Abstract
The ATP-dependent carboxylate-amine/thiol ligase superfamily is known to contain enzymes catalyzing the formation of various types of peptide, such as d-alanyl-d-alanine, polyglutamate, and γ-peptide, but, curiously, no enzyme synthesizing α-dipeptides of l-amino acids is known. We attempted to find such an enzyme. By in silico screening based on the consensus sequence of the superfamily followed by an in vitro assay with purified enzyme to avoid the degradation of the peptide(s) synthesized, ywfE of Bacillus subtilis was found to code for the activity forming l-alanyl-l-glutamine from l-alanine and l-glutamine with hydrolysis of ATP to ADP. No AMP was formed, supporting the idea that the enzyme belongs to the superfamily. Surprisingly, the enzyme accepted a wide variety of l-amino acids. Among 231 combinations of l-amino acids tested, reaction products were obtained for 111 combinations and 44 kinds of α-dipeptides were confirmed by high-performance liquid chromatography analyses, while no tripeptide or longer peptide was detected and the d-amino acids were inert. From these results, we propose that ywfE encodes a new member of the superfamily, l-amino acid ligase.
The formation and degradation of α-peptide bonds are basic functions in living organisms. Both activities are indispensable for cells to proliferate, survive, and adapt to environmental change. Peptide degradation depends on a number of enzymes, such as proteases and peptidases, many of which have broad substrate specificities. By contrast, only one system, the ribosome system, is responsible for the formation of peptide bonds. All cellular organisms are equipped with the ribosome system, in which an amino acid is first ligated to the corresponding tRNA by aminoacyl tRNA synthetase via aminoacyl-AMP, an active intermediate, and next transferred to an elongating peptide by the ribosome (21). The mRNA encodes the amino acid sequence of the peptide to be synthesized; thus this system can form any peptide depending on the information in the mRNA.
While the ribosome system is universal, other machineries specific for certain peptides are also known, including nonribosomal peptide synthetase (NRPS) (4, 5, 26), folylpolyglutamate synthase (23), enzymes involved in bacterial peptidoglycan biosynthesis (17, 27), poly-γ-glutamate synthetase (2, 25), cyanophycin synthetase (1), d-alanine:d-alanine ligase (DDL) (6, 27), and so on.
NRPS has been found in a variety of organisms such as fungi and streptomycetes and other bacteria. In this enzyme, an amino acid is first activated as an aminoacyl-AMP and then bound to a phosphopantetheinyl cofactor, which transfers the amino acid to a synthesizing peptide. Since the domain catalyzing the activation is specific for each amino acid, one NRPS synthesizes only one kind or a few kinds of peptide. While the ribosome and NRPS are completely different systems, they share a common mechanism to activate the substrate amino acids, that is, they take aminoacyl-AMPs as active intermediates (4, 22).
Folylpolyglutamate synthase is distributed in a wide variety of organisms and catalyzes the formation of the poly-γ-glutamate side chain of tetrahydrofolate (23). Peptidoglycan biosynthetic enzymes (in Escherichia coli, enzymes encoded by murC, murD, murE, and murF) are possessed by eubacteria and specifically function in adding a short peptide containing d-amino acids to UDP-N-acetylmuramic acid (17, 27). DDL is also distributed ubiquitously among bacteria and specifically forms the dipeptide d-alanyl-d-alanine (4). Poly-γ-glutamate synthetase and cyanophycin synthetase are found in certain kinds of microorganisms and synthesize specific polymers. The former enzyme synthesizes poly-γ-glutamate in Bacillus spp. (2). The latter synthesizes polymers composed of aspartate and arginine in Cyanobacteriaciae (1). All of these enzymes are categorized into an ATP-dependent carboxylate-amine/thiol ligase superfamily (9, 20), whose members contain an ATP-grasp motif and take acyl-phosphate as an active intermediate.
Despite the versatility of these peptide-forming activities, an enzyme catalyzing the synthesis of α-dipeptide from l-amino acids has not been identified. The enzymes referred to above synthesize a polymer, oligomer, dipeptide of d-amino acids, or a dipeptide with a γ-peptide bond but do not form a dipeptide of l-amino acids with an α-peptide bond. A few biochemical studies with partially purified cell extracts have suggested the existence of a dipeptide-synthesizing enzyme (18, 24), but no gene or purified enzyme for the activity has been reported. We were interested in this point, especially in the curious fact that the ATP-dependent carboxylate-amine/thiol ligase family does not contain such activity though DDL is a member of the family (7). No reason can be given for the absence of an enzyme synthesizing dipeptides from l-amino acids because the ligation reaction between l-amino acids seems to be possible via the same mechanism as that of DDL. We attempted to find this enzyme. In this report, we describe the screening and finding of an enzyme synthesizing dipeptides from l-amino acids. We also present some of the characteristics of the enzyme.
MATERIALS AND METHODS
Materials.
Escherichia coli strain DH5α was purchased from Takara (Kyoto, Japan). Bacillus subtilis (ATCC 15245) was obtained from the American Type Culture Collection. Plasmids pQE-60, pET-28a, and pREP4 were purchased from QIAGEN (Hilden, Germany). Restriction and modification enzymes, DNA ligase, and the PCR kit (LA PCR) were purchased from Takara. His Trap was obtained from Amersham Pharmacia Biotech (Buckinghamshire, England). Dipeptides for standards were purchased from Sigma-Aldrich (St. Louis, MO) or Bachem Feinchemikalien AG (Bubendorf, Switzerland).
