Abstract
In some bacteria, salicylate is synthesized using the enzymes isochorismate synthase and isochorismate pyruvate lyase. In contrast, gene inactivation and complementation experiments with Yersinia enterocolitica suggest the synthesis of salicylate in the biosynthesis of the siderophore yersiniabactin involves a single protein, Irp9, which converts chorismate directly into salicylate. In the present study, Irp9 was for the first time heterologously expressed in Escherichia coli as a hexahistidine fusion protein, purified to near homogeneity, and characterized biochemically. The recombinant protein was found to be a dimer, each subunit of which has a molecular mass of 50 kDa. Enzyme assays, reverse-phase high-pressure liquid chromatography and 1H nuclear magnetic resonance (NMR) spectroscopic analyses confirmed that Irp9 is a salicylate synthase and converts chorismate to salicylate with a Km for chorismate of 4.2 μM and a kcat of 8 min−1. The reaction was shown to proceed through the intermediate isochorismate, which was detected directly using 1H NMR spectroscopy.
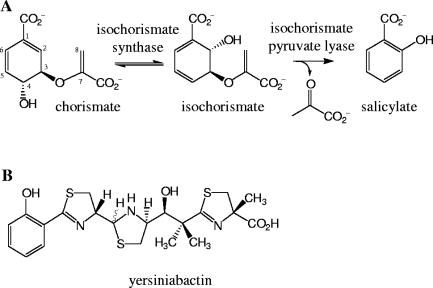
Salicylate is a nonsteroidal anti-inflammatory and is the active component of the analgesic aspirin (12, 31). In some bacteria, salicylate serves as a building block for low-molecular-weight organic Fe3+ chelators (siderophores), such as yersiniabactin in Yersinia pestis and Y. enterocolitica, pyochelin in Pseudomonas aeruginosa, and mycobactin in Mycobacterium tuberculosis (3). In bacteria, salicylate can be derived from chorismate via isochorismate (Fig. 1A). In P. aeruginosa and Pseudomonas fluorescens, two genes coding for an isochorismate synthase and an isochorismate pyruvate lyase were identified as being responsible for salicylate biosynthesis (5, 6, 15). Salicylate is also important in plants, where it induces flowering and is involved in resistance against pathogen infection through the systematic acquired resistance mechanism (4, 29). The transgenic expression of bacterial isochorismate synthase and isochorismate pyruvate lyase genes in tobacco and Arabidopsis thaliana resulted in a constitutive accumulation of salicylate and an enhanced resistance to pathogen attack (14, 32). Recent evidence in A. thaliana suggests that salicylate is synthesized not only through the phenylpropanoid pathway but also, as in bacteria, from chorismate via isochorismate (16, 34).
FIG. 1.
(A) Proposed pathway for salicylate biosynthesis in bacteria. (B) Structure of the siderophore yersiniabactin.
In response to iron deficiency, many bacteria synthesize siderophores that are secreted in the surrounding environment and then reinternalized with bound iron through a specific cell surface receptor (1, 3, 18). An understanding of the mechanism of siderophore biosynthesis is important, because for many pathogenic bacteria the efficiency of iron uptake is directly related to virulence (27, 28). For example, virulence in Y. pestis, the causative agent of bubonic plague, has been correlated with the biosynthesis and transport of the siderophore yersiniabactin, which chelates ferric iron with a dissociation constant of 10−36 M through the nitrogen atoms of its thiazoline rings (21) (Fig. 1B). Formation of yersiniabactin occurs via a hybrid nonribosomal peptide synthetase/polyketide synthase system (33) that assembles the siderophore from the precursor salicylate, a linker group derived from malonyl coenzyme A, three molecules of cysteine, and three methyl groups from S-adenosylmethionine (22).
Pathogenic Yersinia species carry a virulence plasmid (pYV) and a high-pathogenicity island that includes two gene clusters for the biosynthesis, transport, and regulation of yersiniabactin (25). The Y. pestis yersiniabactin genes share some identity with those of Y. enterocolitica, which, unlike the vector-borne pathogen Y. pestis, is an enteric pathogen that infects humans through the ingestion of contaminated food or water and can cause infections such as gastroenteritis (35). The locus involved in the assembly of yersiniabactin in Y. enterocolitica has been genetically characterized and includes nine irp genes (2, 7, 11, 20, 26).
