Abstract
The core oligosaccharide component of the lipopolysaccharide can be subdivided into inner and outer core regions. In Escherichia coli, the inner core consists of two 3-deoxy-d-manno-octulosonic acid and three glycero-manno-heptose residues. The HldE protein participates in the biosynthesis of ADP-glycero-manno-heptose precursors used in the assembly of the inner core. HldE comprises two functional domains: an N-terminal region with homology to the ribokinase superfamily (HldE1 domain) and a C-terminal region with homology to the cytidylyltransferase superfamily (HldE2 domain). We have employed the structure of the E. coli ribokinase as a template to model the HldE1 domain and predict critical amino acids required for enzyme activity. Mutation of these residues renders the protein inactive as determined in vivo by functional complementation analysis. However, these mutations did not affect the secondary or tertiary structure of purified HldE1, as judged by fluorescence spectroscopy and circular dichroism. Furthermore, in vivo coexpression of wild-type, chromosomally encoded HldE and mutant HldE1 proteins with amino acid substitutions in the predicted ATP binding site caused a dominant negative phenotype as revealed by increased bacterial sensitivity to novobiocin. Copurification experiments demonstrated that HldE and HldE1 form a complex in vivo. Gel filtration chromatography resulted in the detection of a dimer as the predominant form of the native HldE1 protein. Altogether, our data support the notions that the HldE functional unit is a dimer and that structural components present in each HldE1 monomer are required for enzymatic activity.
Lipopolysaccharide (LPS), an amphipathic glycolipid in the outer leaflet of the outer membrane of gram-negative bacteria, consists of lipid A and a core oligosaccharide. Some bacteria produce an additional surface-exposed, O-specific polysaccharide attached to the reducing end of the lipid-A core (for a recent review, see reference 22). Lipid A, which is essential for outer membrane stability and bacterial cell viability, consists of two β-1,6-linked glucosamine residues that are phosphorylated and acylated with a variable number of fatty and hydroxy fatty acids (22). The core oligosaccharide can be subdivided into inner and outer core domains. In Escherichia coli K-12 and other enteric bacteria, the outer core usually contains hexoses and hexosamines, while components of the inner core generally include two residues of 3-deoxy-d-manno-octulosonic acid and, depending on the particular bacterial species, two or three residues of l-glycero-d-manno-heptose (10).
LPS contributes to maintaining the structural integrity of the bacterial outer membrane by interacting with outer membrane proteins and divalent cations (8, 9, 21). Phosphate groups covalently attached at various positions on the heptoses facilitate electrostatic interactions with divalent cations, thereby providing for membrane stability and restricted permeability. Thus, E. coli mutants lacking heptoses in the LPS not only exhibit a much shorter core oligosaccharide but also display a wide range of pleiotropic phenotypes due to the reduced stability of the outer membrane. These phenotypes include hypersensitivity to novobiocin and other hydrophobic antibiotics, detergents, and bile salts. Also, these mutants are poor recipients for both plasmid conjugation and generalized transduction (for a recent review, see reference 30). These phenotypes, at least in part, depend on the absence of phosphate groups, since mutations in genes encoding LPS core oligosaccharide kinases also cause pleiotropic properties similar to those found in heptose-deficient mutants but do not affect the formation of a complete core oligosaccharide (32, 33). Heptose-deficient LPS mutants of many bacterial species can survive in the laboratory. However, for some microorganisms, such as Pseudomonas aeruginosa, heptoseless mutants have not been isolated, suggesting that these residues (or the phosphates covalently attached to them) are essential for bacterial survival in vitro (31). Heptose-deficient mutants are also serum sensitive and display reduced virulence in experimental infection models (11, 35). Thus, heptose biosynthetic genes and their products are potentially attractive targets for developing novel antimicrobial compounds.
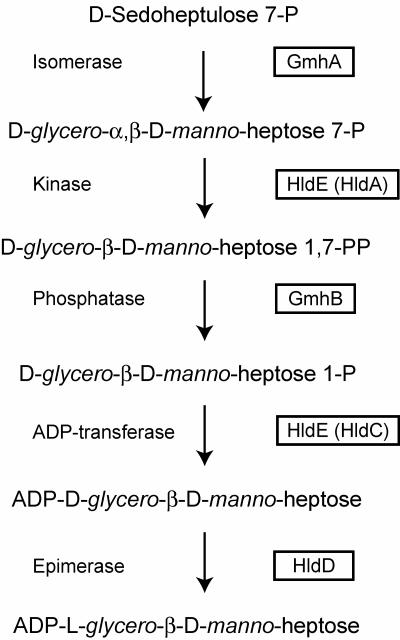
The pathways for the biosynthesis of nucleotide-activated glycero-manno-heptose in gram-positive and gram-negative bacteria have recently been elucidated, revealing the presence in both cases of unique kinase-phosphatase reactions (13, 14). Similar reactions are absent in the classical pathways for the formation of nucleotide-activated sugars, which typically involve a mutase step catalyzing the intramolecular transfer of a phosphate group from the distal carbon to the C-1 position. This phosphate is subsequently modified to form a phosphodiester linkage with the nucleotide by a nucleotidylyltransferase, which results in the synthesis of a nucleotide diphosphate-sugar precursor. Two different biosynthetic pathways, the d-α-d-heptose and the l-β-d-heptose pathways (Fig. 1), can be distinguished on the basis of the specificity of the kinase and the nucleotidylyltransferase steps, while the isomerase and phosphatase enzymes are common to both pathways (30).
FIG. 1.
Biosynthesis pathway of ADP-l-glycero-d-manno-heptose. The protein designations are indicated in the boxes. HldA and HldC are reserved only for those cases where the enzymes are individually encoded by independent genes (30).
In many gram-negative bacteria, the heptokinase and adenylyltransferase activities are contributed by HldE. In the presence of ATP, HldE converts purified glycero-manno-heptose 7-phosphate, prepared from sedoheptulose 7-phosphate and the Aneuribacillus thermoaerophilus GmhA isomerase, into a product consistent with glycero-manno-heptose 1,7-biphosphate (14). HldE is a bifunctional protein with two clearly distinguishable functional domains (29). The N-terminal domain of HldE (HldE1) corresponds to a polypeptide related to the ribokinase superfamily, while the C-terminal domain (HldE2) belongs to the cytidylyltransferase superfamily and presumably carries the ADP transfer activity (28).
