Abstract
Wautersia eutropha H16 (formerly Ralstonia eutropha) mobilizes intracellularly accumulated poly(3-hydroxybutyrate) (PHB) with intracellular poly(3-hydroxybutyrate) depolymerases. In this study, a novel intracellular 3-hydroxybutyrate-oligomer hydrolase (PhaZc) gene was cloned and overexpressed in Escherichia coli. Then PhaZc was purified and characterized. Immunoblot analysis with polyclonal antiserum against PhaZc revealed that most PhaZc is present in the cytosolic fraction and a small amount is present in the poly(3-hydroxybutyrate) inclusion bodies of W. eutropha. PhaZc degraded various 3-hydroxybutyrate oligomers at a high specific activity and artificial amorphous poly(3-hydroxybutyrate) at a lower specific activity. Native PHB granules and semicrystalline PHB were not degraded by PhaZc. A PhaZ deletion mutation enhanced the deposition of PHB in the logarithmic phase in nutrient-rich medium. PhaZc differs from the hydrolases of W. eutropha previously reported and is a novel type of intracellular 3-hydroxybutyrate-oligomer hydrolase, and it participates in the mobilization of PHB along with other hydrolases.
Polyhydroxyalkanoates, which are bacterial polyesters, are a group of storage materials that are used as sources of carbon and energy (1). One of the most abundant polyhydroxyalkanoates is poly(3-hydroxybutyrate) (PHB), a homopolymer of d-(−)-3-hydroxybutyrate (3HB). PHB is synthesized from d-(−)-3-hydroxybutyryl-coenzyme A by PHB synthase (16, 18) and is mobilized by intracellular PHB depolymerases (15, 19).
PHB-producing bacteria contain intracellular PHB depolymerases. Intracellular PHB depolymerases have been found in Rhodospirillum rubrum (15), Paracoccus denitrificans (4), and Wautersia eutropha (19) (formerly Ralstonia eutropha). In W. eutropha H16, a gene encoding an intracellular PHB depolymerases, phaZ1 (19), and a gene encoding a 3HB-oligomer hydrolase, phaZb (formerly phaZ2) (10, 20), have been cloned, and some properties of their products have been reported. Both hydrolases showed hydrolytic activity for amorphous PHB. PhaZb hydrolyzed 3HB oligomers at a high specific activity and degraded PHB into 3HB monomers at a lower specific activity.
The function of PhaZ1 or PhaZb in vivo has been examined by constructing a phaZ1 or phaZb deletion strain (5, 10, 19). The deletion mutants exhibit considerably less degradation of PHB in vivo in some conditions. However, even a PhaZ1-PhaZb double-deletion mutation did not completely inhibit the mobilization of PHB (10). Therefore, it has been suggested that there are other depolymerases or related hydrolases (10).
Since the amino acid sequence of the first intracellular PHB depolymerase, PhaZ1, was reported for W. eutropha H16, several isoenzymes have been found. Recently, the genome sequences of W. eutropha and related bacteria have become available. Four putative PHB depolymerase genes homologous to phaZ1 have been identified using the genome sequence of Wautersia metallidurans (32), the sequence of the megaplasmid of W. eutropha (25), and the incomplete genome sequence of W. eutropha (17).
These putative intracellular PHB depolymerases have been identified based on the amino acid sequence of PhaZ1. In this study, a novel 3HB-oligomer hydrolase of W. eutropha was discovered using the amino acid sequence of an intracellular 3HB-oligomer hydrolase of Acidovorax sp. strain SA1 (27) as a probe. From a BLAST search using this probe, a candidate for an intracellular 3HB-oligomer hydrolase gene was identified in the genome sequence of Ralstonia solanacearum, a close relative of W. eutropha. The counterpart in W. eutropha, designated phaZc, was cloned by colony hybridization and sequenced. The gene product, PhaZc, was characterized, and a deletion mutant was examined to confirm that PhaZc is involved in the mobilization of PHB.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. All Escherichia coli strains were grown aerobically in Luria-Bertani (LB) medium or on solid LB agar (1.5%, wt/vol) plates at 37°C. The following typical concentrations of antibiotics were used: ampicillin, 50 μg/ml; kanamycin, 50 μg/ml; chloramphenicol, 34 μg/ml; and tetracycline, 12.5 μg/ml. All W. eutropha strains were cultivated aerobically at 30°C in a nitrogen-rich medium (N-rich medium) (19, 21) or a minimum salt medium (16, 24). To investigate the accumulation of PHB, W. eutropha was grown in a minimum salt medium containing 2% fructose and 0.1% ammonium sulfate (MSF medium) as described previously (9).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| E. coli JM109 | recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 Δ(lac-proAB)/F′ [traD36 proAB+lacIqlacZΔM15] | Takara |
| E. coli BLR(DE3)/pLysS | F−ompT hsdSB(rB− mB−) gal dcm Δ(srl-recA)306::Tn10(Tcr) (DE3)/pLysS(Cmr) Novagen | |
| W. eutropha H16 | Wild type | ATCC 17699 |
| W. eutropha TK0120 | ΔphaZc, derived from H16 | This study |
| W. eutropha H16DZbc1 | ΔphaZb ΔphaZc, derived from TK0120 | This study |
| W. eutropha OH1 | ΔphaZc, derived from H16 | 10 |
| Plasmids | ||
| pUC19 | Cloning vector; Ampr | Takara |
| pET23b | Expression vector; Ampr | Novagen |
| pE3ReZc | pET23b carrying amplified NdeI-XhoI fragment containing phaZc | This study |
| pETOH | pET23b carrying amplified NdeI-XhoI fragment containing phaZb | 10 |
| pET171H | pET23b carrying amplified NdeI-XhoI fragment containing phaZl | 19 |
| pLO3 | Suicide vector; TcrsacB RP4 oriT ColiE1 ori | 13 |
| pLO3dmZc | pLO3 carrying N-terminal 295 bp and C-terminal 296 bp of phaZc | This study |
| pJPPOH | Suicide vector; carrying 306-bp fragment of phaZb; Kmr | 10 |
Ampr, ampicillin resistant; Cmr, chloramphenicol resistant; Tcr, tetracycline resistant.
