Abstract
The symbiotic interaction between Rhizobium etli and Phaseolus vulgaris, the common bean plant, ultimately results in the formation of nitrogen-fixing nodules. Many aspects of the intermediate and late stages of this interaction are still poorly understood. The R. etli relA gene was identified through a genome-wide screening for R. etli symbiotic mutants. RelA has a pivotal role in cellular physiology, as it catalyzes the synthesis of (p)ppGpp, which mediates the stringent response in bacteria. The synthesis of ppGpp was abolished in an R. etli relA mutant strain under conditions of amino acid starvation. Plants nodulated by an R. etli relA mutant had a strongly reduced nitrogen fixation activity (75% reduction). Also, at the microscopic level, bacteroid morphology was altered, with the size of relA mutant bacteroids being increased compared to that of wild-type bacteroids. The expression of the σN-dependent nitrogen fixation genes rpoN2 and iscN was considerably reduced in the relA mutant. In addition, the expression of the relA gene was negatively regulated by RpoN2, the symbiosis-specific σN copy of R. etli. Therefore, an autoregulatory loop controlling the expression of relA and rpoN2 seems operative in bacteroids. The production of long- and short-chain acyl-homoserine-lactones by the cinIR and raiIR systems was decreased in an R. etli relA mutant. Our results suggest that relA may play an important role in the regulation of gene expression in R. etli bacteroids and in the adaptation of bacteroid physiology.
Rhizobium etli is a gram-negative soil bacterium that elicits nitrogen-fixing nodules on its leguminous host plant Phaseolus vulgaris, the common bean plant. The intracellular bacteria, called bacteroids, are surrounded by a plant plasmalemma-derived membrane and have a physiological state that is different from the free-living form. Bacteroids convert atmospheric nitrogen into ammonia, which is translocated to the plant cell cytoplasm. In return, the bacteria receive a carbon source, typically dicarboxylates, and other nutrients from the plant. Although the precise nutritional conditions under which the bacteroids thrive inside the nodule cells are still not clearly defined, the physiological state of the bacteroids needs to be adapted to the prevailing conditions, such as a microoxic and low-pH environment, specific carbon and nitrogenous compounds, and the presence of oxidative stress (for example, see references 17, 24, 30, and 31).
The rapid adaptation of bacteria to changing environmental conditions is a prerequisite to colonizing novel niches successfully. One of the global regulatory systems in bacteria is the stringent response, which is triggered by various forms of nutritional stress and results in a rapid transcriptional down regulation of ribosomal and tRNA genes and, as a consequence, of ribosome and protein synthesis (3, 7). This process is mediated by the accumulation of the nucleotide alarmones guanosine tetraphosphate (ppGpp) and guanosine pentaphosphate (pppGpp), collectively referred to as (p)ppGpp. In Escherichia coli, these molecules are produced by two enzymes, RelA and SpoT. Under conditions of amino acid starvation, uncharged tRNAs bind to ribosomes and stimulate ribosome-associated RelA to synthesize (p)ppGpp. Recovery from starvation requires the hydrolysis of these molecules, which is primarily catalyzed by the bifunctional SpoT protein. This protein also produces (p)ppGpp in response to glucose starvation (47). Unlike E. coli, a growing number of bacteria, including Bacillus subtilis, possess only a single RelA/SpoT homolog which displays both (p)ppGpp synthase and hydrolase activities (42). Besides the regulation of stable RNA synthesis, (p)ppGpp also influences mRNA transcription. Evidence exists that the regulatory role of (p)ppGpp molecules is exerted through their direct interaction with the β and β′ subunits of core RNA polymerase, thereby possibly affecting the promoter specificity of the RNA polymerase (6, 38).
In recent years, it has become clear that besides its role during nutritional starvation, the stringent response is required in complex physiological processes such as biofilm formation in Listeria monocytogenes, E. coli, and Streptococcus mutans (1, 23, 37), the development of a multicellular fruiting body in Myxococcus xanthus (16), and the development of competence in Bacillus subtilis (19). Also, RelA has been reported to be important for the interaction of bacteria, either pathogenic or beneficial, with their eukaryotic host. For example, ppGpp was shown to accumulate in symbiotic bacteria, including Sinorhizobium meliloti, as a result of amino acid starvation (18, 40). An S. meliloti relA mutant is unable to induce the stringent response and overproduces succinoglycan, an exopolysaccharide that is important for the infection of its host plant, Medicago sativa (40). Moreover, the stringent response is required for nodule formation on its host (40). An analysis of strains that have regained the ability to nodulate revealed the occurrence of two types of suppressor mutations (40). Class I mutations restore nodulation and succinoglycan production to wild-type levels and confer prototrophy. The corresponding mutations map to the rpoB gene, encoding subunit β of RNA polymerase. It was suggested that class I mutants may have a permanent stringent response (41). Class II mutations are found in the rpoC gene, in a region of the β′ subunit that is not conserved, and restore nodulation and succinoglycan production but not prototrophy. These mutations likely reduce the efficiency of transcription of genes in comparison to the levels obtained with a relA mutant strain (41). These findings indicate that an adjustment of rhizobial physiology may be a key process to obtaining a successful symbiosis.
For this work, we identified the R. etli relA gene through a screening of R. etli for symbiotically relevant genes and performed a detailed analysis with respect to its function, regulation, and target genes. Several symbiotically essential genes, including nitrogen fixation and quorum-sensing genes, were shown to be under the control of the relA regulatory circuit. We propose a regulatory model controlled by RelA which is in agreement with the observed symbiotic phenotype of the R. etli relA mutant. Our results indicate a prominent role of the stringent response in R. etli in the adaptation of the bacteria to the endosymbiotic bacteroid state.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used for this work are listed in Table 1. Rhizobium etli strains were cultured in minimal AMS or complex TY medium at 30°C (28). AMS medium was supplemented with various carbon and nitrogen sources at a concentration of 10 mM, unless otherwise indicated, or with Casamino Acids at a final concentration of 0.2%. Escherichia coli strains were routinely grown at 37°C in Luria-Bertani medium. Antibiotics were supplied at the following concentrations: ampicillin, 100 μg ml−1; gentamicin, 50 μg ml−1; kanamycin, 25 μg ml−1; spectinomycin, 50 μg ml−1 (E. coli) or 25 μg ml−1 (R. etli); chloramphenicol, 30 μg ml−1 (E. coli); nalidixic acid, 60 μg ml−1; neomycin, 60 μg ml−1; and tetracycline, 1 μg ml−1. Triparental conjugations and site-directed mutagenesis of R. etli were done as described previously (9).
TABLE 1.
Bacterial strains and plasmids used for this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Escherichia coli strains | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF) recA1 endA1 hsdR17 supE44 thi-1 gyrA96 | Gibco-BRL |
| CF1648 | MG1655 wild type | 47 |
| CF1693 | Kmr Cmr, MG1655 relA251::kan spoT207::cat | 47 |
| Rhizobium etli strains | ||
| CNPAF512 | Nalr, wild type | 28 |
| FAJ1154 | NmrrpoN1::Ω-Km | 28 |
| FAJ1169 | NmrrpoN2::Ω-Km | 27 |
| FAJ1170 | Nmr SprrpoN1::Ω-Sp rpoN2::Ω-Km | 27 |
| FAJ1183 | NmrfnrN::Ω-Km | 29 |
| FAJ4013 | Nmr SprraiI::gusA-Sp cinI::Km (raiI and gusA are in opposite orientations) | 8 |
| Rp1000 | NmrnifA::aphII | 26 |
| CMPG8705 | SprrelA::Ω-Sp, opposite orientation | This work |
| CMPG8706 | SprrelA::Ω-Sp, same orientation | This work |
| CMPG8002 | NmrrelA::mTn5 gusA-oriV | This work |
| Plasmids | ||
| pHP45Ω-Sp | Apr Spr | 14 |
| pJQ200-UC1 | GmrsacB | 32 |
| pBluescript II SK+ | Apr, pUC19 derivative | Stratagene |
| pCRII-TOPO | Apr Kmr, PCR TA cloning vector | Invitrogen |
| pCR4Blunt-TOPO | Apr Kmr, PCR TA cloning vector | Invitrogen |
| pLAFR1 | Tcr, broad-host-range vector | 15 |
| pFAJ1702 | Apr Tcr, stable RK2-derived cloning vector | 12 |
| pFAJ1703 | Apr TcrgusA, stable RK2-derived promoter-probe vector | 12 |
| pCMPG8025 | Tcr, cosmid, pLAFR1 derivative containing the rpoZ relA region | This work |
| pCMPG8705 | Gmr Spr, pJQ200-UC1 containing relA::Ω-Sp (opposite orientation) | This work |
| pCMPG8706 | Gmr Spr, pJQ200-UC1 containing relA::Ω-Sp (same orientation) | This work |
| pCMPG8715 | relA gene in pFAJ1702 | This work |
| pCMPG8711 | relA promoter region and Ω-Sp fragment in pFAJ1703 | This work |
| pCMPG8712 | rpoZ-relA promoter region and Ω-Sp fragment in pFAJ1703 | This work |
| pFAJ4014 | Tcr, cinI-gusA fusion | 8 |
| pFAJ1458 | Tcr, raiI-gusA fusion | E. Luyten, personal communication |
| pFAJ1175 | Tcr, rpoN2 gusA fusion in pLAFR1 | 27 |
| pFAJ1726 | Apr Tcrr, iscN promoter region in pFAJ1703 | 11 |
Cloning of R. etli relA and construction of mutants.