Genetic manipulation.
DNA manipulations were basically performed according to Sambrook et al. (19). The ywfE gene was amplified from the B. subtilis (ATCC 15245) genome by PCR using the primer 5′-TTAACCATGGAGAGAAAAACAGTATTG-3′ to introduce an NcoI site and 5′-ATATGGATCCTACTGGCAGCACATACTTTG-3′ to introduce a BamHI site. The 1.4-kb PCR product was digested with NcoI and BamHI and inserted between the NcoI and BamHI sites of pQE-60. The resulting plasmid, designed to express YwfE with a histidine (His) tag at its C terminal under the control of the T5 promoter and lac operator, was named pQE60ywfE.
In silico screening.
Searches for proteins containing an ATP-grasp motif and homology searches were conducted by using functions provided by ExPASy (expert protein analysis system) available at http://kr.expasy.org/. The BLAST search was done with the blastp program against the complete database of the ExPASy server.
Biochemical procedures.
DH5α/pQE60ywfE cells cultivated in Luria-Bertani medium (10 g/liter Bacto tryptone, 5 g/liter yeast extract, and 5 g/liter NaCl) containing 50 mg/liter of ampicillin at 30°C for 16 h were transferred to fresh TB medium (12 g/liter Bacto tryptone, 24 g/liter yeast extract, 4 ml/liter glycerol, 17 mM KH2PO4, and 72 mM K2HPO4) and incubated at 30°C with vigorous shaking. Three hours after the transfer, 1 mM of IPTG (isopropyl-β-d-thiogalactopyranoside; final concentration) was added, and cultivation was continued a further 4 h. Cells were harvested by centrifugation, resuspended with buffer A (100 mM of Tris-HCl [pH 8.0]) and disrupted by sonication at 4°C. Cellular debris was removed by centrifugation (12,000 × g for 40 min), and the supernatant was subjected to further purification using His Trap (Amersham Pharmacia Biotech) following the instructions of the manufacturer.
The dipeptide-synthesizing activity of YwfE protein was assayed as follows unless otherwise specified. The standard reaction mixture (total volume was 1 ml) contained 50 mg/liter of purified His-tagged YwfE protein, 60 mM ATP, 30 mM MgSO4, and 30 mM substrate amino acid(s) in 50 mM Tris-HCl buffer (pH 9.0). The reaction was carried out at 37°C. A portion of the reaction mixture was taken at each sampling time and immediately subjected to derivatization with fluorenylmethyl chloroformate (FMOC-Cl) (see “Analyses”). For matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOFMS) analysis, a reaction mixture at 4 h was used without derivatization. For determining optimal pH, borate-KCl buffer (pH 9.0 to 11.0) as well as Tris-HCl buffer (pH 7.0 to 10.0) was used. For investigating substrate specificity, reactions were performed under standard assay conditions for 12 h. ADP and the dipeptides formed were analyzed by high-performance liquid chromatography (HPLC). Every combination of one or two amino acids selected from the following 21 kinds of l-amino acids was tested (231 combinations in total): glycine (Gly), l-alanine (Ala), l-serine (Ser), l-cysteine (Cys), l-aspartate (Asp), l-asparagine (Asn), l-threonine (Thr), l-methionine (Met), l-lysine (Lys), l-isoleucine (Ile), l-valine (Val), l-leucine (Leu), l-glutamate (Glu), l-glutamine (Gln), l-proline (Pro), l-arginine (Arg), l-histidine (His), l-tryptophan (Trp), l-phenylalanine (Phe), l-tyrosine (Tyr), and trans-4-hydroxy-l-proline. To test the reactivity with d-amino acids, a reaction with d-alanine (d-Ala), d-serine, d-Ala plus d-Ser, d-Ala plus d-phenylalanine, or d-Ala plus Gln was conducted. For an assay with crude cell extracts, the purified His-tagged YwfE protein in the above assay was replaced with the centrifuged supernatant of cell extracts not yet subjected to His Trap.
Analyses. (i) HPLC analyses.
Amino acids and dipeptides were derivatized with FMOC-Cl (3) and measured by HPLC. A sample (30 μl) was mixed with 270 μl of 0.1 M borate buffer (pH 9.0 with NaOH), and 300 μl of 1.5 mg/ml FMOC-Cl solution in acetone was added. After the mixture had stood for 40 min at room temperature, 600 μl of 25% (vol/vol) CH3CN in 0.25 M borate buffer (pH 5.5) was added. An aliquot (50 μl) of this solution was injected into the HPLC system (LC-10 system; Shimazu, Kyoto, Japan) with a Develosil ODS-HG-5 column (Nomura Kagaku, Tokyo, Japan). For elution, basically, a gradient of solvent A [20 mM (NH4)H2PO4 (pH 6.5 with NH4OH):CH3OH (85:15)] and solvent B (CH3CN:H2O [9:1]) was used with the following program: 0 to 2 min, A:B at 75:25; 2 to 21 min, a linear increase in solvent B to A:B of 55:45; 21 to 36 min, a linear increase to A:B of 45:55; 36 to 37 min, a linear increase to A:B of 1:99; 37 to 39 min, held at A:B of 1:99; 39 to 44 min, a linear decrease in solvent B to A:B of 82:18; 44 to 50 min, A:B of 75:25. The flow rate was maintained at 1.0 ml/min, and the column temperature was kept at 35°C. After separation, fluorescence was monitored at 254 and 630 nm for excitation and emission, respectively. In some cases, the conditions for elution were modified slightly (pH of solvent A or time program of the gradient). ATP, ADP, and AMP were measured using HPLC analyses with absorption at 254 nm after separation with a Develosil C30-UG-5 column (Nomura Kagaku, Tokyo, Japan) using 200 mM acetate and 200 mM triethylamine (pH 6.6) as a mobile phase at 25°C.