DNA preparation and manipulation.
Standard methods were used for preparation and manipulation of DNA, Southern hybridization, and colony hybridization (23). Sequencing was performed with a SEQ-4 × 4 system and Thermosequenase Cy 5.5 (Amersham Biosciences, Tokyo, Japan) as recommended by the manufacturer. Sequences were processed using the program GENETYX-MAC/ATSQ, version 4.2.0 (Software Development Co., Ltd., Tokyo, Japan).
Design of probes to identify the 3HB-oligomer hydrolase gene.
A candidate 3HB-oligomer hydrolase gene was identified by a BLAST comparison using a 3HB-oligomer hydrolase of Acidovorax sp. strain SA1 (accession number AB044565) (27) and the genome sequence of R. solanacearum, which has recently become available in its entirety (accession number NC_003295) (22). A putative hydrolase (protein ID number CAD15276.1) of R. solanacearum exhibited a high level of similarity to the 3HB-oligomer hydrolase. The DNA sequence 400 bp upstream and C terminal of the putative hydrolase gene was used to design a set of primers, forward primer 5′-GCGCATCATGTCCCTCGATCTCGGCAGC-3′ and reverse primer 5′-CGCGCTTCAGGGTGTTGGTGCTCATG-3′. With chromosomal DNA of W. eutropha as the template, these primers were used to amplify an approximately 1.3-kbp fragment that included the equivalent gene, phaZc. The PCR product was 32P labeled and used as a probe in Southern hybridization and colony hybridization. W. eutropha genomic DNA was completely digested with BamHI, EcoRI, KpnI, SalI, SmaI, or XbaI. The resulting fragments were subjected to Southern hybridization.
Construction of pE3ReZc.
To express phaZc, the gene was amplified by PCR with a pair of primers, primers phaZcf (5′-CCGGAATTCATATGTCTGCCAGTCCGCGTCTCGG-3′) and phaZcr (5′-CGAATTCTCGAGTCAGGCCCCGGTAAAGAACTGCGAGG-3′). The products were digested with NdeI and XhoI and inserted into corresponding sites of pET23b (Novagen, Madison, WI). The resulting plasmid was designated pE3ReZc.
Purification of PhaZc from Escherichia coli.
Expression of phaZc was induced by isopropyl-β-d-thiogalactopyranoside as described previously (10) with E. coli BLR(DE3)/pLysS transformed with pE3ReZc. Bacteria were harvested and resuspended in 20 mM Tris-HCl (pH 8.0). The cell suspension was sonicated and centrifuged at 10,000 × g for 20 min. The supernatant (crude extract) was mixed with glycerol at a final concentration of 50% (vol/vol) and stored at −20°C prior to use.
The enzyme was purified in two steps: hydrophobic column chromatography and ion-exchange column chromatography. Ammonium sulfate was added to the crude extract at a final concentration of 1 M, and the mixture was centrifuged at 10,000 × g for 20 min. The supernatant was loaded onto a Toyopearl ether-650M column (15 by 170 mm) that was preequilibrated with 10 mM Tris-HCl (pH 8.0) containing 1 M ammonium sulfate. After one wash with the same buffer, the enzyme was eluted with a linear gradient of ammonium sulfate (200 ml, 1 to 0 M). The active fractions were collected and dialyzed against 10 mM Tris-HCl (pH 8.0). The sample was further purified on a Toyopearl DEAE-650M column (15 by 60 mm) that was preequilibrated with 10 mM Tris-HCl (pH 8.0). The active fractions that were eluted with a linear gradient of NaCl (200 ml, 0 to 0.5 M) were collected and stored at −20°C in 50% (vol/vol) glycerol.
Preparation of substrates.
Semicrystalline PHB was isolated from W. eutropha H16 and purified by hypochlorite treatment (26). Artificial amorphous PHB granules were prepared from purified semicrystalline PHB granules described by Horowitz and Sanders (6, 10). Native PHB was isolated from W. eutropha H16 and purified by glycerol density gradient (5). The 3HB oligomers were prepared as described previously (28).