An mTn5gusA-oriV mutant library of R. etli CNPAF512 was constructed and screened for genes induced under conditions mimicking symbiosis, essentially as described by Xi et al. (44, 45). To clone the genomic region carrying the transposon insertion in the R. etli mutant CMPG8002, genomic DNA was digested with XhoI, NdeI, or AscI, ligated, and transformed into E. coli. The DNA sequence of the genomic region flanking the 5′ end of the gusA gene was determined by using the gusA primer RHI128 (5′-CGGTACCTGACTAGCTAAGGAG-3′). On the basis of this partial DNA sequence, the primers RHI321 (5′-AGCGCCTATGTCAAGGGCCGGC-3′) and RHI322 (5′-TCTTCCGGGCTGTCGCCTTCGG-3′), amplifying a fragment of approximately 0.45 kb, were designed. These primers were used to screen a genomic library of R. etli CNPAF512, which was previously constructed in pLAFR1 (9). Briefly, this library was pooled in groups of 16 clones which were used to prepare plasmid DNAs. The purified DNAs were analyzed by PCR using Taq polymerase and primers RHI321 and RHI322. If a PCR fragment of the correct size was obtained, the clones of the positive pool were individually tested for the presence of relA. One such positive clone, pCMPG8025, carrying the complete relA gene locus, was selected for further analysis. The relA gene region was sequenced by using universal sequencing primers on cloned restriction fragments or by primer walking. DNA sequences were run on an ABI 3100-Avant apparatus (Applera Belgium).
To mutate the R. etli relA gene, a 3.3-kb fragment containing relA was amplified by PCR using pfx DNA polymerase and the primers RHI435 (5′-ACTGGCGGCCGCCGCCGCAACATGGAGACAGCC-3′) and RHI436 (5′-ACTGGCGGCCGCAATAGGCGCGCAGGAGGTAGC-3′), which carried NotI recognition sites at their 5′ ends (in italics), with pCMPG8025 used as a template. This fragment was cloned into pCR4Blunt-TOPO (Invitrogen), digested with HindIII, and ligated to the 2.1-kb HindIII fragment from pHP45Ω carrying a polar Ω-Spr cassette flanked by transcription termination signals, thereby interrupting relA between codons 137 and 138. From plasmids carrying the cassette either in the same or the opposite orientation with respect to relA, a NotI fragment was removed and subsequently cloned into the NotI site of the suicide plasmid pJQ200-UC1, yielding pCMPG8705 (opposite orientation) and pCMPG8706 (same orientation). These constructs were introduced into R. etli. The corresponding R. etli relA mutants CMPG8705 (opposite) and CMPG8706 (same orientation) were selected as described by D'hooghe et al. (9) and verified by appropriate Southern blot hybridization.
Constructs for complementation analysis.
To complement the R. etli relA mutants, a fragment was amplified by a PCR using pfx, with pCMPG8025 used as a template. A 2.8-kb fragment containing only relA was produced using RHI435 and RHI437 (5′-ACTGGCGGCCGCTGGTTCTGCTCGCCAGCCACC-3′; the NotI site is indicated in italics). This fragment was first cloned into pCR4Blunt-TOPO and subsequently transferred as a NotI fragment to the broad-host-range plasmid pFAJ1702, generating pCMPG8715.
Construction of relA-gusA fusions.
Two relA promoter-gusA fusions were constructed. For their construction, promoter fragments were amplified by PCRs using pfx DNA polymerase, with pCMPG8025 as a template and either primers RHI433 (5′-CGATTCTAGAATGGACGATACCGAGCGCACG-3′; the XbaI restriction site is shown in italics) and RHI434 (5′-ATCGCCCGGGTGGTTCTGCTCGCCAGCCACC-3′; the SmaI restriction site is shown in italics), yielding a 1.3-kb fragment, or primers RHI433 and RHI432 (ATCGCCCGGGAATAGGCGCGCAGGAGGTAGC; the SmaI restriction site is shown in italics), yielding a 1.8-kb fragment (Fig. 1). Both fragments were cloned into pCRII-TOPO and verified by DNA sequence analysis. To facilitate subsequent cloning steps, the constructs were digested with SmaI and, after being blunted with Klenow, ligated to a 2.1-kb ClaI-SmaI fragment carrying an Ω-Spr cassette. This Ω-Spr cassette was originally removed from pHP45Ω as a HindIII fragment and cloned into pBluescript II SK(+), from which it was removed as a ClaI-SmaI fragment. In the resulting pCRII-TOPO constructs, plasmids were identified in which the promoter fragment and the Ω-Spr cassette were in opposite orientations. Finally, two relAp-Spr cassette fragments were cloned as XbaI-SmaI fragments into XbaI-HpaI-digested pFAJ1703, a broad-host-range plasmid, thereby fusing relAp to a promoterless gusA gene. The resulting promoter-gusA fusion plasmids were named pCMPG8711 (1.3-kb promoter) and pCMPG8712 (1.8-kb promoter).
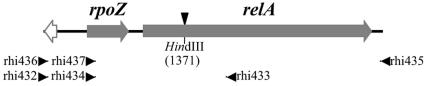
FIG. 1.
Schematic representation of R. etli relA gene region (3,305 bp). The primers used to amplify (parts of) relA are indicated. Filled arrows show the lengths of the relA and rpoZ genes. The white arrow represents an incomplete open reading frame. The HindIII restriction site used to mutagenize relA is indicated with an arrow.
Determination of (p)ppGpp content.
The method used to determine the cellular ppGpp content was adapted from the work of Cashel et al. (4). Bacteria were grown overnight in TY medium at 30°C (R. etli strains) or in morpholinepropanesulfonic acid (MOPS) medium containing 0.4% glucose and 0.2% Casamino Acids (E. coli strains). The overnight cultures were diluted to an optical density at 600 nm (OD600) of 0.6, centrifuged, and washed with 10 mM MgSO4. E. coli cells were resuspended in M56CP medium and R. etli cells were resuspended in AMS medium supplemented with 10 mM NH4Cl and 0.02 g/liter Casamino Acids to induce the stringent response. The cultures were grown for 1 h at 30°C. Next, carrier-free [32P]phosphoric acid (Amersham Biosciences) was added to a final concentration of 100 μCi ml−1. The cultures (250 μl) were further incubated for 3.5 h in a heat block at 30°C before being sampled. After incubation, 50-μl aliquots were taken and an equal volume of formic acid (2 M) was added. Samples were incubated on ice for 15 min and centrifuged for 5 min at maximum speed. Thirty microliters of each supernatant was spotted on a polyethyleneimine-cellulose thin-layer chromatography (TLC) sheet and developed in 1.5 M KH2PO4 for 2.5 h. The (p)ppGpp was visualized by autoradiography at room temperature for at least 2 weeks for Rhizobium strains and overnight for E. coli strains.
