Abstract
We have cloned and functionally characterized a Na+-coupled dicarboxylate transporter, SdcS, from Staphylococcus aureus. This carrier protein is a member of the divalent anion/Na+ symporter (DASS) family and shares significant sequence homology with the mammalian Na+/dicarboxylate cotransporters NaDC-1 and NaDC-3. Analysis of SdcS function indicates transport properties consistent with those of its eukaryotic counterparts. Thus, SdcS facilitates the transport of the dicarboxylates fumarate, malate, and succinate across the cytoplasmic membrane in a Na+-dependent manner. Furthermore, kinetic work predicts an ordered reaction sequence with Na+ (K0.5 of 2.7 mM) binding before dicarboxylate (Km of 4.5 μM). Because this transporter and its mammalian homologs are functionally similar, we suggest that SdcS may serve as a useful model for DASS family structural analysis.
Na+-coupled transport is a common method of substrate uptake in which the movement of Na+ down its electrochemical gradient facilitates the accumulation of small solutes within the cell (14, 32, 37, 39). Mammalian members of the divalent anion/Na+ symporter (DASS) family mediate the transport of dicarboxylates and inorganic anions across the cellular membrane in such a Na+-dependent manner (24, 27, 28, 31). Functionally characterized proteins of this group include the Na+/dicarboxylate cotransporters NaDC-1 and NaDC-3 (5, 16, 26), as well as the Na+-coupled SO4 transporters NaSi-1 and SUT-1 (9, 23). While study of these proteins has led to a detailed understanding of their transport mechanism, information concerning their structure remains limited.
To address this problem, we sought to identify a bacterial homolog of these mammalian transporters that can serve as a model for their structural analysis. The DASS family (also referred to as the SLC13 gene family in the human gene nomenclature) is a collection of evolutionarily related transport proteins that spans all three kingdoms of life, with representatives from prokaryotes comprising the majority of its membership (31). However, to date no bacterial member of this family has been shown to be mechanistically similar to its mammalian counterparts. Here, we report the cloning and functional expression of SdcS, a Staphylococcus aureus DASS transporter that shares sequence similarity with a number of mammalian members of this family. Our findings indicate that SdcS is a Na+/dicarboxylate symporter with transport properties similar to those of NaDC-1 and NaDC-3. This represents the first characterization of a bacterial Na+-coupled DASS transporter and suggests that structural studies of SdcS will provide insight into the architecture of mammalian proteins in this family.
MATERIALS AND METHODS
Bacterial strains.
Escherichia coli strain XL1-Blue (recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10]) (Stratagene) was used for all cloning steps. E. coli strain BL21 [F− ompT hsdSB(rB− mB−) gal dcm] (Novagen) served as host for tests of expression and function of plasmid-encoded SdcS.
Cloning.
The sdcS gene (GenBank accession number BA000017; locus SAV1916) was amplified from S. aureus genomic DNA (ATCC 700699D) by PCR and cloned into plasmid pQE-80L (bla lacIq) (QIAGEN) via restriction sites (BamHI and HindIII) created with the primers 5′-GCATGGATCCATGGCTTATTTCAATCAACATC-3′ and 5′-CGCTAAGCTTACTATTTCAATGGCAGTGGTTG-3′ (restriction sites are underlined). This recombinant plasmid (pQE-80L/SdcS) encodes SdcS with an N-terminal MRGS(H)6GS amino acid extension and places SdcS expression under control of a T5 promoter/lac operator element. To construct an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible C-terminal MRGS(H)6GS-tagged SdcS, a pair of primers (5′-CATGGAATTCAGAGGATCCCATCACCATCACCATCACTA-3′ and 5′-AGCTTAGTGATGGTGATGGTGATGGGATCCTCTGAATTC-3′) encoding the MRGS(H)6GS epitope were annealed to one another and inserted between the NcoI and HindIII sites of plasmid pTrc99A (Amersham Biosciences). The resulting plasmid was digested with NcoI and BamHI, ligated to the sdcS gene amplified as described above using the primers 5′-GGGAAATCCATGGCTTATTTCAATCAACATC-3′ and 5′-GATGGGATCCTCTTTTCAATGGCAGTGGTTG (restriction sites are underlined), and named pTrc99A/SdcS. Constructs were sequenced across the sdcS gene at the University of Texas Medical Branch Protein Chemistry Laboratory to ensure that they were identical to the published sequence (19).
Transport assays.
