Abstract
Sensory adaptation in bacterial chemotaxis is mediated by covalent modification of chemoreceptors. Specific glutamyl residues are methylated and demethylated in reactions catalyzed by methyltransferase CheR and methylesterase CheB. In the well-characterized chemosensory systems of Escherichia coli and Salmonella spp., efficient modification by either enzyme is dependent on a conserved pentapeptide sequence, NWETF or NWESF, present at the extreme carboxyl terminus of high-abundance chemoreceptors. To what extent is position at the extreme carboxyl terminus important for pentapeptide-mediated enhancement of adaptational modification? Is this position equally important for enhancement of both enzyme activities? To address these questions, we created forms of high-abundance receptor Tsr or Tar carrying one, six, or eight additional amino acids extending beyond the pentapeptide at their carboxyl termini and assayed methylation, demethylation, deamidation, and ability to mediate chemotaxis. In vitro and in vivo, all three carboxyl-terminal extensions reduced pentapeptide-mediated enhancement of rates of adaptational modification. CheB-catalyzed reactions were more affected than CheR-catalyzed reactions. Effects were less severe for the complete sensory system in vivo than for the minimal system of receptor and modification enzymes in vitro. Notably, extended receptors mediated chemotaxis as efficiently as wild-type receptors, providing a striking example of robustness in chemotactic systems. This could reflect compensatory reductions of rates for both modification reactions, mitigation of effects of slower reactions by the intertwined circuitry of signaling and adaptation, or tolerance of a range of reactions rates for adaptational modification. No matter what the mechanism, the observations provide a challenging test for mathematical models of chemotaxis.
Sensory cells adapt to the continued presence of a stimulus. In bacterial chemotaxis the molecular basis of this adaptation is covalent modification of chemoreceptors, methylation, and demethylation of specific glutamyl residues in the cytoplasmic domains of these transmembrane proteins (9, 10, 19). A dedicated methyltransferase, CheR, uses S-adenosylmethionine to create carboxyl methyl esters at methyl-accepting glutamyl residues. A dedicated methylesterase, CheB, hydrolyzes those esters, recreating the original carboxylate and releasing methanol. CheB is also a deamidase. Two of the four to six methyl-accepting sites in the receptors of Escherichia coli are created by CheB-catalyzed deamidation. In the well-characterized chemotaxis systems of E. coli and Salmonella enterica serovar Typhimurium, the efficiency of adaptational modification catalyzed by CheR and CheB is strongly dependent on a specific amino acid sequence present as the final five, carboxyl-terminal residues of one class of chemoreceptors, the high-abundance receptors (5, 38). In the current study, we have investigated the consequences of perturbing one feature of this sequence: its position at the very end of the receptor.
Chemoreceptors form complexes with the chemotaxis-specific histidine kinase CheA and a Src homology domain 3-related coupling protein CheW, enhancing an otherwise low rate of kinase autophosphorylation and providing phosphoryl groups for transfer to two response regulators, CheY and CheB (9, 10, 19). Phospho-CheY binds the flagellar motor and determines the direction of rotation. CheB is phosphorylated on its regulatory domain, activating its catalytic domain. Binding of chemoattractant to receptor reduces kinase activity and thus cellular levels of phospho-CheY, resulting in altered rotational bias and altered swimming. However, these changes are short-lived, because an increase in ligand occupancy also initiates the feedback loop of adaptation. Stimulated receptors accumulate methyl groups through a combination of a reduced cellular level of phospho-CheB and occupancy-specific activation of methyl-accepting sites. Increased methylation creates a compensatory change in the receptor-kinase complex that restores CheA activity to its null, receptor-activated state even though the increased level of attractant persists. Control of receptor methylation and demethylation determines steady-state cellular behavior, the ability of the chemosensory system to provide exact adaptation over a wide dynamic range, and the process of molecular memory. Thus, the means by which receptor modifications are modulated are of great interest.
Efficient adaptational modification of chemoreceptors depends on the presence of a conserved pentapeptide sequence, asparagine-tryptophan-glutamate-threonine-phenylalanine (NWETF in the single-letter code), or its variant NWESF (5, 38). These sequences are found in natural receptors only at the extreme carboxyl terminus, a position distant from sites of modification (Fig. 1), and occur in the high-abundance receptors, Tsr, Tar, and Tcp, but not the low-abundance receptors, Trg and Tap. Low-abundance receptors as well as high-abundance receptors with the pentapeptide sequence eliminated by truncations or altered by mutations are inefficiently methylated, demethylated, and deamidated (5, 17, 23, 25, 30) and are ineffective on their own at mediating tactic response and directed movement (16, 17, 20, 36, 39). Such receptors mediate effective taxis only with the assistance of NWETF-containing receptors (16, 17, 20, 36). Adding a sequence ending in the pentapeptide to the carboxyl terminus of low-abundance receptor Trg greatly enhances modification by both CheR and CheB in vivo (17) and in vitro (4, 26).