(ii) MALDI-TOFMS analysis.
The reaction mixture was diluted 50-fold with water and spread on a target plate coated with 2,5-dihydroxy benzoic acid (DHB) as a matrix. The plate was dried and loaded into a MALDI-TOF mass spectrometer (REFLEX mass spectrometer; Bruker Daltonics).
(iii) Phosphate.
The concentration of phosphate was determined using a spectrophotometric assay utilizing the formation of phosphomolybdic acid (10) with a kit (Phospha-C Test Wako; Wako, Osaka, Japan).
RESULTS
Screening of l-amino acid ligase.
An activity-based screening of dipeptide-forming enzymes using living cells or cell extracts is practically impossible because all living organisms possess dipeptide-degrading activities. Therefore in silico screening was conducted. Three keys were set up to screen for enzymes catalyzing the formation of dipeptides from l-amino acids. First, the enzyme would have an ATP-grasp motif, which is a signature of the ATP-dependent carboxylate-amine/thiol ligase family. Second, the open reading frame of the gene would be unassigned; that is, the function of the gene would be unknown. Third, the enzyme would have slight homology with DDL. The screening was carried out using a public database and its functions.
Proteins containing an ATP-grasp domain (accession number PS 50975) in PROSITE (http://kr.expasy.org/prosite/) were nominated from the Swiss-Prot database (first key). More than 300 proteins were listed, but most were assigned a function: acetyl-coenzyme A (CoA) carboxylase, carbamoyl-phosphate synthase, DDL, phosphoribosylamine-glycine ligase, etc. After eliminating the proteins of known function (second key), 13 proteins remained. To examine their similarity to DDL (third key), a BLAST search was done using the amino acid sequence of each protein as a query. One protein, Y4SG, has strong homology with DDL and thus was judged not to meet the third key. The other 10 had no similarity to DDL (we set the limit of similarity at an E value of <e−3); MG011, MG012, MJ0620, MJ1001, MPN015, and MPN016 showed high levels of similarity to ribosomal protein; MJ0776, MJ0136, and yfiQ had homology to carbamoyl-phosphate synthase, phosphoribosylamine-glycine ligase, and acetyl-CoA synthetase, respectively; and MJ0815 had no homology to a function-known protein. Only two sequences, ywfE (E value, 2e−4) and Y4RH (E value, 4e−6), remained. As the latter was assumed to be a homolog of biotin carboxylase due to its strong homology to carboxylases from various organisms (8), ywfE was chosen as the most promising candidate.
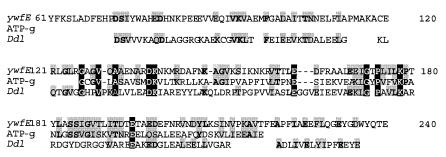
ywfE, an open reading frame in the Bacillus subtilis genome, has a clear ATP-grasp fold and encodes a protein with homology with DDL (Fig. 1). Its function was unknown, but it is part of the bacilysin biosynthetic gene cluster (12). Bacilysin is an antibiotic dipeptide, l-alanyl-anticapsin, synthesized from Ala and an unusual amino acid, anticapsin, in the presence of ATP. The protein is encoded by the gene cluster though the exact function of each gene has not been elucidated (12, 18, 29).
FIG. 1.
Multiple alignment of the regions conserved between YwfE protein (the upper line), ATP-grasp motif (pfam0222, the middle line), and the conserved sequence of DDL (pfam01820, the lower line). The numbers in the upper line indicate distances from the N-terminal amino acid of YwfE protein. Homologous or identical amino acids are shaded. Identical amino acids are in boldface, and amino acids conserved among the three sequences are in white with black shading.
Confirmation of YwfE protein activity.
Since anticapsin, a substrate for bacilysin-synthesizing activity, was not available, other amino acids were used for checking the activity. Ala and Gln were selected, because l-alanyl-l-alanine (Ala-Ala) was reported to be formed by the activity (18) and anticapsin was reported to act as an analog of Gln in vivo (13).
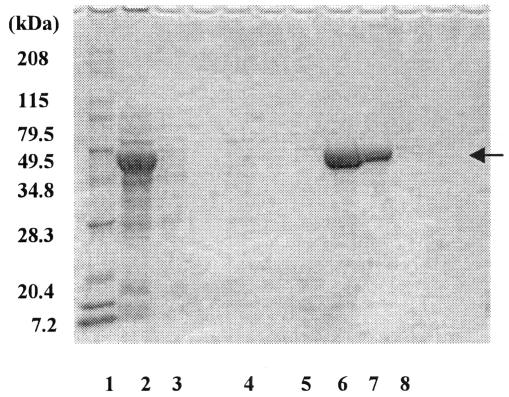
The gene was amplified with PCR, and expression plasmid pQE60ywfE was constructed. In this plasmid, the ywfE gene was driven by the T5 promoter to yield YwfE protein with a His tag at its C terminus. Expression in the host, E. coli strain DH5α, was confirmed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis analysis (Fig. 2). The molecular mass of the expressed protein was estimated at 53 kDa (Fig. 2, lane 8), which is fairly consistent with that estimated from the amino acid sequence of the His-tagged YwfE protein (53.1 kDa).