Overnight cultures were diluted 200-fold into Luria-Bertani broth (containing 100 μg/ml ampicillin and 150 μM IPTG). Cells were grown at 37°C to a density of 5 × 108 to 1 × 109 cells/ml (approximately 3 h), harvested by centrifugation, and then washed twice and resuspended in assay buffer (50 mM MOPS [morpholinepropanesulfonic acid], 5 mM NaCl, 95 mM choline chloride, pH 7 [pH adjusted with Tris base]) at an optical density at 660 nm of 1.4, equivalent to about 1 × 109 to 2 × 109 cells/ml. After equilibration at room temperature, tests of substrate transport were initiated by adding a 1/20 volume of labeled substrate to a final concentration of 100 μM. At indicated times, aliquots were removed for filtration on Millipore filters (0.45-μm pore size, type HAWP), rinsed twice with 5 ml wash buffer (50 mM MOPS-Tris, 100 mM choline chloride, pH 7), and counted by liquid scintillation using Econo-Safe (Research Products International Corp.) as scintillant. Unless otherwise indicated, SdcS transport activity is reported after subtracting background uptake values from strain BL21 housing plasmid pQE-80L.
Apparent kinetic constants (Km, Vmax) for SdcS dicarboxylate substrates were determined by fitting initial transport rates (measured at 1 min) to the Michaelis-Menten equation {v = (Vmax[S])/(Km + [S]) } using nonlinear regression analysis. The apparent half-saturation constant (K0.5) values for Na+ and Li+ were estimated by fitting initial cation-dependent dicarboxylate transport rates (1 min) assayed at subinhibitory cation concentrations (≤Vmax) to the Hill equation {v = (Vmax[S]n)/(K0.5n +[S]n), where n represents the Hill coefficient } using nonlinear regression analysis. True Km (for succinate) and K0.5 (for Na+) constants were calculated from a replot of the apparent Km of succinate as a function of Na+, with the assumption that Na+ binding to the SdcS active site precedes dicarboxylate binding (35).
Immunoblot analysis.
Cells prepared for transport were resuspended in sample buffer, loaded without preheating, and separated by sodium dodecyl sulfate-polyacrylamide (11%) gel electrophoresis (20). Protein was transferred to nitrocellulose and probed with a mouse monoclonal antibody reactive to the SdcS N- and C-terminal RGS(H)4 epitope tags (QIAGEN). To evaluate expression, Western blots were developed by the chemiluminescence method (SuperSignal West Pico Chemiluminescent Substrate; Pierce) using a horseradish peroxidase-conjugated anti-mouse immunoglobulin G antibody (Amersham Biosciences).
Chemicals.
Unlabeled substrates and [14C]fumarate (6.3 mCi/mmol) were from Sigma Chemical Company. [35S]Na2SO4 (1,500 Ci/mmol) and [14C]malate (52 mCi/mmol) were obtained from PerkinElmer Life Sciences and Amersham Biosciences, respectively. [14C]citrate (55 mCi/mmol) and [14C]succinate (44 mCi/mmol) were from Moravek Biochemicals.
RESULTS
Cloning and functional expression of SdcS.
In the present study our objective was to identify and characterize a bacterial homolog of the mammalian members of the DASS family. To do this, a protein-versus-protein similarity search was performed with FASTA3 (default settings) from the European Bioinformatics Institute (http://www.ebi.ac.uk/fasta33/) using the low-affinity Na+/dicarboxylate cotransporter from human kidney, NaDC-1, as query (25). This search resulted in the identification of the putative SAV1916 gene from S. aureus (strain Mu50), whose protein product is approximately 35% identical in sequence to that of human NaDC-1.
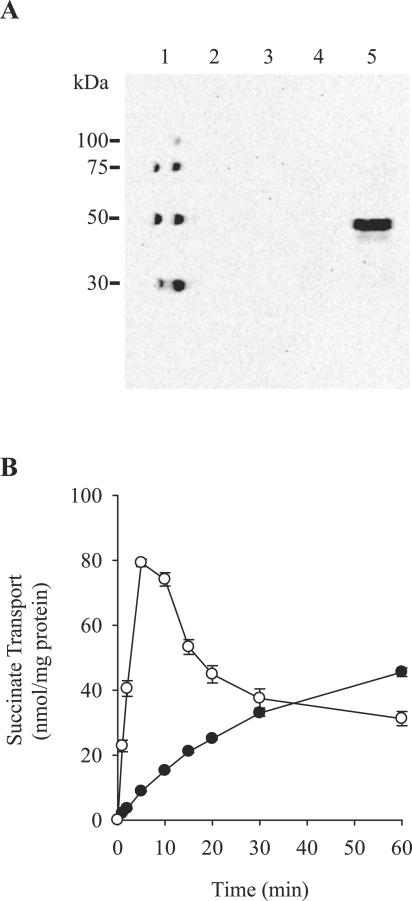
The SAV1916 gene, renamed sdcS (for sodium/dicarboxylate symporter), encodes a hypothetical protein of 520 residues having a molecular mass of 57.2 kDa. To test whether SdcS is functionally expressed, we cloned sdcS into plasmid pQE-80L and monitored its expression and transport activity in E. coli. As illustrated in Fig. 1A, N -terminal histidine-tagged SdcS is expressed only in IPTG-induced cells housing plasmid pQE-80L/SdcS, resulting in a protein band corresponding to 45 to 50 kDa. Placement of the histidine tag at the C terminus of SdcS identified a protein band of the same molecular mass, indicating that the fast migration of N-terminal histidine-tagged SdcS on sodium dodecyl sulfate-polyacrylamide gels was not due to premature termination of translation or proteolytic cleavage of the C-terminal region of this protein (data not shown). Rather, the increased mobility of SdcS is likely due to its hydrophobic nature which, as is frequently exhibited by membrane transport proteins, results in an apparent molecular mass lower than that predicted from its protein sequence (18, 34). Parallel experiments designed to evaluate transport activity demonstrated that only the SdcS-expressing strain rapidly accumulated succinate (Fig. 1B). Uptake in this strain was followed by a slow fall to the equilibrium level, an observation that we attribute to both the leakage and metabolism of internalized substrate. That succinate transport, albeit slow, also occurred in the absence of SdcS is likely due to the action of the endogenous E. coli dicarboxylate transporter DctA. This transporter catalyzes H+/dicarboxylate cotransport and is expressed at a low level under aerobic growth conditions such as that used here (6, 13).