FIG. 1.
Extensions beyond the carboxyl-terminal, modification-enhancing pentapeptide. A. Cartoon of a chemoreceptor dimer (receptor dimer), the methyltransferase (CheR), and methylesterase (CheB). The molecules are shown as ribbon diagrams based on three-dimensional structures of CheR (15) and CheB (13) and on a model of a chemoreceptor based on three-dimensional structures of receptor fragments (22). The modification-enhancing pentapeptide sequence at the carboxyl termini of receptors is marked by an oval and arrows from the aligned sequences in panel B. The two horizontal lines indicate the cytoplasmic membrane. Sites of adaptational modification on the chemoreceptor are marked by balls. B. Aligned carboxyl-terminal amino acid sequences of E. coli Tar (TarE) and Tsr (TsrE) and of Salmonella Tar (TarS), Tsr (TsrS), and Tcp (TcpS). The modification-enhancing pentapeptide sequences at the carboxyl termini of each high-abundance receptor are marked by a shaded box. Other sequence identities are marked by a black background and sequence similarities are boxed. C. Diagrams of sequences of wild-type chemoreceptors (Tar/Tsr), vari-ants deleted of the pentapeptide (TarΔpp/TsrΔpp), and variants with carboxyl-terminal extensions (Tsr +1, Tar +6, Tsr +8). The added amino acids are indicated with the one-letter code. Membrane-spanning segments are marked by hatched boxes and sites of adaptational modification by black dots.
The pentapeptide interacts with and enhances the enzymatic action of methyltransferase CheR and methylesterase/deamidase CheB in distinctly different ways. In the case of methylation, the sequence binds with a ∼2 μM Kd (38) to a small β-sheet subdomain of the carboxyl-terminal, catalytic domain (14) and enhances methylation activity by serving as a docking site that increases enzyme concentration near the methyl-accepting sites (38). In contrast, binding of pentapeptide to CheB has a ∼150 μM Kd (3), occurs at the junction of the amino-terminal domain and the linker between domains (6), and enhances methylesterase activity allosterically, apparently by an effect on the chemoreceptor substrate (3).
To what extent is the position of the pentapeptide at the extreme carboxyl terminus of a chemoreceptor important for enhancement of adaptational modification? Is this location equally important for enhancement of both enzyme activities? To address these questions, we created forms of high-abundance receptors Tsr and Tar carrying extensions beyond their natural carboxyl termini and tested methylation, demethylation, and deamidation in vitro and in vivo, as well as the ability to mediate chemotaxis as the sole chemoreceptor species in the cell.
(Initial aspects of this work were submitted in partial fulfillment of the requirements for an M.S. in Biochemistry at Washington State University by W.-C. Lai.)
MATERIALS AND METHODS
Strains and plasmids.
E. coli K-12 strains CP362 (32), CP553 (11), and RP3098 (33) are deleted, respectively, of the genes for all four methyl-accepting chemotaxis proteins, those chemoreceptor genes plus cheR and cheB or flhA through flhD, thus eliminating expression of all chemoreceptor and che genes. Plasmids pCT1 (18) and pNT201 (8) carry the tsr and tar genes, respectively, under the control of modified lac promoters and also carry the lacIq gene, providing a means of controlled expression of the chemoreceptor genes. pAL79 and pAL61 (5), carrying tsrΔpp and tarΔpp, respectively, were constructed by introducing a stop codon by PCR-based mutagenesis at the fifth-to-last codon of the receptor gene in pCT1 and pNT201, respectively. We used PCR-based mutagenesis of pCT1 and pNT201 to create plasmids pAL41 and pAL67, coding, respectively, for Tsr+1, the receptor with a single cysteine added to its carboxyl terminus, and Tar+6, with six histidines added to the receptor carboxyl terminus. In an initial attempt to create pAL41, the mutagenic oligonucleotide mistakenly lacked a “T”, creating a frame shift that created pAL40, coding for Tsr+8, Tsr with a carboxyl-terminal extension of eight amino acids (VNRHENAR in the single-letter code).
In vitro assays of modification.