Monitoring bacterial growth.
R. etli cultures were grown overnight in TY medium, washed with 10 mM MgSO4, brought to equal cell densities, and subsequently diluted 100 times in 10 mM MgSO4. Five microliters of each suspension was inoculated into 295 μl of growth medium. Five replicates of each 300-μl culture were inoculated into the wells of sterile Honeycomb plates. These plates were incubated at 30°C with continuous shaking, and the OD600 was measured every 30 min in a Bioscreen C microbiology workstation (Labsystems Oy, Zellik, Belgium).
Isolation and detection of autoinducer molecules.
R. etli cultures grown overnight were diluted 100-fold in 500 ml TY medium. The cultures were incubated aerobically at 30°C. Autoinducer molecules were isolated from 50 ml of culture every 6 h by extraction with acidified dichloromethane, thereby avoiding the occurrence of open ring structures as a result of alkalinization of the medium. Autoinducer molecules were detected on C18 reversed-phase TLC plates using the Agrobacterium tumefaciens tra reporter system as described elsewhere (34, 35).
Plant experiments and acetylene reduction assay.
Common bean plants (Phaseolus vulgaris cv. Limburgse vroege) were cultivated and assayed for nitrogen fixation capacity using the acetylene reduction method (ARA) as described previously (11). The isolation of bacteroids was performed essentially as detailed by Michiels et al. (28).
β-Glucuronidase assay.
Unless otherwise specified in the text, R. etli cells were grown overnight in TY medium, washed in 10 mM MgSO4, and resuspended in defined AMS or TY medium with a 50-fold dilution. Cultures were incubated for 12 h either aerobically or microaerobically in stoppered tubes in the presence of an oxygen concentration of 0.3% as previously described (27). GusA expression assays were carried out using p-nitrophenyl-β-d-glucuronide as a substrate for β-glucuronidase as described earlier (27). Experiments were carried out at least in triplicate and confirmed independently. To take into account the differences in cell size between the wild type and the mutants, the β-glucuronidase activity was normalized using OD600 values.
Electron and light microscopy.
Transmission electron micrographs were obtained from thin sections of 3-week-old nodules and analyzed in a Zeiss EM 900 electron microscope as described by Xi et al. (46). Light microscopic examinations of bacteria were done with a Nikon Optiphot-2 microscope equipped with a fluorescence unit after staining of the bacteria with toluidine blue or with the LIVE/DEAD BacLight bacterial viability assay. Images were taken using a digital DS-5 M (Nikon) camera head controlled by a DS-L1 camera control unit (Nikon).
Nucleotide sequence accession number.
The nucleotide sequence of the R. etli relA gene locus has been deposited in the DDBJ-EMBL-GenBank nucleotide sequence database under accession no. DQ010057.
RESULTS
Sequence of R. etli relA.
To obtain novel symbiotic R. etli mutants, an R. etli transposon mutant collection was screened essentially as described by Xi et al. (44). The details of this genome-wide screening will be presented elsewhere. From this screening, the R. etli mutant strain CMPG8002, displaying a strongly reduced symbiotic nitrogen fixation activity (see below), was selected for an in-depth structural and functional analysis.
A complete sequence analysis of the gene inactivated by the transposon insertion in mutant CMPG8002 revealed that it codes for a protein of 744 amino acids (aa) with a predicted molecular mass of 83,690 Da. This protein displays high similarity to RelA proteins. A search using the Pfam HMM database (Pfam 14.0) revealed the presence of four conserved domains in the R. etli RelA protein sequence, namely, the HD, Rela_SpoT, TGS, and ACT domains. The HD domain (PF01966) extends from aa 45 to 144 and is also found in several phosphohydrolases. The catalytic histidine (H) and aspartate (D) residues are also found in the E. coli SpoT protein, but not in E. coli RelA, and are therefore likely involved in (p)ppGpp degradation. Although experimental evidence is currently lacking, these results confirm previous suggestions stating that unlike other gram-negative bacteria, α-proteobacteria only possess one relA/spoT homologue, coding for a dual-function enzyme displaying both (p)ppGpp synthetase and hydrolase activities (40, 48).
One hundred forty-two nucleotides upstream of the relA gene, a gene homologous to E. coli rpoZ was identified (Fig. 1). R. etli rpoZ codes for a protein of 134 aa with a deduced molecular mass of 14,724 Da. A similar rpoZ-relA organization is conserved in other members of the Rhizobiales order.
Symbiotic phenotype.
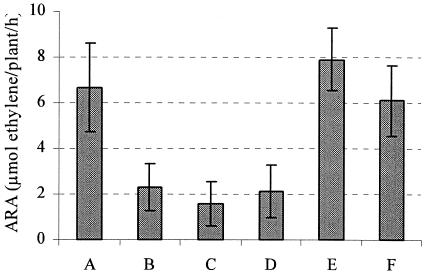
To study the role of relA during symbiosis and to confirm the observed reduction of nitrogenase activity in plants nodulated by the relA mutant CMPG8002, additional R. etli relA mutants (CMPG8705 and CMPG8706) were constructed by site-specific cassette mutagenesis as detailed in Materials and Methods. In addition, plasmids carrying either a genomic region of approximately 20 kb flanking the relA gene or solely the relA gene located on a 2.8-kb fragment (pCMPG8715) were used to complement these mutants. The results of the ARA measurements of the nodules induced by these strains are presented in Fig. 2. The nitrogenase activities of nodules induced by CMPG8705 and CMPG8706 were approximately 25% that of the wild-type nodules. The ARA measurements were confirmed at different time points during symbiosis. Also, the leaves of plants nodulated by the relA mutants were yellowish, indicating a clear deficiency in available nitrogen. This observation confirms the symbiotic phenotype of the initial transposon insertion mutant CMPG8002. The nitrogenase activities of the relA mutant strains containing the different complementation constructs did not differ significantly from the wild-type level, indicating full complementation of the phenotype (Fig. 2).
FIG. 2.
Nitrogenase activities of R. etli strains during symbiosis with P. vulgaris. Bean plants nodulated by the R. etli wild type and relA mutants were analyzed for acetylene reduction activity 3 weeks after inoculation. The R. etli strains tested were the wild type (A), relA mutants CMPG8002 (B), CMPG8705 (C), and CMPG8706 (D), and complemented relA mutants CMPG8002/pCMPG8025 (E) and CMPG8705/pCMPG8715 (F). The data are means of at least five replicates. Bars represent standard deviations.
To further investigate the symbiotic role of relA in more detail, sections of mature nodules were prepared and examined by transmission electron microscopy. This analysis demonstrated an aberrant shape of relA bacteroids, which were enlarged compared to wild-type bacteroids (Fig. 3). A quantitative analysis of bacteroid lengths demonstrated that the mean length of wild-type bacteroids (1.79 μm; standard deviation [SD], 0.51 μm) was significantly different (P < 0.0001; Student's t test) from those of the relA mutant bacteroids CMPG8705 (mean, 2.12 μm; SD, 0.65 μm) and CMPG8002 (mean, 2.01 μm; SD, 0.54 μm) but not from those of the complemented strains CMPG8705 carrying pCMPG8715 (mean, 1.72 μm; SD, 0.55 μm) and CMPG8002 containing pCMPG8025 (mean, 1.78 μm; SD, 0.46 μm). Moreover, the relA mutant bacteroids were generally individually enclosed in a symbiosome membrane with little symbiosome space, whereas symbiosomes from nodules formed by the wild type were composed of multiple bacteroids with a larger symbiosome space than that observed for the relA mutants. These results indicate that in addition to the reduced nitrogen fixation activity of the mutants, structural and morphological differences exist between wild-type and relA mutant bacteroids and symbiosomes. In contrast, the wild-type and relA mutant strains were similar in their nodulation of bean roots, although the colors of the mature nodules were slightly different (data not shown). These results indicate that in the R. etli-bean symbiosis, symbiotic defects of a relA mutant are obvious only at the intermediate and/or late stages of the interaction.
FIG. 3.