Recently, Pelludat et al. (19) reported genetic evidence that Irp9 may function as a salicylate synthase by converting chorismate directly into salicylate, the precursor of yersiniabactin. It was demonstrated that both enzymes from P. fluorescens, isochorismate synthase (PmsC), which catalyzes the conversion of chorismate into isochorismate, and isochorismate pyruvate lyase (PmsB), which converts isochorismate into salicylate, were required for the complementation of a Y. enterocolitica irp9 mutant. Also, irp9 was not able to complement an Escherichia coli isochorismate synthase entC mutant for the production of enterobactin, which requires isochorismate as a precursor. These results suggested that Irp9 might carry out the direct conversion of chorismate to salicylate. This is further supported by the fact that no isochorismate pyruvate lyase homolog can be found in the recently annotated Y. enterocolitica genome (http://www.sanger.ac.uk/Projects/Y_enterocolitica/).
Protein sequence alignment reveals that Irp9 has significant sequence similarity with the bacterial enzymes isochorismate synthase, 4-amino-4-deoxychorismate synthase (which catalyzes the reaction of chorismate and ammonia to aminodeoxychorismate), and anthranilate synthase component I (which catalyzes the related conversion of chorismate to anthranilate, the corresponding amino analogue of salicylate).
To gain insight into the function of Irp9, the corresponding gene was cloned and heterologously expressed in E. coli. This paper presents the first biochemical characterization of the bacterial enzyme Irp9 and the first 1H nuclear magnetic resonance (NMR) spectroscopic studies that elucidate the steps in the Irp9 catalytic sequence and define Irp9 as a bifunctional salicylate synthase.
MATERIALS AND METHODS
Materials.
Restriction enzymes were obtained from New England Biolabs. Herculase Hotstart DNA polymerase was from Stratagene. 2′-Deoxynucleoside 5′-triphosphates (dNTPs) were from Invitrogen. The oligonucleotide primers were synthesized by Sigma-Genosys. The rapid DNA ligation kit from Roche was employed for all ligations. The NovaBlue and XL1-Blue competent cells and the pET-28a vector were obtained from Novagen. The pGEM-T Easy Vector System I for cloning PCR products was purchased from Promega. PCR products or DNA fragments after restriction digestions were purified using a QIAquick PCR purification kit (QIAGEN). Plasmid DNA was isolated using a QIAprep spin plasmid kit (QIAGEN). Culture medium components were obtained from the Sigma-Aldrich Company. The protein centrifugal concentrator (cutoff molecular weight, 10,000) was obtained from Vivascience. DNA sequencing was performed at the Department of Biochemistry, University of Cambridge.
PCR and cloning.
The irp9 gene (1,305 bp) from Y. enterocolitica 8081 and the entC gene (1,176 bp) from E. coli K-12 were amplified by PCR using the forward primers 5′-CATATGAAAATCAGCGAATTT-3′ and 5′-CATATGGATACGTCACTGGCT-3′, which introduced an NdeI site (bold), and the reverse primers 5′-CCTCGAGCTACTACACCATTAAATA-3′ and 5′-CCTCGAGTCATTAATGCAATCC-3′, which introduced an XhoI site (bold). All PCRs contained 1× Herculase buffer, 2.5 units of Herculase Hotstart DNA polymerase, a 0.2 mM concentration of each dNTP, 0.4 μM primers, and 10 ng of genomic DNA from Y. enterocolitica 8081 (irp9) or from E. coli K-12 (entC). At the beginning of each PCR cycle, an initial PCR activation at 95°C was carried out for 2 min. The PCR cycles consisted of a denaturation period of 30 s at 95°C, annealing at 55°C for 1 min, and extension at 72°C for 1 min. Thirty-five cycles were run, followed by 2 min at 72°C for a final extension. PCR products were subcloned into the plasmid PGEM-T Easy Vector System I; the resulting clones were screened by restriction digestion. Plasmids with the correct insert were isolated and digested, and the resulting gene was cloned into the pET-28a vector for expression in order to place the gene in frame with an N-terminal His6-tagged sequence cleavable by thrombin. The identities of the constructs were confirmed by DNA sequencing.
Overexpression and purification of His6-Irp9 and His6-EntC.
An overnight culture (100 ml) of E. coli C41(DE3) competent cells (17) transformed with either plasmid pET-28a/irp9 or pET-28a/entC was added to 1 liter of fresh 2× YT medium supplemented with 30 μg ml−1 kanamycin and divided into two 500-ml cultures. Expression of His6-Irp9 and His6-EntC was induced at an optical density at 600 nm of 0.6 by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM. After 4 h shaking at 37°C, the IPTG-induced cells were harvested by centrifuging at 6,084 × g for 15 min at 4°C. Cell pellets were stored at −20°C.