Methylation, demethylation, deamidation, and receptor-mediated CheA kinase activity were assayed in vitro as described previously (5), with the modifications noted below, using membrane vesicles prepared from CP553 harboring the relevant plasmid and grown in Luria broth plus a concentration of isopropyl-β-d-thiogalactopyranoside that provided maximal production of the receptor, 0.5 mM for plasmids, that coded for Tsr or its derivatives and 1 mM for Tar plasmids. Approximately 50% of vesicle-borne receptors were oriented with their cytoplasmic domains accessible from the exterior, as determined by maximal extents of methylation or deamidation. Methylation and demethylation assays were performed using 0.125 μM modification enzyme. CheA, CheW, and CheB were purified, and lysates containing CheR were prepared as described previously (4, 6). For demethylation experiments, membrane vesicles carrying previously methylated receptors were incubated for 10 min at room temperature, and the reaction was initiated by addition of a solution of phosphoramidate, KCl, MgCl2, and CheB, which had been incubated together for 30 s. Final concentrations were 2 μM [3H]methyl group (∼5 μM receptors), 10 mM KCl, 25 mM MgCl2, 50 mM phosphoramidate, 0.25 μM CheB. Initial rates of methylation and demethylation in vitro or in vivo were determined from fits over periods sufficiently brief that the extent of modification was linear as a function of time or, as necessary, by fits of exponential rises to a maximum or falls to a minimum. Deamidation experiments were performed using 5 μM total receptor in membrane vesicles, 0.5 μM CheB, 10 mM MgCl2, and 15 mM phosphoramidate.
In vivo assays of receptor modification.
For in vivo assays of receptor modification, we used derivatives of CP362 harboring a receptor-encoding plasmid and grown in the absence of inducer in H1 minimal salts medium plus 0.2% ribose (16, 17, 20, 36, 39). Quantitative immunoblotting (27) revealed that in these growth conditions Tar, Tar +6, and TarΔpp were produced at a level ∼75% the content of total receptors in the corresponding strain wild type for chemotaxis. Tsr, Tsr +8, and Tsr +1 were produced at approximately the same level as total receptors in a wild-type cell, and TsrΔpp was present at a twofold higher level. Labeling with [3H]methyl groups was performed as described previously (17), with the following changes. Washed cells were equilibrated with rotary agitation at ∼200 rpm. l-[methyl-3H]methionine was added to a final concentration of 1.5 μM. Samples, including one taken before addition of a stimulant, contained 7.5 × 107 cells and were mixed with ice-cold trichloroacetic acid at a final concentration of ∼10%. After sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and immunoblotting with a receptor-specific antiserum, regions of the immunoblot corresponding to all receptor species were excised, and receptor methylation was quantified by alkali hydrolysis and diffusion of the released methanol (12). For in vivo demethylation, cells were incubated with 1.5 μM l-[methyl-3H]methionine for 15 min, a receptor-specific attractant was added (10 mM α-aminoisobutyrate for Tsr or 25 mM succinate for Tar), incubation was continued for an additional 30 to 60 min, and repellent was added. Exposure to attractant for a time sufficient to allow adaptation provided an initial level of methylation from which repellent-induced reductions could be easily monitored. Samples were taken and modification was assayed as in the assay for methylation.
Behavioral assays.
Assays for formation of chemotactic rings and for accumulation in capillaries containing chemoeffectors were performed as described previously (17, 35), using plasmid-harboring derivatives of CP362 grown as for in vivo assays of receptor modification. Capillary assays for attractants were performed using suspensions at 1 × 106 to 4 × 106 cells/ml, concentrations at which accumulation in a capillary containing a specific attractant concentration is proportional to the cell concentration in the suspension (1). Thus, accumulations in capillaries measured on different days were normalized to a standard cell concentration. Assays of responses to repellents were done as described previously at substantially higher cell concentrations, in the order of 0.5 × 108 to 1 × 108 cells/ml (35), which are required to obtain reliable results but at which accumulation is not proportional to cell concentration. Thus, normalization was not performed.
RESULTS
To investigate the functional importance of the position of the modification-enhancing pentapeptide sequence NWETF at the extreme carboxyl terminus of a chemoreceptor, we created forms of high-abundance receptors Tsr and Tar extended by additional residues after the pentapeptide (Fig. 1C). These were Tsr with an additional cysteine at its carboxyl terminus (Tsr+1), Tsr extended by an arbitrary sequence of eight residues (Tsr+8), and Tar with six histidines added after NWETF (Tar+6). The activities of these receptors were compared those of the respective wild-type receptors (Tsr and Tar), and their truncated derivatives deleted of the pentapeptide (TsrΔpp and TarΔpp).
Adaptational modification in vitro.