In this work, we have compared the HldE1 polypeptide with members of the ribokinase family for which structural information is available. We have modeled the HldE1 protein with the structure of its closest homolog, the E. coli ribokinase, and predicted residues in HldE1 that would be critical for catalysis and ATP binding. We introduced conservative amino acid substitutions in these residues, which abolished enzymatic activity as determined by in vivo complementation of an ΔhldE1 mutant. We also provide direct genetic and biochemical evidence that the HldE heptokinase functions as a dimer.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The properties of the strains and plasmids used in this study are summarized in Table 1. Bacteria were cultured in Luria broth, supplemented when necessary with ampicillin, kanamycin, novobiocin, or trimethoprim at final concentrations of 100 μg ml−1, 25 μg ml−1, 50 μg ml−1, and 50 μg ml−1, respectively. E. coli DH5α was used for maintenance and propagation of plasmids. E. coli strain SØ874 was employed as the parental strain for the construction of deletion mutants FAM2 (ΔhldE), FAM3 (ΔhldE1), and FAM4 (ΔhldE2) by the one-step replacement method described by Datsenko and Wanner (7). The partially or completely deleted hldE gene in these mutants was replaced by the nonpolar kanamycin resistance cassette amplified from pKD4 (7). The gene replacements in the mutants were verified by PCR amplifications. The DNA sequences of the primers used in this study are available from the authors upon request.
TABLE 1.
Characteristics of strains and plasmids used in this study
| Strain or plasmid | Relevant propertiesa | Source or reference |
|---|---|---|
| Strains | ||
| DH5α | F− φ80lacZ M15 endA recA hsdR(rK− mK−) supE thi gyrA relA Δ(lacZYA-argF)U169 | Laboratory stock |
| W3110 | E. coli K-12 rph-1 IN(rrnD-rrnE)1 | Laboratory stock |
| SØ874 | lacZ trp Δ(sbcB-rfb) upp relA rpsL λ− | Laboratory stock |
| FAM2 | SØ874 ΔhldE | This study |
| FAM3 | SØ874 ΔhldE1 (deletion of the heptokinase domain) | This study |
| FAM4 | SØ874 ΔhldE2::Km (deletion of the ADP-transferase domain) | This study |
| Plasmids | ||
| pCP20 | FLP+, λ cI857+, λ pR Repts, Apr, Cmr | 7 |
| pFM1 | pGEX2T, gst-hldE | This study |
| pFM2 | pGEX2T, gst-hldE2 | This study |
| pFM3 | pGEX2T, gst-hldE1 | This study |
| pFM5 | pFM3, gst-hldE1D264N | This study |
| pFM6 | pFM3, gst-hldE1D264E | This study |
| pFM7 | pFM3, gst-hldE1N195E | This study |
| pFM8 | pFM3, gst-hldE1E198D | This study |
| pFM25 | pFM1, gst-hldEN195E | This study |
| pFM34 | pSCRhaB2, hldE1-FLAG | This study |
| pGEX-2T | Cloning vector for the construction and expression of GST-fused proteins | Amersham Biosciences |
| pKD4 | Template plasmid for mutagenesis, Apr, Kmr | 7 |
| pKD46 | γ, β, and exo from γ phage, araC-ParaB, Apr | 7 |
| pSCRhaB2 | pMLBAD (15) with the PRHA (rhamnose-inducible promoter), Tpr | S. T. Cardona |
Km, kanamycin; Tp, trimethoprim; Cm, chloramphenicol.
Construction and purification of HldE1 (heptokinase domain) protein derivatives.
The entire HldE protein, the HldE1 domain, and their mutant protein derivatives were all purified as glutathione S-transferase (GST) fusions. For this purpose, a PCR product spanning the coding region of the hldE gene was cloned into the BamHI and SmaI sites of the GST gene fusion vector pGEX-2T. This experiment resulted in pFM1, which encodes the entire HldE protein N terminally fused to GST and an intervening thrombin protease site. This plasmid was used as a template to amplify only the regions encoding the GST tag and the adenylyltransferase domain in a manner such that the amplification progressed through the backbone of the plasmid, deleting HldE1. BamHI recognition sites were added to each primer to ensure that the GST and hldE2 genes fused in frame. This procedure gave rise to pFM2, which encodes a GST-HldE2 fusion protein. The plasmid pFM3 was made using the same strategy, except that in this case the GST-HldE1 fusion was preserved while the region of the hldE gene encoding the HldE2 domain was deleted. The PCRs were carried out with a long-template PCR kit (Roche), using 2 μM of each primer and 200 μM deoxynucleoside triphosphates. The conditions for amplification were as follows: 93°C for 2 min; 10 cycles of 93°C for 10 s, 63°C for 30 s, and 68°C for 4.5 min; 16 cycles of 93°C for 10 s, 63°C for 30 s, and 68°C initially for 4.5 min and adding 20 s per cycle thereafter; and a final extension at 68°C for 7 min. The plasmid pFM3 was used as a template for the construction of plasmids encoding HldE1 domain mutant proteins with the following amino acid substitutions: D264N (pFM5), D264E (pFM6), N195D (pFM7), and E198D (pFM8). In a similar fashion, pFM1 (hldE) was used as a template to make pFM25, which encodes a protein with the N195D replacement. These amino acid replacements were introduced using a site-directed mutagenesis kit (Stratagene) according to the supplier's instructions. The presence of the correct mutation in each of the plasmids was verified by DNA sequencing of the insert. The plasmid pFM34, encoding HldE1 with a C-terminal FLAG epitope fusion, was constructed using pSCRhaB2, a vector in which the expression of the cloned gene is under the control of a rhamnose-inducible promoter (5).
Recombinant fusion proteins were purified with GSTrap columns (Amersham-Pharmacia) according to the protocol suggested by the supplier. For copurification experiments, bacterial cell pellets were washed with phosphate-buffered saline and resuspended in 5 mM Tris-HCl, pH 7.0. Bacteria were lysed by three passages through a French press. After centrifugation to remove cell debris, the lysate was passed through the GSTrap. The column was washed with 50 column volumes of 5 mM Tris-HCl, pH 7.0, and the protein was eluted in 5 mM Tris-HCl, 10 mM reduced glutathione, pH 8.0.
Protein concentrations were determined by the Bradford assay (Bio-Rad Laboratories), and purity was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining. The GST tag was removed from the rest of the protein by treatment with thrombin.
Analytical methods.
Western blotting was performed using a rabbit anti-GST polyclonal antibody from Molecular Probes and a FLAG M2 monoclonal antibody (Sigma). Reacting bands were visualized by fluorescence with an Odyssey infrared imaging system (LI-COR Biosciences) using IRDye800CW affinity-purified anti-rabbit immunoglobulin G (Rockland Immunochemicals) and Alexa Fluor 700-conjugated anti-mouse immunoglobulin G antibodies (Molecular Probes).
