Abstract
The poly-d-glutamic acid capsule of Bacillus anthracis is essential for virulence. Control of capsule synthesis occurs at the level of transcription and involves positive regulation of the capsule biosynthetic operon capBCAD by a CO2/bicarbonate signal and three plasmid-borne regulators: atxA, acpA, and acpB. Although the molecular mechanism for control of cap transcription is unknown, atxA affects cap expression via positive control of acpA and acpB, two genes with partial functional similarity. Transcriptional analyses of a genetically complete strain indicate that capB expression is several hundred-fold higher during growth in 5% CO2 compared to growth in air. atxA was expressed appreciably during growth in air and induced only 2.5-fold by CO2. In contrast, expression of acpA and acpB was induced up to 23-fold and 59-fold, respectively, by CO2. The 5′-end mapping of gene transcripts revealed atxA-regulated and atxA-independent apparent transcription start sites for capB, acpA, and acpB. Transcripts mapping to all atxA-regulated start sites were increased during growth in elevated CO2. The acpA gene has one atxA-regulated and one atxA-independent start site. acpB lies downstream of capBCAD. A single atxA-independent start site maps immediately upstream of acpB. atxA-mediated control of acpB appears to occur via transcriptional read-through from atxA-dependent start sites 5′ of capB. One atxA-independent and two atxA-regulated start sites map upstream of capB. Transcription from the atxA-regulated start sites of capBCAD was reduced significantly in an acpA acpB double mutant but unaffected in mutants with deletion of only acpA or acpB, in agreement with the current model for epistatic relationships between the regulators.
Control of virulence gene expression in Bacillus anthracis is highly dependent upon atxA, a regulatory gene located on virulence plasmid pXO1. In genetically complete strains harboring pXO1 and the second virulence plasmid, pXO2, atxA acts as a global regulator controlling expression of the pXO2-encoded capsule biosynthetic gene operon, capBCAD (3, 6, 9, 22); the toxin structural genes, pagA, lef, and cya on pXO1 (4, 13, 20); and a number of other genes located on the plasmids and chromosome (3). The mechanism by which atxA exerts its effect on target gene transcription is unknown. A direct effect of atxA on transcription has not been demonstrated for any atxA-controlled gene.
The capBCAD genes are required for virulence in a mouse model for inhalation anthrax (7). The capsule biosynthetic genes capBCA are predicted to encode the proteins responsible for the synthesis, transport and attachment of the poly-d-glutamic acid capsule polymers to the outside of the bacterial cells (14, 15). Enzymatic or structural functions for CapB, CapC, and CapA have not been demonstrated. CapD (formerly Dep) is an enzyme that depolymerizes the large capsule polymers into smaller d-glutamic acid peptide fragments that are released from the surface of the bacterial cells (21). Given the significance of the capsule biosynthetic gene operon in virulence, determining the mode of regulation of these genes is of interest.
In our current model for capsule gene regulation, atxA controls cap gene transcription and capsule synthesis via the positive regulation of two pXO2-encoded regulators, acpA and acpB. The model arose from studies employing a genetically complete (pXO1+ pXO2+) parent strain and isogenic mutants with deletion of atxA, acpA, and/or acpB (6). In pXO1+ pXO2+ strains, while deletion of acpA or acpB alone does not appreciably affect capB transcription or capsule synthesis, an acpA acpB double mutant exhibits drastically reduced capB transcription and is noncapsulated. Thus, acpA and acpB have some functional similarity. The amino acid sequences of the predicted products of these genes are approximately 62% homologous. Moreover, the proteins also share significant amino acid sequence similarity with the predicted product of atxA.
For many atxA-controlled genes, including acpA and acpB, expression is induced during growth in 5% atmospheric CO2 or in media containing bicarbonate (2, 6, 11, 13, 18, 22, 23). CO2-induced transcription of all three toxin genes has been demonstrated in experiments employing promoter-reporter gene fusions (2, 13, 18). RNA slot blot analysis of capB, the first gene of the capsule biosynthetic operon, and acpA transcripts demonstrated an increase in both transcripts during culture in elevated CO2 (22, 23). We recently demonstrated elevated acpB expression during growth in 5% CO2 using reverse transcription-PCR (RT-PCR) (6).
CO2/bicarbonate is likely to be a physiologically significant signal encountered by the bacterium in the host environment. Concentrations of bicarbonate/CO2 (15 to 40 mM) in the bloodstream of the host (5) are comparable to the concentration of bicarbonate/CO2 present in the bicarbonate-supplemented growth media during culture in vitro (48 mM). Although induction of cap gene expression in vivo has not been assessed quantitatively, our recent experiments employing a mouse model for inhalation anthrax demonstrate the importance of the capsule biosynthetic operon and its regulators during infection (7). The noncapsulated acpA acpB mutant is completely attenuated in the mouse model. The 50% lethal dose and mean time to death for the mutant were comparable to those of a mutant with deletion of the entire capsule biosynthetic gene operon, capBCAD, suggesting that the regulators function similarly during in vivo and in vitro growth.
