CF). CF patients manifest a host defense defect localized to the conducting airways of the lung that results in chronic colonization by several bacterial species (21, 26, 37). P. aeruginosa is thought to cause the most clinically important CF airway infections, as P. aeruginosa colonization heralds the onset of chronic pulmonary symptoms and declining lung function (21).
CF P. aeruginosa infections have several remarkable characteristics. In most cases, P. aeruginosa airway colonization occurs after other bacteria, such as Staphylococcus aureus, have infected the patient's airway (13, 21). Once P. aeruginosa infection develops, it often displaces other bacteria and becomes the predominant bacterium in the CF lung (14, 21). P. aeruginosa also routinely reaches very high densities within the respiratory secretions (108 to 1010 CFU/ml) (21). In addition, CF P. aeruginosa infections are thought to involve coordinated bacterial activities facilitated by cell-to-cell communication. These include the formation of multicellular biofilm communities and density-dependent gene regulation (3, 5, 6, 9, 44).
While controversy about the initial steps in the infection pathogenesis remains, consensus is emerging that once chronic P. aeruginosa colonization is established, a large proportion of the infecting bacteria grow within airway sputum (21, 33). Sputum is a complex mixture of airway mucus, inflammatory substances that are induced by infection, and bacteria and bacterial products. The inflammatory components include massive numbers of polymorphonuclear leukocytes as well as antibodies, antimicrobial peptides, dead host cells, and serum components that enter the airway due to vascular leak and pulmonary hemorrhage (21). In addition to providing a physical substrate for bacterial growth, the sputum very likely serves as the nutritional source for the infecting organisms (21, 33).
Many bacterial functions, including virulence determinants central to disease pathogenesis, are known to be influenced by specific nutrients. For example, the production of a biosurfactant by P. aeruginosa is modulated by the particular carbon and nitrogen sources available (11, 29). This surfactant facilitates surface motility and hydrocarbon assimilation. Similarly, induction of the type III secretion system, a key virulence factor, can be regulated by the level of calcium present (54). Many other functions, including biofilm formation, secretion of exoproducts, and the ability to kill predators such as nematodes, are also influenced by the nutrients present in a given environment (15, 24, 34, 47, 50).
The strong influence of nutritional conditions on bacterial functioning led us to hypothesize that growth in sputum from CF patients could profoundly impact P. aeruginosa physiology. To test this, we used Affymetrix GeneChips to globally evaluate gene expression of P. aeruginosa during growth using CF sputum as the sole source of carbon and energy. We found that CF sputum obtained from several different patients supported P. aeruginosa growth to high population densities. Transcriptome analysis revealed that gene expression was markedly affected by growth in CF sputum and suggested that amino acids within sputum were the likely source of carbon for the bacteria. The expression analysis led us to other key aspects of P. aeruginosa physiology that were affected by growth in sputum. These include swimming motility, quorum sensing by the Pseudomonas quinolone signal (PQS) system, and the production of bactericidal factors that enhance the competitiveness of P. aeruginosa during growth with S. aureus. Thus, CF sputum is an excellent growth medium, and the physiological changes it induces in P. aeruginosa could impact the pathogenesis of CF infections.
MATERIALS AND METHODS
Bacterial strains and growth media.
P. aeruginosa strain UCBPP-PA14 (39) and S. aureus strain MN8 (42) were used in these studies. P. aeruginosa was grown in morpholinepropanesulfonic acid (MOPS)-buffered medium (50 mM MOPS [pH 7.2], 93 mM NH4Cl, 43 mM NaCl, 3.7 mM KH2PO4, 1 mM MgSO4, and 3.5 μM FeSO4 · 7H2O). Twenty mM glucose, 6.3 mM glucose, 13 mM succinate, or 0.11% (wt/vol) Casamino Acids were added as the sole carbon and energy source. P. aeruginosa reaches equal maximum population densities when grown in 6.3 mM glucose, 13 mM succinate, and 0.11% Casamino Acids. For routine growth, P. aeruginosa was grown in tryptic soy broth. For in vitro growth of S. aureus or P. aeruginosa/S. aureus binary cultures, brain heart infusion broth was used. For differential isolation of P. aeruginosa and S. aureus in coculture, Pseudomonas isolation agar (PIA) and Baird Parker agar (Remel) were used, respectively. Escherichia coli pKDT17 and E. coli pECP61.5 were used for quantitation of 3OC12-HSL and C4-HSL, respectively, as outlined previously (52).
Sputum sampling and medium preparation.
Sputum samples from CF and non-CF patients were obtained by expectoration into sterile collection tubes. Only sputum samples containing total bacterial titers of ≤108 CFU of P. aeruginosa/ml and devoid of antibiotics were used in this study. Sputum samples were frozen on dry ice immediately after collection and were stored at −80°C until processing. Frozen sputum was thawed, transferred to a 250-ml flask, and lyophilized overnight (VirTis). Fifty ml of CF sputum corresponded to approximately 2 g dry weight. Powdered sputum was stored at −20°C under desiccating conditions.
Prior to medium preparation, powdered sputum was weighed and sterilized in a HybriLinker HL-2000 (UVP) for 10 to 20 min. Sputum was then resuspended in MOPS minimal medium to 10% sputum (vol/vol) and homogenized by sonication with a tip sonicator (Branson Ultrasonics). Samples were sonicated up to 5 times at 40 to 50% output for 30 s, depending on the consistency of the medium. This medium will be referred to below as MOPS-sputum medium. In some cases, MOPS-sputum medium was centrifuged at 16,000 × g for 5 min to remove any insoluble material and then filtered through a 0.45-μm-pore-size filter. Mucus isolated from primary lung epithelia (55) and UV-sterilized bovine mucin (Worthington Biochem) resuspended in MOPS medium were used as a control in some cases.
Growth of P. aeruginosa in CF sputum.
P. aeruginosa was grown with shaking (250 rpm) in MOPS-sputum or MOPS-glucose medium at 37°C. Washed cells from exponentially growing cultures in MOPS minimal medium with glucose (optical density at 600 nm [OD600] of 0.4 to 0.6) were the source of the inoculum, and all cultures were diluted to a starting OD600 of 0.001. Cell density was monitored using serial dilution/plate counts and/or optical density at 600 nm. For optical density measurements of MOPS-sputum medium-grown P. aeruginosa, uninoculated MOPS-sputum medium was used as a blank.
Global expression profiling.
P. aeruginosa growing in MOPS-glucose or MOPS-sputum medium were harvested at an OD600 of 0.1 to 0.2 and mixed 1:1 with the RNA stabilizing agent RNAlater (Ambion). RNA was isolated using RNeasy mini-columns (QIAGEN) and prepared for hybridization to Affymetrix GeneChip microarrays as previously described (43). DNA contamination of RNA samples was assessed by PCR amplification of the P. aeruginosa rplU gene with the primers rplU-for (5′-CGCAGTGATTGTTACCGGTG-3′) and rplU-rev (5′-AGGCCTGAATGCCGGTGATC-3′). Washing, staining, and scanning of the GeneChips was performed by the University of Iowa DNA core facility using an Affymetrix fluidics station. GeneChips were performed in duplicate or triplicate for each condition tested, and data were analyzed using Microarray Suite software.
Semiquantitative RT-PCR.
Semiquantitative reverse transcription (RT)-PCR was performed using Superscript II reverse transcriptase as outlined by the manufacturer (Invitrogen). One hundred ng of P. aeruginosa RNA served as the template for cDNA synthesis using 250 ng of the random primer (NS)5. Five ng of the resulting purified cDNA was used as template in the subsequent PCR. PCR (25-μl reaction volume) was performed with the Expand Long Template PCR system (Roche) with the following conditions: 95°C for 2 min; and 30 cycles of 95°C for 45 s, 60°C for 45 s, and 68°C for 1 min. For visualization, 5 μl of the resulting PCR was subjected to agarose gel electrophoresis and stained with ethidium bromide.
