Abstract
Sha (also known as Mrp/Mnh/Pha) is a Na+/H+ antiporter encoded by a cluster of six or seven genes that probably form a multisubunit transport complex. The Sha system is important for the homeostasis of H+, Na+, and other monovalent cations and plays a critical role in various functions, including alkaliphily, sporulation, and symbiosis. Here, we characterized the sha homologue genes from the opportunistic pathogen Pseudomonas aeruginosa, which exist as a cluster of six genes (PA1054 to PA1059). The gene cluster PA1054 to PA1059, but not the cluster with a deletion of PA1054, complemented a growth defect in the presence of 0.2 M NaCl and a defect in Na+/H+ antiport activity of the Escherichia coli TO114 mutant lacking the three major Na+/H+ antiporters, indicating that genes PA1054 to PA1059 are responsible for Na+/H+ antiport activity. We disrupted PA1054 (a shaA homologue gene) and determined its effect on Na+ tolerance during growth, Na+ efflux, and pathogenicity in mice. Disruption of PA1054 resulted in severe Na+ sensitivity during growth and decreased Na+ efflux activity. In mice, the deletion mutant of PA1054 also exhibited an attenuated virulence in systemic, pulmonary, and urinary tract infections and also a decrease in colonization of the infected organs. From these results, we conclude that the genes PA1054 to PA1059 encode a Na+/H+ antiporter that is largely responsible for Na+ extrusion in P. aeruginosa and has a role in the infection of the pathogen. We propose to designate PA1054 to PA1059 as the sha (sodium hydrogen antiporter) genes, shaABCDEFG.
Na+/H+ antiporters are ubiquitous membrane proteins that are involved in homeostasis of H+ and Na+ throughout the biological kingdoms (16, 19, 21). We have studied multigene-encoded Na+/H+ antiporters called Sha (14, 15, 35) and also known as Mrp (11, 12), Pha (24), and Mnh (9). sha homologue genes are found as a cluster containing six or seven genes on the genomes of various bacteria. Several studies indicate that all genes in the sha cluster are necessary for ion transport and function, and Sha proteins most probably form a multisubunit transport complex (9, 12, 24, 35). It is well mentioned that Sha proteins show similarity to membrane subunits of respiratory NADH:quinone oxydoreductase (complex I); i.e., ShaA, ShaC, and ShaD are related to NuoL, NuoK, and NuoM/NuoN of Escherichia coli complex I, respectively (6, 9, 11, 18, 24). Recently, several exciting studies have implied a relation between the Sha antiporter and complex I. It was shown that Na+ extrusion mediated by Mrp (= Sha) of Bacillus subtilis is more resistant to a protonophore than that mediated by a typical Na+/H+ antiporter, NhaA of E. coli, and the Mrp (= Sha) system is hypothesized to have an additional primary energization mode (10, 33). On the other hand, recent accumulating evidence on complex I suggests that it may be capable of Na+ translocation besides H+ pumping, although it remains unclear whether the mode of Na+ transport is primary or secondary (2, 3, 29-31). These observations provide an interesting basis for understanding the transport mechanism of the Sha system and complex I and the evolutionary relationship between them.
We have studied the physiological significance of the Sha system with different bacteria. It is characteristic that mutations in sha genes result in serious sensitivity to pH, Na+, K+, and Li+, indicating that the Sha system is responsible for the major homeostatic capacity of pH and the monovalent cations (6, 11, 12, 14, 24). The Sha system is necessary for the maintenance of cytoplasmic pH under extreme alkaline conditions in B. halodurans C-125 and is regarded as an alkaliphilic factor (6). The Sha system of B. subtilis is largely responsible for Na+ extrusion, and the function of Sha is required for the initiation of sporulation (14, 15). We recently reported that a loss of Sha function results in an altered induction of alternative sigma factors and, thus, the expression of many genes is affected in the transition phase in B. subtilis (13). A substantial homeostatic capacity is also required for adaptation and resistance to a change in the external environment. Pha, the Sha homologue of Sinorhizobium meliloti, is required for infection of the bacterium into its host plants (24). S. meliloti pha mutants showed sensitivity to K+, but not to Na+, during growth, and the Pha system is suggested to be a K+/H+ antiporter (24). It is thought that the Pha system plays a role in adaptation to the altered pH and/or ionic milieu inside the host plants during infection (24).