Complementation experiments.
Strains FAM2, FAM3, and FAM4, carrying deletions of the hldE gene, were transformed with the various plasmids constructed in this work (Table 1). The presence or absence of the heptoseless phenotype was evaluated by culturing transformants on plates containing 50 μg ml−1 novobiocin. E. coli DH5α was also transformed with pFM3, pFM5, pFM6, pFM7, and pFM8 to assess the effect of mutant proteins on the function of the chromosomally encoded HldE. The optical density at 600 nm of overnight cultures was standardized to 0.1, and 10 μl of serial dilutions was plated on medium containing novobiocin, with and without IPTG (isopropyl-β-d-thiogalactopyranoside), according to the protocol of Jett et al. (12). Lipid A-core LPS was isolated as described elsewhere (19) and analyzed in 16% Tricine gels (16, 23). LPS samples were visualized by silver staining (19).
Characterization of purified proteins by spectrofluorometry, circular dichroism, and gel filtration chromatography.
Spectrofluorometric determinations based on the excitation of a single tryptophan residue in HldE1 were used to assess changes in tertiary structures of wild-type and mutant proteins, by determining the fluorescence emission at 300 to 450 nm following excitation at 295 nm. These experiments were conducted using a Fluorolog-3 spectrofluorometer (Spex Industries) with the excitation slit set to 1 nm and the emission slit set to 2 nm. Secondary structures of the wild-type and mutant proteins were examined by circular dichroism using a JASCO-JA1 spectrometer. The structure of the protein was measured from 200 nm to 250 nm, the range which demonstrates the presence of α-helices, using a 0.1-mm-path-length cell. Data were acquired at 25°C with a 100-nm averaging per minute and a 1-nm band pass. Five replicate samples were averaged for each protein. The helical content was calculated with an optimized self-organizing map algorithm (K2d program [http://www.embl-heidelberg.de/∼andrade/k2d.html]), as described by Andrade et al. (1).
Purified proteins were concentrated using a Vivaspin 6 ml concentrator (Sartorius AG, Goettingen, Germany) with a molecular mass cutoff of 5,000 Da. Concentrated samples were applied to a Supradex 200 10/300 GL column equilibrated with a buffer consisting of 250 mM HEPES and 50 mM sodium phosphate, pH 7.0. The column was connected to an AKTA fast protein liquid chromatography apparatus (Amersham Biosciences Corp., Piscataway, NJ) and eluted with the same buffer. In addition, the column was calibrated using the 12- to 200-kDa gel filtration molecular mass markers (MWGF200; Sigma Chemical Co., St. Louis, Missouri). Fractions containing peaks corresponding to various molecular masses were examined by SDS-PAGE in a 14% acrylamide gel.
Homology modeling.
Cluster analysis was performed with enzymes from the ribokinase superfamily whose structures were deposited in the Protein Data Bank (PDB) (2). Alignment of the sequence of HldE1 to those of ribokinase (PDB entry 1RKA) and human adenosine kinase (PDB entry 1BX4) was performed using ClustalW (27). Homology modeling of HldE1 to the E. coli ribokinase was performed using the MODELLER program (20).
RESULTS
HldE carries two domains that function independently.
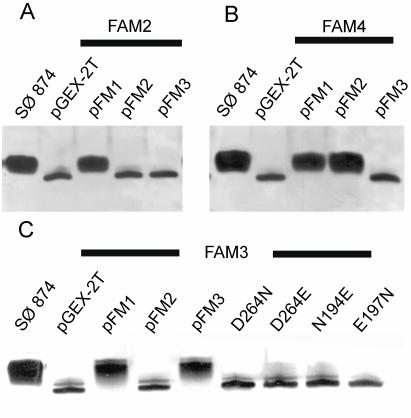
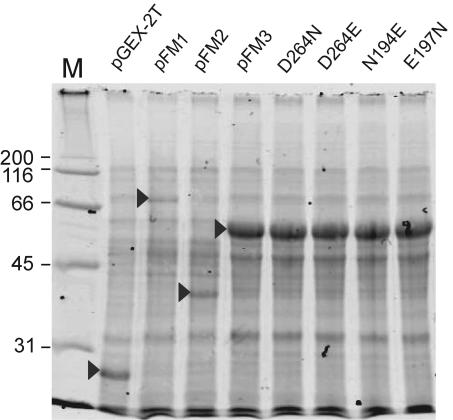
In previous work, we have shown that the E. coli HldE has two distinct domains (29). We also showed that the in vitro-transcribed and -translated HldE polypeptide has an apparent molecular mass of 55 kDa, consistent with the expected mass for HldE predicted from the sequence (29). However, a smaller polypeptide of approximately 35 kDa, corresponding to the predicted mass of the heptokinase (HldE1) domain, was also found, suggesting that the HldE protein could be processed in vivo (29). To investigate this possibility, we cloned the hldE gene in the expression fusion vector pGEXT-2T, resulting in plasmid pFM1, which encodes HldE N terminally fused to GST. Introduction of pFM1 into strain FAM2 (ΔhldE::Km) complemented the heptoseless LPS phenotype as determined by the restoration of novobiocin resistance in this strain and the formation of a wild-type lipid A-core oligosaccharide band (Fig. 2A). Analysis by SDS-PAGE of protein lysates prepared from bacteria containing pFM1 showed the presence of a polypeptide band of approximately 82 kDa (Fig. 3). This band was absent from lysates of cells transformed with the vector pGEXT-2T as a control, which revealed only a prominent 27-kDa polypeptide that was consistent with the expected mass of GST (Fig. 3). Affinity chromatography of the lysates by using GSTrap columns confirmed that the 82-kDa polypeptide was the only GST fusion product recovered (data not shown), indicating that detectable proteolysis of HldE does not occur in vivo.
FIG. 2.
Analysis of lipid A-core from parental strain and hldE mutants. The thick lines indicate the background E. coli strain used to transform recombinant plasmids encoding (as GST-fused proteins) full-length HldE (pFM1), HldE2 (ADP-heptose transferase domain; pFM2), HldE1 (heptokinase domain; pFM3), and the various mutant forms of HldE1. The background strains were SØ874 (wild type) (panels A, B, and C), FAM2 (SØ874 ΔhldE) (panel A), FAM4 (SØ874 ΔhldE2::Km) (panel B), and FAM3 (SØ874 ΔhldE1) (panel C).
FIG. 3.
Analysis of polypeptides by SDS-PAGE. Polypeptides corresponding to full-length HldE (pFM1), HldE2 (ADP-heptose transferase domain; pFM2), HldE1 (heptokinase domain; pFM3), the various mutant forms of HldE, and the GST protein fusion partner (pGEX-2T) are indicated by the arrows. All of the recombinant proteins were detected as GST-fused polypeptides. M, molecular weight markers in thousands.
