Abstract
Staphylococcus aureus expresses various cell wall-associated and extracellular virulence factors, coordinately controlled by different two-component signal transduction systems and transcriptional regulators. In this study, we used microarray technology to identify the genes regulated by ArlR. The microarray data indicate that ArlR functions as a positive regulator and also as a negative repressor to directly and/or indirectly mediate the expression of at least 114 genes involved in different functions, including autolysis, cell division, growth, and pathogenesis.
Staphylococcus aureus is an important human and animal pathogen that causes a wide range of infections, including life-threatening endocarditis and toxic shock syndrome (22, 26). The ability of this organism to cause a variety of diseases is partly due to the expression of different cell wall-associated and secreted virulence factors which enable the bacteria to adhere to and colonize host cells (13) or cause toxic shock syndrome (26). The expression of virulence factors is coordinately controlled by two-component signal transduction systems, such as agr (1), sae (16), arl (14), and srrAB (30), and global regulators, including sar (8, 9), sigB (3, 31), rot (2, 24), and mgr (23). Therefore, the elucidation of the regulons of these regulatory systems is important for better understanding molecular mechanisms of pathogenesis. Recently, S. aureus regulons of agr, sar, sigB, and rot have been revealed by using a microarray-based approach (4, 11, 27). In our studies, we identified target genes controlled by ArlR by a comparison of the transcriptional profile between an arlR mutant and the wild-type strain during the mid-exponential phase of growth by using Affymetrix S. aureus arrays.
Construction of the arlR deletion mutant.
The arlR deletion mutant (Sa316ko) was constructed by bacteriophage φ11-mediated transduction of a cassette containing the tetA gene, flanked by chromosomal fragments upstream and downstream of the alrR, from strain RN4220 into a clinical human isolate strain WCUH29 as described previously (12). Selection for tetracycline resistance and screening for the loss of the erythromycin resistance marker carried by the vector indicated that allelic replacement had occurred and resulted in the arlR mutant strain Sa316ko. The mutation in arlR was verified by PCR and Southern blot analysis (data not shown).
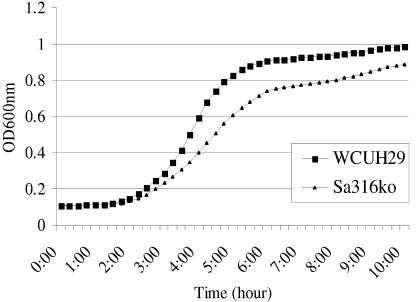
To characterize the arlR mutant strain, we examined the effect of the arlR mutation on the bacterial growth rate, CFU, phenotype, and stress responses to different antibacterial agents, including cell wall synthesis inhibitors such as bacitracin, phosphomycin, and vancomycin. No significant effect of the arlR mutation on stress response to antibacterial agents was observed (data not shown). However, Sa316ko grew slightly slower than WCUH29 between early log and stationary phases (Fig. 1). Growth curves as measured by optical density (OD) were confirmed by determining the effect of arlR mutation on colony size and viable cell counts at intervals during growth. The ArlR mutant strain, Sa316ko, displayed similar-sized colonies on a tryptic soy agar plate and similar morphological features of individual cells under microscopic observation (data not shown) but decreased approximately one log CFU compared to wild-type strain between early log and stationary phases (data not shown). These results suggest that the arlRS regulon may be involved in the modulation of expression of genes associated with growth and cell division. Although it is unlikely that the arlR allelic gene replacement mutation has a polar effect on arlRS downstream gene expression (since arlR and arlS loci are located in a single operon), we examined the transcription level of arlRS downstream gene odhA, encoding 2-oxoglutarate dehydrogenase E1. Our real-time reverse transcriptase (RT)-PCR analyses showed that the arlR mutation had no significant influence on the expression of odhA (see Table 5).
FIG. 1.
Growth curve of the arlR mutant. The arlR mutant strain Sa316ko and parent strain WCUH29 were incubated in TSB at 37°C overnight with shaking; the cultures were inoculated in fresh TSB. The cell growth was monitored at 37°C by a measuring of the OD at 600 nm every 15 min with 1 min of mixing before each reading. These curves represent one of three reproducible experiments.
TABLE 5.