Pseudomonas aeruginosa is a gram-negative bacterium that is found in natural environmental niches, such as soil, and also in hospital environments, where the bacterium is a leading cause of nosocomial infections. The bacterium is an opportunistic pathogen that frequently causes chronic pulmonary infections, as well as severe systemic infections, especially in patients with cystic fibrosis, burns, and immunosuppression (1). During infection, P. aeruginosa, as well as other pathogens, must survive various environmental changes in temperature, pH, ionic and/or osmotic strength, and oxidants. P. aeruginosa has a gene cluster consisting of six genes (PA1054 to PA1059) that encode Sha homologue proteins. Given the case of the Pha system in S. meliloti, we hypothesized that a substantial homeostatic capacity conferred by the sha homologue genes would play a role in the infection of P. aeruginosa. In this study, we characterized the sha homologue genes of P. aeruginosa and focused on its involvement in the virulence of the pathogen.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
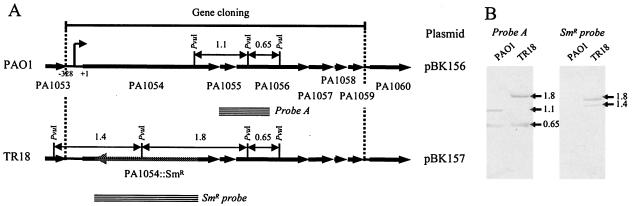
All strains of P. aeruginosa used in this study are derivatives of the standard genetic strain PAO1. TR18 (PA1054::Smr) was constructed by the insertional inactivation of PA1054 by use of a streptomycin resistance (Smr) gene cassette (Fig. 1). Approximately 1 kb of upstream (−985 to +9 positions of PA1054) and downstream (+2791 to +3813 positions of PA1054) regions, which included the first and last three codons of PA1054, respectively, was amplified by PCR. The upstream and downstream fragments were cloned into the EcoRI/BamHI and BamHI/HindIII sites, respectively, of the pUC19 vector. A 2.1-kb Smr gene cassette was excised from the plasmid pHP45Ω (23) and inserted into the BamHI site between the fragments. In this construction, the Smr gene cassette was in the opposite direction to PA1054. The resulting plasmid was introduced into PAO1 by electroporation to generate TR18. Double crossovers were identified as Smr and Cbs. HK3 (nhaP::Cmr) was constructed in the same way as in the case of TR18. A 1.1-kb region of a chloramphenicol resistance (Cmr) gene cassette was prepared from pTn5-CM (27) by PCR amplification. The Cmr gene cassette was inserted between the +9 and +1267 positions of nhaP to inactivate the gene. The Cmr gene cassette was oriented in the same direction as nhaP in HK3. Gene replacements were verified by PCR and Southern blot analysis.
FIG. 1.
(A) Schematic map of the PA1054 to PA1059 region (a sha homologue cluster). The start of the PA1054 gene is shown as +1. In the TR18 mutant, PA1054 is inactivated by insertion of the Smr gene cassette (checked arrow), as described in Materials and Methods. Striped bars show the probes used in Southern blotting, and the predicted size of the PvuI-digested fragment is shown. The region used for gene cloning from the PAO1 and TR18 genomes to construct pBK156 and pBK157, respectively, is shown. (B) Southern blots with PvuI-digested genomes of PAO1 and TR18.
P. aeruginosa was grown at 30°C in LB or LBK (10 g/liter of KCl instead of NaCl in LB). Antibiotic supplements for P. aeruginosa were 200 μg/ml streptomycin, 300 μg/ml carbenicillin (Cb), and 200 μg/ml chloramphenicol. NaCl sensitivity of P. aeruginosa was tested on LB-NaCl plates including 10 g/liter of tryptone, 5 g/liter of yeast extract, and various concentrations of NaCl. E. coli TO114 (nhaA::Kmr, nhaB::Emr, chaA::Cmr), which is deficient in Na+/H+ antiport activity, is a derivative of W3110 (26). TO114 was routinely cultivated at 37°C in LBK supplemented with 30 μg/ml kanamycin (Km), 160 μg/ml erythromycin (Em), 30 μg/ml chloramphenicol, and 100 μg/ml ampicillin, if necessary. NaCl sensitivity of E. coli TO114 was tested in L (K) medium, including 10 g/liter of tryptone, 5 g/liter of yeast extract, 5 g/liter of KCl, and various concentrations of NaCl.
Gene cloning.
DNA regions containing the cluster PA1054 to PA1059 and its putative promoter region (the −328 position of PA1054 to the end of PA1059) were amplified by PCR from the chromosomal DNAs of PAO1 and TR18 (Fig. 1). The amplified fragments were cloned into the EcoRI/KpnI sites of pUC19, resulting in pBK156 (containing the intact cluster) and pBK157 (containing the cluster with a mutation of PA1054::Smr), respectively. The absence of PCR errors was confirmed by sequencing and referring to the whole sequence database of P. aeruginosa PAO1.