All chromatographic purification steps were carried out at 4°C. Harvested cells were suspended in buffer A (0.02 M potassium phosphate, 0.5 M NaCl, 0.02 M imidazole, pH 7.4) containing lysozyme (0.35 mg ml−1) and stirred at room temperature for 30 min. The suspension was homogenized for 20 min by sonication on ice. The crude extract was then centrifuged for 30 min at 38,724 × g. The resulting supernatant was applied at a flow rate of 1.0 ml min−1 to a 5-ml HiTrap chelating column (Amersham Biosciences) previously charged with 0.1 M NiSO4 and equilibrated with buffer A. After a washing with buffer A (10 bed volumes; 2.5 ml min−1), the protein was eluted using a linear gradient of 20 to 500 mM imidazole in 100 ml buffer A (2.5 ml min−1). The elution was followed via sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and fractions containing the protein of interest were concentrated using a protein centrifugal concentrator. The concentrated protein sample was then dialyzed against 100 mM potassium phosphate, pH 7.0, using a PD-10 column containing Sephadex G-25 resin (Amersham Biosciences). The purity of the protein was judged to be ≥95% by SDS-PAGE. Enzyme concentration was evaluated by measuring the absorbance at 280 nm and using the conversion factor (the optical density at 280 nm of 1.0 corresponds to 0.94 mg ml−1) calculated using the software Vector NTI (version 6; Invitrogen). Aliquots of pure His6-tagged recombinant proteins were frozen with liquid nitrogen and stored at −80°C until use. The molecular mass of His6-Irp9 was determined by mass spectrometry using the following conditions: the Irp9 sample was washed into 5 mM Bis-Tris propane buffer (pH 7.0) using a microconcentrator (Vivascience), acetonitrile (50% vol/vol) and acetic acid (1% vol/vol) were added, and 10 μl was injected to a Finnigan MAT triple quadrupole mass spectrometer.
Determination of His6-Irp9 native molecular size by gel filtration.
The molecular size of His6-Irp9 was determined by gel filtration chromatography (HiLoad 26/60 Superdex 200). The column was eluted using 50 mM potassium phosphate and 150 mM NaCl, pH 7.0, at a flow rate of 0.5 ml min−1. A calibration curve was obtained with the following protein standards (by use of gel filtration low-molecular-weight and high-molecular-weight calibration kits from Amersham Biosciences): ferritin from horse spleen (molecular weight, 440), catalase from bovine liver (232), aldolase from rabbit muscle (158), albumin from bovine serum (67), ovalbumin from hen egg (43), chymotrypsinogen A from bovine pancreas (25), and RNase A from bovine pancreas (13.7). The molecular weight calibration curve was calculated using the equation Kav = (Ve − Vo)/(Vt − Vo), where Kav is the partition coefficient, Ve is the elution volume, Vo is the void volume, and Vt is the total volume. For His6-Irp9, Ve equals 206 ml, Vo equals 130 ml, and Vt equals 345 ml. The protein elution was followed by determination of the absorbance at 280 nm.
Enzyme assays.
Steady-state kinetic experiments were performed spectrophotometrically on a microplate reader by monitoring the product salicylate by fluorescence (excitation, 305 nm; emission, 440 nm) as described previously (5, 6). The assays containing 1 μM enzyme were performed in triplicate at 25°C in 100 mM potassium phosphate and 10 mM MgCl2, pH 7.0. The amount of salicylate formed was determined from a standard curve obtained with 0 to 25 μM salicylate. Data were fitted using GraFit (version 5.0.10; Erithacus Software).
Preparation of chorismate for reverse-phase high-pressure liquid chromatography (RP-HPLC) and 1H NMR experiments.
Chorismate (barium salt; Sigma) was purified by recrystallization using the method of Guilford et al. (10). Barium chorismate (50 mg) was suspended in ether (2.5 ml), and 2 N HCl (2.5 ml) was added. After brief shaking, the etheral layer was separated and stirred with activated charcoal (∼100 mg) for 5 min at 0°C and then filtered through Celite and washed with ether (7.5 ml). The filtrate was concentrated in a Craig tube to a volume of ∼0.5 ml under a stream of nitrogen. One volume of cold dichloromethane was added to nucleate the crystallization. This was then followed by the gentle addition of cold hexane (approximately 1 volume), and the crystals left to form at 0°C. The crystals were harvested by centrifugation, dried under vacuum, dissolved in water (20 mM final concentration), and stored at −80°C as 50-μl aliquots.
RP-HPLC assays.
Recombinant protein (approximately 5 μM) was incubated for 30 min at 30°C with 1.5 mM of chorismate in a total volume of 400 μl of 100 mM potassium phosphate and 10 mM MgCl2, pH 7.0. After incubation, the reaction mixture was filtered with a Vivaspin500 instrument (molecular weight cutoff, 10,000) and spun at 1,400 × g for 10 min at ambient temperature. The filtrate was then recovered, and 100 μl was injected immediately for RP-HPLC analysis. Analytical RP-HPLC was performed on a Phenomenex Jupiter 5-μm C18 column (250 mm by 4.6 mm). The flow rate was 1.0 ml min−1. Solvent A was 0.1% aqueous trifluoroacetic acid, and solvent B was 0.1% trifluoroacetic acid in acetonitrile. The following gradient was used for all runs: 0 min, 5% solvent B; 30 min, 95% solvent B; and 35 min, solvent 5% B. The effluent was monitored at 280 nm.