Here we further investigate the expression patterns of capB and the cap gene regulators, acpA and acpB, with respect to the CO2/bicarbonate signal during culture in vitro. We also identify atxA- and CO2-controlled transcripts of capB, acpA, and acpB to further elucidate the relationships between these regulators and this important cue.
MATERIALS AND METHODS
Strains.
Table 1 contains a complete list of strains, including plasmid content and relevant genotypes. Construction of the strains was described previously (3, 6).
TABLE 1.
Strains used in this study
| Strain name | Plasmid content | Genotype | Relevant characteristic(s)a | Reference |
|---|---|---|---|---|
| UT500 | pXO1+ pXO2+ | pXO2 from 6602 transduced into 7702 | 3 | |
| UT501 | pXO1+ pXO2+ | atxA | Kmr | 3 |
| UT502 | pXO1+ pXO2+ | acpA | Spr | 3 |
| UT525 | pXO1+ pXO2+ | acpB | Kmr | 6 |
| UT526 | pXO1+ pXO2+ | acpA acpB | Spr Kmr | 6 |
Abbreviations: Kmr, kanamycin resistance; Spr, spectinomycin resistance.
Media and growth conditions.
For analysis of gene expression during growth in elevated CO2, strains were grown under conditions known to promote capsule synthesis as described previously (6, 8). NBYCO3 medium was nutrient broth yeast medium (8) supplemented with 0.8% sodium bicarbonate (wt/vol). LBgoh was Luria-Bertani (1) broth containing 0.5% glycerol to suppress sporulation. When indicated, media contained kanamycin (50 μg/ml) (Fisher Scientific, Pittsburg, PA) and/or spectinomycin (100 μg/ml) (Sigma-Aldrich, St. Louis, MO). Briefly, 30 ml of LBgoh (plus antibiotics when required) in a 250-ml flask was inoculated with vegetative cells from an NBYCO3 plate. Cultures were incubated at 30°C with agitation (200 rpm) for 12 to 14 h. Cultures were diluted into 50 ml of NBYCO3 broth (without antibiotics) to obtain an initial optical density at 600 nm (OD600) of approximately 0.1. Cultures were grown in 5% CO2 at 37°C with stirring (200 rpm) and sampled at early, mid-exponential, late-exponential, and early-stationary phases. Under these growth conditions, the parent and isogenic mutant strains had similar growth rates. For analysis of gene expression during growth in air, duplicate cultures were grown in unsupplemented NBY broth and incubated in air at 37°C with agitation (200 rpm).
Real-time Q-RT-PCR.
RNA was extracted from cultures using the protocol and reagents of the Ribopure Bacteria kit (Ambion, Austin, TX). Typically, 10 to 30 μg of RNA was obtained from 1 ml of culture. RNA preparations were treated with DNase-Free (Ambion, Austin, TX) according to the protocol of the supplier. The protocol and equipment used for quantitative RT-PCR (Q-RT-PCR) assays were described previously (6). The primer and probe sequences for the assays are shown in Table 2.
TABLE 2.
Primer and probe sequences used in Q-RT-PCR assays
| Gene | Accession no.a | Forward primer (+) and antisense primer (−) | Probe (5′FAM)b |
|---|---|---|---|
| gyrB | NC003997.3:4584-6506 | (+) ACTTGAAGGACTAGAAGCAG | CGAAAACGCCCTGGTATGTATA |
| (−) TCCTTTTCCACTTGTAGATC | |||
| atxA | NC001496.1:150042-151469 | (+) ATTTTTAGCCCTTGCAC | CTTTTATCTCTTGGAAATTCTATTACCACA |
| (−) AAGTTAATGTTTTATTGCTGTC | |||
| acpA | NC002146.1:68909-70360 | (+) ATTATCTTTACCTCAGAATCAG | CAATTTCTGAAGCCATTTCTAATCTT |
| (−) AACGTTAATGATTTCTTCAG | |||
| acpB | NC002146.1:49418-50866 | (+) TTTTTCAATACCTTGGAACT | CTTGAAGAATCATTAGGAATCTCATTACA |
| (−) AATGCCTTTTAGAAACCAC | |||
| capB | NC002146.1:56089-57483 | (+) TTTGATTACATGGTCTTCC | ATAATGCATCGCTTGCTTTAGC |
| (−) CCAAGAGCCTCTGCTAC |
Each accession number is followed by the version number and the nucleotide region for that sequencing project.
FAM, 6-carboxyfluorescein.
Nonquantitative reverse transcription-PCR.
RNA was isolated and DNase treated as described above for Q-RT-PCR. Nonquantitative RT-PCR was performed using the protocol and reagents supplied in the RETROscript kit by Ambion. The protocol has been described previously (6). Primers MD62 and MD129 were employed. Primer sequences are shown in Table 3.