FIG. 2.
SDS-polyacrylamide gel electrophoresis analysis of cell lysates and purified enzyme fractions. Lane 1, marker; lane 2, crude cell extracts; lanes 3, 4, and 5, fractions eluted from the nickel column with an imidazole buffer of 10 mM, 40 mM, and 100 mM, respectively; lanes 6 to 8, fractions eluted with 500 mM imidazole buffer (amounts of protein applied to the SDS gel were different).
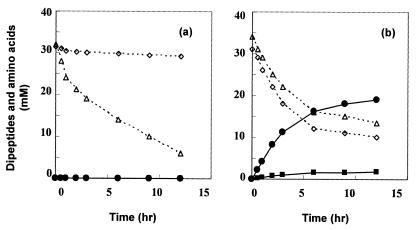
Cell extracts of E. coli expressing the His-tagged YwfE protein were incubated with Ala, Gln, and ATP, but no Ala-Ala, l-alanyl-l-glutamine (Ala-Gln), or l-glutamyl-l-alanine (Gln-Ala) was detected (see Fig. 5a). In this reaction, a decrease of Gln was observed due to glutaminase contained in the extracts. As expected, the extracts had strong dipeptide-degrading activity (data not shown), suggesting the possibility that the synthesizing activity of the ywfE gene product was masked.
FIG. 5.
Time course of reactions with crude extracts (a) and with the purified enzyme fraction (b). Reaction conditions were as described in Materials and Methods except for the enzyme concentration (250 mg/liter). Open diamond; Ala; open triangle, Gln; solid circle, Ala-Gln; solid square, Ala-Ala.
To exclude this possibility, an assay with purified protein was conducted. Cell extract of DH5α/pQE60ywfE was applied to a nickel column, and the gene product was eluted with imidazole buffer and then dialyzed with Tris-HCl buffer. Through these procedures, the gene product was purified to apparent homogeneity (Fig. 2).
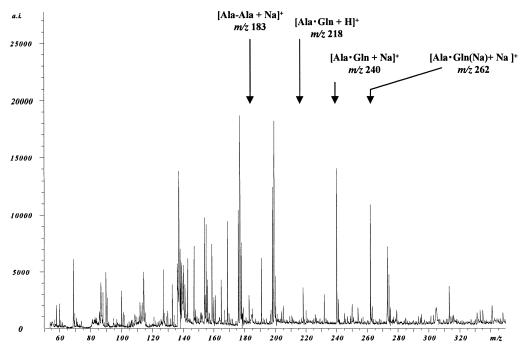
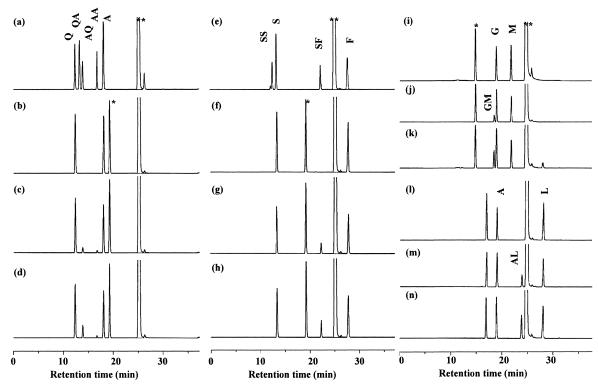
This purified fraction was incubated with Ala, Gln, and ATP for 4 h and analyzed with MALDI-TOFMS and HPLC. In the MALDI-TOFMS analysis, peaks corresponding to [Ala-Ala + Na]+ (m/z 183) and a dipeptide(s) composed of Ala and Gln ([Ala-Gln + H]+ or [Gln-Ala + H]+, m/z 218; [Ala-Gln + Na]+ or [Gln-Ala + Na]+, m/z 240; [Ala-Gln {Na} + Na]+ or [Gln-Ala{Na } + Na]+, m/z 262) were detected (Fig. 3). No peak corresponding to tripeptide (for example, [Ala-Ala-Ala + Na]+, m/z 272, or [Ala-Ala-Gln + H]+, m/z 307) was observed. While the MS analysis could not identify a dipeptide with Ala and Gln as Ala-Gln or Gln-Ala, HPLC analyses clearly revealed that dipeptides formed were Ala-Ala and Ala-Gln but not Gln-Ala (Fig. 4a to d). No unassigned product was found in the HPLC analyses, supporting the absence of tripeptides.
FIG. 3.
MALDI-TOFMS analysis of the reaction catalyzed by YwfE protein. Peaks assigned are as follows: m/z 90, [Ala + H]+; m/z 134, [Ala(Na) + Na]+; m/z 136, [DHB-H2O]+; m/z 137, [DHB-H2O + H]+; m/z 147, [Gln + H]+; m/z 154, [DHB]+; m/z 155, [DHB + H]+; m/z 159, [DHB-H2O + Na]+; m/z 169, [Gln + Na]+; m/z 176, [DHB-H + Na]+; m/z 177, [DHB + Na]+; m/z 183, [Ala-Ala + Na]+; m/z 191, [Gln(Na) + Na]+; m/z 199, [DHB -H + 2Na]+; m/z 218, [Ala · Gln + H]+; m/z 240, [Ala · Gln + Na]+; m/z 262, [Ala · Gln(Na) + Na]+. a.i., absolute intensity.