1H NMR structure determination of the reaction product salicylate and the reaction intermediate isochorismate.
1H NMR spectra were recorded at 27°C or at 10°C on a Bruker Avance 500 MHz spectrometer equipped with a 5-mm triple resonance inverse automatic tuning and matching cryoprobe with Z gradients. Solvent suppression was accomplished by use of a presaturation technique or of the WATERGATE (water suppression by gradient-tailored excitation) gradient spin echo sequence (23). For the direct detection of product salicylate, the sample for analysis (total volume of 0.7 ml) contained 10 μM His6-Irp9 and 2 mM chorismate in 20 mM potassium phosphate (pH 7.0) and 5 mM MgCl2 in D2O. The reaction was followed by NMR spectroscopy over 45 min at 27°C. For the detection of the intermediate isochorismate, a time course experiment was performed by acquiring NMR spectra every 3 min over 90 min at 10°C. In this experiment, the sample contained 5 μM His6-Irp9 and 2 mM chorismate in 20 mM potassium phosphate (pH 7.0) and 5 mM MgCl2 in D2O. For the experiment consisting of analyzing the turnover of isochorismate by Irp9, a reaction volume consisting of 10 μM His6-EntC and 2 mM chorismate in 20 mM potassium phosphate (pH 7.0) and 5 mM MgCl2 in D2O was first prepared. The reaction was monitored by 1H NMR spectroscopy until it reached equilibrium, at which point the solution was filtered using a Vivaspin500 (molecular weight cutoff, 10,000) and spun at 1,400 × g for 10 min at ambient temperature to remove the isochorismate synthase EntC. The filtrate was then recovered, and His6-Irp9 (5 μM) was added in a total volume of 700 μl. The reaction was followed over 90 min at 10°C by acquiring NMR spectra every 3 min. δH (500 MHz, D2O): chorismate, 6.42 (1H, s, H2), 6.17 (1H, d, J 10.0 Hz, H6), 5.82 (1H, dd, J 10.0 and 2.5 Hz, H5), 5.06 (1H, d, J 3.0 Hz, H8b); isochorismate, 6.68 (1H, d, J 5.5 Hz, H6), 6.20 (1H, dd, J 9.5 Hz and 5.5 Hz, H5), 6.06 (1H, dd, J 9.5 and 4.5 Hz, H4), 5.09 (1H, d, J 2.5 Hz, H8b); salicylate, 7.65 (1H, dd, J 7.8 Hz and 1.5 Hz, H6), 7.30 (1H, ddd, J 8.3 Hz, 7.4 Hz and 1.5 Hz, H4), 6.80 (2H, m, H3 and H5) ppm.
RESULTS
Cloning, overexpression, and purification of His6-Irp9.
The irp9 gene was amplified from Y. enterocolitica (strain 8081) genomic DNA and cloned into an N-terminal His6-tagged vector for expression. Standard inducible E. coli overexpression resulted in the isolation of 60 mg purified Irp9 per liter of cell culture after Ni2+-nitrilotriacetic acid affinity chromatography. The molecular mass of His6-Irp9 was 50,033 ± 2 Da, in good agreement with the value from the deduced amino acid sequence of His6-Irp9 lacking the first methionine (calculated as 50,032 Da).
Irp9 is a homodimer.
The molecular subunit arrangement of His6-Irp9 was determined by gel filtration chromatography. The protein was eluted from a HiLoad 26/60 Superdex 200 column with a retention time consistent with a dimer and an apparent molecular size of 100 kDa. SDS-PAGE analysis of His6-Irp9 purified by gel filtration (data not shown) showed the presence of a single band at 50 kDa, suggesting that Irp9 is a homodimer.
Irp9 is a salicylate synthase.
The catalytic activity of the recombinant protein Irp9 was investigated by following the formation of salicylate using fluorescence assay analysis (excitation, 305 nm; emission, 440 nm). The recombinant enzyme Irp9 showed hyperbolic saturation kinetics with its substrate chorismate with an apparent Km of 4.2 μM and a kcat of 8 min−1.