TABLE 3.
Primer sequences used in primer extension and nonquantitative PCR assaysa
| Gene | Primer name | Primer sequence (5′→3′) |
|---|---|---|
| acpA | MD28 | CTGTTTGTCATGTAAATAATTCTT |
| MD33 | AGAAATGGCTTCAGAAATTG | |
| MD34 | TCGGCTAATATCTTTTTCCAT | |
| acpB | MD62 | TGTAAACCATTTTTCTTCGC |
| MD64 | TCCCTTGCTTTTAAGAATGT | |
| MD65 | GTCAGAATCCCTGGTTTGTA | |
| MD108 | GATTCAGCAGTGTTTCCAAT | |
| MD129 | AGGCCTTAATTAAAGACGAGA | |
| capB | PE2 | CCATTTCTATACTAGATGTTGCATG |
DNA oligonucleotides were purchased from Integrated DNA Technologies (Coralville, Iowa). PCRs were performed using a PE-Applied Biosystems (Norwalk, Conn.) PCR system 9700.
Primer extension.
For primer extension analysis, RNA was extracted from cultures using RNAwiz (Ambion, Austin, TX) according to the protocol provided by the supplier. Typically between 20 and 100 μg of RNA was obtained per ml of culture. The method for primer extension analysis has been described previously (13, 19). Briefly, 30 to 50 μg of RNA was incubated in the presence of end-labeled primer [γ-32P]ATP (10mCi/ml). Reverse transcription reactions were carried out with Superscript (Invitrogen, Carlsbad, CA). Primer sequences are listed in Table 3. Primers MD28, MD33, and MD34 were used for analysis of acpA transcripts. Primers MD62, MD64, MD65, and MD108 were used for analysis of acpB transcripts. For analysis of the capB gene, primer PE2 developed by Uchida et al. (22) was employed.
The 5′ ends of the acpA, acpB, and capB genes were sequenced using the fmol Sequencing kit (Promega, Madison, WI) according to the protocol of the supplier. Primers employed in the sequencing reactions were the same primers used for the corresponding primer extension reactions (listed above).
RESULTS
Quantitative and temporal assessment of CO2-enhanced capB, acpA, and acpB expression during culture.
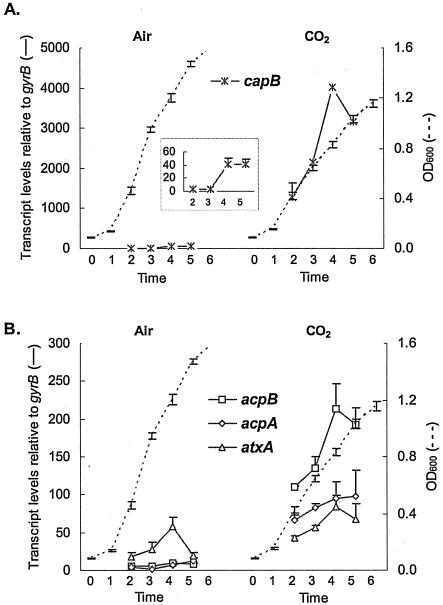
Although CO2-enhanced expression of capB, acpA, and acpB has been reported, the expression patterns of the regulators and the capsule biosynthesis gene during culture were not known. We used Q-RT-PCR (Taqman) to accurately measure capB transcript levels during growth in air and in 5% atmospheric CO2 (Fig. 1A). The capB gene is the first gene in the capsule biosynthetic gene operon. Throughout growth, capB transcript levels were 57- to 448-fold higher when cells were cultured in the presence of 5% CO2, compared to cells grown in air. capB transcription was extremely low during growth in air, but increased 12- to 15-fold as the culture entered the late-exponential growth phase (Fig. 1A, insert). The highest capB transcript levels observed during growth in air were still remarkably less than levels observed at any time throughout growth in elevated CO2.
FIG. 1.
Real-time transcript levels detected during growth in elevated CO2 and during growth in air for (A) capB and (B) atxA, acpA, and acpB. The transcript levels shown represent four different data sets that were normalized to gyrB transcript levels. A representative growth curve is shown for each experiment. The inset in panel A shows capB transcript levels using a different scale.
We used the same method to assess the relative transcript levels of atxA, acpA, and acpB throughout growth in the presence of elevated CO2 and during growth in air (Fig. 1B). Q-RT-PCR results revealed that during growth in air, acpA and acpB expression was relatively unchanged and very low. atxA was expressed at a higher level than either acpA or acpB (two- to fivefold), and expression levels were highest at late-exponential phase under this growth condition. The expression of acpA and acpB was markedly higher in 5% CO2 than during culture in air. Expression of the acpA and acpB genes was induced 16- to 23-fold and 7- to 59-fold, respectively, depending on the growth phase. In contrast, the presence of CO2 only affected atxA gene transcription approximately 2.5-fold.
The capB gene has atxA-regulated and atxA-independent transcriptional start sites.