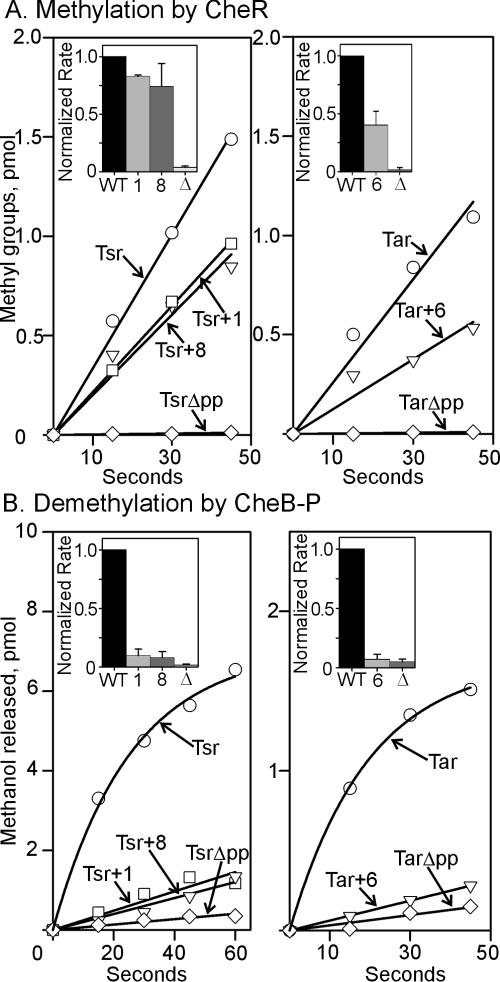
We assayed in vitro methylation, demethylation, and deamidation of wild-type, truncated, and extended forms of Tsr and Tar (Fig. 2 and 3). Initial rates of methylation and demethylation were measured using radiolabeled methyl groups, and deamidation was assessed using the shifts in electrophoretic mobility of a receptor that occur when a glutamine at a methyl-accepting site is deamidated (21, 29). The data in Fig. 2B and 3 are for demethylation and deamidation catalyzed by the phosphorylated form of CheB, generated by the presence of phosphoramidate. We also performed parallel measurements of the two reactions catalyzed by CheB in the absence of phosphorylation. Rates were lower, but similar patterns of differences between the receptor forms were observed (data not shown). For methylation, demethylation, and deamidation, extensions past the pentapeptide sequence reduced rates to values intermediate between the wild-type and pentapeptide-deleted receptor, even if the extension was as short as a single residue. The extent of reduction was different, depending on the extension, receptor, and modification assayed. There was not a consistent relationship between extension length and extent of reduction. However, effects were greater for the CheB-catalyzed reactions of demethylation and deamidation than for CheR-catalyzed methylation and greater for Tar than for Tsr.
FIG. 2.
Methylation and demethylation in vitro. Line graphs show representative time courses and bar graph inserts show mean values of initial rates (three or more independent experiments), normalized to the respective wild-type forms of the receptor, for methylation catalyzed by CheR (A) and demethylation catalyzed by phospho-CheB (B). Assays were performed on high-abundance receptors that were wild-type (Tsr and Tar or WT in bar graphs), deleted of the carboxyl-terminal pentapeptide (TsrΔpp and TarΔpp or Δ in bar graphs), or extended by a single cysteine (Tsr+1 or 1 in bar graphs), a sequence of eight arbitrary residues (Tsr+8 or 8 in bar graphs) or six histidines (Tar+6 or 6 in bar graphs). No other chemotaxis components were present in these assays. Initial rates were determined by linear fits to the data except for demethylation of Tsr and Tar, for which we used an expression for exponential rise to a maximum. Error bars are standard deviations.
FIG. 3.
Deamidation in vitro. The figure shows representative immunoblots of samples of membrane-embedded receptors taken at the indicated times after addition of phospho-CheB. Forms of Tsr and Tar are labeled as in Fig. 2. No other chemotaxis components were present in these assays. Positions of unmodified (U) and deamidated receptors (D) are indicated. Unmodified receptors were in the form encoded by wild-type receptor genes, two glutamines at methyl-accepting sites. Deamidation at one site reduced electrophoretic migration in SDS-polyacrylamide gel electrophoresis, and deamidation at both sites caused a greater reduction, corresponding to the two bands migrating above the unmodified form that is the only species present at time = 0. Unmodified receptors with fewer or more residues migrated slightly more rapidly or slightly more slowly than the wild-type protein.
We assayed our membrane preparations of wild-type, truncated, and extended forms of Tsr and Tar for the ability to activate the kinase activity of CheA and to modulate that activity upon ligand occupancy using the standard assay for formation of phospho-CheY. As reported previously (5, 17, 23, 25, 30), we observed that deletion of the pentapeptide did not significantly alter the ability of either high-abundance receptor to activate kinase activity or to reduce that activity upon attractant binding. This was also the case for the three carboxyl-terminal extensions (data not shown).
Adaptational modification in vivo.