We next determined whether the heptokinase and adenylyltransferase domains can function independently. Strains FAM3 and FAM4, carrying partial deletions of the hldE gene corresponding to the regions encoding the heptokinase (ΔhldE1::Km) and the adenylyltransferase (ΔhldE2::Km) domains, respectively, were constructed. In both strains, the deletions resulted in the replacement of a portion of hldE by a nonpolar kanamycin resistance cassette. Phenotypically, both mutant strains were sensitive to novobiocin and exhibited a fast-migrating core-lipid A band in Tricine-SDS polyacrylamide gels. As expected, pFM1 complemented the heptoseless phenotype in FAM3 and FAM4 (Fig. 2B and C) and also restored resistance to novobiocin. The plasmid pFM3, encoding the heptokinase domain of HldE, could complement only the heptoseless phenotype of strain FAM3 and not that of strains FAM2 and FAM4 (Fig. 2A to C). Conversely, pFM2, encoding the adenylyltransferase C-terminal domain of HldE, complemented only the heptoseless phenotype of FAM4 and did not complement the phenotypes of strains FAM2 and FAM3 (Fig. 2A to C). These results conclusively demonstrated that each domain of the HldE protein can independently function in ADP-heptose synthesis.
Cluster analysis and molecular modeling of the HldE1 (glycero-manno-heptose 7-phosphate kinase) domain.
We have previously shown that HldE1 shows strong homology with members of the ribokinase superfamily, most notably with the E. coli ribokinase (28, 29). The ribokinase superfamily includes a family of small-molecule kinases that phosphorylate carbohydrate and noncarbohydrate substrates (4, 6, 17, 25, 34). The monomeric fold of the ribokinase superfamily is well conserved, yet differences can be observed in the quaternary structures for the active forms of the proteins in this superfamily (34), which range from monomers to tetramers. A cluster analysis involving members of the ribokinase superfamily for which structural information is available showed that HldE1 grouped with enzymes whose active forms are dimers (Fig. 4), and it was most closely associated with the E. coli ribokinase (PDB entry 1RKA) and Mycobacterium adenosine kinase (18). In both enzymes, the active sites are located at the interface of the monomers (25).
FIG. 4.
Cluster tree of various members of the ribokinase superfamily with known crystal structures.
A second branching of dimers included the Salmonella enterica serovar Typhimurium 4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate kinase (6) and the sheep pyridoxal kinase (17). In these proteins, the dimers have independent active sites that are not located at the interface of the monomers. These proteins also were closely associated with a tetrameric hypothetical protein from Bacillus subtilis (PDB entry 1KYH), which has been shown to be a much more primitive member of the ribokinase superfamily (34).
The Thermotoga maritima hypothetical phosphofructokinase Tm0828 (PDB entry 1O14) and 2-keto-3-deoxygluconate kinase Tm0067 (PDB entry 1J5V) are arranged as dimers that exhibit edge-to-edge dimerization similar to that of the ribokinase (25, 34). These proteins also grouped with the trimeric 4-methyl-5-β-hydroxy-ethylthiazole kinase (PDB entry 1EKK) from B. subtilis. In contrast, the human and Toxoplasma eukaryotic adenosine kinases, which are known to function as monomers, also grouped together (Fig. 4). This analysis placed HldE1 closest to the E coli ribokinase (Fig. 4), suggesting that this protein could be the best template to use for structural predictions on the HldE1 domain.
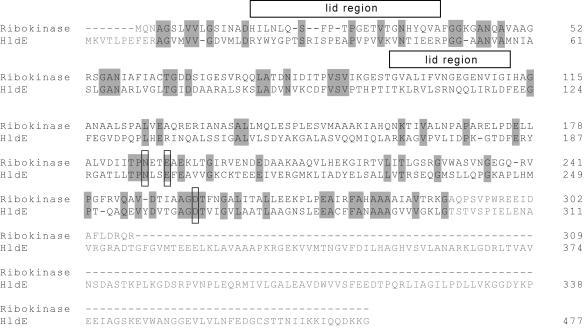
The E. coli ribokinase has two main structural units: the first is a central α/β fold comprised of a twisted nine-stranded β-sheet flanked on both faces by α helices; the second is a protruding β-sheet (“lid region”) formed by two insertions into the sequence of the central α/β fold. Essentially all catalytic components lie within the central α/β fold. X-ray crystallography has revealed that a dimer interface is formed by the interaction of the protruding β-sheets (25). A structural alignment of HldE1 and ribokinase (Fig. 5) showed several regions of homology between these two proteins, consistent with their overall 37% similarity and 20% identity. In contrast, a similar alignment with the human adenosine kinase (monomeric active form) showed a lower degree of sequence homology with HldE1 (data not shown) and could not be used as a template for modeling.
FIG. 5.
Alignment of E. coli ribokinase and HldE1 amino acid sequences. Only residues indicated in black type were used for the analysis. Gray shading indicate the conserved residues. Mutated amino acids in ATP binding and catalytic activity sites are indicated with boxes. The boxes above the alignment indicate the amino acids involved in forming the “lid” region.
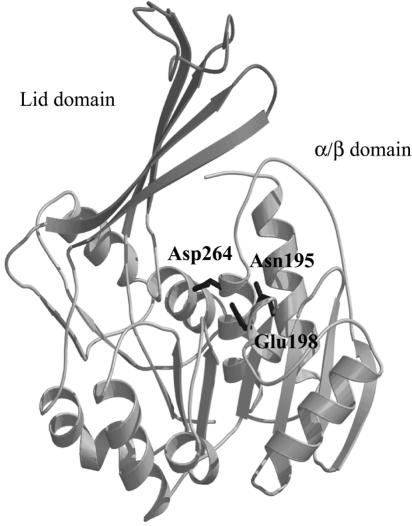
The structural alignment revealed that the critical amino acids in the ribokinase were all conserved in HldE1 and allowed us to identify several conserved amino acids in the α/β unit that could be essential for catalysis and substrate binding within the predicted three-dimensional structure of the heptokinase domain HldE1. These residues were aspartic acid (D) 264 (Fig. 6), which is equivalent to the catalytic D256 in the ribokinase, and glutamine (N) 195 and glutamic acid (E) 198, both of which are part of the NXXE motif for ATP binding (25). The identity between the two proteins was weaker near the ends of the sequences and in the lid region. The differences in the lid region of the proposed heptokinase model probably reflect the larger size of the substrate for this enzyme (the seven-carbon sugar glycero-manno-heptose 7-phosphate) in contrast to that of the ribokinase (the five-carbon sugar ribose). From these results we concluded that HldE1, like the E. coli ribokinase, may form a dimer with the active sites located at the interface of the monomers.