FIG. 1.
Functional expression of SdcS. (A) Western blot of whole cells (optical density at 660 nm of 10; 10 μl/lane): molecular mass standards (lane 1); BL21 housing pQE-80L grown with (lane 3) and without (lane 2) IPTG; BL21 housing pQE-80L/SdcS grown with (lane 5) and without (lane 4) IPTG. Protein was detected using a monoclonal antibody directed against the N-terminal SdcS histidine tag. (B) Succinate transport activity of IPTG-induced strain BL21 housing pQE-80L (•) and pQE-80L/SdcS (○) in the presence of 100 μM succinate and 5 mM NaCl. Cells housing either pQE-80L or pQE-80L/SdcS and grown in the absence of IPTG exhibited succinate transport profiles similar to that of IPTG-induced cells carrying plasmid pQE-80L. Values shown are means ± standard errors for three independent experiments.
Carboxylate specificity.
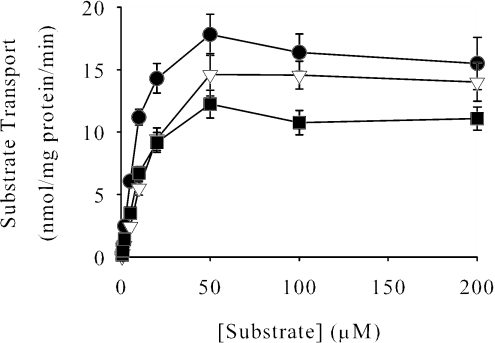
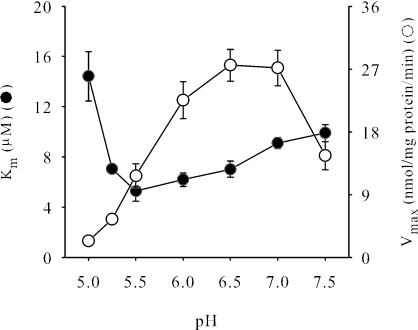
Members of the DASS family have broad anionic substrate specificity, transporting a wide array of di- and tricarboxylates as well as inorganic oxyanions such as sulfate. Analysis of SdcS substrate preference tested directly by using radiolabeled citrate, sulfate, fumarate, malate, and succinate indicated that only the latter three were transported appreciably (Fig. 2 and data not shown). Further study found that other dianionic carboxylates—aspartate, glutamate, glutarate, α-ketoglutarate, and maleate—did not inhibit succinate transport when present at 1 mM, suggesting that these compounds are, at best, poor SdcS substrates (data not shown). Ligands transported by SdcS exhibit similar kinetic profiles, with nearly equivalent maximal velocities, and Km values between 5 and 15 μM (Fig. 2; Table 1). To determine which charged form(s) of carboxylate SdcS was transported, we evaluated succinate uptake at external pH values from pH 5.0 to 7.5, a range that spans the pK2 of this substrate (pK2 = 5.6). We noted considerable changes in both kinetic parameters (Km and Vmax) over this pH range (Fig. 3). In particular, as assay pH fell below the pK2 of succinate, the affinity of this substrate for SdcS decreased quickly, showing a nearly threefold drop from its value of 5.3 μM at pH 5.5 to near 15 μM at pH 5.0. This observation provides a strong indication that SdcS recognizes and transports only the divalent form of its natural substrates.
FIG. 2.
Kinetics of SdcS-mediated dicarboxylate transport. Initial rates of fumarate (▿), malate (▪), and succinate (•) transport were estimated as described in Materials and Methods. Data from three independent trials are shown as means ± standard errors. Km and Vmax values for SdcS under these conditions are shown in Table 1.