Real-time RT-PCR analysis of expression of genes regulated by ArlR in different growth phases
| N315 ORF | N315 gene | N315 description | Change in expression level (n-fold)a in RT-PCR by phase
|
||
|---|---|---|---|---|---|
| Early log | Mid-log | Stationary | |||
| SA0250 | lytS | Two-component sensor kinase | −4.4 | −7 | −6.7 |
| SA1844 | agrA | Two-component response regulator | −2.7 | −2.3 | −3.9 |
| SA1882 | kdpD | Two-component sensor kinase | −2.6 | 13.8 | 1.2 |
| SA1993 | lacF | PTS system, lactose specific IIA component | 6.3 | 4 | 13 |
| SA1245b | odhA | 2-Oxoglutarate dehydrogenase E1 | NC | NC | NC |
Normalized values in the arlR mutant over values in the wild-type strain. Negative numbers denote up-regulation in the wild-type strain.
Downstream gene of arlRS. NC, no detectable change.
Identification of the arlRS regulon using microarray assay.
To better define the arlRS regulon, gene expression profiles of the arlR mutant and parent cells were analyzed by using Affymetrix S. aureus arrays as described previously (11). The S. aureus array (Affymetrix) contains probe sets to over 3,300 S. aureus open reading frames (ORFs) based on the updated S. aureus genomic sequences of N315, Mu50, NCTC 8325, and COL. Total RNA was extracted from S. aureus cells grown to mid-log phase (OD at 600 nm, 0.4) by using the RNAPrep kit (Promega, MI) and treated with a DNA-free kit (Ambion). The RNA (10 μg) was reverse transcribed to cDNA by using Superscript II reverse transcriptase and random primers (Invitrogen). The cDNA was treated with NaOH, purified by using the QIAquick PCR purification kit (QIAGEN), and digested with DNase I. The fragmented cDNAs were then directly labeled with biotin by using a biotin-ddUTP kit (Affymetrix). Biotinylated cDNA (3 μg) was hybridized to the GeneChips. The GeneChips were then washed and subjected to a series of staining procedures as described in the manual for the Affymetrix array. Each GeneChip was washed and scanned at a 570-nm wavelength and a 3-μm resolution in an Affymetrix GeneChip scanner. The Affymetrix Microarray Suite 4.0 algorithms calculated the signal intensities (average differences) and the present or absent determinations for each open reading frame. The GeneChips were then normalized, and their backgrounds were defined by using GeneSpring 4.0 (Silicon Genetics). The GeneSpring software was used to further analyze the transcription patterns of genes. To identify genes with significantly altered expression levels, a series of statistical analyses (filtering) were performed; cutoff values for ratio of expression levels of 1.80 and 0.55 were used to filter genes with expression level changes (n-fold) greater than ±1.8 in all three independent biological samples. Genes with variations (n-fold) of >1.5 across the three samples were excluded. Furthermore, a statistical group comparison using the Student t test/analysis of variance was conducted to compare the mean expression levels of the control and the arlR mutant samples. The genes with significant differential expression levels (P value, <0.05) were selected.
The results of three independent experiments demonstrated that on average, transcripts for 73% of all genes on the arrays were detected by the Affymetrix arrays in the mid-log phase of bacterial cells of WCUH29. A comparison analysis of gene expression levels between wild-type WCUH29 and the arlR mutant revealed that the expression levels of 114 genes were significantly altered in the arlR mutant. Of these, 37 genes showed a decrease (Table 1) and 77 genes showed an increase (Table 2) in expression level after the mutation of arlR.
TABLE 1.