FIG. 6.
Homology model of the E. coli HldE1 domain (heptokinase) based on the crystal structure of ribokinase (PDB entry 1RKA), using MODELLER. Alignment of HldE1 and ribokinase was performed using CLUSTAL W. The first 11 residues of HldE1 and the first 3 residues of the ribokinase were removed. The last 18 residues in HldE1 are not included in this model.
Construction and functional characterization of amino acid substitution mutants in HldE1 domain.
We constructed amino acid replacements in the HldE1 domain to test whether the residues indicated above, which are predicted to be of functional relevance for HldE1 function, were indeed required for enzyme activity. The putative catalytic residue D264 was replaced by glutamic acid (HldE1D264E; pFM6) and asparagine (HldE1D264N; pFM5). Residues N195 and E198, which are proposed to be involved in ATP binding, were replaced by aspartic acid (HldE1N195D [pFM7] and HldE1E198D [pFM8]). We have shown previously that the kinase activity of HldE can be demonstrated biochemically by following the conversion of glycero-manno-hepose 7-phosphate into a product consistent with glycero-manno-heptose 1,7-biphosphate (14). However, only minimal quantities of the carbohydrate substrate can be prepared from sedoheptulose 7-phosphate by using purified A. thermoaerophilus GmhA isomerase (14), thus preventing us from conducting a detailed kinetic analysis of the reaction in vitro. Therefore, we investigated the function of HldE1 and its mutant derivatives by functional complementation in vivo. Without exception, all of the replacements rendered the protein nonfunctional, as demonstrated by the inability of mutant proteins to complement the heptoseless phenotype of strain FAM3 (ΔhldE1::Km) (Fig. 2C). Lack of complementation was not due to the lack of protein expression, since in all cases the polypeptides were detected as GST fusion proteins by SDS-PAGE (Fig. 3).
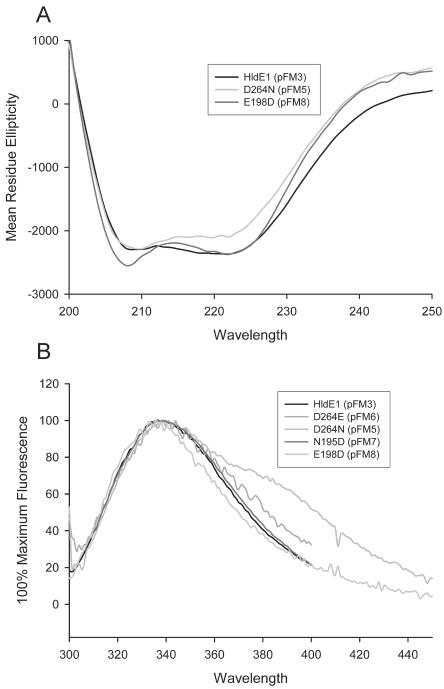
To determine whether the absence of complementation was due to misfolding of the protein or to a true functional defect in HldE1 activity, the structural integrity of the mutant proteins was examined by circular dichroism. Figure 7A shows similar circular dichroism spectra for the parental HldE1 (77% helical content) and the mutants HldE1D264N (72% helical content) and HldE1E198D (80% helical content). These results suggest that the loss of enzymatic activity in the mutant proteins was not caused by drastic alterations in protein structure. We also examined parental and mutant proteins by spectrofluorometric analysis, taking advantage of a single tryptophan residue (W25) that is in the beginning of the first lid segment. Mutant proteins and parental HldE1 showed a maximum absorption at 338 ± 3 nm (Fig. 7B). The protein encoded by pFM5 also had an additional shoulder in the region of 380 nm to 400 nm. This may be due to the presence of protein aggregates (see below). Overall, the spectrofluorometry results indicated that the tryptophan residues in all of these proteins were equally accessible to the solvent and did not show a radical change in their environment upon introduction of the amino acid substitution, suggesting that their tertiary structures remained consistent. We therefore concluded that no significant changes in tertiary protein structure had occurred as a consequence of the amino acid replacements in the mutant forms of the HldE1 domain, although it is possible that the location of the tryptophan in the lid region may be spatially far from potentially minor folding changes that could occur at the point of the mutations. From the combined circular dichroism and spectrofluorometric analyses we concluded that the substitutions in amino acids D264, N195, and N198 directly affect the enzymatic function of HldE1, as predicted from the molecular model.
FIG. 7.
A, circular dichroism spectrum of HldE1, D264N, and E198D mutant proteins. B, spectrofluorometric profile based on the excitation of the HldE tryptophan 25 residue at 295 nm. The excitation slit was set to 1 nm; the emission slit was set to 2 nm. Maximum fluorescence for parental and mutant proteins showed maximum excitation at 338 ± 3 nm.
HldE1 functions as a dimer.
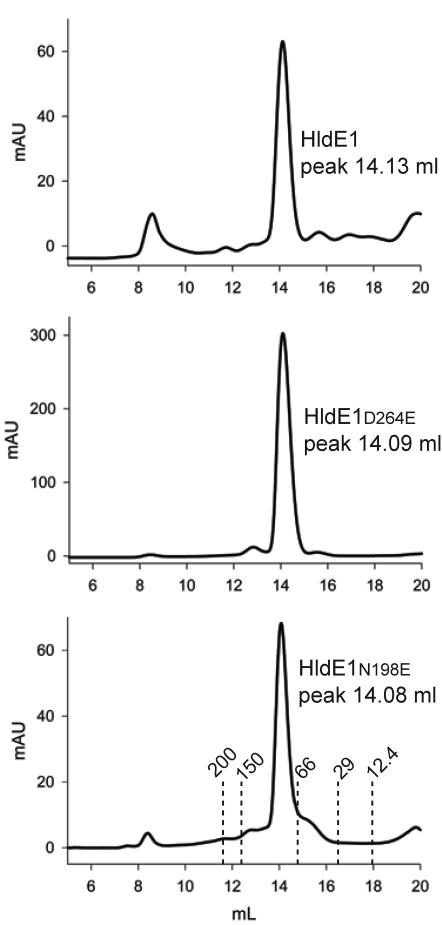
Crystallographic data, supported by size exclusion chromatography, have shown that the active functional unit of the E. coli ribokinase is a dimer whereby an exchange of β strands in the lid allows each subunit to assist in covering the active site of the other (25). The closest homology between HldE1 and E. coli ribokinase (Fig. 5) suggested that HldE1 may also function as a dimer. Therefore, we estimated the native molecular mass of HldE1 by size exclusion chromatography. Figure 8 shows that the predominant form of purified HldE1 and its mutant derivatives eluted in a region corresponding to a molecular mass of 83 kDa, consistent with dimers. A strong band corresponding to a 35-kDa polypeptide (the predicted mass of the HldE1 monomer) and a weak band corresponding to an 80-kDa polypeptide were identified when the fractions from these peaks were run on a denaturing gel (data not shown). The 80-kDa polypeptide is consistent with a partially denatured dimeric form of HldE1. Therefore, the size exclusion chromatography data strongly suggest that native HldE1 is a dimer.