We investigated effects of the carboxyl-terminal extensions on adaptational modification in vivo, measuring methylation after stimulation by an attractant and demethylation after stimulation by a repellent. Cells containing a wild-type, truncated, or extended form of Tar or Tsr as the sole methyl-accepting chemotaxis protein were treated with radiolabeled methionine to establish steady-state labeling of methyl groups on the receptors, stimulated with attractant or repellent, and receptor methylation was monitored over time (Fig. 4) using the vapor diffusion method (12). Carboxyl-terminal extensions reduced in vivo rates of methylation after attractant stimulation and rates of demethylation after repellent stimulation in a pattern parallel to that observed in vitro, but the reductions were less extreme. CheB-catalyzed demethylation was reduced more than CheR-catalyzed methylation, and effects were more extensive for Tar than for Tsr. The combination of these differences resulted in essentially no effect of Tsr extensions on methylation, only on demethylation. As observed in vitro, effects of carboxyl-terminal extensions on in vivo rates of modification did not vary directly with the length of the extension. As would be expected for effects on rate, after sufficient time modification of the extended forms of the high-abundance receptors attained a steady-state level similar to that of the wild-type receptor (data not shown).
FIG. 4.
Methylation and demethylation after stimulation in vivo. Line graphs show representative time courses (methylation) or data from several experiments (demethylation), and bar graph inserts show mean values of initial rates (methylation) or rate constants (demethylation) from three or more independent experiments, normalized to the respective wild-type forms of the receptor. Forms of Tsr and Tar are labeled as in Fig. 2. Methylation (A) was measured after stimulation with a saturating concentration of attractant (10 mM serine for forms of Tsr or 10 mM aspartate for forms of Tar) and demethylation (B) measured after stimulation with a saturating concentration of repellent (70 mM leucine for forms of Tsr or 100 μM nickel sulfate for forms of Tar). The cells contained chromosomally encoded levels of Che proteins and the indicated receptor, as the single kind of receptor in the cell, expressed at a level approximating the cellular content of receptors in a wild-type cell (see Materials and Methods). Initial rates of methylation were determined by linear fits to the data, and rate constants of demethylation were determined by fitting the data to exponential decay to a new steady-state value. Error bars are standard deviations. WT, wild type.
Chemotactic response to spatial gradients.
We tested effects of carboxyl-terminal extensions of Tar and Tsr on the ability of those receptors to mediate chemotaxis by observing the formation of chemotactic rings in semisolid agar plates in spatial gradients of attractants (Fig. 5) or repellents (data not shown). In this commonly used assay, there were only modest or no differences between wild-type and extended forms of the chemoreceptors. Next we utilized the more demanding capillary assay (1). In this assay, microcapillaries containing an attractant were placed in a suspension of cells containing only buffer or microcapillaries containing buffer were placed in a suspension of cells containing a repellent. Thus, cells moving up a gradient of attractant or down a gradient of repellent would both accumulate in the capillary. Carboxyl-terminal extensions past the pentapeptide had no significant effect on chemotactic accumulations in capillaries in response to either attractants or repellents (Fig. 6).
FIG. 5.
Chemotaxis to metabolically generated gradients of attractants. Cells as for Fig. 4 were inoculated into semisolid agar plates containing a limiting amount of a single metabolizable attractant, serine (Ser) for Tsr, aspartate (Asp) or maltose (Mal) for Tar, or a mixture of metabolizable attractants, tryptone (Tryp), containing both serine and aspartate, and formation of chemotactic rings monitored over time of incubation at 35°C. Panel A shows images of plates containing serine (left panel) or aspartate (right panel) after ∼10 h of incubation. Panel B shows plots of mean rates of ring movement (three or more independent measurements) normalized to the rate of cells containing the respective wild-type forms of Tsr (left panel) or Tar (right panel). Error bars are standard deviations. Asterisks indicate that no distinct ring formed (e.g., TarΔpp in panel A). WT, wild type.
FIG. 6.
Chemotaxis to diffusionally generated gradients of attractants and repellents. Cells as for Fig. 4 were tested for accumulation in microcapillaries containing the indicated attractants from a cell suspension in buffer (A) or for accumulation in microcapillaries containing buffer from a cell suspension containing the repellent glycerol (31). Error bars are standard errors for three or more independent experiments.
DISCUSSION
We found that maximal enhancement of the reactions of adaptational modification by the pentapeptide NWETF required that this sequence be the final residues at the carboxyl terminus of a chemoreceptor. Receptors with carboxyl-terminal extensions past the pentapeptide exhibited rates of modification lower than a wild-type receptor but higher than the low rates for receptors deleted of the pentapeptide. The effect was greater for CheB-catalyzed modifications than for CheR-catalyzed methylation, greater for the minimal in vitro system of membrane-embedded receptor plus enzyme than for the complete sensory system in vivo, and greater for receptor Tar than for receptor Tsr. In spite of lower rates of adaptational modification, receptors with carboxyl-terminal extensions mediated chemotaxis as effectively as a wild-type receptor. These observations are discussed below.