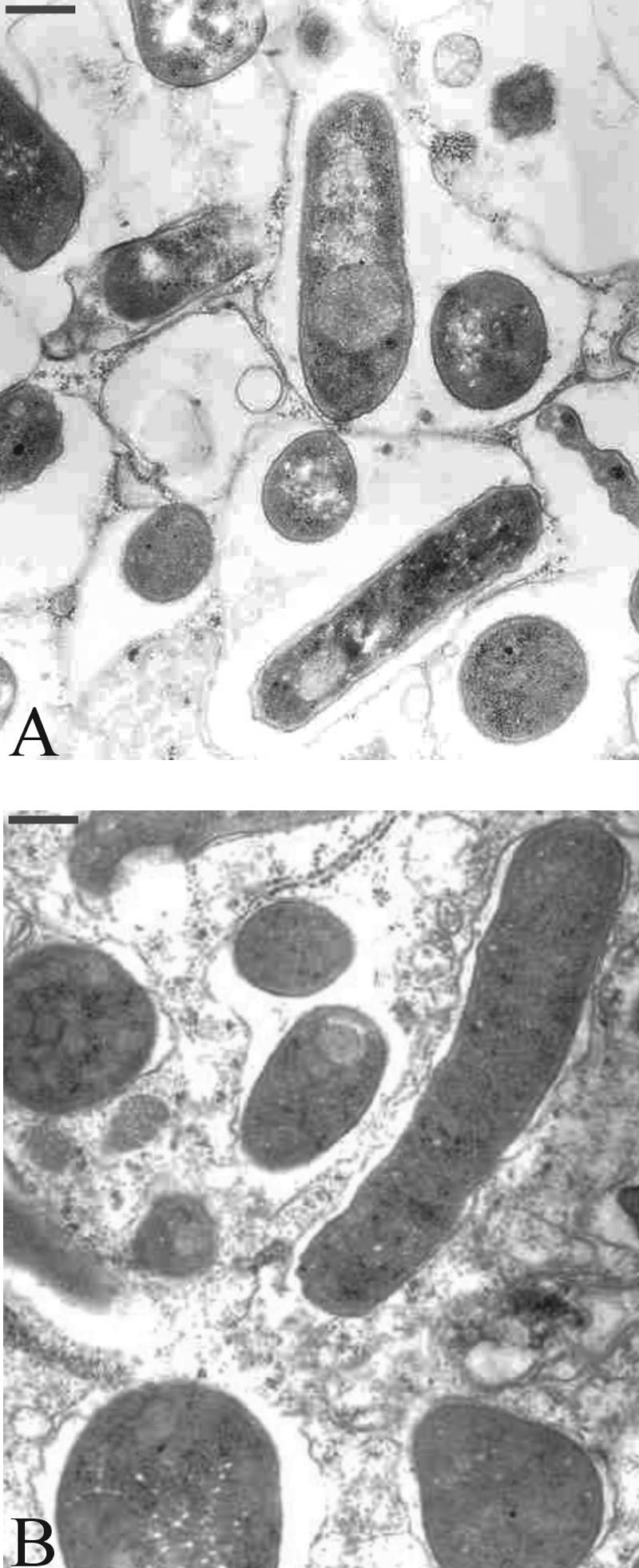
Transmission electron micrographs of sections from nodules colonized by the R. etli wild type (A) and the relA mutant CMPG8002 (B). Mature nodules were harvested 3 weeks after the inoculation of P. vulgaris seedlings. Bars, 0.4 μm.
ppGpp accumulation.
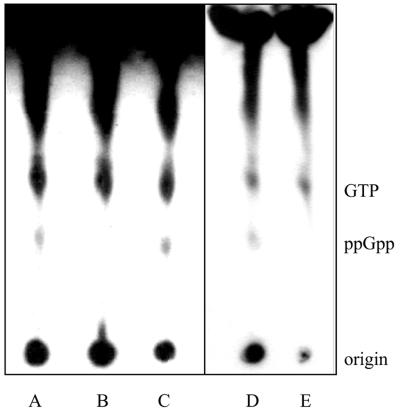
To determine whether the inactivation of the relA gene in R. etli results in the inability to produce ppGpp, the nucleotide pool of the relA knockout strain CMPG8002 was compared with that of the wild-type strain. The stringent response was induced by shifting an exponential culture of R. etli from rich medium to amino acid-limiting conditions, a treatment that also induces the accumulation of ppGpp in other bacteria (40). A TLC analysis was performed after incubation of the cultures with [32P]H3PO4. The E. coli wild-type strain CF1648 and the relA spoT mutant CF1693 were used as control strains. In contrast to the wild type, the R. etli relA mutant strain CMPG8002 failed to accumulate ppGpp after a shift from complex TY medium to conditions of amino acid starvation (Fig. 4). Complementation of the relA mutant restored the ability of the relA mutant to synthesize ppGpp. In the presence of plasmid pCMPG8025, a clear complementation of ppGpp synthesis was observed in the relA mutant. These results indicate that the gene product of relA is the major or only ppGpp synthetase in R. etli.
FIG. 4.
TLC analysis of ppGpp production in R. etli and E. coli. Analyses of the total intracellular nucleotide content were performed as originally described by Cashel et al. (4). The nucleotide pools were labeled with 32P and separated by polyethyleneimine-TLC as described in Materials and Methods. The strains assayed were as follows: lane A, R. etli wild type; lane B, R. etli relA mutant strain CMPG8002; lane C, CMPG8002 complemented with plasmid pCMPG8025; lane D, E. coli wild-type strain CF1648; and lane E, E. coli relA spoT mutant CF1693 (47).
Growth characteristics.
To examine the importance of relA during free-living growth, tests were performed with R. etli CNPAF512 and CMPG8705 (relA) on different growth media. The OD600 was measured with a Bioscreen C apparatus. Several growth defects were observed, depending on the composition of the medium (data not shown). With complex TY medium, the doubling time and lag phase were similar for the wild type and the relA mutant CMPG8705. However, higher optical densities were always reached by the relA mutant CMPG8705 in TY medium than by the wild type (see Fig. 6). This difference in optical density could be fully complemented by pCMPG8715. With minimal AMS medium, the differences in growth kinetics between the wild-type strain and the relA mutant CMPG8705 were dependent on the carbon source used. An extended lag phase was observed for the relA mutant CMPG8705 in defined medium in the presence of all carbon sources used. However, the effect was most obvious in the presence of Casamino Acids (extension of 20.5 h compared to wild-type lag phase) compared to succinate (12.5 h) and mannitol (2.0 h). With this last carbon source, the increase of the lag phase was small, but a strong increase in the optical density reached at stationary phase was observed (OD600, 0.7 for the wild type and 1.2 for CMPG8705). This increase was not observed with AMS supplemented with either succinate or Casamino Acids. These effects were fully complemented by introducing plasmid pCMPG8715 into the relA mutant strain. To explain this difference in OD600 values at stationary phase between the different media, a light microscopic analysis and viable cell counts were performed. These analyses indicated that the same numbers of R. etli wild-type and relA mutant cells per ml or the same CFU were reached in each of the different media at the onset of stationary phase (data not shown). However, a fluorescence microscopic examination of cells stained with the LIVE/DEAD BacLight assay demonstrated that the sizes of the wild-type and mutant bacteria differed. An increase in the length of the relA mutant CMPG8705 compared with the wild type was observed for cells grown to stationary phase in TY medium. The average length of relA mutant bacteria (mean, 1.67 μm; SD, 0.39 μm) was increased by 16% (P < 0.0001; Student's t test) compared to the wild-type bacteria (mean, 1.44 μm; SD, 0.38 μm), which might explain the observed difference in OD600 values. Also, with AMS medium containing mannitol as a carbon source, the average length of relA mutant bacteria was 2.02 μm (SD, 0.27 μm), whereas that of the wild-type bacteria was 1.65 μm (SD, 0.26 μm), indicating an increase of 22% (P < 0.0001; Student's t test) for relA mutant bacteria relative to the wild type.