TABLE 1.
Transport properties of SdcS
| Substrate pair | Dicarboxylate kineticsa
|
Cation activationb
|
||
|---|---|---|---|---|
| Km (μM) | Vmax (nmol/mg protein/min) | K0.5 (mM) | Vmax (nmol/mg protein/min) | |
| Na+/fumarate | 15 ± 0.54 | 16 ± 1.7 | 1.5 ± 0.02 | 11 ± 0.84 |
| Na+/malate | 8.1 ± 0.40 | 12 ± 1.0 | 1.2 ± 0.01 | 14 ± 0.93 |
| Na+/succinate | 6.6 ± 0.75 | 17 ± 2.0 | 1.0 ± 0.04 | 15 ± 3.1 |
| Li+/succinate | NDc | ND | 23 ± 0.60 | 7.2 ± 0.52 |
Rates of transport were measured as described in Materials and Methods, and kinetic constants were determined by fitting initial transport rates to the Michaelis-Menten equation. Km and Vmax values are means ± standard errors for three independent experiments. The concentration range of dicarboxylate used to approximate kinetic constants was 1 to 200 μM; Na+ concentration was fixed at 5 mM.
As in footnote a, except that kinetic constants were determined by fitting transport rates to the Hill equation. K0.5 and Vmax values are means ± standard errors for three independent experiments. The Na+/Li+ concentration range used to approximate kinetic constants was 0.5 to 7.5 mM; dicarboxylate concentration was fixed at 100 μM. Isotonic conditions were maintained at 100 mM by supplementation with choline chloride.
ND, not determined.
FIG. 3.
Effects of pH on kinetic constants for succinate transport. Km and Vmax values for SdcS are as indicated. Assays were performed as described in Materials and Methods except that assay and wash buffers used morpholineethanesulfonic acid (MES)-MOPS-Tris rather than MOPS-Tris. The concentrations of succinate used to estimate kinetic constants were 1 μM to 200 μM. Kinetic constants are reported as means ± standard errors for three independent experiments.
Cation specificity.
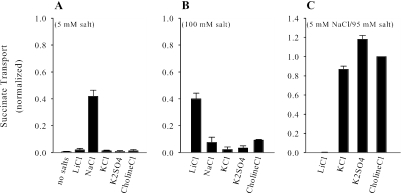
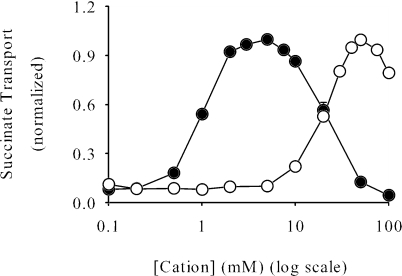
Because members of the DASS family are Na+-coupled transporters, the cation requirements and selectivity of SdcS were examined. We found that at low salt concentrations (5 mM) only Na+ stimulated succinate transport (Fig. 4A). By contrast, at high concentrations of cation (100 mM) Li+ promoted uptake of succinate whereas Na+ had an inhibitory effect (Fig. 4B). We also determined what effect, if any, the addition of various salts had upon Na+/dicarboxylate transport. Interestingly, the presence of either choline chloride, KCl, or K2SO4 in the assay medium increased Na+-dependent succinate uptake roughly two- to threefold (Fig. 4C), indicating that high medium osmolarity may enhance SdcS transport activity. The inclusion of both Na+ and Li+ in the assay medium did not stimulate succinate transport but, instead, completely inhibited it. This result, coupled with the finding that these cations promote dicarboxylate uptake when present individually, leads us to suggest that all cation binding sites must be occupied by the same cationic species for transport to occur.
FIG. 4.
Cation selectivity of SdcS. Rates of cation-dependent succinate transport were measured in the presence of 5 mM salt (A), 100 mM salt (B), and 5 mM NaCl-95 mM salt (C). To maintain isotonicity, K2SO4 was used at one-half the noted concentrations. Transport rates were estimated after a 5-min incubation of cells with labeled substrate as described in Materials and Methods, except that assay and wash buffers contained 50 mM MOPS-Tris, pH 7, plus the noted additives. Data from three independent trials were normalized to the transport value measured in the presence of 5 mM NaCl-95 mM choline chloride (38 ± 5.5 nmol/mg protein) and are shown as means ± standard errors.