Transport assay.
Activity of the Na+/H+ antiporter was measured by a quenching method with everted membrane vesicles, as described previously (4, 25). Cells were grown in LBK medium to the late exponential phase of growth (optical density [OD600] of 1.0 to 1.3). Everted membrane vesicles were prepared in TSCD buffer (10 mM Tris-HCl, 250 mM sucrose, 140 mM choline-Cl, 0.5 mM dithiothreitol, pH 7.4) containing 0.5 mg/ml lysozyme, 0.1 mg/ml DNase, 0.01 mg/ml RNase, and 1 mM phenylmethylsulfonyl fluoride at 4°C. The reaction mixture (2 ml) contained 20 mM bis-Tris propane, 140 mM KCl, and 5 mM MgCl2, adjusted to pH as indicated. After the addition of acridine orange (2 μM) and membrane vesicles (50 μg of protein), 10 mM Tris-lactate was added to induce ΔpH formation that was accompanied by quenching of the fluorescence. Fluorescence of acridine orange was monitored using an F-2500 fluorescence spectrophotometer (HITACHI, Japan) with excitation at 493 nm and emission at 530 nm.
An assay of 22Na+ efflux energized by an inwardly ΔpH gradient was described previously (14). Right-side-out vesicles were prepared in acetate buffer (100 mM potassium acetate, 5 mM MgSO4, 10 mM sodium phosphate, pH 6.0). Downhill 22Na+ efflux was initiated by dilution of the membrane vesicles into gluconate buffer (100 mM potassium gluconate, 5 mM MgSO4, 10 mM potassium phosphate, pH 6.0) or acetate buffer. At given times, the reaction was terminated by filtration through 0.45-mm-size filters. The filter was washed, dried, and counted by liquid scintillation spectrometry.
Virulence assay.
The virulence of P. aeruginosa was tested in 4-week-old male (for systemic and pulmonary infection) or female (for urinary tract infection) ICR mice (SLC, Japan). Overnight cultures of PAO1 and TR18 were grown on LBK agar and LBK, supplemented with streptomycin at 30°C, respectively. Cultures were suspended in 50 mM KCl (for systemic infection) or 150 mM KCl (for pulmonary and urinary tract infection). In the systemic infection model, mice (n = 10 per group) were pretreated to induce neutropenia by intraperitoneal administration of cyclophosphamide at 200 mg/kg of body weight, 4 days before infection. Mice were intravenously challenged with approximately 5 × 104 or 5 × 105 CFU per mouse, and survival rate was monitored for 8 days. In the pulmonary infection model, mice (n = 5 to 10 per group) were pretreated with cyclophosphamide, as described above. Mice were anesthetized by 50 mg/kg of body weight of pentobarbital and challenged intranasally with approximately 1 × 104, 3 × 104, or 1 × 105 CFU per mouse. The survival rate was monitored for 6 days. In parallel, mice were sacrificed 1 and 3 days after infection. The lungs were removed aseptically from surviving mice and homogenized with 150 mM KCl, and the number of viable cells was counted on the LBK plates. In the urinary tract infection model, mice (n = 10 per group) were denied water for a whole day before bacterial challenge. Mice were anesthetized by 50 mg/kg pentobarbital and challenged via uterine insertion of 1.6 × 104 or 105 CFU per mouse, and the ureterostoma was closed with a clip for 30 min. At 1, 3, and 7 days after infection, the kidney and bladder were aseptically removed. The incised bladder was rinsed with 150 mM KCl, the organs were homogenized with 150 mM KCl, and the number of viable bacteria was counted on LBK plates. Comparisons of survival curves of PAO1 and TR18 were performed by Logrank testing. Bacterial counts of PAO1 and TR18 were statistically compared using Student's t test or an Aspin-Welch test.
RESULTS
The cluster PA1054 to PA1059 is responsible for Na+/H+ antiport activity.