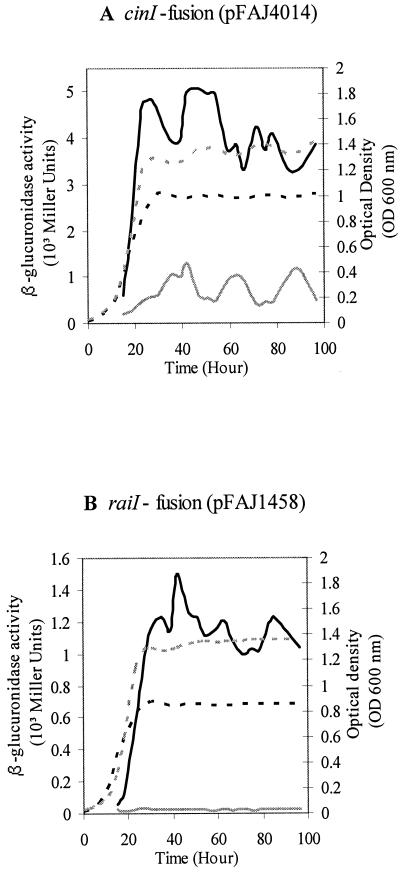
FIG. 6.
Expression of raiI and cinI in R. etli wild type and relA mutant strain. The GusA activities of cinI-gusA (pFAJ4014) (A) and raiI-gusA (pFAJ1458) (B) fusions were monitored in the R. etli wild type (black) and the relA mutant CMPG8705 (gray) during free-living growth in complex TY medium as a function of time. Curves representing the OD600 value of the cultures are represented separately by dashed lines for the wild type (black) and the relA mutant CMPG8705 (gray). Expression is given in Miller units.
Expression patterns of R. etli relA.
To investigate the regulation of relA expression in R. etli, two transcriptional relA-gusA fusions, containing either the rpoZ-relA intergenic region and the 3′ end of rpoZ (pCMPG8711) or a region of approximately 1 kb upstream of relA containing the rpoZ gene (pCMPG8712), were constructed and introduced into R. etli. The β-glucuronidase activity was measured during free-living aerobic and microaerobic growth in TY medium and in bacteroids isolated from 3-week-old nodules. The results for the free-living aerobic and symbiotic expression of pCMPG8712 are shown in Fig. 5. Because of the differences in cell size, the expression levels were normalized by using the OD600 value.
FIG. 5.
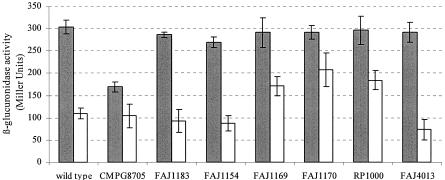
Expression analysis of relA in different R. etli regulatory mutant backgrounds. The expression of relA-gusA (pCMPG8712) was monitored in free-living cultures grown in TY medium aerobically (dark gray) and in bacteroids (white). The R. etli strains tested were the wild-type strain CNPAF512, the relA mutant CMPG8705, the fnrN mutant FAJ1183, the rpoN1 mutant FAJ1154, the rpoN2 mutant FAJ1169, the rpoN1 rpoN2 double mutant FAJ1170, the nifA mutant RP1000, and the raiI cinI double mutant FAJ4013. Expression is given in Miller units. Data are the means of at least three replicates. Bars represent the standard deviations.
During aerobic growth, both relA-gusA fusions were expressed, indicating that the relA gene was actively transcribed. In the relA mutant CMPG8705 grown in complex medium, GusA activity was reproducibly reduced to approximately 55% of the wild-type level for both fusions tested (Fig. 5; data not shown for pCMPG8711). These results indicate that the relA gene is partially positively autoregulated under these conditions. Therefore, it seems that relAp belongs to those promoters that are upregulated during the stringent response. These results were confirmed by measuring the expression of relA as a function of the OD600 value. A twofold increase in expression occurred during the exponential growth phase and reached a maximum expression level in stationary phase (data not shown). These data are consistent with the twofold reduction of relA expression observed with the R. etli relA mutant.
Under microaerobic conditions, depending on the fusion tested, the expression of relA was reduced 3.6-fold (pCMPG8711) or 2-fold (pCMPG8712) compared with the aerobic GusA level in the wild type (data not shown). Under conditions of microaerobiosis, the GusA activities of both relA fusions were not different from that of the relA mutant (data not shown). Finally, in bacteroids, relA expression was even further reduced compared to that under conditions of microaerobiosis. Also, the expression levels were not different between the wild type and the relA mutant strain.
Comparing the expression levels of relA between both relA-gusA fusions, the GusA levels were two- to sevenfold higher in strains carrying pCMPG8712 than in those containing pCMPG8711 (data not shown). At present, it is not clear whether these higher expression levels were caused by an additional promoter or regulatory sequence upstream of rpoZ which also affected relA transcription or by the extra rpoZ gene present on this plasmid. In any case, although the absolute values differed, the relative expression values between both plasmids were comparable among the different strains and growth conditions.
To further analyze the control of relA expression, the relA-gusA fusion pCMPG8712 was introduced into various R. etli regulatory mutants (fnrN, rpoN, nifA, and raiI cinI). For each of the mutants tested, relA expression was assayed during growth under free-living aerobic and microaerobic conditions in rich medium and during symbiosis. Under free-living aerobic conditions, no reduction of relA expression was observed in the mutants compared to the wild type, except for the described autoregulation (Fig. 5). Also, under free-living microaerobic conditions no reduction of relA expression was observed (data not shown). Under symbiotic conditions, the expression of relA was clearly upregulated in the rpoN2 mutant FAJ1169, in the rpoN1 rpoN2 double mutant FAJ1170, and in the nifA mutant Rp1000, indicating a negative regulation of relA expression by the RpoN and NifA proteins in bacteroids (Fig. 5). NifA is an oxygen-dependent enhancer binding-type regulator and a key activator promoting the expression of many nitrogen fixation genes in R. etli. In several control experiments, the expression of gusA fusions was either unaffected or reduced in nifA or rpoN mutant backgrounds, making it unlikely that the observed increase in relA expression is indirectly caused by a defect in nitrogen fixation (data not shown). Also, the inactivation of the second regulatory protein, FnrN, controlling the oxygen-dependent expression of bacteroid genes involved in respiration and leading to a decrease in nitrogen fixation of approximately 80% (29), had no effect on relA expression. An interaction between the quorum-sensing and (p)ppGpp regulatory circuits was previously described for A. tumefaciens (48) and Pseudomonas aeruginosa (39). It was therefore tested whether the quorum-sensing systems of R. etli influence the expression of relA. The N-acyl homoserine lactone (AHL) quorum-sensing system in R. etli consists of two luxIR-like systems, raiIR and cinIR (8, 34). However, the inactivation of raiI and cinI did not affect relA expression in R. etli.
Genes regulated by R. etli relA.
In Pseudomonas putida, the RpoN-dependent Pu promoter of the TOL plasmid is controlled by ppGpp (2). Because of the central role of RpoN in the regulation of nitrogen fixation gene expression in R. etli (27, 28), a possible dependence of promoters regulated by RpoN on the stringent response was studied. For this, the expression of rpoN2, a major symbiotic regulator, and of the nitrogen fixation gene iscN, both previously demonstrated to be part of the RpoN regulon (11, 27), was examined using rpoN2-gusA (pFAJ1175) and iscN-gusA (pFAJ1726) fusions in the wild type and the relA mutant strain CMPG8705. As previously demonstrated, both fusions were not expressed under free-living aerobic conditions in TY medium but were strongly induced during microaerobiosis and in bacteroids (Table 2). Whereas the expression levels of rpoN2 and iscN under free-living conditions were not different in wild-type and relA mutant strains in TY medium as well as in AMS medium supplemented with 10 mM NH4Cl and 10 mM succinate (data not shown), the bacteroid GusA levels of both σN-dependent promoter-gusA fusions were reduced approximately 2.5-fold in the relA mutant. In S. meliloti, the expression of SMb21243, which is positively controlled by RelA, also exhibited a reduction of approximately 2.5-fold (41). These results point to a positive regulation by RelA of rpoN2 and iscN, specifically under symbiotic conditions. Since the expression of iscN is positively regulated by RpoN2 during symbiosis (11), the lower expression of iscN in the relA mutant may be the result of a lower level of rpoN2 expression.
TABLE 2.