Enzyme assays.
3HB-oligomer hydrolase activity and PHB depolymerase activity were assayed based on the amount of 3HB or 3HB oligomers released from the 3HB oligomers or PHB, as described previously (10, 31). The reaction mixture (50 μl) was composed of 100 mM Tris-HCl (pH 8.5), PHB granules (0.5 mg/ml as a solid), and enzyme. The reaction was started by addition of substrate at 30°C. For 3HB-oligomer hydrolase activity, all 3HB oligomers were dissolved in distilled water at a concentration of 10 mM and used as substrates at a concentration of 2 mM instead of PHB granules. The rate of hydrolysis of p-nitrophenyl esters was determined spectrophotometrically, and lipase activity was assayed by titration of the acid released from olive oil, as described previously (33).
Electrophoresis and immunoblot analysis.
Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and immunoblot analysis were performed by the procedure of Laemmli (12) and the method of Towbin et al. (29), respectively. Polyclonal antibody against PhaZc was prepared from rabbits.
Construction of deletion mutants.
A 291-bp deletion of phaZc was generated by PCR using the following primers: ReZc-dmNF (5′-GCTCTAGAATGTCTGCCAGTCCGCGTCTCG), ReZc-dmNR (5′-CGGAATTCGTTGAGCCGCGCAATCAGCGTGAC), ReZc-dmCF (5′-CGGAATTCGCCTGGAATGGGGCTTGCATTACG), and ReZc-dmCR (5′-AACTGCAGTCAGGCCCCGGTAAAGAACTGCG). The truncated gene was treated with XbaI and PstI and inserted into the corresponding site of pLO3 (13). E. coli S17-1 was transformed using the resulting plasmid, pLO3dmZc. pLO3dmZc was transferred to W. eutropha H16 by conjugation, and the transformant was selected based on tetracycline resistance and auxotrophy. Deletion mutants were obtained by selection of sucrose-resistant colonies on LB agar containing 15% sucrose, as described Lenz and Friedrich (13). The mutant was verified by Southern and PCR analyses. The phaZc deletion mutant obtained was designated TK0120. A double-deletion mutant lacking phaZb and phaZc, H16DZbc1, was constructed from the phaZc deletion mutant using pJPPOH, as described previously (10).
Other methods.
PhaZ1 and PhaZb were purified as described previously (10). The protein concentration was determined by the method of Lowry et al. (14) or Bradford (2) with reagents obtained from Bio-Rad Laboratories (Hercules, CA). The N-terminal amino acid sequence was determined with a Procise 491-HT (Applied Biosystems, Tokyo, Japan) used according to the instructions in the manual. The PHB content was quantitated as crotonic acid by high-pressure liquid chromatography as described by Karr et al. (8). Cell dry weight was determined with freeze-dried whole cells.
Nucleotide sequence accession number.
The phaZc sequence has been deposited in the GenBank/EMBL/DDBJ databases under accession no. AB180691.
RESULTS
Identification of the analogue of 3HB-oligomer hydrolase.
The search for amino acid sequences similar to the sequence of the 3HB-oligomer hydrolase of Acidovorax sp. strain SA1 revealed putative sequences in W. metallidurans, R. solanacearum, R. rubrum, Rhodobacter sphaeroides, and other species whose genome sequences are available (Fig. 1). Among these sequences, the amino acid sequence of a putative hydrolase of R. solanacearum exhibited 37% identity to the 3HB-oligomer hydrolase sequence. The DNA sequence of the putative hydrolase gene was used to design primers for use in PCR with W. eutropha genomic DNA. A PCR product of the expected size (about 1.3 kbp) was used as a probe for hybridization.
FIG. 1.
Comparison of PhaZc amino acid sequences. A BLAST search using an intracellular 3HB-oligomer hydrolase amino acid sequence of Acidovorax sp. strain SA1 revealed several putative 3HB-oligomer hydrolases from PHB-accumulating bacteria. (A) Results of a multiple alignment using the Clustal W program (1.8.2). The candidate catalytic center is indicated by boldface type, and the lipase box is underlined. Asterisks indicate residues that are conserved in all proteins; colons indicate substitution of amino acid residues with very similar residues; and periods indicate substitution of amino acid residues with residues that are slightly less similar. (B) Phylogenetic tree. The cluster analysis was based on the neighbor-joining method. The numbers at the nodes are percentages of 1,000 bootstrap resamplings (only values greater than 50% are shown). The following proteins were included in the analyses: 3HB-oligomer hydrolase from Acidovorax sp. strain SA1 (SA1), predicted hydrolases from W. metallidurans (Wmet), R. solanacearum (Rsol), R. rubrum (Rrub), and R. sphaeroides (Rsph), and PhaZc from W. eutropha cloned in this study (Weut). The accession numbers of these proteins are AB044565, ZP_00276649, NP_519695, ZP_00268816, ZP_00007013, and AB180691, respectively.