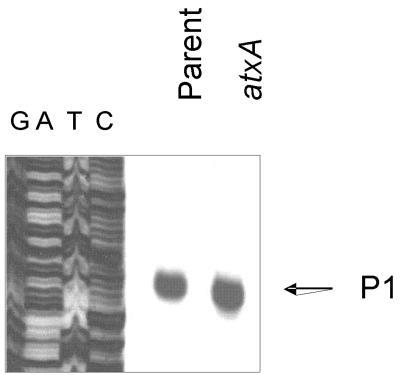
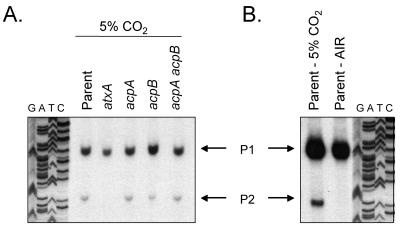
A previous report indicated two major transcriptional start sites for capB, P1 and P2, located 731 and 625 bp upstream of the translational start, respectively (Fig. 2) (22). These results were obtained from experiments employing strains lacking both virulence plasmids and carrying capBCA and acpA or atxA on multicopy vectors. The data indicated that P1 and P2 were atxA and acpA regulated, but acpA had a much larger effect on transcription than atxA (22). We performed capB primer extension reactions using the same primer as Uchida et al. (PE2) (22), and RNA was obtained from the pXO1+ pXO2+ strain UT500 and isogenic mutants deleted for specific regulatory genes. All cultures were grown to mid-exponential phase in air or in elevated CO2.
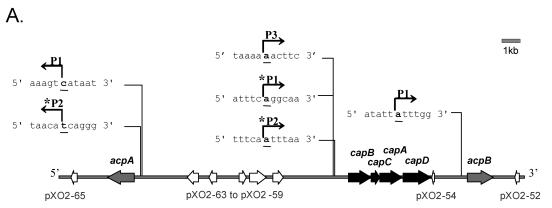
FIG. 2.
(A) Model showing transcriptional start sites for the capsule biosynthetic gene operon (capBCAD) and the cap gene regulators, acpA and acpB. (B) DNA sequence of the region upstream of capB. The predicted translational start site is underlined and capitalized. Nucleotides corresponding to the transcriptional start sites are in bold and capitalized. * denotes atxA/CO2-regulated start site. P1 and P2 upstream of capB were reported previously by Uchida and coworkers (17).
In contrast to the previous report, we found that transcripts mapping to P1 and P2 were reduced in the atxA mutant but unaffected in the acpA mutant (Fig. 3A). Transcription from P1 and P2 was also unaffected in the acpB mutant. Nonetheless, steady-state levels of transcripts originating at P1 and P2 decreased appreciably in the acpA acpB double mutant. These results are in accord with our previously established model for capB gene regulation (6) in which one of the two regulators, acpA or acpB, is required for atxA-mediated trans-activation of capB transcription. Deletion of either acpA or acpB does not significantly affect capB transcripts because the two regulators have some partial functional similarity.
FIG. 3.
Primer extension analysis of capB transcripts. PE2 primer was employed (22). (A) RNA was extracted from cells grown in 5% CO2. Lane 1, UT500 (Parent); lane 2, UT501 (atxA); lane 3, UT502 (acpA); lane 4, UT525 (acpB); lane 5, UT526 (acpA acpB). (B) RNA was extracted from cells as shown. Lane 1, UT500 (Parent) grown in 5% CO2; lane 2, UT500 grown in air.
Our data also revealed a previously unidentified transcriptional start site, P3, that is located 796 bp upstream from the translational start site of capB (Fig. 2). P3 transcription is not affected by acpA or acpB (Fig. 3). We observed a slight but reproducible increase in P3 transcription in the atxA-null mutant, suggesting that other factors may be involved in regulation of transcription from this start site. It is unlikely that P1 and P2 transcripts are the result of atxA-dependent processing of the P3 transcript since P3 transcription remained unchanged in the acpA acpB mutant as P1 and P2 transcripts decreased significantly. Other bands (unlabeled in Fig. 3) were visualized consistently but did not appear to be affected by the capsule gene regulators. It is likely that these minor bands correspond to regions of RNA with secondary structure that lead to pausing of reverse transcriptase. The B. anthracis genome is A-T rich (65% for the chromosome, 67.5% for pXO1, and 67% for pXO2) (17). Alternatively, the bands may represent processed P3 transcripts.
atxA has been linked to CO2-enhanced gene expression in B. anthracis (11, 13). Previous reports of toxin gene expression have indicated atxA/CO2-regulated transcriptional start sites for lef, cya, and pagA (4, 13). The lef and cya genes each have one apparent start site that is atxA dependent and CO2 induced (4). The pagA gene has two apparent start sites; one is constitutively expressed at a low level, while the other is atxA dependent and CO2 induced (13). To determine whether the atxA-regulated transcriptional start sites observed for the capB gene were also influenced by CO2, we compared the primer extension results obtained using RNA from the parent strain cultured in air and RNA isolated from the parent strain grown in elevated CO2 (Fig. 3B). Levels of transcripts corresponding to both atxA-regulated start sites, P1 and P2, were elevated during growth in 5% CO2. Thus, the low level of capB transcript detected during aerobic growth (Fig. 1A) most likely correlates with transcripts initiating at P3.