S. aureus genes up-regulated by ArlR
| N315 ORF | N315 gene | N315 description | Change in expression level (n-fold)a | agr, sar, rot, or sigB effectb |
|---|---|---|---|---|
| SA1844 | agrA | Response regulator | −2.8 | |
| SA1842 | agrB | Accessory gene regulator B | −2.2 | |
| SA1843 | agrC | Sensor histidine kinase | −2.4 | |
| SAS066 | agrD | AgrD protein | −1.6 | |
| SA1248 | arlR | Response regulator | −602.2 | |
| SAS065 | hld | Delta-hemolysin | −2.2 | agr + |
| SA0250 | lytS | Two-component sensor histidine kinase | −3.1 | rot + |
| SA0251 | lytR | Two-component response regulator | −2.2 | |
| SA0144 | cap5A | Capsular polysaccharide synthesis enzyme Cap5A | −1.8 | σB + |
| SA0147 | cap5D | Capsular polysaccharide synthesis enzyme Cap5D | −1.5 | σB + |
| SA0211 | Putative oxidoreductase | −2.1 | ||
| SA0212 | Hypothetical protein | −1.9 | ||
| SA0220 | Hypothetical protein | −2.8 | rot + | |
| SA0252 | lrgA | Holin-like protein LrgA | −16.5 | sar +, sigB − |
| SA0253 | lrgB | Holin-like protein LrgB | −19.7 | sar +, sigB − |
| SA0269 | Hypothetical protein | −2.3 | ||
| SA0270 | ssaA | Secretory antigen precursor SsaA | −2 | sigB |
| SA0271 | Hypothetical protein | −5.9 | agr +, rot +, | |
| SA0272 | Hypothetical protein | −7.4 | sigB − | |
| SA0275 | Hypothetical protein | −4.2 | sigB − | |
| SA0276 | Similar to diarrheal toxin | −4.4 | ||
| SA0417 | Hypothetical protein | −2.5 | ||
| SA0519 | sdrC | Ser-Asp-rich fibrinogen-binding protein SdrC | −2.3 | sar +, rot + |
| SA0520 | sdrD | Ser-Asp-rich fibrinogen-binding protein SdrD | −3.7 | |
| SA0521 | sdrE | Ser-Asp-rich fibrinogen-binding protein SdrE | −2.1 | |
| SA0746 | Staphylococcal nuclease | −1.9 | ||
| SA0893 | Hypothetical protein | −1.9 | ||
| SA1269 | Blt-like protein | −3.1 | ||
| SA1270 | Similar to amino acid pearmease | −1.8 | ||
| SA1271 | IlvA threonine deaminase | −3 | ||
| SA1272 | Alanine dehydrogenase | −2.4 | ||
| SA1305 | Cell-dividion initiation protein | −4.9 | ||
| SA1583 | rot | Repressor of toxin | −1.9 | |
| SA2222 | Bicyclomycin-resistant protein TcaB | −2 | ||
| SA2303 | Hypothetical protein | −11.2 | rot + | |
| SA2486 | 2-Oxoglutarate/malate translocator | −1.8 |
TABLE 2.
S. aureus genes repressed by ArlR
| N315 ORF | N315 gene | N315 description | Change in expression level (n-fold)a | agr, sar, rot, or sigB effectb |
|---|---|---|---|---|
| SA0104 | Hypothetical protein | 1.9 | ||
| SA0123 | Hypothetical protein | 22.1 | rot− | |
| SA0124 | Hypothetical protein | 35 | ||
| SA0125 | Hypothetical protein | 39.6 | ||
| SA0126 | Hypothetical protein | 20.4 | ||
| SA0127 | cpsM | Capsular polysaccharide repeat, Unit transporter cpsM | 4 | rot − |
| SA0135 | Hypothetical protein | 1.6 | ||
| SA0136 | Hypothetical protein | 2.7 | ||
| SA0137 | Hypothetical protein | 3.6 | ||
| SA0138 | Hypothetical protein | 5.6 | ||
| SA0165 | Hypothetical protein | 1.8 | rot − | |
| SA0299 | PfkB family carbohydrate kinase | 2.5 | ||
| SA0304 | nanA | N-Acetylneuraminate lyase | 2.5 | |
| SA0318 | Putative membrane protein | 2.5 | ||
| SA0319 | Hypothetical protein | 4.7 | ||
| SA0320 | Hypothetical protein | 5.1 | ||
| SA0321 | Putative PTS multidomain regulator | 5.5 | ||
| SA0331 | Hypothetical protein | 14.2 | ||
| SA0332 | Hypothetical protein | 8 | ||
| SA0333 | Conserved hypothetical protein | 10.6 | ||
| SA0395 | Hypothetical protein | 2.2 | ||
| SA0710 | Conserved hypothetical protein | 7.4 | rot − | |
| SA0850 | Hypothetical protein | 10.7 | rot − | |
| SA0851 | Oligopeptide ABC transporter, ATP-binding protein | 2.2 | ||
| SA0899 | sspC | Cysteine protease | 3.1 | agr +, rot − |
| SA0904 | Hypothetical protein | 10.7 | rot − | |
| SA0956 | Hypothetical protein | 2.9 | ||
| SA0978 | isdC | Conserved hypothetical protein | 1.9 | |
| SA1090 | lytN | LytN protein | 10.4 | agr +, sar +, rot −, sigB − |
| SA1091 | fmhC | FmhC protein | 3.9 | rot − |