Effects on CheR and CheB actions.
Reductions of rates of pentapeptide-enhanced methylation by carboxyl-terminal receptor extensions (Fig. 2A) were consistent with a deduction from the three-dimensional structure of a CheR-pentapeptide complex (14). In that structure, a glutamine side chain of CheR (Gln-182) formed a hydrogen bond with the carboxyl group of the carboxyl terminus of the pentapeptide. The authors suggested that this bond contributed to binding of CheR and pentapeptide and that it would not form if the receptor were extended past the pentapeptide carboxyl terminus. Thus, carboxyl-terminal extensions should reduce binding affinity and presumably methylation rate. We observed reductions in rates of methylation (Fig. 2A and 4A). The reduced rates were still greater than rates for receptors lacking the pentapeptide, consistent with important contributions to CheR activation and presumably binding energy made by the tryptophan and phenylalanine side chains of the pentapeptide (14, 34), which would still occur in receptors carrying carboxyl-terminal extensions.
Less is known about the details of interaction between CheB and the pentapeptide, but it is notable that carboxyl-terminal receptor extensions resulted in greater reductions in enhancement by the pentapeptide for CheB-catalyzed than for CheR-catalyzed reactions. This differential effect adds to the list of differences between interactions of CheR and CheB with the NWETF sequence. The differential effect might reflect different locations of pentapeptide binding (6, 13) and thus different contributions of features of the pentapeptide to binding interactions, different affinities (3, 38), different mechanisms for enhancing enzyme action (3, 38), or a combination of factors.
A caution about carboxyl-terminal extensions of chemoreceptors.
Studies of chemoreceptors can be facilitated by placing additional residues at their carboxyl termini. Such extensions include a single cysteine to provide a reactive sulfhydryl for receptor-specific labeling, six histidines to provide a nickel-binding site for purification, and large entities like derivatives of green fluorescent protein to monitor cellular localization. We found that extensions of a single cysteine, six histidines, or eight arbitrary residues reduced enhancement of CheR- and CheB-catalyzed rates of modification. The implication is that similar reductions would be caused by extensions we did not test. Furthermore, the most commonly used general test for function of an altered chemotaxis component, mediation of chemotactic ring formation by intact cells, did not reveal the defects we identified by testing for specific aspects of receptor function, and neither did the more demanding capillary assay. Investigators using receptors with carboxyl-terminal extensions need to be aware of the likelihood of reduced rates of adaptational modification and that general assays of chemotactic function will probably not reveal these defects.
Robustness.
Amino acid extensions past the NWETF interaction site reduced the extent to which the pentapeptide enhanced rates of adaptational modification (Fig. 2 to 4) but had little or no effect on receptor-mediated cellular migration in spatial gradients of an attractant or repellent (Fig. 5 and 6). This apparent contradiction can be understood as an example of the robustness in the chemotactic system (2). Several factors, not mutually exclusive, could contribute to this robustness. It might be that the slowed rates of methylation and demethylation are sufficiently compensatory that the sensory system still functions effectively. This is a possible explanation for the performance of Tar +6, for which in vivo rates of both methylation and demethylation were intermediate between the wild-type and pentapeptide-deleted forms of the receptor. It is a less attractive explanation for the performance of the extended forms of Tsr, for which in vivo rates of methylation were approximately the same as for the wild-type receptor, but in vivo rates of demethylation were reduced. For these extended receptors, and perhaps also for the extended form of Tar, the intertwined circuitry of signaling and adaptation in the relevant time frame of a few seconds (7) might mitigate the consequences of slower adaptational modification to provide effective chemotactic migration. In fact, in cells containing the complete circuitry receptor (Fig. 4), carboxyl-terminal extensions had less effect than in mixtures containing only receptor and enzyme (Fig. 2). Also, receptor clusters, which require CheA and CheW (28), might sequester the modification enzymes (24) and thus reduce the impact of lowered affinity at the pentapeptide site. Finally, it is possible that CheR- and CheB-catalyzed reactions can perform their crucial physiological functions over a range of absolute rates, and the reduced rates we observed in vivo are within those windows. For instance, a receptor alternation that resulted in a threefold extension of tumble duration, presumably reflecting reduced rate of demethylation, did not reduce chemotactic efficiency but instead resulted in more efficient migration in an attractant gradient (37). Whatever the contributing factors, the robustness we have documented for rates of adaptational modification provides an interesting and potentially challenging test of mathematical models of bacterial chemotaxis.