FIG. 8.
Size exclusion chromatography profiles obtained with wild-type HldE1, HldE1D264E, and HldE1N198E. The x axis corresponds to milliliters passed over the column, and the y axis indicates the absorption read in the detector at 280 nm. The height of the peak depends on the relative concentration of protein loaded into the column. Standards used to calibrate the column were blue dextran (2000 kDa), β-amylase (200 kDa), alcohol dehydrogenase (150 kDa), bovine serum albumin (66 kDa), carbonic anhydrase (29 kDa), and cytochrome c (12.4 kDa). The predominant peak of elution (14.1 ml) in all cases corresponded to an approximate mass of 83 kDa, which is consistent with expected mass of the dimeric form of HldE1.
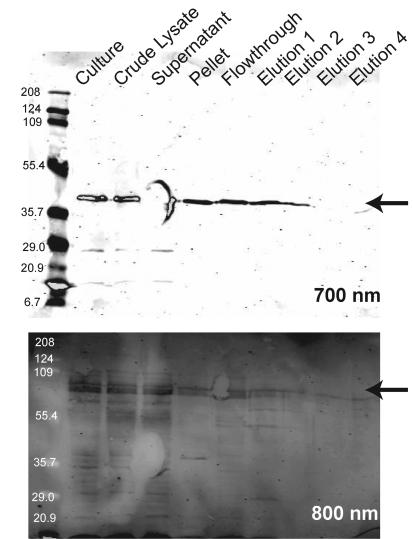
To demonstrate that HldE1 monomers are indeed associated in vivo, we also conducted a coelution experiment. We first constructed the plasmid pFM34, which expresses the HldE1 protein tagged with the FLAG epitope at the C-terminal end. This plasmid is compatible with pFM1, which encodes HldE as a GST fusion protein. Both pFM34 and pFM1 were cotransformed into strain FAM2 (ΔhldE). A cell extract was purified using the GSTrap column, and the eluate was tested by Western blotting with anti-FLAG and anti-GST monoclonal antibodies. Figure 9 shows the two proteins coeluted in the purification by GST affinity chromatography, despite the fact that the protein encoded by pFM34 does not have a GST tag. As a control, the vector pGEX-2T (encoding only GST) and pFM34 were cotransformed into strain FAM2. Although both GST and HldE-FLAG proteins were visible during the protein purification steps, only the GST was recovered in the elution fractions (data not shown). From these results, we conclude that HldE must exist in vivo at least as a dimer.
FIG. 9.
Western blot from a copurification experiment using a strain coexpressing a full-length HldE N terminally fused to GST and an HldE1 domain C terminally fused to the FLAG epitope tag. A cell-free lysate was passed through a GSTrap column to immobilize the GST-tagged protein. After washing with 50 times the column volume, proteins were eluted with reduced glutathione. The various fractions were examined by immunoblotting probed with antibodies against both GST and FLAG tags. The blots were visualized by fluorescence scanning with an Odyssey infrared imaging system (LI-COR Biosciences) using both 700-nm (for detection of the FLAG epitope) and 800-nm (for detection of the GST tag) channels (for more details, see Materials and Methods).
If, as predicted from the model, HldE1 has a lid region on one monomer that assists in covering the active site of the other, we should expect that point mutations affecting the activity of one monomer would have a dominant negative effect on the function of the wild-type protein. In contrast, if the monomers can function enzymatically independently from one another, we should not see any reduction in the enzyme activity of the intact protein. To test these possibilities, we used the strain FAM4, which has a chromosomal deletion of hldE2 and is therefore devoid of ADP-transferase activity while retaining heptokinase activity. FAM4 was transformed with pFM25, which encodes a complete HldEN195D protein with a substitution affecting the predicted ATP binding site of the heptokinase domain HldE1, but with a normal ADP-transferase domain HldE2. Because the heptokinase and ADP-transferase activities are independent, the chromosomal HldE1 should provide the heptokinase activity and the C-terminal domain of HldEN195D should provide the ADP-transferase activity. Since the expression of the pFM25-encoded HldEN195D protein is under the control of the lac promoter, we investigated the sensitivity of FAM4(pFM25) to 50 μg ml−1 novobiocin in the presence and in the absence of the inducer IPTG as a marker for a heptoseless phenotype. We observed that FAM4(pFM25) cells were sensitive to novobiocin when plated in the presence of 1 mM IPTG, but they grew on plates with novobiocin and no IPTG. This result was consistent with the notion of a dominant negative phenotype caused by the mutant HldE protein and could be explained by the overexpression of the HldEN195D, which would lead to greater amounts in the bacterial cell of HldEN195D homodimers and HldEN195D/chromosomally encoded HldE heterodimers than homodimers of the wild-type HldE protein. We reasoned that a similar phenomenon should be seen when expressing mutated proteins in E. coli DH5α, which, unlike FAM4, carries the parental hldE gene. E. coli DH5α transformed with plasmids pFM7 and pFM8, which encode mutant forms of HldE1 with amino acid replacements in the predicted ATP binding site, displayed a novobiocin-sensitive phenotype when transformants were plated in the presence of IPTG. In contrast, bacteria transformed with pFM5 or the control plasmid pFM3 remained resistant to novobiocin (Table 2). The plasmid pFM5 encodes an HldE1 domain with an amino acid replacement in the catalytic site D256, while pFM3 encodes the parental HldE1 protein. Therefore, we concluded from these experiments the dominant negative phenotype caused by the coexpression of mutant and parental HldE forms was manifested only in the case of mutations affecting the ATP binding site and not in the case of mutations affecting catalytic residues. These results also supported the notion that HldE functions as a dimer in vivo.
TABLE 2.