| SA1145 | Host factor-1 protein | 3 | ||
| SA1154 | Conserved hypothetical protein | 9.1 | ||
| SA1266 | Hypothetical protein | 6.4 | ||
| SA1267 | ebhA | Hypothetical protein | 15.1 | rot − |
| SA1268 | ebhB | Hypothetical protein | 28.6 | agr +, rot − |
| SA1552 | Hypothetical protein | 3.7 | ||
| SA1577 | Hypothetical protein | 17.2 | ||
| SA1630 | splB | Serine protease, V8 protease | 4.3 | agr +, sar +, rot −, sigB − |
| SA1637 | lukD | Leukotoxin, LukD | 1.9 | |
| SA1638 | lukE | Leukotoxin, LukE | 1.6 | |
| SA1752 | hlb | Truncated beta-hemolysin | 2.5 | |
| SA1848 | nrgA | Probable ammonium transporter | 1.9 | |
| SA1882 | kdpD | Two-component sensor kinase KdpD | 2.9 | |
| SA1883 | kdpE | Two-component response regulator KdpE | 2 | |
| SA1991 | lacG | 6-Phospho-beta-galactosidase | 4.1 | |
| SA1992 | PTS system, lactose-specific IIBC component | 3.5 | ||
| SA1993 | lacF | PTS system, lactose-specific IIA component | 6.5 | |
| SA1994 | lacD | Tagatose 1,6-diphosphate aldolase | 4.4 | |
| SA1995 | lacC | Tagatose-6-phosphate kinase | 4.9 | |
| SA1996 | lacB | Galactose-6-phosphate isomerase LacB subunit | 7.2 | |
| SA1997 | lacA | Galactose-6-phosphate isomerase LacA subunit | 5.1 | |
| SA2006 | Hypothetical protein | 5.5 | ||
| SA2007 | Hypothetical protein | 2.8 | ||
| SA2008 | alsS | Alpha-acetolactate synthase | 2.4 | |
| SA2081 | Urea transporter | 3.1 | ||
| SA2082 | ureA | Urease gamma subunit | 9.3 | rot − |
| SA2083 | ureB | Urease beta subunit | 12.1 | rot − |
| SA2084 | ureC | Urease alpha subunit | 7 | rot − |
| SA2085 | ureE | Urease accessory protein UreE | 5.2 | rot − |
| SA2086 | ureF | Urease accessory protein UreF | 4.6 | rot − |
| SA2087 | ureG | Urease accessory protein UreG | 3.6 | rot − |
| SA2088 | ureD | Urease accessory protein UreD | 4.5 | rot − |
| SA2091 | Hypothetical protein | 6.7 | ||
| SA2092 | Hypothetical protein | 4.5 | ||
| SA2209 | hlgC | Gamma-hemolysin component C precursor | 3.2 | agr +, rot − |
| SA2287 | Staphylococcal regulator SarH2 | 7.9 | ||
| SA2315 | Putative membrane protein | 2.6 | ||
| SA2319 | Putative l-serine dehydratase | 3.8 | ||
| SA2320 | Putative membrane protein | 4.7 | ||
| SA2329 | Murine hydrose exporter | 3.8 | ||
| SA2337 | feoB | Ferrous iron transporter protein | 4 | |
| SA2338 | Hypothetical protein | 7.3 | ||
| SA2382 | Hypothetical protein | 5.4 | ||
| SA2455 | cap8C | Capsular polysaccharide synthesis enzyme Cap8C | 12.7 | rot −, sigB + |
| SA2456 | cap8B | Capsular polysaccharide synthesis enzyme Cap8B | 18.1 | rot −, sigB + |
| SA2457 | capA | Capsular polysaccharide biosynthesis CapA | 31.2 | rot −, sigB + |
| SA2482 | arcA | Arginine deiminase | 2 | |
| SAV0397 | Hypothetical protein | 12.1 |
The array data indicated that ArlR positively regulates a two-component system, lytR-lytS, which encodes a response regulator and a sensor histidine kinase and is involved in autolysis (6, 14). The positive regulation of lytS expression by ArlR was confirmed by real-time RT-PCR analysis using the Stratagene Mx3000P real-time PCR system. Gene-specific primers were designed to yield ∼100 bp of specific products (Table 3), and the housekeeping gene 16S rRNA was used as an endogenous control (29). All samples were analyzed in triplicate and normalized against 16S rRNA gene expression. The results were statistically analyzed for correlation to the microarray results. Compared to the wild type, the arlR mutant possessed low levels of lytS mRNA in the early log, mid-log, and stationary phases of growth (Tables 4 and 5). We also found that the mutation in arlR significantly down-regulates the expression of lrgA and lrgB, encoding different holin-like proteins involved in murein hydrolase transport and inhibition of murein hydrolase activity (7, 17). These enzymes are involved in the cleavage of specific cell wall components, which are important for cell division and growth (7, 17).