TEM.
Negative staining of P. aeruginosa for transmission electron microscopy (TEM) was performed as described elsewhere using phosphotungstic acid (23). Bacteria were harvested from liquid suspension using a wide-bore pipette tip (3 mm) to minimize flagellar shearing.
PQS extraction and quantitation.
PQS was extracted from exponentially growing (OD600 of 0.1 or of 0.4 to 0.6) P. aeruginosa in MOPS-glucose, MOPS-sputum, MOPS-succinate, or MOPS-succinate medium containing amino acids using acidified ethyl acetate as outlined previously (16, 35). Uninoculated MOPS-sputum medium was also extracted as a control. Ethyl acetate extracts were evaporated under a continuous stream of N2 and resuspended in 50 μl of acetonitrile/acidified ethyl acetate (1:1 ratio). PQS in these extracts (20 to 40 μl) was analyzed using thin-layer chromatography (TLC) and visualized under UV light as described previously (16, 35). Although several solvent systems have been used to evaluate PQS production, it is important to point out that the solvent system used in this study has been shown to separate PQS from other quinolones/quinolines within the culture supernatant (4, 35). Quantitation of PQS on TLC plates was performed using spot densitometry with a Fluorchem 8900 gel imager (Alpha Innotech) with synthetic PQS as a standard.
RESULTS
P. aeruginosa growth in CF sputum.
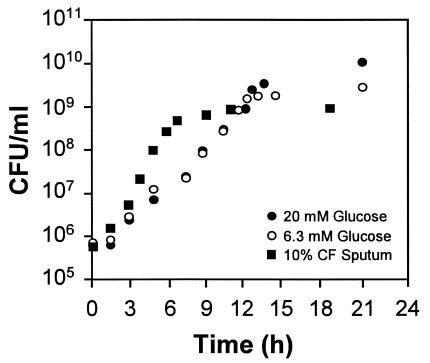
The first step in understanding the physiology of P. aeruginosa during growth in CF sputum was development of an in vitro CF sputum medium. To accomplish this, we used sterile lyophilized CF sputum as the sole source of carbon and energy in a standard MOPS buffer. Similar growth kinetics were observed for P. aeruginosa grown using sputum from 10 different CF patients, and a representative growth curve is shown in Fig. 1. P. aeruginosa grows aerobically using CF sputum as a sole source of carbon and energy with a mean doubling time of approximately 40 min (Fig. 1). Maximum P. aeruginosa growth yields in 10% CF sputum medium are 1 × 109 bacteria/ml. At this concentration of CF sputum, P. aeruginosa growth is limited by the amount of metabolizable carbon, since the addition of glucose to CF sputum medium increased P. aeruginosa growth yields (data not shown).
FIG. 1.
Growth of P. aeruginosa in MOPS medium containing 20 mM glucose (•), 6.3 mM glucose (○), or 10% CF sputum (▪) as the sole source of carbon and energy. Bacteria were grown with shaking (250 rpm) at 37°C and sampled during exponential growth (108 CFU of P. aeruginosa/ml) for Affymetrix GeneChip analysis. Representative growth curves are shown.
Transcriptome analysis of CF sputum-grown P. aeruginosa.
We used Affymetrix GeneChips to examine P. aeruginosa gene expression during growth in 10% CF sputum. Glucose-grown P. aeruginosa were used as the control for these experiments. All cultures were sampled for GeneChip analysis at an OD600 of 0.1 to 0.2, and the maximum doubling times in glucose and CF sputum were similar (50 min for glucose; 40 min for CF sputum). For sputum GeneChip experiments, P. aeruginosa was grown using sputum samples from two different CF patients, and the data were averaged to eliminate patient-specific factors. On average, 70% of the gene-specific tiles present on the Affymetrix GeneChip hybridized at levels sufficient for statistical analysis; thus, we were able to compare expression profiles of approximately 4,000 genes. A total of 147 genes (approximately 3% of all P. aeruginosa genes) were differentially regulated at least fivefold during growth of P. aeruginosa in CF sputum compared to glucose-grown bacteria (Table 1). Of these genes, the majority were up-regulated (113 genes), while a smaller number were repressed (34 genes).
TABLE 1.
P. aeruginosa genes differentially regulated during growth in CF sputum
| Function and ORFa | Genea | Category or classa | Fold regulationb |
|---|---|---|---|
| Amino acid biosynthesis | |||
| PA0035 | trpA | Tryptophan synthase alpha chain | −7 |
| PA0036 | trpB | Tryptophan synthase beta chain | −9 |
| PA0904 | lysC | Aspartate kinase alpha and beta chain | −2.9 |
| PA1326 | ilvA2 | Threonine dehydratase, biosynthetic | −9.1 |
| PA3118 | leuB | 3-Isopropylmalate dehydrogenase | −4.3 |
| PA3120 | leuD | 3-Isopropylmalate dehydratase small subunit | −7 |
| PA3121 | leuC | 3-Isopropylmalate dehydratase large subunit | −7 |
| PA3525 | argG | Argininosuccinate synthase | −2.6 |
| PA3537 | argF | Ornithine carbamoyltransferase, anabolic | −3.7 |
| PA4588 | gdhA | Glutamate dehydrogenasec | −4.2 |
| PA4695 | ilvH | Acetolactate synthase isozyme III small subunit | −2.6 |
| PA4696 | ilvI | Acetolactate synthase large subunit | −2.7 |
| PA5035 | gltD | Glutamate synthase small chainc | −4.8 |
| PA5036 | gltB | Glutamate synthase large chain precursorc | −4.2 |
| Amino acid transport and degradation | |||
| PA0782 | putA | Proline dehydrogenase PutA | 4.3 |
| PA0865 | hpd | 4-Hydroxyphenylpyruvate dioxygenase | 66 |
| PA0866 | aroP2 | Aromatic amino acid transport protein AroP2 | 13 |
| PA0870 | phhC | Aromatic amino acid aminotransferase | 9 |
| PA0871 | phhB | Pterin-4-α-carbinolamine dehydratase | 5 |
| PA0872 | phhA | Phenylalanine-4-hydroxylased | 32 |
| PA0897 | aruG | Arginine/ornithine succinyltransferase AII subunit | 3 |
| PA0898 | aruD | Succinylglutamate-5-semialdehyde dehydrogenase | 2.7 |
| PA2001 | atoB | Acetyl coenzyme A acetyltransferase | 16 |
| PA2007 | maiA | Maleylacetoacetate isomerase | 8 |
| PA2008 | fahA | Fumarylacetoacetase | 9 |
| PA2009 | hmgA | Homogentisate 1,2-dioxygenase | 11 |
| PA2247 | bkdA1 | 2-Oxoisovalerate dehydrogenase, α subunit | 20 |
| PA2248 | bkdA2 | 2-Oxoisovalerate dehydrogenase, β subunit | 19 |
| PA2249 | bkdB | Branched-chain α-keto acid dehydrogenase | 13 |
| PA2250 | lpdV | Lipoamide dehydrogenase-Val | 19 |
| PA3766 | Probable aromatic amino acid transporter | 2.8 | |
| PA4470 | fumC1 | Fumarate hydratase | 6 |
| PA5302 | dadX | Catabolic alanine racemase | 9 |
| PA5304 | dadA | d-amino acid dehydrogenase, small subunit | 20 |
| Glucose transport and metabolism | |||
| PA2322 | Gluconate permease | −5.5 | |
| PA2323 | Probable glyceraldehyde-3-phosphate dehydrogenase | −3.8 | |
| PA3181 | 2-Keto-3-deoxy-6-phosphogluconate aldolase | −3 | |
| PA3186 | oprB | Glucose/carbohydrate outer membrane porin OprB | −2.7 |
| PA3195 | gapA | Glyceraldehyde-3-phosphate dehydrogenase | −3.2 |