Cloning of phaZc.
The 3.5-kbp SalI fragments were inserted into pUC19, and a positive clone was obtained by colony hybridization. The cloned fragment was sequenced, which revealed that it included two genes, bdh [d-(−)-3-hydroxybutyrate dehydrogenase gene] (accession number AF145230) and phaZc. The phaZc gene represented an 898-bp open reading frame, whose translation product was estimated to have a molecular mass of 31,542 Da. The primary structure of PhaZc contained the conserved sequence of the active site or the lipase box pentapeptide (Gly-X1-Ser-X2-Gly) which is characteristic of serine-dependent catalysis (Fig. 1A) (7, 11). Phylogenetic analysis showed that PhaZc proteins from W. eutropha, W. metallidurans, and R. solanacearum are closely related (Fig. 1B).
Properties of PhaZc.
E. coli transformed with pE3ReZc was used to produce PhaZc. SDS-PAGE of the cytosolic fraction of E. coli/pE3ReZc resulted in a thick ∼32-kDa band whose size corresponded to the size of PhaZc deduced from the amino acid sequence (Fig. 2, lane 1). PhaZc was purified with a hydrophobic column and an anion-exchange column. The results of the purification are summarized in Table 2. The final preparation exhibited apparent homogeneity on an SDS-polyacrylamide gel (Fig. 2, lane 1). The N-terminal amino acid sequence of the purified enzyme was determined chemically to be SASPRLGFV, which corresponded to the deduced amino acid sequence except for the first methionine. Immunostaining of the crude extract from W. eutropha showed that the molecular mass was the same as the molecular mass of purified PhaZc from E. coli.
FIG. 2.

SDS-PAGE analysis of purified PhaZc from E. coli harboring pE3ReZc. E. coli overexpressing phaZc was sonicated and centrifuged, and the soluble crude extract (14 μg) (lane 1) was purified on a hydrophobic column (2 μg) (lane 2) and an anion-exchange column (1 μg) (lane 3). The arrow indicates the position of PhaZc. Lane M contained molecular mass markers.
TABLE 2.
Purification of PhaZc from E. coli harboring pE3ReZca
| Step | Protein (mg) | Total activity (U)b | Sp act (U/mg) | Yield (%) |
|---|---|---|---|---|
| Crude extract | 170 | 2,500 | 15 | 100 |
| Toyopearl ether-650M | 7.8 | 1,100 | 140 | 44 |
| Toyopearl DEAE-650M | 2.9 | 900 | 310 | 36 |
Enzyme was purified from 2 g (wet weight) of E. coli cells harboring pE3ReZc as described in Materials and Methods.
3HB pentamer was used as a substrate to measure the activity. One unit of enzyme activity catalyzed the formation of 1 μmol of 3HB per min.
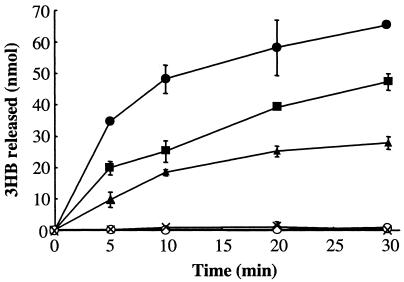
The hydrolytic activity was measured using semicrystalline PHB, artificial amorphous PHB, native PHB inclusion bodies (granules), and several 3HB oligomers (dimer, trimer, tetramer, and pentamer) (Table 3). PhaZc did not hydrolyze semicrystalline PHB or native PHB inclusion bodies, but it hydrolyzed artificial amorphous PHB (Fig. 3) and 3HB oligomers. The larger oligomers were hydrolyzed more rapidly. PhaZc showed slight hydrolytic activity with p-nitrophenyl esters and no lipase activity with olive oil. The enzyme activity was strongly inhibited by 1 mM diisopropylfluorophosphate (2% of residual activity), which suggests that the serine residue is the active center. Because the PHB depolymerase activity of PhaZ1 is synergically enhanced by PhaZb (10), the synergic effect of PhaZ1 or PhaZb on PhaZc was examined. PhaZ1 or PhaZb did not enhance the PHB depolymerase activity of PhaZc (data not shown).
TABLE 3.
Substrate specificity of PhaZc
| Substrate | Sp act (μmol/min/mg)a |
|---|---|
| 3HB oligomers | |
| Dimer | 31 |
| Trimer | 200 |
| Tetramer | 240 |
| Pentamer | 310 |
| PHB | |
| Artificial amorphous PHB | 3.0 |
| Native PHB granules | <0.1 |
| Semicrystalline PHB | <0.1 |
| p-Nitrophenyl esters | |
| Acetate | 1.0 |
| Butyrate | 6.0 |
| Palmitate | <0.01 |
| Olive oil | <0.5 |
Activity was assayed as described in Materials and Methods.
FIG. 3.