TABLE 3.
Primers used in real-time RT-PCR
| Primer | Sequence (5′-3′) |
|---|---|
| SA0123for | ATATTACGGCGAACGGACGAC |
| SA0123rev | TGGCTTGTTATGCTCAAATGAATCG |
| SA0250for | GCATGGTTCTATCGTCGGTACATTG |
| SA0250rev | ACTTACTTTGCGTTTCGGCTTCAC |
| SA0252lrgAfor | TGAAACAACAAAAAGACGCATCAAAACCAG |
| SA0252lrgArev | ACTTCGCCTAACTTAACAGCACCAG |
| SA0270ssaAfor | GGCATCCAAGTCAATTAAACCAAGATAATG |
| SA0270ssaArev | CAGTACGGTAGCTGTTTGTGTTGTAAC |
| SA0319for | GCACCATCTGATATCGAAGTTGAAC |
| SA0319rev | TAGGCGTTCGGCATTTTCAGC |
| SA1248for | TGACAAAGTTGCTGGGCTTGATTAC |
| SA1248rev | TGTGGCTGACGACGTAAAATTGC |
| SA1583for | TCAGCGAGATTGAAAGCGAATAC |
| SA1583rev | CTGTCCATTTCTTTAAGCGTCATAG |
| SA1637for | TGGGGCGGTAAGTATAATGTTTCG |
| SA1637rev | GATCCATTCAATCCACCTGATAAGC |
| SA1882for | GAAAGACAAGCTGGTGCAACAAC |
| SA1882rev | AACGGCGAGAGAAAGTTCATTTAAC |
| SA1844agrAfor | GTGAAATTCGTAAGCATGACCCAGTTG |
| SA1844agrArev | TGTAAGCGTGTATGTGCAGTTTCTAAAC |
| SA1993for | GAAGGAAACAATTGCATTGCTGAAG |
| SA1993rev | ATATCATCACCTTGCGCTTCTTTAG |
| SA1245odhA1665for | GCAATGAACCACCCGTAGAATAGC |
| SA1245odhA1833rev | ACGAGAGCAGCACAAGATGATACAC |
| 16S rRNAfor | CTGTGCACATCTTGACGGTA |
| 16S rRNArev | TCAGCGTCAGTTACAGACCA |
TABLE 4.
Real-time RT-PCR analysis of expression of genes regulated by ArlR
| N315 ORF | N315 gene | N315 description | Change in expression level (n-fold)a
|
|
|---|---|---|---|---|
| RT-PCR | Microarray | |||
| SA0123 | Hypothetical protein | 42.8 | 22.1 | |
| SA0319 | Hypothetical protein | 5.1 | 4.7 | |
| SA1637 | lukD | Leukotoxin LukD | 2 | 1.9 |
| SA1882 | kdpD | Two-component sensor kinase | 3.1 | 2.9 |
| SA1993 | lacF | PTS system, lactose-specific IIA component | 17 | 6.5 |
| SA1844 | agrA | Two-component response regulator | −5.7 | −2.8 |
| SA0250 | lytS | Two-component sensor kinase | −2.8 | −3.1 |
| SA0252 | lrgA | Holin-like protein LrgA | −64.8 | −16.5 |
| SA0270 | ssaA | Secretory antigen precursor SsaA | −1.7 | −2 |
| SA1583 | rot | Repressor of toxin | −4 | −1.9 |
Normalized values in the arlR mutant over values in the wild-type strain. Negative numbers denote up-regulation in the wild-type strain.