Expression analysis of rpoN-dependent genes
| Strain | Plasmid | Activity (U)a
|
||
|---|---|---|---|---|
| Aerobic | Microaerobic | Symbiotic | ||
| Wild type | rpoN2-gusA | 21 (2) | 1,105 (108) | 2,368 (490) |
| relA | rpoN2-gusA | 23 (1) | 1,030 (84) | 934 (276) |
| Wild type | iscN-gusA | 9 (1) | 524 (60) | 5,427 (625) |
| relA | iscN-gusA | 5 (1) | 511 (29) | 2,129 (329) |
GusA activity of R. etli rpoN2- gusA (pFAJ1175) and iscN-gusA (pFAJ1726) in R. etli wild-type and relA mutant (CMPG8705) backgrounds. Expression after growth in complex TY medium is given in Miller units. Free-living expression data are the means of at least four replicates, and symbiotic expression data are the means of at least eight replicates. Standard deviations are in parentheses.
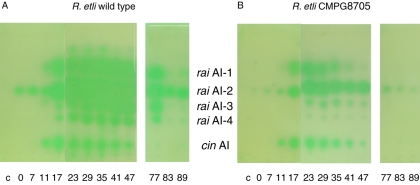
Finally, we examined whether the expression of the cin and rai quorum-sensing systems in R. etli was altered in the relA mutant strain. For this, the expression of raiI-gusA and cinI-gusA fusions was examined as a function of time in cultures of wild-type R. etli and the relA mutant strain CMPG8705 grown in TY medium. It can be seen from Fig. 6 that the expression of both fusions was clearly reduced in the relA background compared to the wild type. Depending on the cell density, the GusA activity of raiI-gusA in the relA mutant strain was reduced approximately 40- to 50-fold compared to the expression level of the wild type, while that of cinI-gusA was reduced 5- to 6-fold. To corroborate these results, the cin- and rai-dependent production of autoinducer molecules was also examined. For this, autoinducers were isolated from wild-type and relA mutant cultures grown aerobically in TY medium as a function of time (Fig. 7). Clearly, for the wild type the autoinducer profile changed as a function of time and cell density. All autoinducer spots were most abundant in our assay between 23 and 47 h of growth for the wild type, and thereafter the amount of autoinducers detected declined. However, the amount of AHLs produced differed significantly between the wild type and the relA mutant CMPG8705. This effect was less obvious for the level of synthesis of the saturated long-chain 3-hydroxy-acyl-homoserine lactone (cin AI in Fig. 7) produced by the CinI synthase (8). The abundance of RaiI-dependent AHLs (as shown for rai AI-1 to 4 in Fig. 7) was at least 10-fold lower in the relA mutant than in the wild-type strain when examined at identical time points in the growth curve. In particular, the production of rai AI-1, comigrating with synthetic 3O,C6-HSL, and rai AI-4, comigrating with synthetic 3O,C8-HSL, was strongly affected. However, this difference may also be due to a differential sensitivity of the A. tumefaciens tra reporter towards different types of AHLs. Since 3O-acyl-HSLs may comigrate with the same 3OH derivatives, it is not possible to discriminate between both types of molecules in a TLC assay. The expression tests as well as the autoinducer profiles indicate that the expression of the R. etli cin and rai systems is positively controlled by relA.
FIG. 7.
Analysis of quorum-sensing molecules in R. etli. Precultures of wild-type R. etli (A) and the relA mutant CMPG8705 (B) were diluted 100-fold and grown further aerobically in TY medium. Cells were harvested at the indicated times. Autoinducer molecules were isolated from the culture supernatant, separated by TLC, and visualized using the A. tumefaciens tra reporter system (35). The stationary phase was reached after approximately 24 h of growth. The positions of some of the rai (rai AI-1 to -4) and cin (cin AI) autoinducers are indicated. In the control lane (c), the extract from an uninoculated culture was spotted.
DISCUSSION
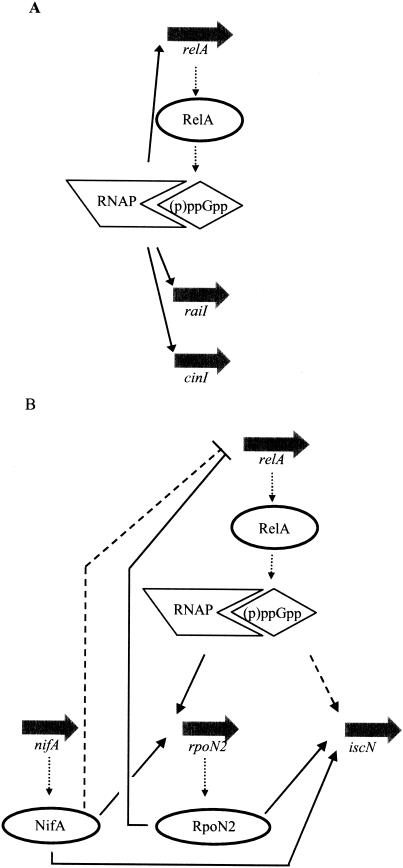
When cells enter the stationary growth phase, the binding of (p)ppGpp to the RNA polymerase reprograms the enzyme and drastically changes the global cellular transcription pattern by inhibiting transcription from stringent promoters (3). Global regulation may be directed at the level of transcription of the target gene and/or involve an altered expression of regulatory genes (5). At the level of activity, besides other models that suggest a direct effect on promoter recognition and kinetics, the σ factor competition model proposes that elevated (p)ppGpp levels may alter the affinity of the RNA polymerase core in favor of alternative sigma factors, such as σS and σH (e.g., see references 20 and 21). It is generally assumed that the available core is limiting for transcription. Under favorable growth conditions, the (p)ppGpp concentration is low and hence favors binding of σ70 to the core enzyme. On the other hand, conditions of growth arrest or stress result in increased (p)ppGpp levels, facilitating the replacement of σ70 by alternative σ factors and the concomitant activation of expression of stress regulons. The expression of most of the nitrogen fixation genes in Rhizobium species is controlled by σN in concert with the transcriptional activator NifA. σN is structurally different from the large σ70 family of sigma factors, including σS and σH, and typically recognizes promoters with a conserved −12/−24 consensus (25). Transcriptional activation of these promoters requires an enhancer binding protein such as NtrC or NifA. In R. etli, σN is encoded by two different rpoN genes. While rpoN1 has important functions during free-living growth, rpoN2 primarily has a symbiotic function (27, 28). The rpoN2 gene is autoregulated by σN/NifA and strongly up-regulated in bacteroids, thereby likely providing the cell with an increased amount of σN to meet the requirement for nif and fix gene activation. Here we demonstrated that (p)ppGpp is required for the high-level expression of two σN-dependent promoters, namely those of the rpoN2 and iscN genes (schematically shown in Fig. 8B). Clearly, there is not an absolute requirement for (p)ppGpp for transcription from these promoters. Possibly, in the presence of (p)ppGpp, σN is more competitive for core RNA polymerase, leading preferentially to the activation of nitrogen fixation genes. In P. aeruginosa, (p)ppGpp allows the transcriptional activation of the σN-dependent Po and Pu promoters, controlling genes involved in the catabolism of aromatic compounds (2, 36). The stimulatory effect of (p)ppGpp on the σN-dependent expression of Po has been attributed to direct effects on transcription and successful competition of σN for the available core RNA polymerase (22, 36). Interestingly, more than 20 years ago a possible role of ppGpp in the derepression of nif genes in the free-living nitrogen-fixing bacterium Klebsiella pneumoniae was already suggested (33). In addition to the effect of (p)ppGpp on σN-dependent gene expression, σN also negatively controls the expression of relA during symbiosis (Fig. 8B). This negative feedback loop possibly avoids the accumulation of too high levels of σN in bacteroids as a result of the positive control of (p)ppGpp on rpoN2 expression. This effect is also observed in the R. etli nifA mutant background, which corroborates the observed effects of an rpoN2 mutation, as the transcription of the latter gene is NifA dependent. In contrast, inactivation of the rpoN1 gene does not affect relA expression, which is in agreement with previous observations that RpoN2, but not RpoN1, fulfills a major function during symbiotic nitrogen fixation. At present, it is not clear how RpoN2 could mechanistically affect relA gene expression, either through a direct interaction of σN with the relA promoter region, which occurs at the rpoN1 gene promoter, or indirectly by competing with the available core RNA polymerase.