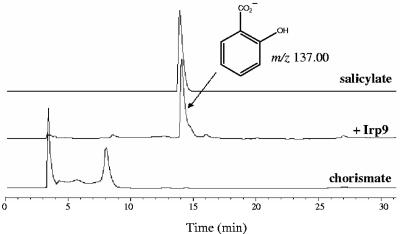
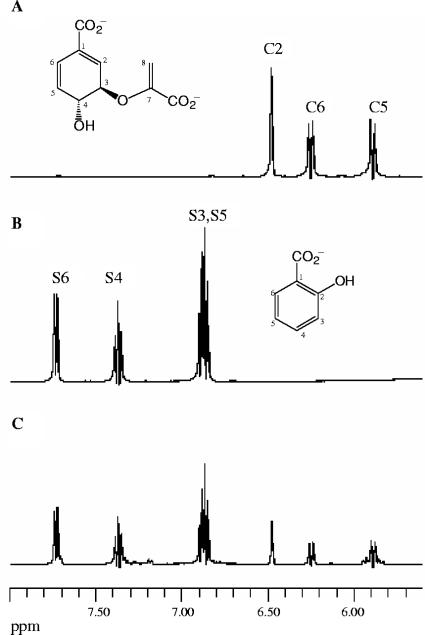
The identity of the salicylate product released during the reaction of Irp9 with chorismate was first confirmed by RP-HPLC analysis and comparison with an authentic standard. The Irp9 enzyme showed no detectable catalytic activity in the absence of Mg2+; hence, MgCl2 was added in all enzyme reactions. This cation is known to be required for the catalytic activity of isochorismate synthases (13). As can be seen in Fig. 2, in the presence of a catalytic amount of recombinant Irp9, chorismate (retention time [tR], 8.0 min) was consumed with the concomitant appearance of salicylate (tR, 14.0 min). The peak at a tR of 14.0 min was characterized by mass spectrometry and was shown to have a mass corresponding to that of salicylate (Fig. 2). To further identify salicylate as the product of the Irp9 reaction, 1H NMR experiments were carried out (Fig. 3). NMR spectra were recorded over 45 min at 27°C for chorismate alone (Fig. 3A), salicylate alone (Fig. 3B), and chorismate in the presence of Irp9 (Fig. 3C). In the latter spectrum, peaks corresponding to salicylate were present, which indicated that Irp9 catalyzes formation of this compound. The inherent reactivity of chorismate leads to the uncatalyzed formation of quantities of 4-hydroxybenzoic acid and prephenic acid, the latter being readily converted into phenylpyruvate under acidic conditions (8, 9). Consequently, the RP-HPLC and NMR experiments were carefully performed using freshly recrystallized chorismate in order to minimize the amount of these impurities.
FIG. 2.
RP-HPLC analysis of the Irp9-catalyzed reaction after incubation for 30 min at 30°C with 5 μM enzyme and 1.5 mM chorismate in 100 mM potassium phosphate and 10 mM MgCl2, pH 7.0. Chorismate was detected at a tR of 8.0 min, and salicylate was detected at a tR of 14 min. Chorismate and salicylate in the absence of enzyme were also incubated separately for 30 min at 30°C and filtered using a Vivaspin500 instrument before being injected for RP-HPLC analysis. The early peak at a tR of 3.3 min represents the solvent front. Detection was carried out at 280 nm.
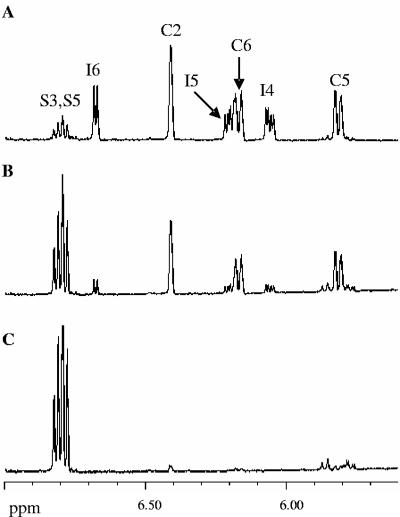
FIG. 3.
1H NMR spectroscopic analysis of the Irp9-catalyzed conversion of chorismate to salicylate. Each spectrum was acquired at 27°C over a period of 45 min in 20 mM potassium phosphate and 5 mM MgCl2, pH 7.0. (A) Substrate control, 2 mM chorismate. (B) Product control, 2 mM salicylate. (C) Irp9-catalyzed reaction, 10 μM Irp9 and 2 mM chorismate. Only the vinylic regions (5.0 to 8.0 ppm) of the NMR spectra are displayed. The abbreviations C and S correspond to protons from chorismate and salicylate, respectively.