Our next tests addressed the relationship between cation concentration and SdcS transport activity. Initial work (Fig. 4) suggested that both Na+ and Li+ stimulated dicarboxylate uptake in a concentration-dependent manner. Further analysis confirmed this supposition. Thus, the K0.5 for Na+ was 1 to 2 mM, while that for Li+ was 20-fold greater (Table 1). Also, the maximal SdcS dicarboxylate transport activity attained with Li+ was only one-half that found with Na+ (Fig. 5; Table 1). This lowered affinity for Li+ and the reduced transport of dicarboxylates in its presence are characteristics of several vertebrate DASS family members (2, 29, 41). The sigmoidal nature of cation-dependent transport (Fig. 5) indicated cooperative binding between SdcS cation binding sites, and Hill coefficients for various cation (Li+ and Na+)/dicarboxylate (fumarate, malate, and succinate) pairings ranged from 2.5 to 3.5. However, unlike other DASS transporters, SdcS exhibited strong cation-mediated inhibition, with Na+-dependent succinate transport having a Ki for Na+ of 26 ± 3.4 mM (Fig. 5). The closeness of the K0.5 and Ki of Na+ prevents SdcS from attaining maximal transport activity (Vmax) (8), resulting in an overestimation of the Hill coefficient. Thus, it is likely that two to three Na+ (or Li+) cations along with a single dicarboxylate must occupy the SdcS active site for transport to occur. Work with permeabilized whole cells indicated that valinomycin had no effect on SdcS transport activity, suggesting an electroneutral symport reaction with a Na+/dicarboxylate ratio of 2:1 (data not shown).
FIG. 5.
Kinetics of cation-dependent succinate transport. Initial rates of succinate transport (100 μM) in the presence of NaCl (•) and LiCl (○). Transport was estimated as described in Materials and Methods except that the assay buffer contained 50 mM MOPS-Tris, pH 7, plus the indicated concentrations of NaCl (or LiCl). Assay buffer isotonicity was maintained at 100 mM by the addition of the necessary amounts of choline chloride. Data from three independent trials were normalized to peak values (16 ± 2.9 nmol/mg protein/min for NaCl; 6.7 ± 0.49 nmol/mg protein/min for LiCl) and are shown as means ± standard errors. K0.5 and Vmax values for SdcS under these conditions are shown in Table 1.
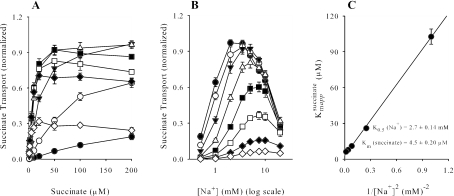
Transport kinetics of SdcS.
The requirement of Na+ (or Li+) for transport function implies that SdcS operates as a symporter. As such, the true affinity of one substrate will be observed only when its cosubstrate is present at saturating concentrations. With this in mind, we generated a series of transport curves in which the concentration of one substrate was varied while the other was held constant (Fig. 6). This experiment not only documented the stimulatory and inhibitory properties of Na+ upon succinate transport (Fig. 6A) but indicated that the K0.5 of Na+ for SdcS decreases as succinate concentration increases (Fig. 6B). Furthermore, at subinhibitory Na+ concentrations, the apparent affinity of succinate (100 μM at 1 mM Na+ decreasing to 6.1 μM at 7.5 mM Na+), but not the maximal velocity (17 to 20 nmol/mg protein/min), was variable. This finding was interpreted as reflecting an ordered bireactant system in which Na+ binds before succinate (35), and a replot of these data gave true Km values for Na+ and succinate of 2.7 ± 0.14 mM and 4.5 ± 0.20 μM, respectively (Fig. 6C). In agreement with work cited above, the finding that linearization of the replot was obtained only when the “Na+” term was raised to the second power reflects an apparent coupling of at least two Na+ ions for each succinate transported.
FIG. 6.
(A) Initial rates (1 min) of succinate transport as a function of succinate at 0.5 (•), 1 (○), 2 (▾), 3.5 (▵), 5 (▪), 7.5 (□), 10 (⧫), and 20 (⋄) mM NaCl. (B) Replots of panel A as a function of Na+ ion concentration: 1 (⋄), 2 (⧫), 5 (□), 10 (▪), 20 (▵), 50 (▾), 100 (○), and 200 (•) μM succinate. Data from three independent trials were normalized to peak values (17 to 19 nmol/mg protein/min) and are shown as means ± standard errors. (C) A replot of apparent Km values was used to derive true K0.5 and Km constants for Na+ and succinate assuming an ordered bireactant system in which KBapp = {(KA)2 KB + KAKB [A]}/[A]2 + KB, where A and B denote Na+ and succinate, respectively.
DISCUSSION
The primary function of proteins of the DASS family is the transport of divalent carboxylates and oxyanions. In multicellular organisms, these transporters provide a mechanism to regulate the extracellular concentration of their respective substrates, and in certain instances their dysfunction can lead to a variety of disorders. For example, mice lacking the Na+/sulfate cotransporter NaSi-1 exhibit growth retardation and spontaneous seizures (7), while Drosophila melanogaster flies heterozygous for the Na+-independent dicarboxylate transporter gene Indy display a near doubling in their average life span (12, 17, 33). Also, due to its role in recovery of citrate from the renal proximal tubule, it has been postulated that improper NaDC-1 function may lead to hypocitraturia and the subsequent development of kidney stones (24, 27, 28). Despite their physiological importance, analysis of members of this transport family has been hindered by the limited information available regarding their structure.