FIG. 8.
Schematic overview of the free-living aerobic (A) and symbiotic (B) regulation of relA and target genes. Under aerobic free-living conditions, relA is positively autoregulated and controls the expression of the QS genes cinI and raiI. The effect is likely mediated by ppGpp and RNA polymerase (RNAP). During symbiosis, the expression of rpoN2, coding for σN, and iscN is induced by relA. Possibly, the effect of relA on iscN expression is indirect (dashed line) via RpoN2. In addition, the expression of relA is negatively controlled by σN and NifA. The effect of NifA on relA expression is either direct or indirect, as NifA also controls rpoN2 transcription (11, 27).
The stringent response is initiated upon metabolic stress. This situation often occurs simultaneously with an increase in the cell density, such as at the transition from the exponential to the stationary growth phase. We therefore examined whether there was an interaction between the relA and quorum-sensing regulons in R. etli. We have previously described two quorum-sensing regulatory systems in R. etli, i.e., the raiIR and cinIR systems. The cin system is involved in the synthesis of saturated long-chain 3-hydroxy-acyl-homoserine lactone [3OH-(slc)-HSL] (8), whereas rai controls the production of multiple short-chain AHLs (34, 43). In this study, we observed an effect of a relA mutation on the production of AHLs in R. etli. The production of AHLs catalyzed by RaiI and CinI in R. etli was shown to be at least 10-fold lower in a relA mutant. The observed reduction in the expression of the synthases of both quorum-sensing systems (Fig. 8A) may account for the decreased autoinducer levels. Links between the relA and quorum-sensing regulatory circuits have also been noticed previously for A. tumefaciens and P. aeruginosa. In the former, relA regulates the stationary-phase-dependent transcription of the attM gene coding for an AHL-lactonase (48). As a result of this induction, the quormone signal 3-oxo-C8-HSL molecules in A. tumefaciens are degraded at stationary phase (48). In P. aeruginosa, relA overexpression results in the premature activation of the lasR and rhlR regulatory genes and the premature production of 3-oxo-C12-AHL and C4-AHL (39). In addition, under low-magnesium growth conditions, RelA contributes to the production of high levels of 3-oxo-C12-AHL (13).
The inactivation of R. etli relA leads to a strongly affected symbiotic phenotype at the late stages of the interaction. Also, as in other bacterial species, the stringent response in R. etli is an important metabolic regulator, as the inactivation of relA affects growth in several media. This points to a central function of RelA in bacteroid physiology. One may therefore speculate that at least part of the bacteroid metabolism may resemble that of free-living bacteria subjected to a metabolic stress. In S. meliloti, the stringent response was previously found to control the nodulation of S. meliloti on its leguminous host, alfalfa, as the nodule number was strongly decreased (40). A detailed study of relA suppressor strains that have wild-type nodulation and succinoglycan production clearly indicated that careful control of the bacterial metabolic demand is a key factor controlling a successful symbiosis (41). Besides an overall effect on physiology, the strongly decreased nitrogen fixation ability of the R. etli relA mutant may result from a decreased expression of specific (symbiotic) target genes, including σN-dependent genes such as the rpoN2 gene and other nitrogen fixation genes. It will be important in future experiments to address the question of whether (p)ppGpp may redirect gene expression in bacteroids, favoring the transcription of σN-dependent genes. Indeed, it was recently shown using an in silico approach for the detection of the −24/−12 type of promoters in Rhizobiales that the σN regulon may control more genes than traditionally assumed (10). Interestingly, S. meliloti class I suppressors carry mutations in a region previously implicated in σ factor recognition (41). The fine-tuning of gene expression in bacteroids by the stringent response may therefore be a system to obtain a tight coupling between bacterial metabolism and symbiotic nitrogen fixation.
Acknowledgments
M.M. was a recipient of a fellowship from IWT-Flanders, and K.B. is an aspirant of the Fund for Scientific Research-Flanders. This work was supported by grants from the Research Council of the K.U. Leuven (GOA/2003/09) and from the Fund for Scientific Research-Flanders (G.0108.01 and G.0287.04).
We thank M. Cashel and D. Vinella for providing the E. coli strains CF1648 and CF1693.
REFERENCES
- 1.Balzer, G. J., and R. J. McLean. 2002. The stringent response genes relA and spoT are important for Escherichia coli biofilms under slow-growth conditions. Can. J. Microbiol. 48:675-680. [DOI] [PubMed] [Google Scholar]
- 2.Carmona, M., M. J. Rodriguez, O. Martinez-Costa, and V. de Lorenzo. 2000. In vivo and in vitro effects of (p)ppGpp on the sigma(54) promoter Pu of the TOL plasmid of Pseudomonas putida. J. Bacteriol. 182:4711-4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In R. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology Press, Washington, D.C.
- 4.Cashel, M., R. A. Lazzarini, and B. Kalbacher. 1969. An improved method for thin-layer chromatography of nucleotide mixtures containing 32P-labelled orthophosphate. J. Chromatogr. 40:103-109. [DOI] [PubMed] [Google Scholar]
- 5.Chang, D. E., D. J. Smalley, and T. Conway. 2002. Gene expression profiling of Escherichia coli growth transitions: an expanded stringent response model. Mol. Microbiol. 45:289-306. [DOI] [PubMed] [Google Scholar]
- 6.Chatterji, D., N. Fujita, and A. Ishihama. 1998. The mediator for stringent control, ppGpp, binds to the beta-subunit of Escherichia coli RNA polymerase. Genes Cells 3:279-287. [DOI] [PubMed] [Google Scholar]
- 7.Chatterji, D., and A. K. Ojha. 2001. Revisiting the stringent response, ppGpp and starvation signaling. Curr. Opin. Microbiol. 4:160-165. [DOI] [PubMed] [Google Scholar]
- 8.Daniels, R., D. E. De Vos, J. Desair, G. Raedschelders, E. Luyten, V. Rosemeyer, C. Verreth, E. Schoeters, J. Vanderleyden, and J. Michiels. 2002. The cin quorum sensing locus of Rhizobium etli CNPAF512 affects growth and symbiotic nitrogen fixation. J. Biol. Chem. 277:462-468. [DOI] [PubMed] [Google Scholar]
- 9.D'hooghe, I., J. Michiels, K. Vlassak, C. Verreth, F. Waelkens, and J. Vanderleyden. 1995. Structural and functional analysis of the fixLJ genes of Rhizobium leguminosarum biovar phaseoli CNPAF512. Mol. Gen. Genet. 249:117-126. [DOI] [PubMed] [Google Scholar]
- 10.Dombrecht, B., K. Marchal, J. Vanderleyden, and J. Michiels. 2002. Prediction and overview of the RpoN-regulon in closely related species of the Rhizobiales. Genome Biol. 3:RESEARCH0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dombrecht, B., M. Z. Tesfay, C. Verreth, C. Heusdens, M. C. Napoles, J. Vanderleyden, and J. Michiels. 2002. The Rhizobium etli gene iscN is highly expressed in bacteroids and required for nitrogen fixation. Mol. Genet. Genomics 267:820-828. [DOI] [PubMed] [Google Scholar]
- 12.Dombrecht, B., J. Vanderleyden, and J. Michiels. 2001. Stable RK2-derived cloning vectors for the analysis of gene expression and gene function in gram-negative bacteria. Mol. Plant-Microbe Interact. 14:426-430. [DOI] [PubMed] [Google Scholar]
- 13.Erickson, D. L., J. L. Lines, E. C. Pesci, V. Venturi, and D. G. Storey. 2004. Pseudomonas aeruginosa relA contributes to virulence in Drosophila melanogaster. Infect. Immun. 72:5638-5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fellay, R., J. Frey, and H. Krisch. 1987. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of gram-negative bacteria. Gene 52:147-154. [DOI] [PubMed] [Google Scholar]