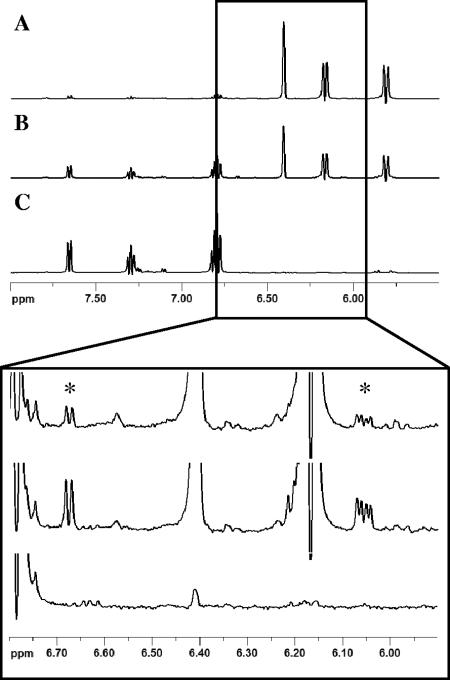
The existence of isochorismate as a reaction intermediate was confirmed in 1H NMR experiments with the Irp9 reaction carried out at 10°C (Fig. 4). Irp9 was incubated with chorismate, and NMR spectra were recorded approximately every 3 min in a time course experiment under steady-state conditions over 90 min. Concomitant with the decrease of the chorismate peaks and the increase of the salicylate peaks (Fig. 4A to C) peaks corresponding to isochorismate could be observed (Fig. 4, lower panel). These NMR signals correspond to unbound isochorismate released in solution. Isochorismate was found to accumulate up to a constant level of 4% with respect to the initial level of substrate and then slowly disappear. This is consistent with isochorismate being formed as an intermediate in the Irp9-catalyzed reaction (Fig. 1A).
FIG. 4.
1H NMR spectroscopic time course analysis of the Irp9 reaction at 10°C using 5 μM Irp9 and 2 mM chorismate after 3 min (A), after 22 min (B), and at completion after 90 min (C). Panels A to C show chorismate turnover into salicylate. The 5.9- to 6.8-ppm region is highlighted in the lower panel to show the formation of isochorismate. Peaks for isochorismate are indicated by asterisks.
Additional evidence that isochorismate is an intermediate of the Irp9 reaction came from a second 1H NMR spectroscopic time course experiment starting with an equilibrium mixture of chorismate and isochorismate. This equilibrium mixture (at a ratio of approximately 60:40 for chorismate:isochorismate) (30) was generated by the reaction of chorismate with the isochorismate synthase EntC. EntC was then removed by ultrafiltration. The addition of Irp9 to this solution mixture led to the initial turnover of isochorismate (Fig. 5), which appeared to be faster than the consumption of chorismate.
FIG. 5.
1H NMR spectroscopic time course analysis of the Irp9 reaction at 10°C using 5 μM Irp9 acting on an equilibrium mixture of chorismate and isochorismate formed from chorismate (2 mM) by the action of EntC, which was removed prior to the addition of Irp9 after 3 min (A), after 40 min (B), and at completion after 90 min (C). The signals for isochorismate in solution disappear faster than those for chorismate. Eventually, both are converted to salicylate. The abbreviations C, I, and S indicate protons from chorismate, isochorismate, and salicylate, respectively.
DISCUSSION
Irp9 is homologous to the monofunctional isochorismate synthases which convert chorismate to isochorismate by an initial addition of water to the C-2 of chorismate with a concomitant loss of the C-4 hydroxyl group. Irp9 is also closely related to the bifunctional anthranilate synthases, which initially convert chorismate to 2-amino-2-deoxyisochorismate (the amino analogue of isochorismate) and then catalyze the elimination of the enolpyruvyl side chain to form anthranilate (the amino analogue of salicylate). Based on these comparisons, a plausible mechanism for the reaction catalyzed by Irp9 would involve the initial conversion of chorismate to isochorismate and the subsequent elimination of the enolpyruvyl side chain in a lyase reaction to give salicylate.
The irp9 gene from the bacterium Y. enterocolitica was cloned into an overexpression system, and an N-terminal His6-tagged form of the encoded protein was purified. We have confirmed that Irp9 converts chorismate into salicylate (19). This is the first in vitro experimental demonstration for such a salicylate synthase. Furthermore, 1H NMR spectroscopic studies provided direct evidence for the existence of isochorismate as an intermediate reaction product during the catalytic cycle of Irp9. This demonstrates that Irp9 is a bifunctional enzyme which catalyzes the initial isomeric conversion of chorismate to isochorismate and the subsequent formation of salicylate (Fig. 1A).