| Flagellar synthesis and chemotaxis | |||
| PA1092 | fliC | Flagellin type B | −21 |
| PA2867 | Probable chemotaxis transducer | −8 | |
| PA4307 | pctC | Chemotactic transducer PctC | −8 |
| PA4310 | pctB | Chemotactic transducer PctB | −23 |
| Pseudomonas quinolone signaling | |||
| PA0996 | pqsA | Probable coenzyme A ligase | 18 |
| PA0997 | pqsB | Homologous to β-keto-acyl-acyl-carrier protein synthase | 17 |
| PA0998 | pqsC | Homologous to β-keto-acyl-acyl-carrier protein synthase | 19 |
| PA0999 | pqsD | 3-Oxoacyl-(acyl carrier protein) synthase III | 17 |
| PA1000 | pqsE | Quinolone signal response protein | 19 |
| PA1001 | phnA | Anthranilate synthase component I | 22 |
| PA1002 | phnB | Anthranilate synthase component II | 14 |
| Other genes | |||
| PA0034 | Probable two-component response regulator | −7 | |
| PA0435 | Hypothetical protein | 9 | |
| PA0672 | hemO | Heme oxygenase | 6 |
| PA0730 | Probable transferase | −7 | |
| PA0781 | Hypothetical protein | 36 | |
| PA1093 | Hypothetical protein | −20 | |
| PA1300 | Probable σ70 factor, ECF subfamily | 6 | |
| PA1325 | Conserved hypothetical protein | −11 | |
| PA1892 | Hypothetical protein | −5 | |
| PA1894 | Hypothetical protein | −13 | |
| PA1895 | Hypothetical protein | −9 | |
| PA1896 | Hypothetical protein | −15 | |
| PA1897 | Hypothetical protein | −23 | |
| PA1922 | Probable TonB-dependent receptor | 9 | |
| PA1924 | Hypothetical protein | 22 | |
| PA1925 | Hypothetical protein | 6 | |
| PA1981 | Hypothetical protein | −14 | |
| PA1982 | exaA | Quinoprotein alcohol dehydrogenase | −18 |
| PA1983 | exaB | Cytochrome c550 | −16 |
| PA1999 | Probable coenzyme A transferase, subunit A | 28 | |
| PA2000 | Probable coenzyme A transferase, subunit B | 22 | |
| PA2006 | Probable MFS transporter | 7 | |
| PA2027 | Hypothetical protein | −25 | |
| PA2194 | hcnB | Hydrogen cyanide synthase HcnB | 5 |
| PA2384 | Hypothetical protein | 12 | |
| PA2385 | pvdQ | PvdQ | 15 |
| PA2386 | pvdA | l-Ornithine-N5-oxygenase | 24 |
| PA2392 | pvdP | PvdP | 9 |
| PA2393 | Probable dipeptidase precursor | 19 | |
| PA2394 | pvdN | PvdN | 15 |
| PA2395 | pvdO | PvdO | 15 |
| PA2396 | pvdF | Pyoverdine synthetase F | 14 |
| PA2397 | pvdE | Pyoverdine biosynthesis protein PvdE | 12 |
| PA2399 | pvdD | Pyoverdine synthetase D | 10 |
| PA2400 | pvdJ | PvdJ | 28 |
| PA2401 | pvdJ | PvdJ | 17 |
| PA2402 | Probable nonribosomal peptide synthetase | 15 | |
| PA2405 | Hypothetical protein | 5 | |
| PA2411 | Probable thioesterase | 80 | |
| PA2412 | Conserved hypothetical protein | 32 | |
| PA2413 | Probable class III aminotransferase | 21 | |
| PA2424 | pvdL | PvdL | 16 |
| PA2425 | pvdG | PvdG | 12 |
| PA2426 | pvdS | σ factor PvdS | 10 |
| PA2427 | Hypothetical protein | 8 | |
| PA2451 | Hypothetical protein | 6 | |
| PA2452 | Hypothetical protein | 46 | |
| PA2807 | Hypothetical protein | −28 | |
| PA2862 | lipA | Lactonizing lipase precursor | −7 |
| PA2911 | Probable TonB-dependent receptor | 6 | |
| PA2912 | Probable ATP-binding component of ABC transporter | 5 | |
| PA3281 | Hypothetical protein | 16 | |
| PA3282 | Hypothetical protein | 10 | |
| PA3283 | Conserved hypothetical protein | 23 | |
| PA3284 | Hypothetical protein | 48 | |
| PA3407 | hasAp | Heme acquisition protein HasAp | 13 |
| PA3444 | Conserved hypothetical protein | −8 | |
| PA3526 | Probable outer membrane protein precursor | −8 | |
| PA3598 | Conserved hypothetical protein | 6 | |
| PA3600 | Conserved hypothetical protein | 171 | |
| PA3601 | Conserved hypothetical protein | 80 | |
| PA3662 | Hypothetical protein | −6 | |
| PA3749 | Probable MFS transporter | −10 | |
| PA3757 | Probable transcriptional regulator | 5 | |
| PA3758 | Probable N-acetylglucosamine-6-phosphate deacetylase | 6 | |
| PA3759 | Probable aminotransferase | 6 | |
| PA3789 | Hypothetical protein | 12 | |
| PA3790 | oprC | Outer membrane porin OprC | 14 |
| PA3922 | Conserved hypothetical protein | −5 | |
| PA3936 | Probable permease of ABC taurine transporter | −8 | |
| PA3938 | Probable periplasmic taurine-binding protein precursor | −5 | |
| PA4063 | Hypothetical protein | 23 | |
| PA4064 | Probable ATP-binding component of ABC transporter | 18 | |
| PA4065 | Hypothetical protein | 10 | |
| PA4066 | Hypothetical protein | 8 | |
| PA4131 | Probable iron-sulfur protein | 7 | |
| PA4170 | Hypothetical protein | 36 | |
| PA4171 | Probable protease | 8 | |
| PA4218 | Probable transporter | 23 | |
| PA4219 | Hypothetical protein | 66 | |
| PA4220 | Hypothetical protein | 104 | |
| PA4221 | fptA | Fe(III)-pyochelin outer membrane receptor precursor | 44 |
| PA4222 | Probable ATP-binding component of ABC transporter | 88 | |
| PA4223 | Probable ATP-binding component of ABC transporter | 41 | |
| PA4224 | pchG | Pyochelin biosynthetic protein PchG | 96 |
| PA4225 | pchF | Pyochelin synthetase | 59 |
| PA4226 | pchE | Dihydroaeruginoic acid synthetase | 75 |
| PA4228 | pchD | Pyochelin biosynthesis protein PchD | 81 |
| PA4229 | pchC | Pyochelin biosynthetic protein PchC | 80 |
| PA4230 | pchB | Salicylate biosynthesis protein PchB | 139 |
| PA4231 | pchA | Salicylate biosynthesis isochorismate synthase | 121 |
| PA4471 | Hypothetical protein | 5 | |
| PA4498 | Probable metallopeptidase | 5 | |
| PA4514 | Probable outer membrane receptor for iron transport | −10 | |
| PA4570 | Hypothetical protein | 5 | |
| PA4633 | Probable chemotaxis transducer | −5 | |
| PA4770 | lldP | l-Lactate permease | 35 |
| PA4771 | lldD | l-Lactate dehydrogenase | 9 |
| PA4772 | Probable ferredoxin | 9 | |
| PA4811 | fdnH | Nitrate-inducible formate dehydrogenase, β subunit | 6 |
| PA4834 | Hypothetical protein | 80 | |
| PA4835 | Hypothetical protein | 45 | |
| PA4836 | Hypothetical protein | 57 | |
| PA4837 | Probable outer membrane protein precursor | 43 | |
| PA4838 | Hypothetical protein | 8 | |
| PA4929 | Hypothetical protein | −6 | |
| PA5303 | Conserved hypothetical protein | 21 | |
| PA5532 | Hypothetical protein | 8 | |
| PA5534 | Hypothetical protein | 42 | |
| PA5535 | Conserved hypothetical protein | 43 | |
| PA5536 | Conserved hypothetical protein | 74 | |
| PA5538 | amiA | N-acetylmuramoyl-l-alanine amidase | 38 |
| PA5539 | Hypothetical protein | 27 | |
| PA5540 | Hypothetical protein | 28 | |
| PA5541 | Probable dihydroorotase | 40 | |
| tRNA-Arg | Noncoding RNA gene | 16 |
From P. aeruginosa genome website, http://www.pseudomonas.com.