To determine whether the growth defect of the mutant is attributable to increased susceptibility to cell lysis, we examined the effect of ArlR on autolysis induced by Triton X-100 and detected the cell wall murein hydrolase activity by using a zymographic assay as described previously (19). Consistent with a previous report (14), the arlR mutant cells displayed increased lysis in the presence of 0.01% Triton and showed enhanced peptidoglycan hydrolase activity compared to the parent control (unpublished data). These results indicated that increased autolysis in the arlR mutant may result from the significant down-regulation of the lytSR and lrgAB operons and, in turn, may partially affect bacterial growth. Although it has been reported that the mutation in rat, another autolysis regulator, exhibited a growth defect and enhanced autolysis partly due to increased murein hydrolase activity (19), the reason why the mutation in arlR led to a slight impact on growth remains undefined, since the lrgAB mutation did not show significant impact on the cell shape and growth rate (17).
Also, our microarray data showed that ArlR positively regulates virulence factor genes, such as sdrC, sdrD, and sdrE, encoding different Ser-Asp-rich bone sialoprotein-binding proteins (Table 1) (28). The result for sdrD was confirmed by using real-time RT-PCR and demonstrated that the level of sdrD mRNA in the arlR mutant strain is significantly decreased compared with that in the wild-type strain (Table 4). The up-regulation of sdrC by ArlR may indirectly function via up-regulated rot expression, since Rot positively regulates the expression of sdrC (11). In addition, ArlR positively regulates the tcaB gene (which encodes a bicyclomycin-resistant protein), the secretory antigen precursor ssaA, and toxin genes, such as hld and SA0276, encoding delta-hemolysin and diarrheal toxin, respectively (Table 1).
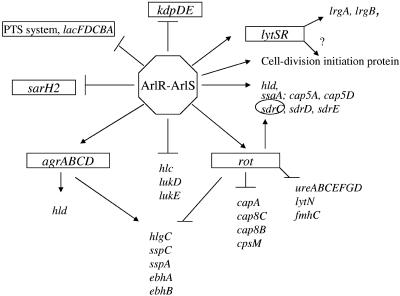
Moreover, the array data showed that ArlR positively regulates the accessory gene regulator (agr) (Table 1). A real-time RT-PCR was employed to validate this result and demonstrated that the mutation in arlR significantly decreased the level of agrA expression (Table 4). The expression of agrA was constantly up-regulated by ArlR at different times of growth (Table 5). These results are inconsistent with the finding that the mutation in arlS leads to the overexpression of Agr (15). This controversy may be due to different sensitivities, culture conditions, and time points of sampling between different assays. In addition, the array results indicated that ArlR also positively regulates the expression of the repressor of toxins (rot). This result was confirmed by real-time RT-PCR and demonstrated that the mutation in arlR causes a decrease in rot expression (Table 4). Our findings are consistent with previous reports that the expression of certain secreted enzymes, toxins, and ureases are repressed by Rot (9, 27). Most virulence factors negatively regulated by ArlR were also repressed by Rot but were up-regulated by Agr (5, 25). Therefore, the down-regulation of these toxins, proteases, and adhesins may be mediated directly by ArlR or indirectly controlled via Rot or Agr (Fig. 2).
FIG. 2.
Schematic figure showing how the staphylococcal two-component signal transduction regulatory system, ArlRS, directly and/or indirectly modulates gene expression. →, positive regulation; ⊥, negative regulation.
To determine the role of the ArlRS regulatory system in pathogenesis, we examined the effect of the arlR mutation on virulence by using a murine hematogenous pyelonephritis model as described previously (20). The virulence of the arlR mutant was significantly attenuated compared to that of the parent control (unpublished data). Collectively, these findings suggest that ArlRS is a global two-component virulence regulatory system which can interact with other regulators to modulate the expression of virulence factors.
On the other hand, ArlR functions as a repressor of virulence factors. Our microarray data showed that ArlR negatively regulates some toxin genes, lukD, lukE, phlC (hlb), and hlgC, which encode leukotoxin D, leukotoxin E, beta-hemolysin component C, and gamma-hemolysin component C, respectively (Table 2). Gamma-hemolysin is an S. aureus virulence factor that has been shown to play a role in S. aureus endophthalmitis and corneal pathogenesis (10, 21). The microarray result for lukD was confirmed by real-time RT-PCR. As shown in Table 4, the lukD transcript was present at higher levels in the arlR mutant strain than in the wild-type strain. The array results also demonstrated that ArlR negatively regulates splB (encoding V8 protease), sspC (encoding cysteine protease), and ebhA and ebhB (encoding adhesins) (Table 2). The negative regulation of ebhB expression by ArlR was confirmed by a real-time RT-PCR (Table 4).