We searched for consensus sequences upstream of the capB transcriptional start sites to reveal putative promoter regions. Canonical −10 and −35 sequences typical of B. subtilis σA promoters (10) were not apparent upstream of the P3 start site (Fig. 2B), suggesting that additional regulatory mechanisms may be involved in controlling transcription at this start site. We noted a potential −10 but not a −35 consensus sequence upstream of P1. Prototypical −10 and −35 sequences were not readily identifiable for the P2 start site, indicating that P2 may be a breakdown product of P1, as suggested previously by Uchida (22).
acpB is cotranscribed with capD.
acpB transcript levels decrease significantly (15-fold) in the absence of atxA (6). We performed 5′-end mapping experiments using RNA isolated from the parent strain and the atxA-null mutant to map atxA-regulated and atxA-independent transcriptional start sites for acpB and revealed potential promoter sequences. Using various primers (Table 3), we identified a single transcriptional start site, P1, located 310 bp upstream from the translational start codon (Fig. 2A and Fig. 4). A weak −10 promoter sequence (GATAAT) was identified upstream of this start site, but a canonical −35 sequence was not readily discernible. Surprisingly, the steady-state level of this transcript was unchanged in the atxA mutant.
FIG. 4.
Primer extension analysis of acpB transcripts. MD65 primer was employed (See Materials and Methods). RNA was extracted from cells grown in 5% CO2. Lane 1, UT500; lane 2, UT501 (atxA).
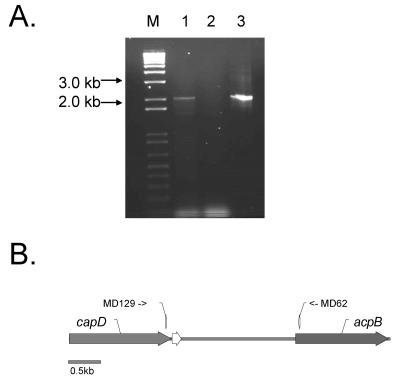
The acpB gene is located approximately 2 kb downstream from and in the same orientation as the cap gene operon (17) (Fig. 2A). The inability to identify an atxA-regulated transcriptional start site in the region between capD, the last gene of the capsule biosynthetic gene operon and acpB suggested that acpB may be cotranscribed with capD and that atxA-regulation of acpB results from transcription associated with the capBCAD operon. We employed RT-PCR to test for transcripts extending from capD to acpB. Using primers corresponding to sequences of the capD and acpB genes we obtained the amplicon shown in Fig. 5, indicating cotranscription of the two genes. Primer extension experiments employing various primers corresponding to the capBCAD coding sequence did not reveal transcripts with 5′ ends mapping within the region (data not shown). Taken together, the data indicate that acpB is cotranscribed with capD and suggest that atxA-mediated regulation of acpB occurs via the atxA-dependent P1 and P2 start sites of capB (Fig. 2).
FIG. 5.
RT-PCR of the capD-acpB region. (A) capD-acpB transcript was detected. Lane M, DNA markers with sizes as indicated; lane 1, cDNA template plus reverse transcriptase; lane 2, cDNA template minus reverse transcriptase; lane 3, DNA template, PCR control. (B) Illustration showing location of primers MD62 and MD129 employed in the PCRs used in panel A. The open arrow indicates the position of pXO2-54.
The acpA gene has atxA-regulated and atxA-independent transcriptional start sites
. We performed a similar analysis of the other atxA-controlled cap regulator, acpA. Primer extension analysis of the acpA gene revealed two major transcriptional start sites, P1 and P2, located 379 and 353 bp, respectively, upstream from the translational start (Fig. 2A and 2B and Fig. 6A). P1 expression was unaffected by atxA, yet P2 expression was atxA-regulated since P2 transcripts were undetectable in the atxA-null mutant (Fig. 6A). There was no evidence of acpA autoregulation as P1 and P2 transcription did not vary in the acpA mutant samples. Similarly, in an acpB mutant and an acpA acpB double mutant, the acpA P1 and P2 transcript levels did not change, indicating that atxA-mediated regulation of acpA does not appear to require any of these factors. Similar to the results obtained for the capB gene, transcripts from the atxA-regulated start site of acpA, P2, but not the atxA-independent start site, P1, were increased during growth in elevated CO2 (Fig. 6B). Canonical −10 and −35 promoter sequences could not be readily identified upstream of either the P1 or P2 site, indicating that the expression of acpA is most likely sigma A independent.
FIG. 6.