FIG. 4.
HPLC analysis of the reaction catalyzed by YwfE protein. (a) Authentic samples of Ala (A), Gln (Q), Ala-Ala (AA), Ala-Gln (AQ), and Gln-Ala (QA). (b to d) Reaction with Ala and Gln at time zero (b) and 4 h (c) and reaction in which the sample at 4 h was coinjected with FMOC-derivatized Ala-Gln (d). (e) Authentic samples of Ser (S), Phe (F), Ser-Ser (SS), Ser-Phe (SF), and Phe-Ser (FS). (f to h) Reaction with Ser and Phe at time zero (f) and 4 h (g) and reaction in which the sample at 4 h was coinjected with FMOC-derivatized Ser-Phe (h). (i to k) Reaction with Gly (G) and Met (M) at time zero (i) and 4 h (j) and reaction in which the sample at 4 h was coinjected with FMOC-derivatized Gly-Met (GM) (k). (l to n) Reaction with Ala (A) and Leu (L) at time zero (l) and 4 h (m) and reaction in which the sample at 4 h was coinjected with FMOC-derivatized Ala-Leu (AL). Peaks indicated by single and double asterisks are derived from Tris buffer used for the enzyme reactions and FMOC reagent remaining in the reaction mixture, respectively.
The time course of the reaction was monitored with HPLC (Fig. 5b). As the reaction proceeded, Ala and Gln levels decreased and Ala-Gln and Ala-Ala were found to be accumulated in the reaction mixture. After 12 h, 20.5 mM of Gln and 21 mM of Ala were consumed and 19 mM of Ala-Gln and 1.8 mM of Ala-Ala were formed. The amounts of amino acids used were almost equal to the amounts of dipeptides formed, indicating that the purified fraction contained no substantial dipeptide-degrading activity or glutaminase activity. At the end of the reaction, residual ATP (28 mM), ADP (30 mM), and phosphate (29 mM) were detected but not AMP. When ATP was omitted in the reaction, no dipeptide was detected. These results clearly demonstrated that YwfE protein catalyzes the ligation reaction of Ala and Gln with the decomposition of ATP to ADP.
Characterization of YwfE protein.
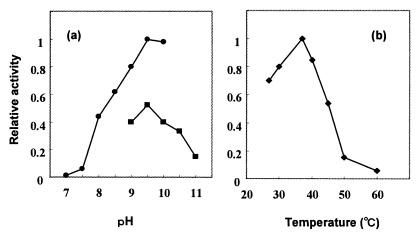
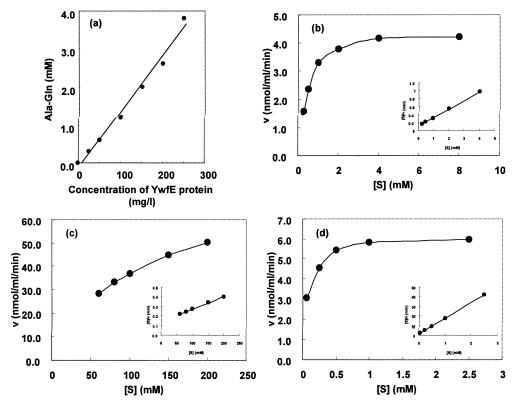
Using Ala and Gln as substrates, some of the characteristics of the enzyme were examined. The optimal pH and temperature were around 9.5 and 37°C, respectively (Fig. 6). GTP did not substitute for ATP. Mg or Mn was essential for the activities. Dipeptide formation was completely abolished by the addition of 2 mM of CdSO4 or ZnSO4 to the standard reaction mixture, indicating strong inhibitory effects of these metal ions. No coenzyme, such as NAD(P), NAD(P)H, pyridoxal phosphate, CoA, or dihydrofolate was required. Apparent Kms for Ala, Gln, and ATP were 0.42 (±0.07), 105.0 (±17.9), and 0.07 (±0.1) mM, respectively. The Vmax for Ala-Gln synthesis was 764.6 ± 17.9 nmol · min−1 · mg−1 protein (Fig. 7).
FIG. 6.
Effects of pH (a) and temperature (b) on YwfE protein activity. (a) Purified His-tagged YwfE protein (50 mg/liter) was incubated with 10 mM ATP, 5 mM each of Ala and Gln, and 10 mM MgSO4 in 100 mM Tris-HCl (pH 7.0 to 10.0) or 100 mM borate-carbonate (pH 9.0 to 11.0) at 37°C for 1 h and the amount of Ala-Gln formed was measured. The amount relative to that formed at pH 9.5 with Tris-HCl was expressed as relative activity. (b) Purified His-tagged YwfE protein (50 mg/liter) was incubated with 10 mM ATP, 5 mM each of Ala and Gln, and 10 mM MgSO4 in 100 mM Tris-HCl (pH 9.5) at various temperatures. The amount of Ala-Gln relative to that formed at 37°C was expressed.
FIG. 7.