Furthermore, the array results showed that ArlR negatively regulates the expression of genes involved in different PTS systems, such as the lacG, lacF, lacD, lacC, lacB, and lacA operons, as well as hypothetical proteins. To validate these results, real-time RT-PCR was performed, and it demonstrated that the mutation of arlR increases the expression of hypothetical proteins (SA0123 and SA0319) and constantly up-regulates lacF expression at different phases of growth (Tables 4 and 5). Also, we found that in the arlR mutant, the levels of kdpD and kdpE transcripts (which encode a two-component sensor kinase and response regulator and involve K+ transport [18]) were obviously higher than those in the wild-type strain. To confirm this result, real-time RT-PCR was performed, and the results demonstrated that the mutation of arlR increases kdpD expression in the mid-log phase of growth but decreases kdpD expression in the early log phase of growth (Tables 4 and 5). These results suggest that ArlR differentially regulates kdpD expression at different times of growth.
Conclusion.
The regulon of ArlRS has been identified by employing transcriptome technology using Affymetrix S. aureus arrays. The results demonstrate that ArlRS is a global transcriptional regulator which directly and/or indirectly interacts with other regulators in regulatory networks and modulates the expression of genes involved in autolysis, cell growth, and pathogenesis (Fig. 2). Some genes/operons mediated by ArlRS may be missed, due to their lack of stability and/or kinetic regulation as well as low detection sensitivity. Further studies to investigate which genes identified using microarray assays are directly regulated by ArlR and are involved in autolysis and/or pathogenesis are in progress.
Acknowledgments
We thank Junsong Sun for his assistance in the real-time RT-PCR analysis, Aaron Becker for his assistance in the microarray analysis, and Doug Weiss for his critical reading of the manuscript and for his helpful suggestions.
This work was supported by grant O3-O2 from the Academic Health Center at the University of Minnesota. This research was also supported in part by grant AI057451 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Abdelnour, A., S. Arvidson, T. Bremell, C. Ryden, and A. Tarkowski. 1993. The accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine arthritis model. Infect. Immun. 61:3879-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arvidson, S., and K. Tegmark. 2001. Regulation of virulence determinants in Staphylococcus aureus. Int. J. Med. Microbiol. 291:159-170. [DOI] [PubMed] [Google Scholar]
- 3.Bischoff, M., J. M. Entenza, and P. Giachino. 2001. Influence of a functional sigB operon on the global regulators sar and agr in Staphylococcus aureus. J. Bacteriol. 183:5171-5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bischoff, M., P. Dunman, J. Kormanec, D. Macapagal, E. Murphy, W. Mounts, B. Berger-Bächi, and S. Projan. 2004. Microarray-based analysis of the Staphylococcus aureus σB regulon. J. Bacteriol. 186:4085-4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bronner, S., H. Monteil, and G. Prevost. 2004. Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol. Rev. 28:183-200. [DOI] [PubMed] [Google Scholar]
- 6.Brunskill, E. W., and K. W. Bayles. 1996. Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J. Bacteriol. 178:611-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brunskill, E. W., and K. W. Bayles. 1996. Identification of LytSR-regulated genes from Staphylococcus aureus. J. Bacteriol. 178:5810-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung, A. L., J. M. Koomey, C. A. Butler, S. J. Projan, and V. A. Fischetti. 1992. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc. Natl. Acad. Sci. USA 89:6462-6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung, A. L., K. Schmidt, B. Bateman, and A. C. Manna. 2001. SarS, a SarA homolog repressible by agr, is an activator of protein A synthesis in Staphylococcus aureus. Infect. Immun. 69:2448-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dajcs, J. J., M. S. Austin, G. D. Sloop, J. M. Moreau, E. B. Hume, H. W. Thompson, F. M. McAleese, T. J. Foster, and R. J. O'Callaghan. 2002. Corneal pathogenesis of Staphylococcus aureus strain Newman. Investig. Ophthalmol. Vis. Sci. 43:1109-1115. [PubMed] [Google Scholar]