As a first step in addressing structural aspects of DASS transporters, we have cloned and functionally expressed a bacterial member of this family, the sdcS gene product from S. aureus. In work described here, this transporter was found to exhibit many of the functional properties characteristic of mammalian Na+/dicarboxylate symporters. Thus, SdcS mediates the Na+-dependent uptake of substrates similar to those transported by its eukaryotic counterparts. These substrates are recognized with affinities comparable to those found for the high-affinity Na+/dicarboxylate cotransporter NaDC-3 (Km for substrate ranging from 2 to 30 μM) (5, 16, 36). Our kinetic studies showed that SdcS affinity for succinate, but not its transport velocity, was dependent on activating concentrations of Na+ (Fig. 6A). Moreover, the apparent affinity of Na+ for SdcS increased as succinate concentration increased (Fig. 6B). The competitive activation of succinate transport by Na+ and the uncompetitive nature of the effect of succinate on Na+ affinity are symptomatic of an ordered transport mechanism (35). Therefore, we suggest a kinetic model for SdcS that is in good agreement with those proposed for various mammalian DASS transporters (21, 42): Na+/dicarboxylate symport requires that the binding of Na+ at the extracellular surface precede the binding of dicarboxylate.
The presence of Na+ (or Li+) is essential to the function of both SdcS and its mammalian homologs. However, our work highlights two differences in how these transporters couple the cation to dicarboxylate uptake. First, among these cotransporters, only with SdcS does Na+ have closely linked stimulatory and inhibitory activities (Fig. 5); other DASS family members have K0.5 activation values that range from 10 to 50 mM (5, 16, 23, 26) but show no dicarboxylate transport inhibition at Na+ concentrations below 100 mM. While we can offer no definitive answer for this unusual behavior at the present time, this finding does suggest that SdcS activity in S. aureus is tightly regulated by the extracellular Na+ concentration. Second, while functionally characterized NaDC-1 and NaDC-3 orthologs are electrogenic (24, 27, 28)—coupling three Na+ ions to the transport of a single divalent carboxylate—our preliminary work regarding the electrical character of SdcS indicates that this transporter facilitates the electroneutral transport of Na+ and dicarboxylate with a stoichiometry of 2:1. Such differences in coupling stoichiometry are not unusual in functionally (and evolutionarily) related transporters. For instance, the human Na+/glucose transporter (hSGLT1) and the E. coli Na+/proline transporter (PutP), both members of the Na+/substrate symporter family (15, 40), catalyze uptake with a Na+:substrate ratio of 2:1 and 1:1, respectively (4, 38). In spite of this and other dissimilarities, these two proteins, as well as other Na+/substrate symporter family members, share a common structural theme (15, 40).
Structural information gleaned from the study of bacterial transporters has often been applied to the analysis of their eukaryotic homologs. For instance, the solved structures of the glycerol 3-phosphate antiporter (GlpT) and lactose permease (LacY) of E. coli and the oxalate/formate exchange protein (OxlT) from Oxalobacter formigenes provide a framework for the architecture of eukaryotic members of the major facilitator superfamily (1, 10, 11). Similarly, structure and function studies of eukaryotic ATP-binding cassette transporters have been guided by the known structures of the MsbA lipid and BtuCD vitamin B12 transporters of E. coli (3, 22). Within the DASS family, to our knowledge only two prokaryotic proteins have been cloned and functionally characterized, SdcS and the E. coli citrate carrier CitT (30). Of these two proteins, only SdcS has a transport mechanism similar to that of its mammalian counterparts; whereas CitT catalyzes dicarboxylate exchange, SdcS and its eukaryotic homologs function as Na+-dependent cotransporters. Comparison of the SdcS sequence with those of mammalian DASS transporters such as human NaDC-1 and human NaDC-3 showed that these proteins share significant amino acid identity (∼35% for human NaDC-1; ∼33% for human NaDC-3), with conserved residues distributed across their entire lengths. This similarity extends to the structural level where, using membrane topology prediction algorithms, we have recently found that in many cases putative SdcS transmembrane segments are predicted to be present in regions where conserved residues are clustered. Taken together, these findings suggest that SdcS can serve as a bacterial paradigm for the functional and structural properties of its eukaryotic counterparts.
Acknowledgments
This work was supported by grant DK46269 from the National Institutes of Health (to A.M.P.).
We thank Wolfgang Epstein and Peter Maloney for helpful discussions.