Regulation (n-fold) of genes differentially expressed during P. aeruginosa growth in 10% CF sputum compared to growth in glucose; a positive number indicates an up-regulation of the gene during growth in sputum.
Involved in synthesis of glutamate and degradation of glutamate and glutamine.
Involved in synthesis of tyrosine and degradation of phenylalanine.
P. aeruginosa metabolism in CF sputum.
To chronically colonize the CF lung, P. aeruginosa must be able to proliferate within the thick respiratory sputum of the CF lung. The composition of CF sputum is complex, and it is clear that CF sputum supports growth of large numbers of P. aeruginosa in vivo (21, 33). The growth substrate(s) within the CF lung is not known, but high concentrations of proteins and amino acids have been found in CF sputum (2, 33, 48). Since glucose-grown bacteria served as the control comparison in the GeneChip experiments and glucose metabolism is well understood in P. aeruginosa (7, 19, 22, 36, 41), we hypothesized that our array data would provide information regarding carbon metabolism in CF sputum. Eleven genes involved in branch chain and aromatic amino acid catabolism were highly up-regulated during growth in CF sputum (Table 1), and genes involved in biosynthesis of these amino acids were repressed. Genes involved in transport and metabolism of glucose were also repressed during growth in CF sputum (Table 1).
Flagellar motility in CF sputum.
A recent study reported that P. aeruginosa is nonmotile during growth in the presence of dialyzed CF sputum (53). This loss of motility was attributed to repression of fliC, which encodes the major flagellar filament component and is required for flagellum biosynthesis. Analyses of our array results confirmed the repression of fliC during growth in CF sputum (Table 1), and microscopic examination revealed that >95% of P. aeruginosa growing in filtered and nonfiltered CF sputum media are nonmotile. An examination of negatively stained CF sputum-grown P. aeruginosa using transmission electron microscopy revealed that approximately 40% of these bacteria did not possess flagella. Also as previously shown, loss of motility is not specific to CF sputum, as non-CF sputum also inhibited motility (reference 53 and Table 2).
TABLE 2.
Impact of carbon source on P. aeruginosa swimming motility
| Growth substratea | Swimming motilityb |
|---|---|
| Glucose | + |
| Succinate | + |
| Casamino Acids | + |
| Bovine mucin | + |
| Primary lung cell line mucus | + |
| CF sputum (n = 10) | − |
| Non-CF sputum (n = 2) | − |
| Heat-inactivated CF sputum | + |
Carbon and energy source supplied for growth in a MOPS minimal base.
+, >90% of P. aeruginosa examined exhibited swimming motility; −, <1% exhibited swimming motility by phase contrast microscopy.
To determine if motility was repressed by airway mucins or other components of sputum, we grew P. aeruginosa in medium made from mucus collected from primary cultures of lung epithelial cells grown at the air-liquid interface. Epithelium cultures grown in this manner differentiate and secrete mucus on their apical surface, and these secretions should be devoid of substances induced by inflammation as well as material contributed by submucosal glands. Neither mucus collected from primary cells nor bovine submaxillary mucin blocked motility of P. aeruginosa (Table 2). Normal motility also was observed after sputum was heated to 95°C for 10 min prior to growth of P. aeruginosa (Table 2). Taken together, these data suggest that some heat-labile factor in sputum, other than mucus, is responsible for inhibition of motility.
Quorum sensing in CF sputum.
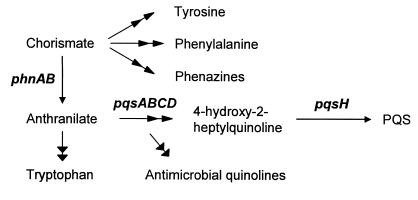
P. aeruginosa uses cell-to-cell communication (quorum sensing) to control the expression of 5 to 10% of its genes, many of which are involved in virulence (43, 51). Quorum sensing in P. aeruginosa is complex, involving the use of at least three signal molecules, the most recently discovered of which being 2-heptyl-3-hydroxy-4-quinolone (designated the Pseudomonas quinolone signal [PQS]) (35). The biosynthesis of PQS likely involves several gene products (Fig. 2) including proteins involved in the biosynthesis of aromatic amino acids and synthesis of the precursor quinoline 4-hydroxy-2-heptylquinoline (HHQ) (10). HHQ is hypothesized to be the immediate precursor of PQS as well as several other quinolones/quinolines, many of which have significant antimicrobial activity (10). Our transcriptome analysis of CF sputum-grown P. aeruginosa revealed that genes involved in quinolone/quinoline biosynthesis are induced during growth in CF sputum (Table 1). Genes involved in the biosynthesis of anthranilate (phnAB) and the conversion of anthranilate to HHQ (pqsA-E) exhibited the highest induction (14- to 30-fold). PA2587 which is hypothesized to perform the final step in PQS synthesis reproducibly showed a twofold induction. Induction of these genes had a significant impact on PQS production, resulting in approximately fivefold higher levels of PQS in the culture supernatants of CF sputum medium-grown bacteria (Fig. 3).
FIG. 2.
Proposed pathway for aromatic amino acid and quinolone/quinoline biosynthesis in P. aeruginosa. Dual arrowheads indicate multiple steps. Genes encoding proteins critical for quinolone/quinoline production are shown in boldface type.
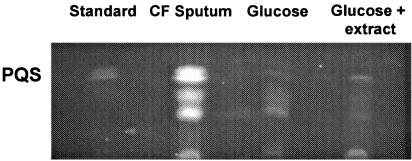
FIG. 3.
Increased production of PQS during growth in CF sputum. TLC was used to monitor PQS production by P. aeruginosa grown with CF sputum, glucose, or glucose containing a CF sputum ethyl acetate extract (see Materials and Methods). Growth in glucose containing an ethyl acetate extract of CF sputum medium was used to test if CF sputum medium contained sufficient P. aeruginosa quorum sensing signals to induce PQS production. Synthetic PQS (150 ng) was included as a reference. Sampling occurred during late exponential phase (approximately 6 × 108 CFU of P. aeruginosa/ml), and similar results were obtained for mid-exponential phase bacteria. Spot densitometry of triplicate experiments revealed that CF sputum-grown P. aeruginosa produced 4.9 ± 0.56 times more PQS than glucose-grown bacteria.
PQS induction is not due to acyl-HSLs endogenous to CF sputum.
Regulation of the pqsA-E genes is complex and is mediated by the ratio of the two primary acyl-homoserine lactone (acyl-HSL) quorum sensing molecules, butyryl-homoserine lactone (C4-HSL) and 3-oxo-dodecanoyl-homoserine lactone (3OC12-HSL) (30). All of the CF sputum samples used in this study contained P. aeruginosa; thus, it is possible that acyl-HSLs produced by the resident bacteria within the CF sputum may account for the increase in PQS production observed during growth in CF sputum medium. To examine this possibility, we extracted and quantitated the levels of C4-HSL and 3OC12-HSL in CF sputum medium. Our results indicate that only low levels of C4-HSL (<10 nM) and 3OC12-HSL (3 nM) are present in CF sputum medium.