Characterization of Ala-Gln-synthesizing reaction by YwfE protein. (a) The linear relationship between the amount of Ala-Gln formed and the concentration of YwfE protein. The reaction was carried out under standard conditions with different amounts of YwfE protein for 1 h. (b to d) The reaction velocity (v) versus substrate concentration ([S]) curves (large graphs) and [S]/v versus [S] curves (Hanes-Woolf plot; insets) were plotted. The reactions were performed by varying the concentration of Ala (b), Gln (c), or ATP (d) under the standard conditions, except for the concentration of YwfE protein. The enzyme concentration used was 100 mg/liter for panels b and c or 40 mg/liter for panel d. The reaction times was 60 min for panels b and c and 15 min for panel d.
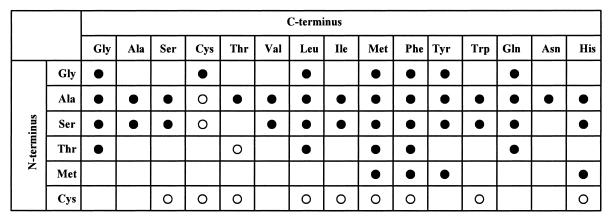
Next, combinations of amino acids were tested as substrates under conditions of pH 9.0 and 37°C. The reaction mixture was analyzed with three methods: by measuring the amount of phosphate formed and amounts of ATP and ADP to check whether the reaction proceeds and by analyzing the dipeptide formed by HPLC to confirm the product. Surprisingly, it was found that YwfE protein accepted a wide variety of amino acids as substrates and various dipeptides were formed. Among all combinations of 21 kinds of l-amino acids tested (231 combinations in total), reaction products were obtained in 111 reactions (see examples in Fig. 4). In some reactions, more than two dipeptides were formed. By comparing the dipeptide standard sample in the HPLC analysis, 44 kinds of dipeptides were confirmed to be produced (Fig. 8).
FIG. 8.
Product spectrum of YwfE protein. Amino acids able to be accepted at the N terminus are listed vertically. The ones able to be accepted at the C terminus are listed horizontally. Arg, Lys, Glu, Asp, Pro, and trans-4-hydroxy-l-proline were inert. A filled circle indicates that the formation of the corresponding dipeptide was confirmed by HPLC analysis. An open circle indicates that synthesis of the corresponding dipeptide was suggested by the formation of ADP and phosphate and by the appearance of a new HPLC peak but could not be confirmed due to the unavailability of standard dipeptide. Reaction conditions were as described in Materials and Methods.
While the enzyme has extremely broad substrate specificity, some preferences were observed; the enzyme did not accept highly charged amino acids such as Lys, Arg, Glu, and Asp; the enzyme did not react with secondary amines such as Pro; the N-terminal residue of the dipeptide formed seemed to be limited to Ala, Gly, Ser, Thr, and Met whereas the C-terminal residue seemed to allow for a wider array of amino acids; and Ala and Ser seemed to be most preferred for the N-terminal residue while Met and Phe seemed to be preferred for the C terminus. The enzyme did not react with d-Ala, d-serine, or d-phenylalanine, confirming that it was not DDL. From these results, it was revealed that YwfE protein had the ability to synthesize various dipeptides from corresponding combinations of l-amino acids in the presence of ATP.
DISCUSSION
We report here that the protein encoded by ywfE, which was found in the B. subtilis genome by in silico screening, catalyzes the formation of α-dipeptides from l-amino acids in an ATP-dependent manner. This is the first gene for an enzyme synthesizing dipeptides from l-amino acids. The most outstanding feature of the enzyme is its broad substrate specificity. Since all other peptide bond-forming enzymes except for the ribosome have very restricted substrate specificities, YwfE protein is unique not only in forming a dipeptide of l-amino acids but also in the wide spectrum of its products. Based on these characteristics, we propose to call the enzyme l-amino acid ligase.
YwfE protein apparently belongs to the ATP-dependent carboxylate-amine/thiol ligase superfamily for the following reasons. (i) It has a clear ATP-grasp sequence (Fig. 1) but shares no homology with a motif of aminoacyl-tRNA synthetase or the adenylation domain of NRPS. (ii) ADP, but not AMP, and phosphate were formed in the reaction catalyzed by the enzyme. Since an enzyme that belongs to the ATP-dependent carboxylate-amine/thiol ligase superfamily transfers phosphate from ATP to its acyl substrate, it forms ADP and phosphate as reaction products (9, 14), whereas aminoacyl-tRNA synthetase or the adenylation domain of NRPS forms acyl-AMP as a reaction intermediate, and thus AMP and pyrophosphate are the reaction products (5). A previous report has suggested that the bacilysin-synthesizing reaction was carried out in an NRPS-like manner (29). The discrepancy with our results may arise from a difference in enzyme purity; the former study used a partially purified fraction whereas we used a fraction purified to apparent homogeneity (Fig. 2).
The substrate specificity of YwfE was unexpectedly relaxed (Fig. 8). For the N-terminal residue, Ala or Ser seems to be most preferred. Following these amino acids, Gly is preferred, suggesting that nonbulky and neutral amino acids tend to be chosen for the N terminus. For the C-terminal residue, Phe and Met are most reactive. Leu is next. Bulky and neutral amino acids seem to be preferred for the C terminus. Due to the unavailability of Cys-X and X-Cys peptides except for Gly-Cys, we could not confirm whether they or part of them are synthesized by the enzyme. However, the consumption of ATP and formation of ADP and phosphate were found with various combinations of amino acids containing Cys. As the reaction with Cys alone formed ADP, Cys is apparently taken as the N-terminal amino acid and, at the very least, Cys-Cys must be formed.