REFERENCES
- 1.Abramson, J., I. Smirnova, V. Kasho, G. Verner, H. R. Kaback, and S. Iwata. 2003. Structure and mechanism of the lactose permease of Escherichia coli. Science 301:610-615. [DOI] [PubMed] [Google Scholar]
- 2.Bai, L., and A. M. Pajor. 1997. Expression cloning of NaDC-2, an intestinal Na+- or Li+-dependent dicarboxylate transporter. Am. J. Physiol. 273:G267-G274. [DOI] [PubMed] [Google Scholar]
- 3.Chang, G., and C. B. Roth. 2001. Structure of MsbA from E. coli: a homolog of the multidrug resistance ATP binding cassette (ABC) transporters. Science 293:1793-1800. [DOI] [PubMed] [Google Scholar]
- 4.Chen, C. C., and T. H. Wilson. 1986. Solubilization and functional reconstitution of the proline transport system of Escherichia coli. J. Biol. Chem. 261:2599-2604. [PubMed] [Google Scholar]
- 5.Chen, X., H. Tsukaguchi, X. Z. Chen, U. V. Berger, and M. A. Hediger. 1999. Molecular and functional analysis of SDCT2, a novel rat sodium-dependent dicarboxylate transporter. J. Clin. Investig. 103:1159-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies, S. J., P. Golby, D. Omrani, S. A. Broad, V. L. Harrington, J. R. Guest, D. J. Kelly, and S. C. Andrews. 1999. Inactivation and regulation of the aerobic C4-dicarboxylate transport (dctA) gene of Escherichia coli. J. Bacteriol. 181:5624-5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dawson, P. A., L. Beck, and D. Markovich. 2003. Hyposulfatemia, growth retardation, reduced fertility, and seizures in mice lacking a functional NaSi-1 gene. Proc. Natl. Acad. Sci. USA 100:13704-13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dixon, M., and E. C. Webb. 1979. Enzymes. Academic Press, New York, N.Y.
- 9.Girard, J. P., E. S. Baekkevold, J. Feliu, P. Brandtzaeg, and F. Amalric. 1999. Molecular cloning and functional analysis of SUT-1, a sulfate transporter from human high endothelial venules. Proc. Natl. Acad. Sci. USA 96:12772-12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heymann, J. A., R. Sarker, T. Hirai, D. Shi, J. L. Milne, P. C. Maloney, and S. Subramaniam. 2001. Projection structure and molecular architecture of OxlT, a bacterial membrane transporter. EMBO J. 20:4408-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang, Y., M. J. Lemieux, J. Song, M. Auer, and D. N. Wang. 2003. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science 301:616-620. [DOI] [PubMed] [Google Scholar]
- 12.Inoue, K., Y. J. Fei, W. Huang, L. Zhuang, Z. Chen, and V. Ganapathy. 2002. Functional identity of Drosophila melanogaster Indy as a cation-independent, electroneutral transporter for tricarboxylic acid-cycle intermediates. Biochem. J. 367:313-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janausch, I. G., E. Zientz, Q. H. Tran, A. Kroger, and G. Unden. 2002. C4-dicarboxylate carriers and sensors in bacteria. Biochim. Biophys. Acta 1553:39-56. [DOI] [PubMed] [Google Scholar]
- 14.Jung, H. 2001. Towards the molecular mechanism of Na+/solute symport in prokaryotes. Biochim. Biophys. Acta 1505:131-143. [DOI] [PubMed] [Google Scholar]
- 15.Jung, H. 2002. The sodium/substrate symporter family: structural and functional features. FEBS Lett. 529:73-77. [DOI] [PubMed] [Google Scholar]
- 16.Kekuda, R., H. Wang, W. Huang, A. M. Pajor, F. H. Leibach, L. D. Devoe, P. D. Prasad, and V. Ganapathy. 1999. Primary structure and functional characteristics of a mammalian sodium-coupled high affinity dicarboxylate transporter. J. Biol. Chem. 274:3422-3429. [DOI] [PubMed] [Google Scholar]
- 17.Knauf, F., B. Rogina, Z. Jiang, P. S. Aronson, and S. L. Helfand. 2002. Functional characterization and immunolocalization of the transporter encoded by the life-extending gene Indy. Proc. Natl. Acad. Sci. USA 99:14315-14319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knol, J., L. Veenhoff, W. J. Liang, P. J. Henderson, G. Leblanc, and B. Poolman. 1996. Unidirectional reconstitution into detergent-destabilized liposomes of the purified lactose transport system of Streptococcus thermophilus. J. Biol. Chem. 271:15358-15366. [DOI] [PubMed] [Google Scholar]