Degradation of artificial amorphous PHB granules by PhaZc. The 3HB released during PHB degradation was measured enzymatically. Symbols: ×, no PhaZc; ▴, 0.46 μg of PhaZc; ⧫, 1.2 μg of PhaZc; •, 4.6 μg of PhaZc; ○, 4.6 μg of boiled PhaZc (100°C, 5 min). The error bars indicate standard deviations of the means based on three independent measurements.
Growth phase dependence and cell localization of PhaZc.
W. eutropha cells were grown in N-rich medium or MSF medium, and the expression of phaZc in the cells at different growth phases was examined by Western blotting. phaZc seemed to be expressed constitutively both in the N-rich medium (cell growth conditions) and in the MSF medium (PHB accumulation conditions) (data not shown).
After cultivation for 2 days in the MSF medium, the PHB-accumulating cells were harvested and sonicated. The resultant disrupted cells were fractionated with a linear sucrose gradient (1 to 2 M). The fractions were analyzed by immunostaining (Fig. 4). Antisera against PhaZ1 and 3HB dehydrogenase used as controls for the protein in PHB inclusion bodies and in the cytosol of the cell, respectively. PhaZc was found mainly in the cytosolic fraction, but some PhaZc was also detected in the fraction containing PHB inclusion bodies. The subcellular localization of PhaZc is different from that of PhaZ1 or PhaZb (10).
FIG. 4.
Subcellular localization of PhaZc determined by using a sucrose density gradient. Sonicates from PHB-rich cells were fractionated by ultracentrifugation in a sucrose gradient (1 to 2 M; total volume, 11 ml). Each fraction (1.1 ml) was collected and analyzed by Western blotting and immunostaining with antiserum against PhaZc, PhaZ1, or 3HB dehydrogenase (3HBDH); 100% corresponded to PHB and protein concentrations of 65 and 3.7 mg/ml, respectively.
PHB mobilization in phaZ deletion strains.
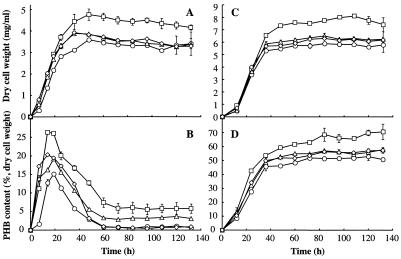
To determine if PhaZc is involved in the mobilization of PHB, phaZc (ΔphaZc) and phaZb (ΔphaZb) deletion mutants and a phaZb-phaZc (ΔphaZbZc) double-deletion mutant were examined for the accumulation of PHB when they were grown in N-rich or MSF medium for several days (Fig. 5). When cultured in N-rich medium, the wild type and all of the mutants temporarily accumulated PHB in the logarithmic phase and then rapidly degraded it, consistent with previous reports (19, 20). During the logarithmic phase, the PHB content of all mutants was higher than that of the wild type, and in the stationary phase, all mutants except the ΔphaZbZc mutant showed elevated levels of PHB. In the logarithmic phase, the ΔphaZb mutant accumulated the largest amount of PHB, and the ΔphaZc and ΔphaZbZc mutants deposited the least. In the stationary phase, the amount of PHB in the ΔphaZbZc mutant was almost the same as the amount in the wild type (Fig. 5B). When cultured in MSF medium (PHB accumulation conditions), all mutants contained higher PHB levels than the wild type. Although the tendency of PHB to accumulate in each mutant was similar to the results obtained in N-rich medium, the difference between the PHB content of each mutant and that of the wild type was less than difference in N-rich medium. In these conditions, all strains accumulated PHB with growth, and temporary accumulation of PHB was not observed in the logarithmic phase (Fig. 5D).
FIG. 5.
Growth and accumulation of PHB in various W. eutropha phaZ deletion mutants in the nutrient-rich medium and the PHB accumulation medium. (A and B) Nutrient-rich medium. (C and D) PHB accumulation medium (mineral salt medium containing 2% [wt/vol] fructose and 0.1% [wt/vol] ammonium sulfate). (A and C) Cell dry weight. (B and D) PHB content. Symbols: ○, wild type; ▵, TK0120 (ΔphaZc mutant); □, OH1 (ΔphaZb mutant); ⋄, H16DZbc1 (ΔphaZb ΔphaZc mutant). The data are means for three independent measurements. The error bars indicate standard deviations.
DISCUSSION
It has been reported that W. eutropha has different types of intracellular PHB depolymerases, PhaZ1 and a 3HB-oligomer hydrolase, PhaZb (formerly PhaZ2). In the present study we showed that there is a second type of 3HB-oligomer hydrolase, PhaZc. PhaZ1 and its homologs, PhaZb and PhaZc, are different in terms of overall amino acid sequence and molecular mass. PhaZ1 is a distinctive type of PHB depolymerase. It does not contain the lipase box pentapeptide (Gly-X1-Ser-X2-Gly) which all extracellular PHB depolymerases contain. Instead of the serine residue, PhaZ1 has a cysteine residue at the active site (11). PhaZb and PhaZc have the lipase box sequence at their active sites. PhaZb is a 78-kDa protein and is similar to the extracellular 3HB-oligomer hydrolase of Ralstonia pickettii A1 (33). The molecular mass of PhaZc (32 kDa) is about one-half that of PhaZb, and the amino acid sequence of PhaZc resembles the amino acid sequence of an intracellular 3HB-oligomer hydrolase of Acidovorax sp. strain SA1 (27). Therefore, PhaZ1 and its homologs, PhaZb and PhaZc, belong to distinct groups.