- 11.Dunman, P. M., E. Murphy, S. Haney, D. Palacios, G. Tucker-Kellogg, S. Wu, E. L. Brown, R. J. Zagursky, D. Shlaes, and S. J. Projan. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183:7341-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan, F., R. D. Lunsford, D. Sylvester, J. Fan, H. Celesnik, S. Iordanescu, M. Rosenberg, and D. McDevitt. 2001. Regulated ectopic expression and allelic-replacement mutagenesis as a method for gene essentiality testing in Staphylococcus aureus. Plasmid 46:71-75. [DOI] [PubMed] [Google Scholar]
- 13.Foster, T. J, and M. Höök. 1998. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 6:484-488. [DOI] [PubMed] [Google Scholar]
- 14.Fournier, B., and D. C. Hooper. 2000. A new two-component regulatory system involved in adhesion, autolysis, and extracellular proteolytic activity of Staphylococcus aureus. J. Bacteriol. 182:3955-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fournier, B., A. Klier, and G. Rapoport. 2001. The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 41:247-261. [DOI] [PubMed] [Google Scholar]
- 16.Giraudo, A. T., A. L. Cheung, and R. Nagel. 1997. The sae locus of Staphylococcus aureus controls exoprotein synthesis at the transcriptional level. Arch. Microbiol. 168:53-58. [DOI] [PubMed] [Google Scholar]
- 17.Groicher, K. H., B. A. Firek, D. F. Fujimoto, and K. W. Bayles. 2000. The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J. Bacteriol. 182:1794-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heermann, R., K. Altendorf, and K. Jung. 2003. The N-terminal input domain of the sensor kinase KdpD of Escherichia coli stabilizes the interaction between the cognate response regulator KdpE and the corresponding DNA-binding site. J. Biol. Chem. 278:51277-51284. [DOI] [PubMed] [Google Scholar]
- 19.Ingavale, S. S., W. Van Wamel, and A. L. Cheung. 2003. Characterization of Rat, an autolysis regulator in Staphylococcus aureus. Mol. Microbiol. 48:1451-1466. [DOI] [PubMed] [Google Scholar]
- 20.Ji, Y., A. Marra, M. Rosenberg, and G. Woodnutt. 1999. Regulated antisense RNA eliminates alpha-toxin virulence in Staphylococcus aureus infection. J. Bacteriol. 181:6585-6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karlsson, A., P. Saravia-Otten, K. Tegmark, E. Morfeldt, and S. Arvidson. 2001. Decreased amounts of cell wall-associated protein A and fibronectin-binding proteins in Staphylococcus aureus sarA mutants due to up-regulation of extracellular proteases. Infect. Immun. 69:4742-4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 23.Luong, T. T., S. W. Newell, and C. Y. Lee. 2003. mgr, a novel global regulator in Staphylococcus aureus. J. Bacteriol. 185:3703-3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McNamara, P. J., K. C. Milligan-Monroe, S. Khalili, and R. A. Proctor. 2000. Identification, cloning, and initial characterization of rot, a locus encoding a regulator of virulence factor expression in Staphylococcus aureus. J. Bacteriol. 182:3197-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429-1449. [DOI] [PubMed] [Google Scholar]
- 26.Projan, S., and R. Novick. 1997. The molecular basis of pathogenicity, p. 55-81. In G. Archer and K. Crossley (ed.), Staphylococci in human diseases. Churchill Livingstone, New York, N.Y.
- 27.Saïd-Salim, B., P. M. Dunman, F. M. McAleese, D. Macapagal, E. Murphy, P. J. McNamara, S. Arvidson, T. J. Foster, S. J. Projan, and B. N. Kreiswirth. 2003. Global regulation of Staphylococcus aureus genes by Rot. J. Bacteriol. 185:610-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tung, H., B. Guss, U. Hellman, L. Persson, K. Rubin, and C. Ryden. 2000. A bone sialoprotein-binding protein from Staphylococcus aureus: a member of staphylococcal Sdr family. Biochem. J. 345:611-619. [PMC free article] [PubMed] [Google Scholar]
- 29.Yarwood, J. M., J. K. McCormick, M. L. Paustian, V. Kapur, and P. M. Schlievert. 2002. Repression of the Staphylococcus aureus accessory gene regulator in serum and in vivo. J. Bacteriol. 184:1095-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yarwood, J. M., J. K. McCormick, and P. M. Schlievert. 2001. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J. Bacteriol. 183:1113-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ziebandt, A. K., H. Weber, J. Rudolph, R. Schmid, D. Hoper, S. Engelmann, and M. Hecker. 2001. Extracellular proteins of Staphylococcus aureus and the role of SarA and sigma B. Proteomics 1:480-493. [DOI] [PubMed] [Google Scholar]