The substrate specificity of an enzyme is not simply defined as a matrix of N terminus-acceptable and C terminus-acceptable amino acids, but rather is determined by the combination of substrate amino acids (Fig. 8). For example, Ser is acceptable for the N terminus and Thr is acceptable for the C terminus, but Ser-Thr was not formed. Given that the enzyme belongs to the ATP-dependent carboxylate-amine/thiol ligase superfamily, the acylphosphate derivative of the N-terminal amino acid would be formed first. Thus, the amino acid accepted as the N-terminal residue may determine the possible range of C-terminal amino acids.
While the apparent Km was lower for Ala than Gln, 10 times more Ala-Gln than Ala-Ala was formed (Fig. 5b). One possibility is that the Vmax value for the synthesis of Ala-Gln is much higher. Another possibility is that the Km value of the N-terminal substrate binding site and that of the C-terminal substrate binding site are different, and the apparent Km for Ala was the sum of these values. This is supported by the fact that DDL has two subsites for binding d-Ala and the Kms of the two sites are different (16). Clarifying how the enzyme recognizes its substrate and what factor affects the reaction rate is important for further application of this enzyme. Since DDL, a related enzyme, has been studied extensively, comparing the secondary structure of YwfE with that of DDL may reveal the relevant residues for substrate recognition.
The physiological significance of YwfE protein other than the participation in bacilysin biosynthesis is unclear. The following points seem to suggest that the enzyme has no additional role in B. subtilis. First, the synthesized dipeptides, except for bacilysin, would be decomposed rapidly by cellular peptidases (see below). This means the formation of a futile cycle between YwfE protein and peptidases that is meaningless for the cells except in some specific circumstances in which ATP is in excess. Second, the expression of ywfE is strictly controlled and coordinated with that of the bacilysin biosynthetic gene cluster (15, 29). Third, a ywfE-deficient mutant was reported to grow normally (11, 28). To elucidate the function of YwfE protein in more detail, examining the effect on cell growth of each dipeptide which can be synthesized by YwfE protein would be helpful.
The distribution of this new type of enzyme in nature is interesting, but no obvious homolog has been found in public databases. As we cannot assign any specific motif to l-amino acid ligase at present, screening with the same procedure used for ywfE would be useful for finding the next example of this type of enzyme. We adapted in silico screening plus an assay with purified enzyme as the screening strategy to avoid the degradation of dipeptides by cellular peptidases. When cell extract of DH5α/pQE60ywfE was reacted with Ala and Gln in the presence of ATP, no dipeptide was detected (Fig. 4a), probably because of the dipeptide-degrading activity of the extracts. These results strongly support that our strategy was effective. In this context, the genes of unknown function we selected may contain other l-amino acid ligases.
Acknowledgments
We thank Akio Ozaki, Makoto Yagasaki, and Yoshiyuki Yonetani for helpful discussions and Aya Kubota-Hada, Reiko Kouda, and Mayumi Fukano for technical assistance. We thank Yoshiko Matsumoto for skillful MS analysis.
REFERENCES
- 1.Aboulmagd, E., F. B. Oppermann-Sanio, and A. Steinbuchel. 2000. Molecular characterization of the cyanophycin synthetase from Synechocystis sp. strain PCC6308. Arch. Microbiol. 174:297-306. [DOI] [PubMed] [Google Scholar]
- 2.Ashiuchi, M., and H. Misono. 2002. Biochemistry and molecular genetics of poly-γ-glutamate synthesis. Appl. Microbiol. Biotechnol. 59:9-14. [DOI] [PubMed] [Google Scholar]
- 3.Bank, R. A., E. J. Jansen, B. Beekman, and J. M. Te Koppele. 1996. Amino acid analysis by reverse-phase high-performance liquid chromatogtaphy: improved derivatization and detection conditions with 9-fluorenylmethyl chloroformate. Anal. Biochem. 240:167-176. [DOI] [PubMed] [Google Scholar]
- 4.Cane, D. E. 1997. Polyketide and nonribosomal polypeptide biosynthesis. Chem. Rev. 97:2463-2705. [DOI] [PubMed] [Google Scholar]
- 5.Cane, D. E., and C. T. Walsh. 1999. The parallel and convergent universes of polyketide syntheses and nonribosomal peptide synthetases. Chem. Biol. 6:R319-R325. [DOI] [PubMed] [Google Scholar]
- 6.Evers, S., B. Casadewall, M. Charles, S. Dutka-Malen, M. Galimand, and P. Courvalin. 1996. Evolution of structure and substrate specificity in D-alanine:D-alanine ligases and related enzymes. J. Mol. Evol. 42:706-712. [DOI] [PubMed] [Google Scholar]