Besides PhaZ1, PhaZ2 to PhaZ5 have been identified as intracellular PHB depolymerases in W. eutropha (17, 25, 32). These depolymerases, however, are actually PhaZ1 homologs; they resemble PhaZ1 in amino acid sequence, the active site residues, and molecular mass (45 to 47 kDa) (except PhaZ4, which has a molecular mass of 27 kDa). PhaZ4 exhibits similarity with the C-terminal side of PhaZ1, but it lacks the N-terminal side consisting of about 150 amino acid residues compared with the other homologs. These homologs should be classified as type 1 intracellular PHA depolymerases. In this study, to avoid confusion over the nomenclature, the 3HB-oligomer hydrolase (formerly PhaZ2) (10, 20) was renamed PhaZb. The 3HB-oligomer hydrolase found in this study was designated PhaZc. In the future, new types of enzymes involved in the degradation of PHB may be designated PhaZd, PhaZe, and so on because it is reasonable to classify enzymes based on type or properties (amino acid sequence, molecular mass, structure, activity, etc.). It is preferable that PhaZ1 homologs be designated PhaZa1 to PhaZa5. The type “a” enzyme (PhaZ1) hydrolyzes PHB but is not active with 3HB oligomers. The type “b” enzyme (PhaZb), which has a high molecular mass (78 kDa), efficiently hydrolyzes 3HB oligomers and degrades amorphous PHB at a lower rate. The b type enzyme has been found only in members of the genus Wautersia, such as W. eutropha or W. metallidurans, so far. The type “c” enzyme (molecular mass, about 30 kDa) exhibits strong 3HB-oligomer hydrolase activity compared with other PHB depolymerases or 3HB-oligomer hydrolases. Some enzymes (PhaZc in this study, PhaZcWeu) hydrolyze the 3HB pentamer most efficiently and exhibit weak hydrolytic activity with amorphous PHB, but another PhaZc in Acidovorax sp. strain SA1 (PhaZcAsp) (27) hydrolyzes the 3HB dimer at a high rate with no PHB-hydrolyzing activity. The type c enzyme was found in many other PHB-producing or PHB-degrading bacteria based on the BLAST search. In addition, three proteins whose molecular masses are about 30 kDa were reported to be 3HB-oligomer hydrolases in Pseudomonas lemoignei (3), Zoogloea ramigera (28), and Pseudomonas denitrificans (30). These enzymes also could be homologous to PhaZc. Although the substrate specificities of PhaZc homologs vary, the widespread distribution may reflect the importance of the PhaZc-like enzymes in the mobilization of PHB.
Some of the enzymatic properties of PhaZc in vitro clearly suggest that the enzyme is a specific 3HB-oligomer hydrolase, not a lipase or nonspecific esterase, and it may function partly as a PHB depolymerase. Although the enzyme hydrolyzed 3HB oligomers efficiently, it also degraded artificial amorphous PHB (Table 3 and Fig. 3), and a certain amount of PhaZc was found in native PHB inclusion bodies (Fig. 2). The PHB depolymerase activity with artificial amorphous PHB observed in vivo does not necessarily mean that PhaZc has weak activity with native PHB in the cell; the amorphous PHB in native PHB inclusion bodies probably differs from the artificial amorphous PHB in terms of accessibility and degradability. In a previous report, we showed that PhaZ1 and PhaZb synergistically hydrolyze PHB (10). For these reasons, PhaZ1, PhaZb, and PhaZc may work together and hydrolyze PHB efficiently, as well as 3HB oligomers in the cell.