Although more experiments are necessary to fully define the catalytic mechanism, the principles learned from Irp9 are likely to be applicable to the homologous proteins MbtI, which is involved in the biosynthesis of the siderophore mycobactin produced by M. tuberculosis (24, 27), and YbtS, involved in yersiniabactin biosynthesis in Y. pestis (7). By analogy, MbtI and YbtS also appear to be bifunctional salicylate synthases. The salicylate synthase Irp9 from Y. enterocolitica provides a convenient model system for the homologous enzyme from Y. pestis. A better understanding of its mechanism may aid the design of potential inhibitors against plague. Furthermore, the biochemical characterization of the first salicylate synthase is an important step forward in our understanding of the diverse roles salicylate plays in plant and microbial metabolism.
Acknowledgments
This work was supported by the Biotechnology and Biological Sciences Research Council, United Kingdom. A.C. also thanks Astex Technology Ltd. and the Gates Cambridge Trust for financial support.
We are very grateful to Brendan Wren, Department of Infectious & Tropical Diseases, London School of Hygiene & Tropical Medicine, United Kingdom, for providing us with genomic DNA from Y. enterocolitica (strain 8081). We also thank Esther M. M. Bulloch for assistance with mass spectrometry.
REFERENCES
- 1.Braun, V., and H. Killmann. 1999. Bacterial solutions to the iron supply problem. Trends Biochem. Sci. 24:104-109. [DOI] [PubMed] [Google Scholar]
- 2.Brem, D., C. Pelludat, A. Rakin, C. A. Jacobi, and J. Heesemann. 2001. Functional analysis of yersiniabactin transport genes of Yersinia enterocolitica. Microbiology 147:1115-1127. [DOI] [PubMed] [Google Scholar]
- 3.Crosa, J. H., and C. T. Walsh. 2002. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev. 66:223-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Durrant, W. E., and X. Dong. 2004. Systemic acquired resistance. Annu. Rev. Phytopathol. 42:185-209. [DOI] [PubMed] [Google Scholar]
- 5.Gaille, C., P. Kast, and D. Haas. 2002. Salicylate biosynthesis in Pseudomonas aeruginosa. Purification and characterization of PchB, a novel bifunctional enzyme displaying isochorismate pyruvate-lyase and chorismate mutase activities. J. Biol. Chem. 277:21768-21775. [DOI] [PubMed] [Google Scholar]
- 6.Gaille, C., C. Reimmann, and D. Haas. 2003. Isochorismate synthase (PchA), the first and rate-limiting enzyme in salicylate biosynthesis of Pseudomonas aeruginosa. J. Biol. Chem. 278:16893-16898. [DOI] [PubMed] [Google Scholar]
- 7.Gehring, A. M., E. DeMoll, J. D. Fetherston, I. Mori, G. F. Mayhew, F. R. Blattner, C. T. Walsh, and R. D. Perry. 1998. Iron acquisition in plague: modular logic in enzymatic biogenesis of yersiniabactin by Yersinia pestis. Chem. Biol. 5:573-586. [DOI] [PubMed] [Google Scholar]
- 8.Gibson, F. 1964. Chorismic acid: purification and some chemical and physical studies. Biochem. J. 90:256-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibson, F. 1964. Preparation of chorismic acid. Methods Enzymol. 17A:362-364. [Google Scholar]
- 10.Guilford, W. J., S. D. Copley, and J. R. Knowles. 1987. On the mechanism of the chorismate mutase reaction. J. Am. Chem. Soc. 109:5013-5019. [Google Scholar]
- 11.Guilvout, I., O. Mercereaupuijalon, S. Bonnefoy, A. P. Pugsley, and E. Carniel. 1993. High-molecular-weight protein 2 of Yersinia enterocolitica is homologous to Angr of Vibrio anguillarum and belongs to a family of proteins involved in nonribosomal peptide synthesis. J. Bacteriol. 175:5488-5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeffreys, D. 2004. Aspirin: the remarkable story of a wonder drug. Bloomsbury, London, United Kingdom.