Although only low levels of acyl-HSLs are present in CF sputum medium, it is possible that these concentrations of signaling molecules are sufficient for PQS induction. To examine this possibility, we extracted CF sputum medium with acidified ethyl acetate to remove all known P. aeruginosa quorum-sensing molecules (PQS and acyl-HSLs). This extract, which contains any acyl-HSLs present in CF sputum medium, was dried and reconstituted in MOPS-glucose medium, and the PQS production by P. aeruginosa in this medium was then compared to MOPS-glucose medium (no extract added) and CF sputum medium. No difference in PQS production was observed after growth of P. aeruginosa in MOPS-glucose medium and MOPS-glucose medium plus CF sputum extract (Fig. 3), indicating that the addition of CF sputum extract (i.e., acyl-HSL molecules in CF sputum) to MOPS-glucose medium had no effect on PQS production. This lack of PQS induction is not likely due to catabolite repression by glucose, since the addition of glucose to CF sputum medium had no effect on the increased PQS production normally observed (data not shown).
Induction of PQS during growth with amino acids.
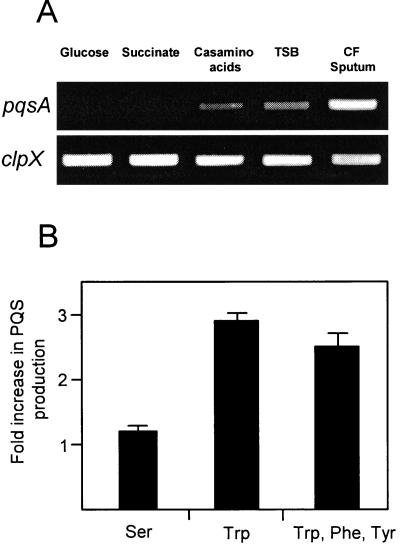
Because quorum sensing signaling molecules within CF sputum do not induce PQS, we next evaluated the impact of various carbon sources on the expression of pqsA to pqsE and production of PQS. RT-PCR was used to evaluate pqsA mRNA levels for P. aeruginosa grown using different carbon sources. The pqsA gene is the first gene in the pqsABCDE operon and serves as a marker gene for expression of this operon in these experiments. As predicted by the microarray results, higher levels of pqsA mRNA were present in CF sputum-grown P. aeruginosa than glucose-grown bacteria (Fig. 4A). Replacing glucose with succinate had no detectable effect on pqsA transcription. Since CF sputum contains high amounts of amino acids, we next evaluated the impact of amino acids on pqsA expression. Growth with Casamino Acids or in the complex medium tryptic soy broth (which contains high concentrations of amino acids) increased expression of pqsA relative to glucose- and succinate-grown P. aeruginosa but not to the level observed for CF sputum-grown bacteria (Fig. 4A).
FIG. 4.
Induction of pqsA and increased production of PQS during growth with amino acids. (A) Semiquantitative RT-PCR was used to examine pqsA transcript levels during growth with glucose, succinate, Casamino Acids, tryptic soy broth, or CF sputum. Bacteria were grown to identical densities, and the constitutively expressed clpX gene was used as the control. (B) P. aeruginosa was grown in MOPS succinate (as the control); MOPS succinate with 1 mM serine (Ser); MOPS succinate with 1 mM tryptophan (Trp); and MOPS succinate with Trp, phenylalanine (Phe), and tyrosine (Tyr) (0.33 μM each). PQS was extracted and quantitated as outlined in Materials and Methods. Data are expressed as increases (n-fold) in PQS production during growth with amino acids compared to growth in succinate alone (PQS produced in MOPS succinate with amino acids/PQS produced in MOPS succinate). Bacteria were sampled at identical densities in late exponential phase.
Since pqsA expression is increased during growth using Casamino Acids and precursors of aromatic amino acid biosynthesis are necessary for PQS production (Fig. 2), we hypothesized that the increased production of PQS observed in CF sputum is a result of the presence of aromatic amino acids in CF sputum. To test this hypothesis, we evaluated production of PQS for P. aeruginosa grown in MOPS succinate medium, MOPS succinate containing the nonaromatic amino acid serine (as a control), MOPS succinate containing the aromatic amino acid tryptophan, and MOPS succinate containing a combination of three aromatic amino acids (tryptophan, tyrosine, and phenylalanine) (Fig. 4B). The addition of tryptophan or a combination of aromatic amino acids significantly increased production of PQS by P. aeruginosa; however, the addition of serine had little effect on PQS production. The observed increase in PQS production during growth in the presence of aromatic amino acids was not a result of increased growth yields, as P. aeruginosa grown with serine reached identical densities (data not shown).
Induction of PQS-controlled genes in CF sputum.
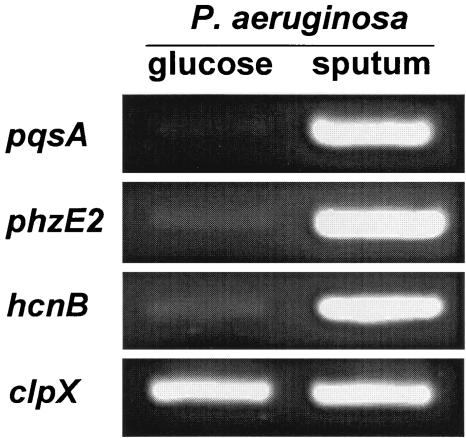
PQS controls the expression of many genes in P. aeruginosa, including genes encoding proteins involved in the production of hydrogen cyanide and pyocyanin (12, 16). Proteins important for the production of hydrogen cyanide and pyocyanin are encoded in part by the hcnABC and phzABCDE genes, respectively. Since PQS controls expression of these genes, we hypothesized that their transcription would be increased during growth in CF sputum. This was not apparent from the microarray data, since many of these genes did not hybridize at levels sufficient for statistical analysis. To test this hypothesis, we used RT-PCR to evaluate the levels of hcnB and phzE mRNA in CF sputum and glucose-grown P. aeruginosa. As expected, these transcripts were present in higher levels in CF sputum-grown bacteria than in glucose-grown bacteria (Fig. 5), indicating that the downstream targets of PQS signaling are induced during growth in CF sputum.
FIG. 5.
Induction of PQS-controlled genes during growth in CF sputum. Semiquantitative RT-PCR using RNA harvested from P. aeruginosa grown with glucose or CF sputum as the sole source of carbon and energy. Genes encoding proteins important for quinoline production (pqsA) and production of the PQS-controlled virulence factors pyocyanin (phzE2) and hydrogen cyanide (hcnA) were tested. For visualization, 5 μl of the resulting PCR was separated by agarose gel electrophoresis and stained with ethidium bromide. The constitutively expressed clpX gene was used as the control.
Dual species cultivation in CF sputum.