There was a significant difference in the accumulation of PHB between various mutants, including the ΔphaZc mutant and the wild type. Under both nitrogen-rich and PHB accumulation conditions, all deletion mutants showed higher PHB levels than the wild type, and the maximal level was observed in the ΔphaZb mutant. The effect of the mutation on accumulation indicates that PhaZc and PhaZb contribute to the mobilization of PHB. It has been reported that accumulation and degradation of PHB occur at the same time (9). It seems that deletion of phaZc and phaZb leads to an imbalance in accumulation and degradation, lowering the degradative activity for PHB or 3HB oligomers in the cell, with the result that a large amount of PHB accumulates. The difference between the ΔphaZb and ΔphaZc mutants in terms of PHB content may be caused by the difference in subcellular localization. Whereas PhaZc mainly localizes in the cytosol, PhaZb is present in both the cytosol and PHB inclusion bodies (10). Although the substrate specificities of PhaZc and PhaZb are similar in vitro, it seems that PhaZb is more concerned with polymer degradation than PhaZc in the cell. Interestingly, the ΔphaZc mutant and the ΔphaZbZc mutant showed similar accumulations of PHB when they were cultured in MSF medium and during the logarithmic phase in N-rich medium. Furthermore, the ΔphaZbZc mutant deposited an amount of PHB similar to the amount deposited by the wild type in the stationary phase in nitrogen-rich conditions. It is not clear why the ΔphaZbZc mutant contains less PHB than the ΔphaZb mutant and shows rapid loss of PHB after accumulation. However, it is possible that the mobilization of PHB is regulated by the concentration of 3HB in the cell. Because both PhaZb and PhaZc efficiently hydrolyze 3HB oligomers, it is clear that the concentration of the 3HB monomer in the cell is changed by these enzymes. The proportions of 3HB, 3HB oligomers, and PHB in deletion mutants should be different from those in the wild type. For these reasons, not only the carbon or nutrient concentration in the culture but also the 3HB oligomers or 3HB concentration in the cell may regulate the mobilization of PHB. The low concentration of 3HB in the cell may be one of the signals for mobilization. In the ΔphaZbZc mutant, the PHB content decreased to almost zero, like the content in the wild type, after temporal accumulation of PHB in N-rich medium. Given that the PHB assay used in this study cannot distinguish PHB, 3HB oligomers, and 3HB, the decrease in the PHB content in the ΔphaZbZc mutant may have been due to a decrease in the 3HB oligomer content caused by an unknown hydrolase(s).
The PHB mobilization system in W. eutropha H16 is very complex and still unclear. Certainly more study is needed.
Acknowledgments
This study was supported by a grant-in-aid for a high-tech research center project (2002) from The Ministry of Education, Culture, Sports, Science and Technology of Japan.
We thank A. Sugiyama and M. Takanashi for providing 3HB oligomers and 3HB dehydrogenase, respectively.
REFERENCES
- 1.Anderson, A. J., and E. A. Dawes. 1990. Occurrence, metabolism, metabolic roles, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol. Rev. 54:450-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Delafield, F. P., K. E. Cooksey, and M. Doudoroff. 1965. β-Hydroxybutyric dehydrogenase and dimer hydrolase of Pseudomonas lemoignei. J. Biol. Chem. 240:4023-4028. [PubMed] [Google Scholar]
- 4.Gao, D., A. Maehara, T. Yamane, and S. Ueda. 2001. Identification of the intracellular polyhydroxyalkanoate depolymerase gene of Paracoccus denitrificans and some properties of the gene product. FEMS Microbiol. Lett. 196:159-164. [DOI] [PubMed] [Google Scholar]
- 5.Handrick, R., S. Reinhardt, and D. Jendrossek. 2000. Mobilization of poly(3-hydroxybutyrate) in Ralstonia eutropha. J. Bacteriol. 182:5916-5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horowitz, D. M., and J. K. M. Sanders. 1994. Amorphous, biomimetic granules of polyhydroxybutyrate: preparation, characterization, and biological implications. J. Am. Chem. Soc. 116:2695-2702. [Google Scholar]
- 7.Jendrossek, D., and R. Handrick. 2002. Microbial degradation of polyhydroxyalkanoates. Annu. Rev. Microbiol. 56:403-432. [DOI] [PubMed] [Google Scholar]
- 8.Karr, D. B., J. K. Waters, and D. W. Emerich. 1983. Analysis of poly-β-hydroxybutyrate in Rhizobium japonicum bacteroids by ion-exclusion high-pressure liquid chromatography UV detection. Appl. Environ. Microbiol. 46:1339-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawaguchi, Y., and D. Yoshiharu. 1992. Kinetics and mechanism of synthesis and degradation of poly(3-hydroxybutyrate) in Alcaligenes eutropus. Macromolecules 25:2324-2329. [Google Scholar]
- 10.Kobayashi, T., M. Shiraki, T. Abe, A. Sugiyama, and T. Saito. 2003. Purification and properties of an intracellular 3-hydroxybutyrate-oligomer hydrolase (PhaZ2) in Ralstonia eutropha H16 and its identification as a novel intracellular poly(3-hydroxybutyrate) depolymerase. J. Bacteriol. 185:3485-3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi, T., and T. Saito. 2003. Catalytic triad of intracellular poly(3-hydroxybutyrate) depolymerase (PhaZ1) in Ralstonia eutropha H16. J. Biosci. Bioeng. 96:487-492. [DOI] [PubMed] [Google Scholar]
- 12.Laemmli, U. K. 1971. Cleavage of structural proteins during the assembly of the head bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 13.Lenz, O., and B. Friedrich. 1998. A novel multicomponent regulatory system mediates H2 sensing in Alcaligenes eutrophus. Proc. Natl. Acad. Sci. USA 95:12474-12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 15.Merrick, J. M., and M. Doudoroff. 1964. Depolymerization of poly-β-hydroxybutyrate by an intracellular enzyme system. J. Bacteriol. 88:60-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peoples, P. O., and A. J. Sinskey. 1989. Poly-β-hydroxybutyrate (PHB) biosynthesis in Alcaligenes eutrophus H16. J. Biol. Chem. 264:15298-15303. [PubMed] [Google Scholar]
- 17.Pötter, M., H. Müller, F. Reinecke, R. Wieczorek, F. Fricke, B. Bowien, B. Friedrich, and A. Steinbüchel. 2004. The complex structure of polyhydroxybutyrate (PHB) granules: four orthologous and paralogous phasins occur in Ralstonia eutropha. Microbiology 150:2301-2311. [DOI] [PubMed] [Google Scholar]
- 18.Rehm, B. H. A., and A. Steinbüchel. 2002. PHA synthases: the key enzyme of PHA synthesis, p. 173-215. In Y. Doi and A. Steinbüchel (ed.), Biopolymers: polyesters I. Wiley-VCH, Weinheim, Germany.