- 13.Kozlowski, M. C., N. J. Tom, C. T. Seto, A. M. Sefler, and P. A. Bartlett. 1995. Chorismate-utilizing enzymes isochorismate synthase, anthranilate synthase, and p-aminobenzoate synthase—mechanistic insight through inhibitor design. J. Am. Chem. Soc. 117:2128-2140. [Google Scholar]
- 14.Mauch, F., B. Mauch-Mani, C. Gaille, B. Kull, D. Haas, and C. Reimmann. 2001. Manipulation of salicylate content in Arabidopsis thaliana by the expression of an engineered bacterial salicylate synthase. Plant J. 25:67-77. [DOI] [PubMed] [Google Scholar]
- 15.Mercado-Blanco, J., K. van der Drift, P. E. Olsson, J. E. Thomas-Oates, L. C. van Loon, and P. Bakker. 2001. Analysis of the pmsCEAB gene cluster involved in biosynthesis of salicylic acid and the siderophore pseudomonine in the biocontrol strain Pseudomonas fluorescens WCS374. J. Bacteriol. 183:1909-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Metraux, J. P. 2002. Recent breakthroughs in the study of salicylic acid biosynthesis. Trends Plant Sci. 7:332-334. [DOI] [PubMed] [Google Scholar]
- 17.Miroux, B., and J. E. Walker. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260:289-298. [DOI] [PubMed] [Google Scholar]
- 18.Neilands, J. B. 1995. Siderophores: structure and function of microbial iron transport compounds. J. Biol. Chem. 270:26723-26726. [DOI] [PubMed] [Google Scholar]
- 19.Pelludat, C., D. Brem, and J. Heesemann. 2003. Irp9, encoded by the high-pathogenicity island of Yersinia enterocolitica, is able to convert chorismate into salicylate, the precursor of the siderophore yersiniabactin. J. Bacteriol. 185:5648-5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pelludat, C., A. Rakin, C. A. Jacobi, S. Schubert, and J. Heesemann. 1998. The yersiniabactin biosynthetic gene cluster of Yersinia enterocolitica: organization and siderophore-dependent regulation. J. Bacteriol. 180:538-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perry, R. D., P. B. Balbo, H. A. Jones, J. D. Fetherston, and E. DeMoll. 1999. Yersiniabactin from Yersinia pestis: biochemical characterization of the siderophore and its role in iron transport and regulation. Microbiology 145:1181-1190. [DOI] [PubMed] [Google Scholar]
- 22.Pfeifer, B. A., C. C. C. Wang, C. T. Walsh, and C. Khosla. 2003. Biosynthesis of yersiniabactin, a complex polyketide-nonribosomal peptide, using Escherichia coli as a heterologous host. Appl. Environ. Microbiol. 69:6698-6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piotto, M., V. Saudek, and V. Sklenar. 1992. Gradient tailored excitation for single quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR 2:661-665. [DOI] [PubMed] [Google Scholar]
- 24.Quadri, L. E. N., J. Sello, T. A. Keating, P. H. Weinreb, and C. T. Walsh. 1998. Identification of a Mycobacterium tuberculosis gene cluster encoding the biosynthetic enzymes for assembly of the virulence-conferring siderophore mycobactin. Chem. Biol. 5:631-645. [DOI] [PubMed] [Google Scholar]
- 25.Rakin, A., C. Noelting, S. Schubert, and J. Heesemann. 1999. Common and specific characteristics of the high-pathogenicity island of Yersinia enterocolitica. Infect. Immun. 67:5265-5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rakin, A., E. Saken, D. Harmsen, and J. Heesemann. 1994. The pesticin receptor of Yersinia enterocolitica: a novel virulence factor with dual function. Mol. Microbiol. 13:253-263. [DOI] [PubMed] [Google Scholar]
- 27.Ratledge, C. 2004. Iron, mycobacteria and tuberculosis. Tuberculosis 84:110-130. [DOI] [PubMed] [Google Scholar]
- 28.Ratledge, C., and L. G. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54:881-941. [DOI] [PubMed] [Google Scholar]
- 29.Shah, J. 2003. The salicylic acid loop in plant defense. Curr. Opin. Plant Biol. 6:365-371. [DOI] [PubMed] [Google Scholar]
- 30.Tewari, Y. B., A. M. Davis, P. Reddy, and R. N. Goldberg. 2000. A thermodynamic study of the conversion of chorismate to isochorismate. J. Chem. Thermodyn. 32:1057-1070. [Google Scholar]
- 31.Vane, J. R., and R. M. Botting. 1992. The history of aspirin, p. 3-34. In J. R. Vane and R. M. Botting (ed.), Aspirin and other salicylates. Chapman and Hall, London, United Kingdom.
- 32.Verberne, M. C., R. Verpoorte, J. F. Bol, J. Mercado-Blanco, and H. J. M. Linthorst. 2000. Overproduction of salicylic acid in plants by bacterial transgenes enhances pathogen resistance. Nat. Biotechnol. 18:779-783. [DOI] [PubMed] [Google Scholar]
- 33.Walsh, C. T. 2004. Polyketide and nonribosomal peptide antibiotics: modularity and versatility. Science 303:1805-1810. [DOI] [PubMed] [Google Scholar]
- 34.Wildermuth, M. C., J. Dewdney, G. Wu, and F. M. Ausubel. 2001. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414:562-565. [DOI] [PubMed] [Google Scholar]
- 35.Wren, B. W. 2003. The yersiniae—a model genus to study the rapid evolution of bacterial pathogens. Nat. Rev. Microbiol. 1:55-64. [DOI] [PubMed] [Google Scholar]