Primer extension analysis of acpA transcripts. MD28 primer was employed. (A) RNA was extracted from cells grown in 5% CO2. Lane 1, UT500 (Parent); lane 2, UT501 (atxA); lane 3, UT502 (acpA); lane 4, UT525 (acpB); lane 5, UT526 (acpA acpB). (B) RNA was extracted from cells as shown. Lane 1, UT500 (Parent) grown in 5% CO2; lane 2, UT500 grown in air.
DISCUSSION
In the work described here, we characterized transcription of capB, the first gene of the B. anthracis capsule biosynthetic operon; acpA and acpB, functionally similar cap regulators; and atxA, a master regulator controlling acpA and acpB expression. In light of the complex regulation of capsule synthesis, we wanted to (i) assess temporal expression of capB and the regulators in batch culture, (ii) examine expression of the genes in response to elevated CO2, a signal considered to be important in the host; and (iii) establish the apparent transcriptional start sites for these genes, distinguishing regulated and nonregulated transcripts.
Capsule synthesis and capB gene expression correlate directly. When B. anthracis is grown in nutrient broth yeast medium (NBY) supplemented with 0.8% bicarbonate in an atmosphere of 5% CO2, capsule is first observed during the mid-exponential phase and cells with the largest capsule diameter are visualized at the early stationary phase (6). The quantitiative RT-PCR analyses performed here indicate that capB gene expression was several hundred-fold higher during growth in elevated CO2 than that observed during growth in air. The highest level of capB transcript observed during growth in 5% CO2 was detected at mid- to late-exponential phase, just before cells become fully capsulated. capB transcription is detected throughout growth in air, yet microscopic examination of India ink preparations of the parent strain do not show capsule on the surface of cells grown in this environment. This suggests that a minimal level of cap transcript must be attained before capsule can be readily identified on the bacterial cell surface. Temporal expression patterns of acpA and acpB indicated that the transcript levels for both regulators in cultures grown in 5% CO2 were relatively high at early exponential phase and increased further as the culture grew. These data suggest that the CO2 signal is sensed quickly by the bacterium, leading to a rapid increase in expression of both acpA and acpB that subsequently leads to induction of capB.
Two apparent transcriptional start sites for capB were identified previously (22). However, the regulation of these start sites did not completely agree with the current model for capsule gene regulation in a genetically complete strain. In the work described here, we have established the apparent transcriptional start sites for capB, distinguishing regulated and nonregulated transcripts, in a pXO1+ pXO2+ strain. Results of primer extension of capB transcripts confirmed two atxA-regulated transcriptional start sites, P1 and P2, as reported previously by Uchida (22). However, in contrast to the previous report, P1 and P2 transcript levels were unaffected in the acpA mutant. It is likely that our findings differ from previously published data because the steady-state levels of atxA and acpA are extremely important for regulation of capB expression. In the previous study (22), elevated P1 and P2 transcription was observed in a pXO1− pXO2− strain containing the capBCA genes and atxA or acpA cloned on high-copy vectors. Taken together, the data suggest that overexpression of acpA can lead to an increase in P1 and P2 transcription in the absence of atxA. In addition, overexpression of atxA may bypass the requirement for acpA or acpB for cap gene activation. We determined that in the pXO1+ pXO2+ acpA acpB strain, cap gene expression is significantly reduced and the strain is noncapsulated, indicating that the levels of atxA normally present in a genetically complete strain are not sufficient for positive regulation of P1 and P2 transcription in the absence of acpA and acpB. The low levels of P1 and P2 transcripts detected in the atxA mutant are likely the result of positive regulation by the low levels of acpA and acpB transcripts present in an atxA mutant. Finally, the nonregulated capB transcription start site P3, was not noted previously by Uchida and coworkers (22). The basal level of capB transcription observed in the noncapsulated acpA acpB mutant and during growth in air most likely results from transcription at this site. A slight increase in P3 transcript was observed in the atxA mutant suggesting an additional level of control.
Primer extension of acpA transcripts revealed an atxA/CO2-regulated transcript (P2) and an atxA-independent transcript (P1). P1 transcription is evident during growth in air and in the absence of atxA. atxA-regulated P2 transcription is induced in the presence of elevated CO2. atxA-independent expression of acpB results from transcription initiating at a start site (P1) immediately upstream of acpB, whereas atxA-mediated regulation of acpB expression occurs via read-through transcription from capD. Using primer extension analysis, we were unable to demonstrate mRNAs with 5′ ends mapping within the capBCAD region (data not shown), indicating that atxA-regulated acpB expression may be attributed to the atxA-regulated promoters P1 and P2 of capB. A capBCAD-acpB transcript would result in a positive feedback loop for acpB expression. Nevertheless, data supporting the existence of a 9-kb capBCAD-acpB mRNA molecule are lacking. Northern hybridization experiments reported by Makino et al. (15) reveal a 6-kb transcript associated with capBCAD. Our quantitative reverse transcription-PCR analyses of UT500 show that capB expression is 20- to 50-fold higher than expression of acpB (Fig. 1) (6), but analysis of sequences in the capBCADacpB region does not reveal potential transcription terminators. Further work will address the stability of a possible mRNA corresponding to capBCAD-acpB.