- 19.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 21.Li, H., and A. M. Pajor. 2003. Serines 260 and 288 are involved in sulfate transport by hNaSi-1. J. Biol. Chem. 278:37204-37212. [DOI] [PubMed] [Google Scholar]
- 22.Locher, K. P., A. T. Lee, and D. C. Rees. 2002. The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science 296:1091-1098. [DOI] [PubMed] [Google Scholar]
- 23.Markovich, D., J. Forgo, G. Stange, J. Biber, and H. Murer. 1993. Expression cloning of rat renal Na+/SO42− cotransport. Proc. Natl. Acad. Sci. USA 90:8073-8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Markovich, D., and H. Murer. 2004. The SLC13 gene family of sodium sulphate/carboxylate cotransporters. Pflugers Arch. 447:594-602. [DOI] [PubMed] [Google Scholar]
- 25.Pajor, A. M. 1996. Molecular cloning and functional expression of a sodium-dicarboxylate cotransporter from human kidney. Am. J. Physiol. 270:F642-F648. [DOI] [PubMed] [Google Scholar]
- 26.Pajor, A. M. 1995. Sequence and functional characterization of a renal sodium/dicarboxylate cotransporter. J. Biol. Chem. 270:5779-5785. [DOI] [PubMed] [Google Scholar]
- 27.Pajor, A. M. 1999. Sodium-coupled transporters for Krebs cycle intermediates. Annu. Rev. Physiol. 61:663-682. [DOI] [PubMed] [Google Scholar]
- 28.Pajor, A. M. 2000. Molecular properties of sodium/dicarboxylate cotransporters. J. Membr. Biol. 175:1-8. [DOI] [PubMed] [Google Scholar]
- 29.Pajor, A. M., B. A. Hirayama, and D. D. Loo. 1998. Sodium and lithium interactions with the Na+/dicarboxylate cotransporter. J. Biol. Chem. 273:18923-18929. [DOI] [PubMed] [Google Scholar]
- 30.Pos, K. M., P. Dimroth, and M. Bott. 1998. The Escherichia coli citrate carrier CitT: a member of a novel eubacterial transporter family related to the 2-oxoglutarate/malate translocator from spinach chloroplasts. J. Bacteriol. 180:4160-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prakash, S., G. Cooper, S. Singhi, and M. H. Saier, Jr. 2003. The ion transporter superfamily. Biochim. Biophys. Acta 1618:79-92. [DOI] [PubMed] [Google Scholar]
- 32.Reizer, J., A. Reizer, and M. H. Saier, Jr. 1994. A functional superfamily of sodium/solute symporters. Biochim. Biophys. Acta 1197:133-166. [DOI] [PubMed] [Google Scholar]
- 33.Rogina, B., R. A. Reenan, S. P. Nilsen, and S. L. Helfand. 2000. Extended life-span conferred by cotransporter gene mutations in Drosophila. Science 290:2137-2140. [DOI] [PubMed] [Google Scholar]
- 34.Ruan, Z. S., V. Anantharam, I. T. Crawford, S. V. Ambudkar, S. Y. Rhee, M. J. Allison, and P. C. Maloney. 1992. Identification, purification, and reconstitution of OxlT, the oxalate:formate antiport protein of Oxalobacter formigenes. J. Biol. Chem. 267:10537-10543. [PubMed] [Google Scholar]
- 35.Segel, I. H. 1993. Enzyme kinetics: behavior and analysis of rapid equilibrium and steady-state enzyme systems. Wiley-Interscience, New York, N.Y.
- 36.Steffgen, J., B. C. Burckhardt, C. Langenberg, L. Kuhne, G. A. Muller, G. Burckhardt, and N. A. Wolff. 1999. Expression cloning and characterization of a novel sodium-dicarboxylate cotransporter from winter flounder kidney. J. Biol. Chem. 274:20191-20196. [DOI] [PubMed] [Google Scholar]
- 37.Wilson, T. H., and P. Z. Ding. 2001. Sodium-substrate cotransport in bacteria. Biochim. Biophys. Acta 1505:121-130. [DOI] [PubMed] [Google Scholar]
- 38.Wright, E. M. 2001. Renal Na+-glucose cotransporters. Am. J. Physiol. Renal Physiol. 280:F10-F18. [DOI] [PubMed] [Google Scholar]
- 39.Wright, E. M., K. M. Hager, and E. Turk. 1992. Sodium cotransport proteins. Curr. Opin. Cell Biol. 4:696-702. [DOI] [PubMed] [Google Scholar]
- 40.Wright, E. M., and E. Turk. 2004. The sodium/glucose cotransport family SLC5. Pflugers Arch. 447:510-518. [DOI] [PubMed] [Google Scholar]
- 41.Wright, E. M., S. H. Wright, B. Hirayama, and I. Kippen. 1982. Interactions between lithium and renal transport of Krebs cycle intermediates. Proc. Natl. Acad. Sci. USA 79:7514-7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wright, S. H., B. Hirayama, J. D. Kaunitz, I. Kippen, and E. M. Wright. 1983. Kinetics of sodium succinate cotransport across renal brush-border membranes. J. Biol. Chem. 258:5456-5462. [PubMed] [Google Scholar]