Phenotypes of E. coli DH5α derivatives carrying each of the plasmids encoding wild-type and point mutations of heptokinases under induced and noninduced conditions
| Plasmid (mutation) | No. of colonies (105)
|
|
|---|---|---|
| Without IPTG | With IPTG | |
| pFM3 (wild type) | 300 | 131 |
| pFM5 (D264N) | 342 | 266 |
| pFM7 (N195D) | 81 | 0 |
| pFM8 (E198D) | 144 | 20 |
DISCUSSION
Phylogenetic analyses conducted in this study with members of the ribokinase superfamily that have known structures revealed that the HldE1 heptokinase was more closely related to the E. coli ribokinase than to any other member of this diverse superfamily. The E. coli ribokinase has been well characterized, including the identification of cofactors, crystal structure, and enzyme kinetics. This enabled us to use the ribokinase as a model for the structure and function of the heptokinase despite the poor relationship in the lid region covering the active site. Differences in the lid region of the HldE1 heptokinase can be explained by a difference in size of the substrate. The ribose sugar that serves as a substrate for the ribokinase is a five-carbon molecule, whereas the heptokinase functions on a seven-carbon sugar, thus requiring a larger lid region. Both the heptokinase and ribokinase share a region of hydrophobic residues corresponding to the region of lid interface, suggesting that the heptokinase, like the ribokinase, may undergo an exchange of strands contributing to the formation of the lid region. Although we could not model well the lid region in the heptokinase, the presence of a GG signature in this region suggests that its putative lid undergoes a conformational change. A GG dipeptide was found to undergo a conformational change after substrate binding and is believed to play a significant role in forming the close conformation of the enzyme (24). A GG dipeptide is found in all other members of the ribokinase superfamily where conformational changes have been demonstrated (25, 26, 34).
The mutant enzymes expressed by the plasmids pFM5, pFM6, pFM7, and pFM8 were designed on the basis of the homology model. All were inactive, as evidenced by their inability to restore a complete core-lipid A in a strain lacking the parental HldE1. Tricine SDS-PAGE demonstrated that these proteins were expressed, while circular dichroism and fluorescence spectroscopy revealed that they folded correctly. These data support the idea that the loss of activity is caused by the loss of substrate binding or catalytic activity in the mutant proteins and not by other effects of the mutations on protein structure or stability. However, biochemical assays to determine the kinetics of parental and mutated enzyme forms need to be performed to unequivocally probe the function of the mutated residues. These studies await the synthesis of glycero-manno-heptose 7-phosphate, which currently is not available from commercial sources.
Our phylogenetic analysis also showed that the heptokinase clusters well with other members of the ribokinase superfamily whose active forms are dimers, most specifically with the E. coli ribokinase and the mycobacterial adenosine kinase. Bork et al. (3) suggested an evolutionary pathway in which the most primitive members of this superfamily function as trimers and lack well defined lids. The most advanced members of this superfamily function as monomers, having evolved a lid that is able to cover its own active site without the contribution from the lid of another monomer. The clustering of the HldE1 heptokinase with other proteins known to form functional dimers suggests the possibility that the heptokinase is also a functional dimer and is also in agreement with recent work by Zhang et al. (34) based on the comparisons of quaternary structures among several members of the ribokinase superfamily.
Biochemical evidence supporting that HldE1 is a functional dimer was obtained from size exclusion chromatography and by a coelution experiment that demonstrated a complex formation, as judged by the copurification of both FLAG-tagged and GST-tagged constructs using a GST affinity column under low-salt conditions. Furthermore, genetic evidence for a functional dimer was obtained by the observation that mutant proteins with amino acid substitutions in the ATP binding site display a dominant negative effect on parental HldE. This dominant negative phenotype was observed only with amino acid substitutions mutations in the predicted ATP binding site of HldE1, suggesting that a conformational change might take place upon binding of ATP, as was proposed previously for the ribokinase (26). It is likely that this conformational change involves the lid region, as the conserved GG dipeptide is also found in predicted lid region of HldE1. The GG dipeptide is present in ribokinase superfamily members where a conformational change has been shown to occur (34).
In summary, we provide new genetic and biochemical evidence demonstrating that the HldE1 heptokinase appears to be a functional dimer and that this protein exhibits structural features in common with those of the E. coli ribokinase. Further work to characterize the kinase reaction of HldE1 in detail using chemically synthesized substrates and to conduct a refined structural characterization by crystallization and X-ray diffraction analysis is under way in our laboratories.
Acknowledgments
This work was supported by grants from the Natural Sciences and Engineering Research Council (to M.A.V.) and the Swedish Natural Sciences Research Council (to S.L.M.). F.M. was a recipient of an Ontario Graduate Scholarship in Science and Technology.
M.A.V. holds a Canada Research Chair in Infectious Diseases and Microbial Pathogenesis.
REFERENCES
- 1.Andrade, M. A., P. Chacón, J. J. Merelo, and F. Morán. 1993. Evaluation of secondary structure of proteins from UV circular dichroism using an unsupervised learning neural network. Protein Eng. 6:383-390. [DOI] [PubMed] [Google Scholar]
- 2.Berman, H. M., J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov, and P. E. Bourne. 2000. The Protein Data Bank. Nucleic Acids Res. 28:235-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bork, P., C. Sander, and A. Valencia. 1993. Convergent evolution of similar enzymatic function on different protein folds: the hexokinase, ribokinase, and galactokinase families of sugar kinases. Protein Sci. 2:31-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campobasso, N., Mathews, I. I., T. P. Begley, and S. E. Ealick. 2000. Crystal structure of 4-methyl-5-β-hydroxyethylthiazole kinase from Bacillus subtilis at 1.5 Å resolution. Biochemistry 39:7868-7877. [DOI] [PubMed] [Google Scholar]
- 5.Cardona, S. T., and M. A. Valvano. An expression vector containing a rhamnose-inducible promoter provides tightly regulated gene expression in Burkholderia cenocepacia. Plasmid, in press. [DOI] [PubMed]
- 6.Cheng, G., E. M. Bennett, T. P. Begley, and S. E. Ealick. 2002. Crystal structure of 4-amino-5-hydroxymethyl-2-methylpyrimidine phosphate kinase from Salmonella typhimurium at 2.3 Å resolution. Structure 10:225-235. [DOI] [PubMed] [Google Scholar]
- 7.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferguson, A. D., W. Welte, E. Hofmann, B. Lindner, O. Holst, J. W. Coulton, and K. Diederichs. 2000. A conserved structural motif for lipopolysaccharide recognition by procaryotic and eucaryotic proteins. Struct. Fold Des. 8:585-592. [DOI] [PubMed] [Google Scholar]
- 9.Hancock, R. E. W., D. N. Karunaratne, and C. Bernegger-Egli. 1994. Molecular organization and structural role of outer membrane macromolecules, p. 263-279. In J. M. Ghuysen and R. Hackenbeck (ed.), Bacterial cell wall, vol. 27. Elsevier Science, Amsterdam, The Netherlands.