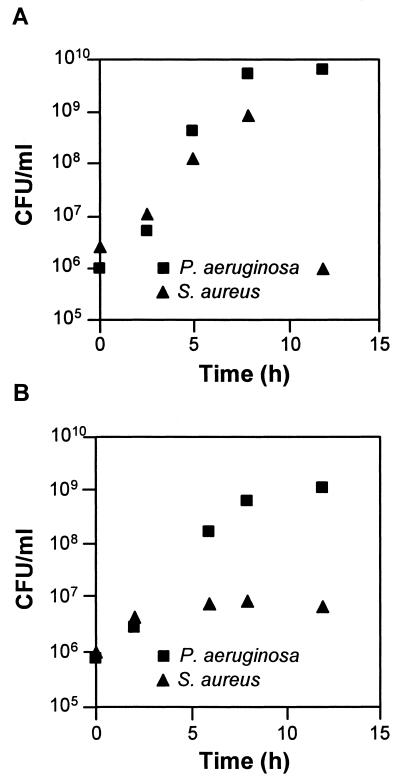
The CF lung is normally colonized by a consortium of bacteria, including P. aeruginosa and S. aureus (21, 25). S. aureus is often the initial colonizer of the CF lung and is later displaced as the primary CF lung inhabitant by P. aeruginosa (8). The mechanism of displacement is unknown but has been hypothesized to be due in part to lysis of S. aureus by P. aeruginosa (27). P. aeruginosa produces several factors that mediate lysis of S. aureus (1, 18, 27, 28), many of which were induced during growth in CF sputum (Table 1 and Fig. 5). The induction of staphylolytic factors during growth in CF sputum leads to the hypothesis that P. aeruginosa-dependent lysis of S. aureus would occur earlier in growth during coculture in CF sputum than in normal laboratory medium. Coculture experiments revealed that within 5 h of binary growth in CF sputum, S. aureus numbers level off and then decrease (Fig. 6). This is in contrast to the timing of lysis observed with laboratory medium, where detectable lysis was not evident until 12 h (Fig. 6). The earlier lysis did not occur because the transition to stationary phase was advanced in CF sputum medium-grown P. aeruginosa (Fig. 6); in fact, lysis in CF sputum is detectable during exponential growth. It should be noted that P. aeruginosa and S. aureus have similar doubling times and maximum growth yields in MOPS-sputum medium (data not shown).
FIG. 6.
Lysis of S. aureus by P. aeruginosa during planktonic growth in brain heart infusion (A) and MOPS-CF sputum (B) media. P. aeruginosa and S. aureus were inoculated to equal densities in test tubes and grown with shaking (250 rpm) at 37°C. Bacterial numbers were determined by differential plating (see Materials and Methods). For all time points, standard deviations were less than 10% of the mean. P. aeruginosa and S. aureus have similar growth rates and maximum cell densities in these media (data not shown).
DISCUSSION
The goal of this study was to evaluate the physiology of P. aeruginosa during growth using CF sputum as the sole source of carbon and energy. Growth of P. aeruginosa was examined using CF sputum obtained from 13 different patients, with 10 of these samples supporting growth kinetics similar to those shown in Fig. 1. These results indicate that the concentrations of metabolizable carbon in the CF sputum samples obtained from these 10 patients are similar. This is an important observation and increases the practicality of these studies, since samples provided by multiple CF patients may be used. The remarkable similarity in P. aeruginosa growth observed with these 10 CF sputum samples may be explained, in part, by our omission of CF sputum samples containing ≥108 CFU of P. aeruginosa/ml of sputum. The reason for poor growth in the other three CF sputum samples is unknown, but it may be a result of the presence of residual antibiotics within the sputum samples. Although we used P. aeruginosa PA14 in these studies, growth of the laboratory strain PAO1 and of a CF isolate of P. aeruginosa in CF sputum were similar (data not shown).
Our transcriptome results indicate that amino acids are a likely growth substrate for P. aeruginosa in CF sputum, because genes involved in catabolism of branch chain and aromatic amino acids were induced during growth in CF sputum. This is not surprising, since high levels (15 to 20 mM) of total amino acids have been observed with CF sputum (2, 48), and amino acids have been shown to be a substrate for P. aeruginosa in respiratory secretions (33). The origin of these amino acids is unclear. A significant fraction of CF sputum is protein (33), and P. aeruginosa produces several extracellular proteases which may liberate amino acids from resident proteins (49). These enzymes could originate from resident P. aeruginosa within the CF sputum before processing or from the bacterial inoculum. However, it is clear that P. aeruginosa is not capable of catabolizing all of the carbon within CF sputum, because P. aeruginosa grows to higher densities in sterilized CF sputum medium which has been stored at 37°C for 7 to 14 days prior to inoculation (data not shown). Whether this increase in metabolizable carbon is due to resident enzymes within CF sputum (either bacterial or host derived) or chemical hydrolysis which act to liberate carbon is unknown but indicates that not all carbon in CF sputum is available for P. aeruginosa growth in our system.
As previously described, P. aeruginosa growing in CF sputum is nonmotile (53). This loss of motility is not specific to CF sputum and does not require the presence of bacteria within sputum since sputum collected from non-CF patients also caused a loss in motility (Table 2). However, motility was not affected by growth on bovine mucin or mucus collected from in vitro-grown human primary lung epithelial cells (55), suggesting that the factor(s) important for loss of motility may be specific to in vivo-collected sputum. In vivo-collected CF sputum is likely very different from in vitro-grown primary lung cell mucus, particularly in regard to the presence of host immunity factors; therefore, it is difficult to speculate on the identity of this signal. Environmental parameters, including subinhibitory concentrations of macrolide antibiotics and antibodies to P. aeruginosa flagellin, have been shown to affect motility (31, 32). These factors are unlikely to affect motility in CF sputum, since the non-CF sputum samples did not contain P. aeruginosa or antibiotics. These data implicate a novel factor affecting motility in sputum, and the observation that this factor is heat labile suggests a proteinaceous component.
It has been proposed that the loss of motility in P. aeruginosa is due to repression of the fliC gene (encoding flagellin) in CF sputum-grown bacteria (53). We also observed repression of fliC during growth in CF sputum (Table 1); however, although over 95% of CF sputum-grown P. aeruginosa cells observed by phase microscopy were nonmotile, approximately 60% have an intact flagellum, as seen upon examination by TEM. The existence of intact flagellum on many of these nonmotile bacteria and the observation that loss of motility by P. aeruginosa occurs very quickly (within 30 min) upon addition of CF sputum suggest that repression of fliC may not be solely responsible for the loss of motility.
P. aeruginosa uses quorum sensing to control the expression of a large number of genes, many of which are important for virulence (43, 51). PQS is the most recently described signaling molecule included in the P. aeruginosa quorum sensing cascade and has been implicated as important in CF disease. Increased production of PQS is observed in early P. aeruginosa colonizers of the CF lung, and PQS has been purified from CF sputum (4, 17). The observation that PQS is induced during growth in CF sputum medium has several implications for CF disease. PQS controls the expression of genes encoding proteins critical for production of the virulence factors hydrogen cyanide and pyocyanin. These factors have been shown to be important for P. aeruginosa pathogenesis in several virulence models (15, 38, 39, 46). Increased expression of virulence factors during growth in CF sputum may be beneficial to P. aeruginosa by liberating nutrients via host cell lysis and by increasing competitiveness in multispecies environments.
Although the las and rhl quorum sensing systems control expression of PQS, our results indicate that quorum sensing signals produced by the resident bacteria within CF sputum do not induce expression of PQS in MOPS-sputum medium. This is not surprising, given that we are using 10% CF sputum and only low levels of quorum sensing molecules are present in CF sputum medium. The PQS-inducing signal(s) in CF sputum medium are also distinct from the signal(s) inhibiting motility, because boiling CF sputum did not alter PQS levels (data not shown). Instead, our data indicate that the induction of PQS during growth within CF sputum is due, in part, to the high concentrations of amino acids in CF sputum (2, 48), specifically, aromatic amino acids. The mechanism of PQS induction by aromatic amino acids may be a result of substrate competition. The biosynthetic pathways for PQS and aromatic amino acids share the precursor molecules chorismate and anthranilate (Fig. 2), which suggests that the presence of aromatic amino acids may reduce competition for these substrates, allowing for increased biosynthesis of PQS. This increase in PQS synthesis is not a general response to all amino acids, since addition of a nonaromatic amino acid had little effect on PQS biosynthesis (Fig. 4B). This hypothesis is predicated on understanding the regulatory mechanisms of amino acid biosynthesis in P. aeruginosa and amino acid transport. Further work utilizing mutants in aromatic amino acid transport and biosynthesis is necessary to test this hypothesis, but it is clear from these data that aromatic amino acids influence PQS levels.