- 19.Saegusa, H., M. Shiraki, C. Kanai, and T. Saito. 2001. Cloning of an intracellular poly[d-(−)-3-hydroxybutyrate] depolymerase gene from Ralstonia eutropha H16 and characterization of the gene product. J. Bacteriol. 183:94-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saegusa, H., M. Shiraki, and T. Saito. 2002. Cloning of an intracellular d(−)-3-hydroxybutyrate-oligomer hydrolase gene from Ralstonia eutropha H16 and identification of the activity site serine residue by site-directed mutagenesis. J. Biosci. Bioeng. 94:106-112. [DOI] [PubMed] [Google Scholar]
- 21.Saito, T., K. Takizawa, and H. Saegusa. 1995. Intracellular poly(3-hydroxybutyrate) depolymerase in Alcaligenes eutrophus. Can. J. Microbiol. 41:187-191. [Google Scholar]
- 22.Salanoubat, M., S. Genin, F. Artiguenave, J. Gouzy, S. Mangenot, M. Arlat, A. Billault, P. Brottier, J. C. Camus, L. Cattolico, M. Chandler, N. Choisne, C. Claudel-Renard, S. Cunnac, N. Demange, C. Gaspin, M. Lavie, A. Moisan, C. Robert, W. Saurin, T. Schiex, P. Siguier, P. Thebault, M. Whalen, P. Wincker, M. Levy, J. Weissenbach, and C. A. Boucher. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415:497-502. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook, J., and D. W. Russell. 2001. Molecular cloning, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Schlegel, H. G., G. Gottschalk, and R. von Bartha. 1961. Formation and utilization of poly-β-hydroxybutyric acid by Knallgas bacteria (Hydrogenomoas). Nature 191:463-465. [DOI] [PubMed] [Google Scholar]
- 25.Schwartz, E., A. Henne, R. Cramm, T. Eitinger, B. Friedrich, and G. Gottschalk. 2003. Complete nucleotide sequence of pHG1: a Ralstonia eutropha H16 megaplasmid encoding key enzymes of H2-based lithoautotrophy and anaerobiosis. J. Mol. Biol. 332:369-383. [DOI] [PubMed] [Google Scholar]
- 26.Smibert, R. M., and N. R. Krieg. 1994. Phenotypic characterization, p. 650. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular bacteriology. American Society for Microbiology, Washington, D.C.
- 27.Sugiyama, A., M. Shiraki, T. Kobayashi, G. Morikawa, M. Yamamoto, M. Yamaoka, and T. Saito. 2002. Cloning and sequencing of an intracellular d(−)-3-hydroxybutyrate oligomer hydrolase from Acidovorax sp. strain SA1 and purification of the enzyme. Curr. Microbiol. 45:123-127. [DOI] [PubMed] [Google Scholar]
- 28.Tanaka, Y., T. Saito, T. Fukui, T. Tanio, and K. Tomita. 1981. Purification and properties of d(−)-3-hydroxybutyrate-dimer hydrolase from Zoogloea ramigera I-16-M. Eur. J. Biochem. 118:177-182. [DOI] [PubMed] [Google Scholar]
- 29.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ueda, S., K. Sano, D. Gao, N. Tomihari, T. Yamane, and I. Endo. 2002. Purification and properties of d-(−)-3-hydroxybutyrate oligomer hydrolase of Paracoccus denitrificans. FEMS Microbiol. Lett. 10:179-184. [DOI] [PubMed] [Google Scholar]
- 31.Williamson, D. H., J. Mellanby, and H. A. Krebs. 1962. Enzymatic determination of d(−)-β-hydroxybutyric acid and acetoacetic acid in blood. Biochem. J. 82:90-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.York, G. M., J. Lupberger, J. Tian, A. G. Lawrence, J. A. Stubbe, and A. J. Sinskey. 2003. Ralstonia eutropha H16 encodes two and possibly three intracellular poly[d-(−)-3-hydroxybutyrate] depolymerase genes. J. Bacteriol. 185:3788-3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, K., M. Shiraki, and T. Saito. 1997. Purification of an extracellular d(−)-3-hydroxybutyrate oligomer hydrolase from Pseudomonas sp. strain A1 and cloning and sequencing of its gene. J. Bacteriol. 179:72-77. [DOI] [PMC free article] [PubMed] [Google Scholar]