- 10.Heinrichs, D. E., M. A. Valvano, and C. Whitfield. 1999. Biosynthesis and genetics of lipopolysaccharide core, p. 305-330. In H. Brade, D. C. Morrison, S. Vogel, and S. Opal (ed.), Endotoxin in health and disease. Marcel Dekker, Inc., New York, N.Y.
- 11.Helander, I. M., B. Lindner, H. Brade, K. Altmann, A. A. Lindberg, E. T. Rietschel, and U. Zahringer. 1988. Chemical structure of the lipopolysaccharide of Haemophilus influenzae strain I-69 Rd−/B+: description of a novel deep-rough chemotype. Eur. J. Biochem. 177:483-492. [DOI] [PubMed] [Google Scholar]
- 12.Jett, B. D., K. L. Hatter, M. M. Huycke, and M. S. Gilmore. 1997. Simplified agar plate method for quantifying viable bacteria. BioTechniques 23:648-650. [DOI] [PubMed] [Google Scholar]
- 13.Kneidinger, B., M. Graninger, M. Puchberger, P. Kosma, and P. Messner. 2001. Biosynthesis of nucleotide-activated d-glycero-d-manno-heptose. J. Biol. Chem. 276:20935-20944. [DOI] [PubMed] [Google Scholar]
- 14.Kneidinger, B., C. L. Marolda, M. Graninger, A. Zamyatina, F. McArthur, P. Kosma, M. A. Valvano, and P. Messner. 2002. Biosynthesis pathway of ADP-d-glycero-l-manno-heptose in Escherichia coli. J. Bacteriol. 184:363-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lefebre, M. D., and M. A. Valvano. 2002. Construction and evaluation of plasmid vectors optimized for constitutive and regulated gene expression in Burkholderia cepacia complex isolates. Appl. Environ. Microbiol. 68:5956-5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lesse, A. J., A. A. Campagnari, W. E. Bittner, and M. A. Apicella. 1990. Increased resolution of lipopolysaccharides and lipooligosaccharides utilizing tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis. J. Immunol. Methods 126:109-117. [DOI] [PubMed] [Google Scholar]
- 17.Li, M. H., F. Kwok, W. R. Chang, C. K. Lau, J. P. Zhang, S. C. Lo, T. Jiang, and D. C. Liang. 2002. Crystal structure of brain pyridoxal kinase, a novel member of the ribokinase superfamily. J. Biol. Chem. 277:46385-46390. [DOI] [PubMed] [Google Scholar]
- 18.Long, M. C., V. Escuyer, and W. B. Parker. 2003. Identification and characterization of a unique adenosine kinase from Mycobacterium tuberculosis. J. Bacteriol. 185:6548-6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marolda, C. L., J. Welsh, L. Dafoe, and M. A. Valvano. 1990. Genetic analysis of the O7-polysaccharide biosynthesis region from the Escherichia coli O7:K1 strain VW187. J. Bacteriol. 172:3590-3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marti-Renom, M. A., A. C. Stuart, A. Fiser, R. Sánchez, F. Melo, and A. Sali. 2000. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 29:291-325. [DOI] [PubMed] [Google Scholar]
- 21.Nikaido, H. 1994. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science 264:382-388. [DOI] [PubMed] [Google Scholar]
- 22.Raetz, C. R. H., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 24.Schumacher, M. A., D. M. Scott, I. I. Mathews, S. E. Ealick, D. S. Roos, B. Ullman, and R. G. Brennan. 2000. Crystal structures of Toxoplasma gondii adenosine kinase reveal a novel catalytic mechanism and prodrug binding. J. Mol. Biol. 298:875-893. [DOI] [PubMed] [Google Scholar]
- 25.Sigrell, J. A., A. D. Cameron, T. A. Jones, and S. L. Mowbray. 1998. Structure of Escherichia coli ribokinase in complex with ribose and dinucleotide determined to 1.8 Å resolution: insights into a new family of kinase structures. Structure 6:183-193. [DOI] [PubMed] [Google Scholar]
- 26.Sigrell, J. A., A. D. Cameron, and S. L. Mowbray. 1999. Induced fit on sugar binding activates ribokinase. J. Mol. Biol. 290:1009-1018. [DOI] [PubMed] [Google Scholar]
- 27.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valvano, M. A. 1999. Biosynthesis and genetics of ADP-heptose. J. Endotoxin. Res. 5:90-95. [Google Scholar]
- 29.Valvano, M. A., C. L. Marolda, M. Bittner, M. Glaskin-Clay, T. L. Simon, and J. D. Klena. 2000. The rfaE gene from Escherichia coli encodes a bifunctional protein involved in the biosynthesis of the lipopolysaccharide core precursor ADP-l-glycero-d-manno-heptose. J. Bacteriol. 182:488-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valvano, M. A., P. Messner, and P. Kosma. 2002. Novel pathways for biosynthesis of nucleotide-activated glycero-manno-heptose precursors of bacterial glycoproteins and cell surface polysaccharides. Microbiology 148:1979-1989. [DOI] [PubMed] [Google Scholar]
- 31.Walsh, A. G., M. J. Matewish, L. L. Burrows, M. A. Monteiro, M. B. Perry, and J. S. Lam. 2000. Lipopolysaccharide core phosphates are required for viability and intrinsic drug resistance in Pseudomonas aeruginosa. Mol. Microbiol. 35:718-727. [DOI] [PubMed] [Google Scholar]
- 32.Yethon, J. A., E. Vinogradov, M. B. Perry, and C. Whitfield. 2000. Mutation of the lipopolysaccharide core glycosyltransferase encoded by waaG destabilizes the outer membrane of Escherichia coli by interfering with core phosphorylation. J. Bacteriol. 182:5620-5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yethon, J. A., and C. Whitfield. 2001. Purification and characterization of WaaP from Escherichia coli, a lipopolysaccharide kinase essential for outer membrane stability. J. Biol. Chem. 276:5498-5504. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, Y., M. Dougherty, D. M. Downs, and S. E. Ealick. 2004. Crystal structure of an aminoimidazole riboside kinase from Salmonella enterica; implications for the evolution of the ribokinase superfamily. Structure 12:1809-1821. [DOI] [PubMed] [Google Scholar]
- 35.Zwahlen, A., L. G. Rubin, C. J. Connelly, T. J. Inzana, and E. R. Moxon. 1985. Alteration of the cell wall in Haemophilus influenzae type b by transformation with cloned DNA: association with attenuated virulence. J. Infect. Dis. 152:485-492. [DOI] [PubMed] [Google Scholar]