Aromatic amino acids may not be the only signal regulating PQS levels in CF sputum, since growth with amino acids does not increase PQS to levels observed during growth in CF sputum. Whether these signals are also in non-CF sputum is also unclear, since growth yields in non-CF sputum were insufficient for comparisons of PQS levels. However, it is likely that increased PQS biosynthesis in response to amino acids will be important in clinical and nonclinical environments.
Interspecies interactions likely shape community structure during infection, particularly in chronic infections, such as CF. The observation that P. aeruginosa induces several distinct staphylolytic factors and preemptively lyses S. aureus during growth in CF sputum indicates that growth in CF sputum may impact community structure. In many cases, it is likely that P. aeruginosa will encounter an established bacterial community (often including S. aureus) upon entering the CF lung; thus, P. aeruginosa must be able to survive and compete in this environment. While it is likely that being a successful competitor in the CF lung involves several factors, the induction of multiple bactericidal factors during growth in CF sputum may increase the competitiveness of P. aeruginosa with the resident CF lung microflora. Although we evaluated interactions between P. aeruginosa and S. aureus, P. aeruginosa is capable of lysing many gram-positive bacteria, including other common inhabitants of the CF lung (e.g., Streptococcus pneumoniae). Whether lysis of resident bacteria is critical for P. aeruginosa colonization of the CF lung is unknown; however, it is clear that CF sputum significantly impacts polymicrobial interactions.
This overview of P. aeruginosa growth in CF sputum indicates that CF sputum contains multiple factors which influence motility and cell-to-cell communication in P. aeruginosa. It is important to understand how P. aeruginosa grows in the CF lung, and our CF sputum medium provides an in vitro model to study growth and metabolism. Factors critical for pathogenesis, including the formation of antibiotic-resistant biofilms, are regulated by the growth substrate (20, 40); thus, elucidation of the nutrient conditions in the CF lung will provide a better understanding of CF pathogenesis. This study indicates that the growth environment is critical for understanding P. aeruginosa cell-to-cell signaling and pathogenesis and illustrates the importance of using appropriate in vivo growth substrates to evaluate P. aeruginosa pathogenesis.
Acknowledgments
We thank Greg Strout for experimental assistance.
This work was supported by a grant from the NIH (1P20RR15564-01 to M.W.) and the Oklahoma Center for the Advancement of Science and Technology (HR03-137S to M.W.). K.L.P. is a University of Oklahoma Graduate Foundation Predoctoral Fellow.
REFERENCES
- 1.Barequet, I. S., G. J. Ben Simon, M. Safrin, D. E. Ohman, and E. Kessler. 2004. Pseudomonas aeruginosa LasA protease in treatment of experimental staphylococcal keratitis. Antimicrob. Agents Chemother. 48:1681-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barth, A. L., and T. L. Pitt. 1996. The high amino-acid content of sputum from cystic fibrosis patients promotes growth of auxotrophic Pseudomonas aeruginosa. J. Med. Microbiol. 45:110-119. [DOI] [PubMed] [Google Scholar]
- 3.Burns, J. L., B. W. Ramsey, and A. L. Smith. 1993. Clinical manifestations and treatment of pulmonary infections in cystic fibrosis. Adv. Pediatr. Infect. Dis. 8:53-66. [PubMed] [Google Scholar]
- 4.Collier, D. N., L. Anderson, S. L. McKnight, T. L. Noah, M. Knowles, R. Boucher, U. Schwab, P. Gilligan, and E. C. Pesci. 2002. A bacterial cell to cell signal in the lungs of cystic fibrosis patients. FEMS Microbiol. Lett. 215:41-46. [DOI] [PubMed] [Google Scholar]
- 5.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 6.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 7.Cuskey, S. M., J. A. Wolff, P. V. Phibbs, Jr., and R. H. Olsen. 1985. Cloning of genes specifying carbohydrate catabolism in Pseudomonas aeruginosa and Pseudomonas putida. J. Bacteriol. 162:865-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cystic Fibrosis Foundation. 1999. Patient registry 1998 annual data report. Cystic Fibrosis Foundation, Bethesda, Md.
- 9.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 10.Deziel, E., F. Lepine, S. Milot, J. He, M. N. Mindrinos, R. G. Tompkins, and L. G. Rahme. 2004. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc. Natl. Acad. Sci. USA 101:1339-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deziel, E., F. Lepine, S. Milot, and R. Villemur. 2003. rhlA is required for the production of a novel biosurfactant promoting swarming motility in Pseudomonas aeruginosa: 3-(3-hydroxyalkanoyloxy) alkanoic acids (HAAs), the precursors of rhamnolipids. Microbiology 149:2005-2013. [DOI] [PubMed] [Google Scholar]
- 12.Diggle, S. P., K. Winzer, S. R. Chhabra, K. E. Worrall, M. Camara, and P. Williams. 2003. The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol. Microbiol. 50:29-43. [DOI] [PubMed] [Google Scholar]
- 13.Di Sant'Agnese, P. A., and D. H. Andersen. 1946. Celiac syndrome: chemotherapy in infections of the respiratory tract associated with cystic fibrosis of the pancreas; observations with penicillin and drugs of the sulfonamide group, with special reference to penicillin aerosol. Am. J. Dis. Child. 72:17-61. [PubMed] [Google Scholar]
- 14.FitzSimmons, S. C. 1993. The changing epidemiology of cystic fibrosis. J. Pediatr. 122:1-9. [DOI] [PubMed] [Google Scholar]
- 15.Gallagher, L. A., and C. Manoil. 2001. Pseudomonas aeruginosa PAO1 kills Caenorhabditis elegans by cyanide poisoning. J. Bacteriol. 183:6207-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallagher, L. A., S. L. McKnight, M. S. Kuznetsova, E. C. Pesci, and C. Manoil. 2002. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J. Bacteriol. 184:6472-6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guina, T., S. O. Purvine, E. C. Yi, J. Eng, D. R. Goodlett, R. Aebersold, and S. I. Miller. 2003. Quantitative proteomic analysis indicates increased synthesis of a quinolone by Pseudomonas aeruginosa isolates from cystic fibrosis airways. Proc. Natl. Acad. Sci. USA 100:2771-2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haba, E., A. Pinazo, O. Jauregui, M. J. Espuny, M. R. Infante, and A. Manresa. 2003. Physicochemical characterization and antimicrobial properties of rhamnolipids produced by Pseudomonas aeruginosa 47T2 NCBIM 40044. Biotechnol. Bioeng. 81:316-322. [DOI] [PubMed] [Google Scholar]
- 19.Hester, K. L., J. Lehman, F. Najar, L. Song, B. A. Roe, C. H. MacGregor, P. W. Hager, P. V. Phibbs, Jr., and J. R. Sokatch. 2000. Crc is involved in catabolite repression control of the bkd operons of Pseudomonas putida and Pseudomonas aeruginosa. J. Bacteriol. 182:1144-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersboll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 21.Hoiby, N. 1998. Pseudomonas in cystic fibrosis: past, present, and future. Cystic Fibrosis Trust, London, United Kingdom.