The data described here and work published previously suggest that acpA and acpB levels are limiting for capB transcription in the absence of atxA and/or elevated CO2 (6, 22, 23). B. anthracis strains cultured in air are normally noncapsulated, but overexpression of acpA in a pXO1− pXO2+ strain leads to capsule production during growth in air (22). Additionally, capB primer extension experiments employing RNA from a pXO1− pXO2− strain with the capBCA genes and acpA cloned on high-copy vectors revealed high levels of transcripts mapping to P1 and P2 in the absence of atxA (22).
Transcriptional regulation of atxA is unlike that observed for acpA and acpB. We only observed a small increase (2.5-fold) in atxA transcript levels during growth in elevated CO2 compared to that observed during growth in air. atxA has a single transcriptional start site, and a previous study employing primer extension reactions and Western hybridizations indicated no difference in atxA expression during growth in elevated CO2 versus growth in air (4). Consensus sequences for promoter recognition by sigma A RNA polymerase (−10 and −35 sequences) have been noted upstream of atxA. Such sequences have not been found upstream of acpA, acpB, or capBCAD. Finally, some data suggest that, in contrast to acpA and acpB, atxA levels are not limiting for target gene expression. Dai et al. (4) demonstrated that overexpression of atxA actually leads to a decrease in expression of pagA and decreased toxin synthesis, and Sirard et al. (18) showed that a second copy of the pagA promoter cloned on pXO1did not affect expression of the pagA gene at the normal pXO1 locus.
The complex regulation of capsule gene expression in B. anthracis, which includes the CO2 signal, a regulatory cascade with functionally similar regulators, multiple regulated and nonregulated transcription start sites, and a potential positive feedback loop, is made even more intriguing by the amino acid sequence similarity of the three regulatory proteins. The master regulator, AtxA, is approximately 50% similar to the cap regulators AcpA and AcpB. AcpA and AcpB are 62% similar to each other. In all cases, the amino acid sequence similarity is throughout the proteins and not limited to a specific region or predicted domain. Secondary and tertiary protein structure predictions suggest that the proteins are soluble, basic proteins. They do not contain strong motifs indicative of nucleic acid binding; however, BLAST results indicate some amino acid homology to the transcriptional regulator Mga (40 to 45% similar) of Streptococcus pneumoniae and BglG (40 to 45% similar), an antiterminator protein in Escherichia coli. Mga has been shown to bind directly to the promoter sequences of the genes that it activates (16), while BglG is an antiterminator protein that binds to the leader sequence of mRNA to allow read-through of its target genes (12). Neither DNA nor RNA binding activities have been ascribed to the B. anthracis proteins. Further investigations will be directed toward discovery of the molecular functions of these unique regulators and the mechanism by which their expression and/or function is linked to the CO2/bicarbonate signal.
Acknowledgments
The assays, reactions, and initial data analysis for real-time PCR were performed by the Quantitative Genomics Core Laboratory in the Department of Integrative Biology and Pharmacology at the University of Texas Health Science Center—Houston.
This work was supported by Public Health Service grant AI33537 from the National Institutes of Health and the Department of Army grant DAMD 17-01-2-0047.
REFERENCES
- 1.Ausubel, F. M. 1993. Current protocols in molecular biology. Greene Publishing Associates and Wiley-Interscience, New York, N.Y.
- 2.Bartkus, J. M., and S. H. Leppla. 1989. Transcriptional regulation of the protective antigen gene of Bacillus anthracis. Infect. Immun. 57:2295-2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bourgogne, A., M. Drysdale, S. G. Hilsenbeck, S. N. Peterson, and T. M. Koehler. 2003. Global effects of virulence gene regulators in a Bacillus anthracis strain with both virulence plasmids. Infect. Immun. 71:2736-2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dai, Z., J. C. Sirard, M. Mock, and T. M. Koehler. 1995. The atxA gene product activates transcription of the anthrax toxin genes and is essential for virulence. Mol. Microbiol. 16:1171-1181. [DOI] [PubMed] [Google Scholar]
- 5.Davson, H., and M. B. Segal. 1975. Introduction to physiology, p. 81-112. Academic Press Inc., London, United Kingdom.