- 15.Friedman, A. M., S. R. Long, S. E. Brown, W. J. Buikema, and F. M. Ausubel. 1982. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene 18:289-296. [DOI] [PubMed] [Google Scholar]
- 16.Harris, B. Z., D. Kaiser, and M. Singer. 1998. The guanosine nucleotide (p)ppGpp initiates development and A-factor production in Myxococcus xanthus. Genes Dev. 12:1022-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hérouart, D., E. Baudouin, P. Frendo, J. Harrison, R. Santos, A. Jamet, G. Van de Sype, D. Touati, and A. Puppo. 2002. Reactive oxygen species, nitric oxide and glutathione: a key role in the establishment of the legume-Rhizobium symbiosis? Plant Physiol. Biochem. 40:619-624. [Google Scholar]
- 18.Howorth, S. M., and R. R. England. 1999. Accumulation of ppGpp in symbiotic and free-living nitrogen-fixing bacteria following amino acid starvation. Arch. Microbiol. 171:131-134. [DOI] [PubMed] [Google Scholar]
- 19.Inaoka, T., and K. Ochi. 2002. RelA protein is involved in induction of genetic competence in certain Bacillus subtilis strains by moderating the level of intracellular GTP. J. Bacteriol. 184:3923-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jishage, M., K. Kvint, V. Shingler, and T. Nystrom. 2002. Regulation of sigma factor competition by the alarmone ppGpp. Genes Dev. 16:1260-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kvint, K., A. Farewell, and T. Nystrom. 2000. RpoS-dependent promoters require guanosine tetraphosphate for induction even in the presence of high levels of sigma(s). J. Biol. Chem. 275:14795-14798. [DOI] [PubMed] [Google Scholar]
- 22.Laurie, A. D., L. M. Bernardo, C. C. Sze, E. Skarfstad, A. Szalewska-Palasz, T. Nystrom, and V. Shingler. 2003. The role of the alarmone (p)ppGpp in sigma N competition for core RNA polymerase. J. Biol. Chem. 278:1494-1503. [DOI] [PubMed] [Google Scholar]
- 23.Lemos, J. A., T. A. Brown, Jr., and R. A. Burne. 2004. Effects of RelA on key virulence properties of planktonic and biofilm populations of Streptococcus mutans. Infect. Immun. 72:1431-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lodwig, E., and P. Poole. 2003. Metabolism of Rhizobium bacteroids. Crit. Rev. Plant Sci. 22:37-78. [Google Scholar]
- 25.Merrick, M. J. 1993. In a class of its own—the RNA polymerase sigma factor sigma 54 (sigma N). Mol. Microbiol. 10:903-909. [DOI] [PubMed] [Google Scholar]
- 26.Michiels, J., I. D'hooghe, C. Verreth, H. Pelemans, and J. Vanderleyden. 1994. Characterization of the Rhizobium leguminosarum biovar phaseoli nifA gene, a positive regulator of nif gene expression. Arch. Microbiol. 161:404-408. [DOI] [PubMed] [Google Scholar]
- 27.Michiels, J., M. Moris, B. Dombrecht, C. Verreth, and J. Vanderleyden. 1998. Differential regulation of Rhizobium etli rpoN2 gene expression during symbiosis and free-living growth. J. Bacteriol. 180:3620-3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michiels, J., T. Van Soom, I. D'hooghe, B. Dombrecht, T. Benhassine, P. de Wilde, and J. Vanderleyden. 1998. The Rhizobium etli rpoN locus: DNA sequence analysis and phenotypical characterization of rpoN, ptsN, and ptsA mutants. J. Bacteriol. 180:1729-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moris, M., B. Dombrecht, C. Xi, J. Vanderleyden, and J. Michiels. 2004. Regulatory role of Rhizobium etli CNPAF512 fnrN during symbiosis. Appl. Environ. Microbiol. 70:1287-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oke, V., and S. R. Long. 1999. Bacteroid formation in the Rhizobium-legume symbiosis. Curr. Opin. Microbiol. 2:641-646. [DOI] [PubMed] [Google Scholar]
- 31.Patriarca, E. J., R. Tate, and M. Iaccarino. 2002. Key role of bacterial NH4+ metabolism in Rhizobium-plant symbiosis. Microbiol. Mol. Biol. Rev. 66:203-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 33.Riesenberg, D., S. Erdei, E. Kondorosi, and C. Kari. 1982. Positive involvement of ppGpp in derepression of the nif operon in Klebsiella pneumoniae. Mol. Gen. Genet. 185:198-204. [DOI] [PubMed] [Google Scholar]
- 34.Rosemeyer, V., J. Michiels, C. Verreth, and J. Vanderleyden. 1998. luxI- and luxR-homologous genes of Rhizobium etli CNPAF512 contribute to synthesis of autoinducer molecules and nodulation of Phaseolus vulgaris. J. Bacteriol. 180:815-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaw, P. D., G. Ping, S. L. Daly, C. Cha, J. E. Cronan, Jr., K. L. Rinehart, and S. K. Farrand. 1997. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc. Natl. Acad. Sci. USA 94:6036-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sze, C. C., and V. Shingler. 1999. The alarmone (p)ppGpp mediates physiological-responsive control at the sigma 54-dependent Po promoter. Mol. Microbiol. 31:1217-1228. [DOI] [PubMed] [Google Scholar]
- 37.Taylor, C. M., M. Beresford, H. A. Epton, D. C. Sigee, G. Shama, P. W. Andrew, and I. S. Roberts. 2002. Listeria monocytogenes relA and hpt mutants are impaired in surface-attached growth and virulence. J. Bacteriol. 184:621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toulokhonov, I. I., I. Shulgina, and V. J. Hernandez. 2001. Binding of the transcription effector ppGpp to Escherichia coli RNA polymerase is allosteric, modular, and occurs near the N terminus of the beta′-subunit. J. Biol. Chem. 276:1220-1225. [DOI] [PubMed] [Google Scholar]
- 39.van Delden, C., R. Comte, and A. M. Bally. 2001. Stringent response activates quorum sensing and modulates cell density-dependent gene expression in Pseudomonas aeruginosa. J. Bacteriol. 183:5376-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wells, D. H., and S. R. Long. 2002. The Sinorhizobium meliloti stringent response affects multiple aspects of symbiosis. Mol. Microbiol. 43:1115-1127. [DOI] [PubMed] [Google Scholar]
- 41.Wells, D. H., and S. R. Long. 2003. Mutations in rpoBC suppress the defects of a Sinorhizobium meliloti relA mutant. J. Bacteriol. 185:5602-5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wendrich, T. M., and M. A. Marahiel. 1997. Cloning and characterization of a relA/spoT homologue from Bacillus subtilis. Mol. Microbiol. 26:65-79. [DOI] [PubMed] [Google Scholar]
- 43.Wisniewski-Dye, F., J. Jones, S. R. Chhabra, and J. A. Downie. 2002. raiIR genes are part of a quorum-sensing network controlled by cinI and cinR in Rhizobium leguminosarum. J. Bacteriol. 184:1597-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xi, C., G. Dirix, J. Hofkens, F. C. Schryver, J. Vanderleyden, and J. Michiels. 2001. Use of dual marker transposons to identify new symbiosis genes in Rhizobium. Microb. Ecol. 41:325-332. [DOI] [PubMed] [Google Scholar]
- 45.Xi, C., M. Lambrecht, J. Vanderleyden, and J. Michiels. 1999. Bi-functional gfp- and gusA-containing mini-Tn5 transposon derivatives for combined gene expression and bacterial localization studies. J. Microbiol. Methods 35:85-92. [DOI] [PubMed] [Google Scholar]
- 46.Xi, C., E. Schoeters, J. Vanderleyden, and J. Michiels. 2000. Symbiosis-specific expression of Rhizobium etli casA encoding a secreted calmodulin-related protein. Proc. Natl. Acad. Sci. USA 97:11114-11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao, H., M. Kalman, K. Ikehara, S. Zemel, G. Glaser, and M. Cashel. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266:5980-5990. [PubMed] [Google Scholar]
- 48.Zhang, H. B., C. Wang, and L. H. Zhang. 2004. The quormone degradation system of Agrobacterium tumefaciens is regulated by starvation signal and stress alarmone (p)ppGpp. Mol. Microbiol. 52:1389-1401. [DOI] [PubMed] [Google Scholar]