- 22.Hunt, J. C., and P. V. Phibbs, Jr. 1983. Regulation of alternate peripheral pathways of glucose catabolism during aerobic and anaerobic growth of Pseudomonas aeruginosa. J. Bacteriol. 154:793-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackson, B. E., V. K. Bhupathiraju, R. S. Tanner, C. R. Woese, and M. J. McInerney. 1999. Syntrophus aciditrophicus sp. nov., a new anaerobic bacterium that degrades fatty acids and benzoate in syntrophic association with hydrogen-using microorganisms. Arch. Microbiol. 171:107-114. [DOI] [PubMed] [Google Scholar]
- 24.Klausen, M., A. Heydorn, P. Ragas, L. Lambertsen, A. Aeas-Jorgensen, S. Molin, and T. Tolker-Nielsen. 2003. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 48:1511-1524. [DOI] [PubMed] [Google Scholar]
- 25.Koch, C., and N. Hoiby. 1993. Pathogenesis of cystic fibrosis. Lancet 341:1065-1069. [DOI] [PubMed] [Google Scholar]
- 26.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2002. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15:194-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Machan, Z. A., G. W. Taylor, T. L. Pitt, P. J. Cole, and R. Wilson. 1992. 2-Heptyl-4-hydroxyquinoline N-oxide, an antistaphylococcal agent produced by Pseudomonas aeruginosa. J. Antimicrob. Chemother. 30:615-623. [DOI] [PubMed] [Google Scholar]
- 28.Mansito, T. B., M. A. Falcon, J. Moreno, A. Carnicero, and A. M. Gutierrez-Navarro. 1987. Effects of staphylolytic enzymes from Pseudomonas aeruginosa on the growth and ultrastructure of Staphylococcus aureus. Microbios 49:55-64. [PubMed] [Google Scholar]
- 29.Mata-Sandoval, J. C., J. Karns, and A. Torrents. 2001. Effect of nutritional and environmental conditions on the production and composition of rhamnolipids by P. aeruginosa UG2. Microbiol. Res. 155:249-256. [DOI] [PubMed] [Google Scholar]
- 30.McGrath, S., D. S. Wade, and E. C. Pesci. 2004. Dueling quorum sensing systems in Pseudomonas aeruginosa control the production of the Pseudomonas quinolone signal (PQS). FEMS Microbiol. Lett. 230:27-34. [DOI] [PubMed] [Google Scholar]
- 31.Molinari, G., C. A. Guzman, A. Pesce, and G. C. Schito. 1993. Inhibition of Pseudomonas aeruginosa virulence factors by subinhibitory concentrations of azithromycin and other macrolide antibiotics. J. Antimicrob. Chemother. 31:681-688. [DOI] [PubMed] [Google Scholar]
- 32.Ochi, H., H. Ohtsuka, S. Yokota, I. Uezumi, M. Terashima, K. Irie, and H. Noguchi. 1991. Inhibitory activity on bacterial motility and in vivo protective activity of human monoclonal antibodies against flagella of Pseudomonas aeruginosa. Infect. Immun. 59:550-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohman, D. E., and A. M. Chakrabarty. 1982. Utilization of human respiratory secretions by mucoid Pseudomonas aeruginosa of cystic fibrosis origin. Infect. Immun. 37:662-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 35.Pesci, E. C., J. B. Milbank, J. P. Pearson, S. McKnight, A. S. Kende, E. P. Greenberg, and B. H. Iglewski. 1999. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:11229-11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phibbs, P. V., Jr., and R. G. Eagon. 1970. Transport and phosphorylation of glucose, fructose, and mannitol by Pseudomonas aeruginosa. Arch. Biochem. Biophys. 138:470-482. [DOI] [PubMed] [Google Scholar]
- 37.Pier, G. B. 2002. CFTR mutations and host susceptibility to Pseudomonas aeruginosa lung infection. Curr. Opin. Microbiol. 5:81-86. [DOI] [PubMed] [Google Scholar]
- 38.Rahme, L. G., F. M. Ausubel, H. Cao, E. Drenkard, B. C. Goumnerov, G. W. Lau, S. Mahajan-Miklos, J. Plotnikova, M. W. Tan, J. Tsongalis, C. L. Walendziewicz, and R. G. Tompkins. 2000. Plants and animals share functionally common bacterial virulence factors. Proc. Natl. Acad. Sci. USA 97:8815-8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rahme, L. G., E. J. Stevens, S. F. Wolfort, J. Shao, R. G. Tompkins, and F. M. Ausubel. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899-1902. [DOI] [PubMed] [Google Scholar]
- 40.Ramsey, M. M., and M. Whiteley. 2004. Pseudomonas aeruginosa attachment and biofilm development in dynamic environments. Mol. Microbiol. 53:1075-1088. [DOI] [PubMed] [Google Scholar]
- 41.Sage, A. E., W. D. Proctor, and P. V. Phibbs, Jr. 1996. A two-component response regulator, gltR, is required for glucose transport activity in Pseudomonas aeruginosa PAO1. J. Bacteriol. 178:6064-6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schlievert, P. M., and D. A. Blomster. 1983. Production of staphylococcal pyrogenic exotoxin type C: influence of physical and chemical factors. J. Infect. Dis. 147:236-242. [DOI] [PubMed] [Google Scholar]
- 43.Schuster, M., C. P. Lostroh, T. Ogi, and E. P. Greenberg. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185:2066-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 45.Smith, R. S., and B. H. Iglewski. 2003. P. aeruginosa quorum-sensing systems and virulence. Curr. Opin. Microbiol. 6:56-60. [DOI] [PubMed] [Google Scholar]
- 46.Tan, M. W., and F. M. Ausubel. 2000. Caenorhabditis elegans: a model genetic host to study Pseudomonas aeruginosa pathogenesis. Curr. Opin. Microbiol. 3:29-34. [DOI] [PubMed] [Google Scholar]
- 47.Tan, M. W., S. Mahajan-Miklos, and F. M. Ausubel. 1999. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc. Natl. Acad. Sci. USA 96:715-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas, S. R., A. Ray, M. E. Hodson, and T. L. Pitt. 2000. Increased sputum amino acid concentrations and auxotrophy of Pseudomonas aeruginosa in severe cystic fibrosis lung disease. Thorax 55:795-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Toder, D. S., S. J. Ferrell, J. L. Nezezon, L. Rust, and B. H. Iglewski. 1994. lasA and lasB genes of Pseudomonas aeruginosa: analysis of transcription and gene product activity. Infect. Immun. 62:1320-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Visca, P., L. Leoni, M. J. Wilson, and I. L. Lamont. 2002. Iron transport and regulation, cell signaling and genomics: lessons from Escherichia coli and Pseudomonas. Mol. Microbiol. 45:1177-1190. [DOI] [PubMed] [Google Scholar]
- 51.Wagner, V. E., D. Bushnell, L. Passador, A. I. Brooks, and B. H. Iglewski. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185:2080-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whiteley, M., M. R. Parsek, and E. P. Greenberg. 2000. Regulation of quorum sensing by RpoS in Pseudomonas aeruginosa. J. Bacteriol. 182:4356-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolfgang, M. C., J. Jyot, A. L. Goodman, R. Ramphal, and S. Lory. 2004. Pseudomonas aeruginosa regulates flagellin expression as part of a global response to airway fluid from cystic fibrosis patients. Proc. Natl. Acad. Sci. USA 101:6664-6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yahr, T. L., L. M. Mende-Mueller, M. B. Friese, and D. W. Frank. 1997. Identification of type III secreted products of the Pseudomonas aeruginosa exoenzyme S regulon. J. Bacteriol. 179:7165-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zabner, J., L. A. Couture, A. E. Smith, and M. J. Welsh. 1994. Correction of cAMP-stimulated fluid secretion in cystic fibrosis airway epithelia: efficiency of adenovirus-mediated gene transfer in vitro. Hum. Gene Ther. 5:585-593. [DOI] [PubMed] [Google Scholar]