- 6.Drysdale, M., A. Bourgogne, S. G. Hilsenbeck, and T. M. Koehler. 2004. atxA controls Bacillus anthracis capsule synthesis via acpA and a newly discovered regulator, acpB. J. Bacteriol. 186:307-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drysdale, M., S. Heninger, J. Hutt, Y. Chen, C. Lyons, and T. M. Koehler. 2005. Capsule synthesis by Bacillus anthracis is required for dissemination in a murine inhalation anthrax. EMBO J. 12:221-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Green, B. D., L. Battisti, T. M. Koehler, C. B. Thorne, and B. E. Ivins. 1985. Demonstration of a capsule plasmid in Bacillus anthracis. Infect. Immun. 49:291-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guignot, J., M. Mock, and A. Fouet. 1997. AtxA activates the transcription of genes harbored by both Bacillus anthracis virulence plasmids. FEMS Microbiol. Lett. 147:203-207. [DOI] [PubMed] [Google Scholar]
- 10.Helmann, J. D., and C. P. Moran, Jr. 2002. RNA polymerase and sigma factors, p. 289-312. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 11.Hoffmaster, A. R., and T. M. Koehler. 1997. The anthrax toxin activator gene atxA is associated with CO2-enhanced non-toxin gene expression in Bacillus anthracis. Infect. Immun. 65:3091-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Houman, F., M. R. Diaz-Torres, and A. Wright. 1990. Transcriptional antitermination in the bgl operon of E. coli is modulated by a specific RNA binding protein. Cell 62:1153-1163. [DOI] [PubMed] [Google Scholar]
- 13.Koehler, T. M., Z. Dai, and M. Kaufman-Yarbray. 1994. Regulation of the Bacillus anthracis protective antigen gene: CO2 and a trans-acting element activate transcription from one of two promoters. J. Bacteriol. 176:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Makino, S.-I., I. Uchida, N. Terakado, C. Sasakawa, and M. Yoshikawa. 1989. Molecular characterization and protein analysis of the cap region, which is essential for encapsulation in Bacillus anthracis. J. Bacteriol. 171:722-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makino, S.-I., M. Watarai, H. I. Cheun, T. Shirahata, and I. Uchida. 2002. Effect of the lower molecular capsule released from the cell surface of Bacillus anthracis on the pathogenesis of anthrax. J. Infect. Dis. 186:227-233. [DOI] [PubMed] [Google Scholar]
- 16.Perez-Casal, J., M. G. Caparon, and J. R. Scott. 1991. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J. Bacteriol. 173:2617-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baillie, I. T. Paulsen, K. E. Nelson, H. Tettelin, D. E. Fouts, J. A. Eisen, S. R. Gill, E. K. Holtzapple, O. A. Okstad, E. Helgason, J. Rilstone, M. Wu, J. F. Kolonay, M. J. Beanan, R. J. Dodson, L. M. Brinkac, M. Gwinn, R. T. DeBoy, R. Madpu, S. C. Daugherty, A. S. Durkin, D. H. Haft, W. C. Nelson, J. D. Peterson, M. Pop, H. M. Khouri, D. Radune, J. L. Benton, Y. Mahamoud, L. Jiang, I. R. Hance, J. F. Weidman, K. J. Berry, R. D. Plaut, A. M. Wolf, K. L. Watkins, W. C. Nierman, A. Hazen, R. Cline, C. Redmond, J. E. Thwaite, O. White, S. L. Salzberg, B. Thomason, A. M. Friedlander, T. M. Koehler, P. C. Hanna, A. B. Kolsto, and C. M. Fraser. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423:81-86. [DOI] [PubMed] [Google Scholar]
- 18.Sirard, J.-C., M. Mock, and A. Fouet. 1994. The three Bacillus anthracis toxin genes are coordinately regulated by bicarbonate and temperature. J. Bacteriol. 176:5188-5192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsui, H. C. T., A. J. Pease, T. M. Koehler, and M. E. Winkler. 1994. Detection and quantitation of RNA transcribed from bacterial chromosomes and plasmids, p. 179-204. In K. W. Adolph (ed.), Molecular microbiology techniques, part A. Academic Press Inc., San Diego, Calif.
- 20.Uchida, I., J. M. Hornung, C. B. Thorne, K. R. Klimpel, and S. H. Leppla. 1993. Cloning and characterization of a gene whose product is a trans-activator of anthrax toxin synthesis. J. Bacteriol. 175:5329-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uchida, I., S. Makino, C. Sasakawa, M. Yoshikawa, C. Sugimoto, and N. Terakado. 1993. Identification of a novel gene, dep, associated with depolymerization of the capsular polymer in Bacillus anthracis. Mol. Microbiol. 9:487-496. [DOI] [PubMed] [Google Scholar]
- 22.Uchida, I., S. Makino, T. Sekizaki, and N. Terakado. 1997. Cross-talk to the genes for Bacillus anthracis capsule synthesis by atxA, the gene encoding the trans-activator of anthrax toxin synthesis. Mol. Microbiol. 23:1229-1240. [DOI] [PubMed] [Google Scholar]
- 23.Vietri, N. J., R. Marrero, T. A. Hoover, and S. L. Welkos. 1995. Identification and characterization of a trans-activator involved in the regulation of encapsulation by Bacillus anthracis. Gene 152:1-9. [DOI] [PubMed] [Google Scholar]