Acknowledgments
We thank Angela Lilly for construction of pAL40, pAL41, pAL61, pAL67, and pAL79, Alexander Barnakov and Ludmilla Barnakova for purified CheA, CheB, CheW, and CheY, and Thomas Boldog for phosphoramidate.
This work was supported in part by research grant GM29963 to G.L.H. from the National Institutes of General Medical Science.
REFERENCES
- 1.Adler, J. 1973. A method for measuring chemotaxis and use of the method to determine optimum conditions for chemotaxis by Escherichia coli. J. Gen. Microbiol. 74:77-91. [DOI] [PubMed] [Google Scholar]
- 2.Alon, U., M. G. Surette, N. Barkai, and S. Leibler. 1999. Robustness in bacterial chemotaxis. Nature 397:168-171. [DOI] [PubMed] [Google Scholar]
- 3.Barnakov, A. N., L. A. Barnakova, and G. L. Hazelbauer. 2002. Allosteric enhancement of adaptational demethylation by a carboxyl-terminal sequence on chemoreceptors. J. Biol. Chem. 277:42151-42156. [DOI] [PubMed] [Google Scholar]
- 4.Barnakov, A. N., L. A. Barnakova, and G. L. Hazelbauer. 1998. Comparison in vitro of a high- and a low-abundance chemoreceptor of Escherichia coli: similar kinase activation but different methyl-accepting activities. J. Bacteriol. 180:6713-6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnakov, A. N., L. A. Barnakova, and G. L. Hazelbauer. 1999. Efficient adaptational demethylation of chemoreceptors requires the same enzyme-docking site as efficient methylation. Proc. Natl. Acad. Sci. USA 96:10667-10672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnakov, A. N., L. A. Barnakova, and G. L. Hazelbauer. 2001. Location of the receptor-interaction site on CheB, the methylesterase response regulator of bacterial chemotaxis. J. Biol. Chem. 276:32984-32989. [DOI] [PubMed] [Google Scholar]
- 7.Block, S. M., J. E. Segall, and H. C. Berg. 1982. Impulse responses in bacterial chemotaxis. Cell 31:215-226. [DOI] [PubMed] [Google Scholar]
- 8.Borkovich, K. A., N. Kaplan, J. F. Hess, and M. I. Simon. 1989. Transmembrane signal transduction in bacterial chemotaxis involves ligand-dependent activation of phosphate group transfer. Proc. Natl. Acad. Sci. USA 86:1208-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bourret, R., and A. Stock. 2002. Molecular information processing: lessons from bacterial chemotaxis. J. Biol. Chem. 277:9625-9628. [DOI] [PubMed] [Google Scholar]
- 10.Bren, A., and M. Eisenbach. 2000. How signals are heard during bacterial chemotaxis: protein-protein interactions in sensory signal propagation. J. Bacteriol. 182:6865-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burrows, G. G., M. E. Newcomer, and G. L. Hazelbauer. 1989. Purification of receptor protein Trg by exploiting a property common to chemotactic transducers of Escherichia coli. J. Biol. Chem. 264:17309-17315. [PubMed] [Google Scholar]
- 12.Chelsky, D., N. I. Gutterson, and D. E. Koshland, Jr. 1984. A diffusion assay for detection and quantitation of methyl-esterified proteins on polyacrylamide gels. Anal. Biochem. 141:143-148. [DOI] [PubMed] [Google Scholar]
- 13.Djordjevic, S., P. N. Goudreau, Q. Xu, A. M. Stock, and A. H. West. 1998. Structural basis for methylesterase CheB regulation by a phosphorylation-activated domain. Proc. Natl. Acad. Sci. USA 95:1381-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Djordjevic, S., and A. M. Stock. 1998. Chemotaxis receptor recognition by protein methyltransferase CheR. Nat. Struct. Biol. 5:446-450. [DOI] [PubMed] [Google Scholar]
- 15.Djordjevic, S., and A. M. Stock. 1997. Crystal structure of the chemotaxis receptor methyltransferase CheR suggests a conserved structural motif for binding S-adenosylmethionine. Structure 5:545-558. [DOI] [PubMed] [Google Scholar]
- 16.Feng, X., J. W. Baumgartner, and G. L. Hazelbauer. 1997. High- and low-abundance chemoreceptors in Escherichia coli: differential activities associated with closely related cytoplasmic domains. J. Bacteriol. 179:6714-6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng, X., A. A. Lilly, and G. L. Hazelbauer. 1999. Enhanced function conferred on low-abundance chemoreceptor Trg by a methyltransferase-docking site. J. Bacteriol. 181:3164-3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gegner, J. A., D. R. Graham, A. F. Roth, and F. W. Dahlquist. 1992. Assembly of an MCP receptor, CheW, and kinase CheA complex in the bacterial chemotaxis signal transduction pathway. Cell 70:975-982. [DOI] [PubMed] [Google Scholar]
- 19.Hazelbauer, G. L. 2004. Bacterial chemotaxis: a model for sensory receptor systems. .In G. Adelman and B. H. Smith (ed.), Encyclopedia of neuroscience, 3rd ed. [CD-ROM.] Elsevier, Amsterdam, The Netherlands.