- 7.Fan, C., P. C. Moews, Y. Shi, C. T. Walsh, and J. R. Knox. 1995. A common fold for peptide synthetases cleaving ATP to ADP: glutathione synthetase and D-alanine:D-alanine ligase of Escherichia coli. Proc. Natl. Acad. Sci. USA 92:1172-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freiberg, C. A., R. Fellay, A. Bairoch, W. J. Broughton, A. Rosenthal, and X. Perret. 1997. Molecular basis of symbiosis between Rhizobium and legumes. Nature 387:394-401. [DOI] [PubMed] [Google Scholar]
- 9.Galperin, M. Y., and E. V. Koonin. 1997. A diverse superfamily of enzymes with ATP-dependent carboxylate-amine/thiol ligase activity. Protein Sci. 6:2639-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hertha, H. T., S. Ephraim, and K. Gloria. 1953. A microcolorimetric method for the determination of inorganic phosphorus. J. Biol. Chem. 202:675-685. [PubMed] [Google Scholar]
- 11.Hilton, M., N. G. Alaeddinoglu, and A. L. Demain. 1988. Bacillus subtilis mutant deficient in the ability to produce the dipeptide antibiotic bacilysin: isolation and mapping of the mutation. J. Bacteriol. 170:1018-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inaoka, T., K. Takahashi, M. Ohnishi-Kameyama, M. Yoshida, and K. Ochi. 2003. Guanine nucleotides guanosine 5′-diphosphate 3′-diphosphate and GTP co-operatively regulate the production of an antibiotic bacilysin in Bacillus subtilis. J. Biol. Chem. 278:2169-2176. [DOI] [PubMed] [Google Scholar]
- 13.Kenig, M., E. Vandamme, and E. P. Abraham. 1976. The mode of action of bacilysin and anticapsin and biochemical properties of bacilysin-resistant mutants. J. Gen. Microbiol. 94:46-54. [DOI] [PubMed] [Google Scholar]
- 14.Mullins, L., L. E. Zawadzke, C. T. Walsh, and F. M. Raushe. 1990. Kinetic evidence for the formation of D-alanyl phosphate in the mechanism of D-alanyl-D-alanine ligase. J. Biol. Chem. 265:8993-8998. [PubMed] [Google Scholar]
- 15.Ozcengiz, G., N. G. Alaeddinoglu, and A. L. Demain. 1990. Regulation of biosynthesis of bacilysin by Bacillus subtilis. J. Ind. Microbiol. 6:91-100. [DOI] [PubMed] [Google Scholar]
- 16.Park, I., and C. T. Walsh. 1997. D-alanyl-D-lactate and D-alanyl-D-alanine synthesis by D-alanyl-D-alanine ligase from vancomycin-resistant Leuconostoc mesenteroides. J. Biol. Chem. 272:9210-9214. [DOI] [PubMed] [Google Scholar]
- 17.Park, J. T. 1987. Murein synthesis, p. 663-671. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, D.C.
- 18.Sakajoh, M., N. A. Solomon, and A. L. Demain. 1987. Cell-free synthesis of the dipeptide antibiotic bacilysin. J. Ind. Microbiol. 2:201-208. [Google Scholar]
- 19.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Sheng, Y., X. Sun, Y. Shen, A. L. Bognar, E. N. Baker, and C. A. Smith. 2000. Structural and functional similarities in the ADP-forming amide bond ligase superfamily: implications for a substrate-induced conformational change in folylpolyglutamate synthetase. J. Mol. Biol. 302:427-440. [DOI] [PubMed] [Google Scholar]
- 21.Soell, D., and P. R. Schimmel. 1974. Aminoacyl-tRNA synthetases. Enzymes 10:489-536. [Google Scholar]
- 22.Stein, T., J. Vater, V. Kruft, A. Otto, B. Wittermann-Liebold, P. Franke, M. Panico, R. MaCowell, and H. R. Morris. 1996. The multiple carrier model of nonribosomal peptide biosynthesis at modular multienzymatic templates. J. Biol. Chem. 271:15428-15435. [DOI] [PubMed] [Google Scholar]
- 23.Sun, X., A. L. Bognar, E. N. Baker, and C. A. Smith. 1998. Structural homologies with ATP- and folate-binding enzymes in the crystal structure of folylpolyglutamate synthetase. Proc. Natl. Acad. Sci. USA 95:6647-6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ueda, H., Y. Yoshihara, N. Fukushima, H. Shiomi, A. Nakamura, and H. Takagi. 1987. Kyotorphin (tyrosine-arginine) synthetase in rat brain synaptosomes. J. Biol. Chem. 262:8165-8173. [PubMed] [Google Scholar]
- 25.Urushibata, Y., S. Tokuyama, and Y. Tahara. 2002. Characterization of the Bacillus subtilis ywsC gene, involved in γ-polyglutamic acid production. J. Bacteriol. 184:337-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Velkov, T., and A. Lawen. 2003. Non-ribosomal peptide synthetases as technological platforms for the synthesis of highly modified peptide bioeffectors—cyclosporin synthetase as a complex example. Biotechnol. Annu. Rev. 9:151-197. [DOI] [PubMed] [Google Scholar]
- 27.Walsh, C. T. 1989. Enzymes in the D-alanine branch of bacterial cell wall peptidoglycan assembly. J. Biol. Chem. 264:2393-2396. [PubMed] [Google Scholar]
- 28.Yazgan, A., G. Ozcengiz, and M. A. Marahiel. 2001. Tn 10 insertional mutations of Bacillus subtilis that block the biosynthesis of bacilysin. Biochim. Biophys. Acta 1518:87-94. [DOI] [PubMed] [Google Scholar]
- 29.Yazgan, A., G. Ozcengiz, E. Ozcengiz, K. Kilinc, M. A. Marahiel, and N. G. Alaeddinoglu. 2001. Bacilysin biosynthesis by a partially-purified enzyme fraction from Bacillus subtilis. Enzyme Microb. Technol. 29:400-406. [Google Scholar]