- 20.Hazelbauer, G. L., and P. Engström. 1980. Parallel pathways for transduction of chemotactic signals in Escherichia coli. Nature 283:98-100. [DOI] [PubMed] [Google Scholar]
- 21.Kehry, M. R., M. W. Bond, M. W. Hunkapiller, and F. W. Dahlquist. 1983. Enzymatic deamidation of methyl-accepting chemotaxis proteins in Escherichia coli catalyzed by the cheB gene product. Proc. Natl. Acad. Sci. USA 80:3599-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, K. K., H. Yokota, and S.-H. Kim. 1999. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature 400:787-792. [DOI] [PubMed] [Google Scholar]
- 23.Le Moual, H., T. Quang, and D. E. Koshland, Jr. 1997. Methylation of the Escherichia coli chemotaxis receptors: intra- and interdimer mechanisms. Biochemistry 36:13441-13448. [DOI] [PubMed] [Google Scholar]
- 24.Levin, M. D., T. S. Shimizu, and D. Bray. 2002. Binding and diffusion of CheR molecules within a cluster of membrane receptors. Biophys. J. 82:1809-1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, J., G. Li, and R. M. Weis. 1997. The serine chemoreceptor from Escherichia coli is methylated through an inter-dimer process. Biochemistry 36:11851-11857. [DOI] [PubMed] [Google Scholar]
- 26.Li, M., and G. L. Hazelbauer. 2005. Adaptational assistance in clusters of bacterial chemoreceptors. Mol. Microbiol. 56:1617-1626. [DOI] [PubMed]
- 27.Li, M., and G. L. Hazelbauer. 2004. Cellular stoichiometry of the components of the chemotaxis signaling complex. J. Bacteriol. 186:3687-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maddock, J. R., and L. Shapiro. 1993. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science 259:1717-1723. [DOI] [PubMed] [Google Scholar]
- 29.Nowlin, D. M., J. Bollinger, and G. L. Hazelbauer. 1987. Sites of covalent modification in Trg, a sensory transducer of Escherichia coli. J. Biol. Chem. 262:6039-6045. [PubMed] [Google Scholar]
- 30.Okumura, H., S. Nishiyama, A. Sasaki, M. Homma, and I. Kawagishi. 1998. Chemotactic adaptation is altered by changes in the carboxy-terminal sequence conserved among the major methyl-accepting chemoreceptors. J. Bacteriol. 180:1862-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oosawa, K., and Y. Imae. 1984. Demethylation of methyl-accepting chemotaxis proteins in Escherichia coli induced by the repellents glycerol and ethylene glycol. J. Bacteriol. 157:576-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park, C., D. P. Dutton, and G. L. Hazelbauer. 1990. Effects of glutamines and glutamates at sites of covalent modification of a methyl-accepting transducer. J. Bacteriol. 172:7179-7187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parkinson, J. S., and S. E. Houts. 1982. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J. Bacteriol. 151:106-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shiomi, D., H. Okumura, M. Homma, and I. Kawagishi. 2000. The aspartate chemoreceptor Tar is effectively methylated by binding to the methyltransferase mainly through hydrophobic interaction. Mol. Microbiol. 36:132-140. [DOI] [PubMed] [Google Scholar]
- 35.Tso, W. W., and J. Adler. 1974. Negative chemotaxis in Escherichia coli. J. Bacteriol. 118:560-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weerasuriya, S., B. M. Schneider, and M. D. Manson. 1998. Chimeric chemoreceptors in Escherichia coli: signaling properties of Tar-Tap and Tap-Tar hybrids. J. Bacteriol. 180:914-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weis, R. M., and D. E. Koshland, Jr. 1990. Chemotaxis in Escherichia coli proceeds efficiently from different initial tumble frequencies. J. Bacteriol. 172:1099-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu, J., J. Li, G. Li, D. G. Long, and R. M. Weis. 1996. The receptor binding site for the methyltransferase of bacterial chemotaxis is distinct from the sites of methylation. Biochemistry 35:4984-4993. [DOI] [PubMed] [Google Scholar]
- 39.Yamamoto, K., R. M. Macnab, and Y. Imae. 1990. Repellent response functions of the Trg and Tap chemoreceptors of Escherichia coli. J. Bacteriol. 172:383-388. [DOI] [PMC free article] [PubMed] [Google Scholar]