The genes PA1054 to PA1059, homologous to sha genes, are found as a six-gene cluster on the PAO1 genome (Fig. 1). In the adjacent region, PA1053 (similar to SlyB of E. coli) and PA1060 (similar to YbfH of B. subtilis) exist 329 bp upstream and 114 bp downstream of the cluster, respectively. The gene products of PA1054 to PA1059 show significant similarity to Pha proteins of S. meliloti, Sha/Mrp proteins of B. subtilis, and Mnh proteins of Staphylococcus aureus (Table 1). The N-terminal region (approximately 1 to 780 residues) and C-terminal region (781 to 933 residues) of the PA1054 product showed similarity to ShaA/MrpA/MnhA and ShaB/MrpB/MnhB, respectively, suggesting that the A and B genes are fused in P. aeruginosa. It was reported that the B. subtilis homologues mrp(= sha)ABCDEFG are transcribed as an operon (11), a possibility that may apply to the genes PA1054 to PA1059. The 328-bp upstream region of PA1054 allowed the complementation of E. coli TO114 without a vector-derived lac promoter (see below), suggesting that the region contains a promoter that is functional in E. coli.
TABLE 1.
The predicted proteins encoded by the sha homologue genes from P. aeruginosa and their similarity to Sha homologue proteins
| PA no. (no. of amino acid residues of the predicted proteins) | Homologous proteins (no. of amino acid residues) [% identity/% similarity]
|
|||
|---|---|---|---|---|
|
S. meliloti
|
B. subtilis | S. aureus | ||
| SMc03179-03184a | SMc00051-00057 | |||
| PA1054 (933) | PhaA1 (999) [46/78] | PhaA2 (791) [39/77] | ShaA/MrpA (774) [43/80] | MnhA (801) [39/78] |
| PhaA1 (999) [46/78] | PhaB2 (139) [40/71] | ShaB/MrpB (143) [47/79] | MnhB (142) [29/70] | |
| PA1055 (112) | PhaC1 (115) [58/85] | PhaC2 (125) [39/85] | ShaC/MrpC (113) [39/86] | MnhC (113) [46/80] |
| PA1056 (499) | PhaD1 (539) [40/74] | PhaD2 (521) [37/78] | ShaD/MrpD (493) [37/77] | MnhD (498) [32/73] |
| PA1057 (174) | PhaE1 (161) [32/73] | PhaE2 (158) [35/71] | ShaE/MrpE (158) [27/70] | MnhE (159) [26/70] |
| PA1058 (89) | PhaF1 (93) [40/85] | PhaF2 (123) [37/78] | ShaF/MrpF (94) [35/81] | MnhF (97) [30/76] |
| PA1059 (115) | PhaG1 (121) [33/73] | PhaG2 (116) [28/73] | ShaG/MrpG (124) [29/73] | MnhG (118) [29/72] |
The loci involved in an Inf− Fix− phenotype (24).
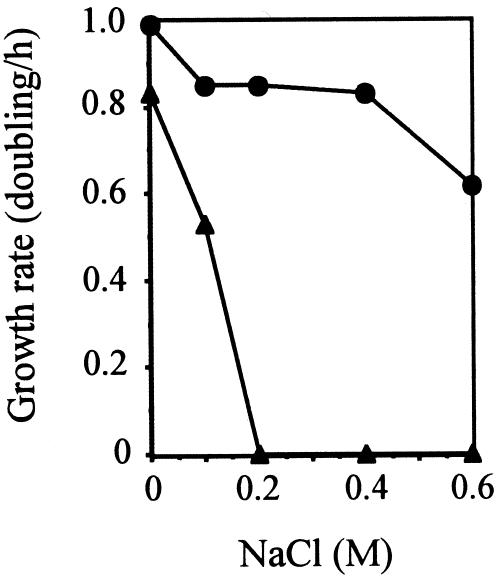
The Sha system is reported to be a Na+/H+ antiporter in B. halodurans (6), B. subtilis (11, 12, 14), and S. aureus (9). We examined Na+/H+ antiport activity of the cluster PA1054 to PA1059 by complementation of E. coil TO114, which lacks three major Na+/H+ antiporters and shows undetectable Na+/H+ antiport activity (26). This mutant is unable to grow in the presence of 0.2 M NaCl at pH 7 (26). We cloned and expressed the PA1054-to-PA1059 cluster under its original promoter in pUC19 (Fig. 1). As shown in Fig. 2, TO114 cells transformed with pBK156 containing the intact cluster restored growth in the presence of up to 0.6 M NaCl, although TO114 with pUC19 (vector) did not grow in the presence of NaCl at concentrations of 0.2 M and above. TO114 with pBK157, containing the cluster with a deletion of PA1054 (see below), also did not grow at 0.2 M NaCl.
FIG. 2.
The restoration of the growth defect of E. coli TO114 by the sha homologue cluster. Cells of TO114 with pBK156 (containing the PA1054-to-PA1059 cluster) (circles) and TO114 with pUC19 (vector only) (triangles) were grown in L (K) medium containing the indicated concentrations of NaCl.
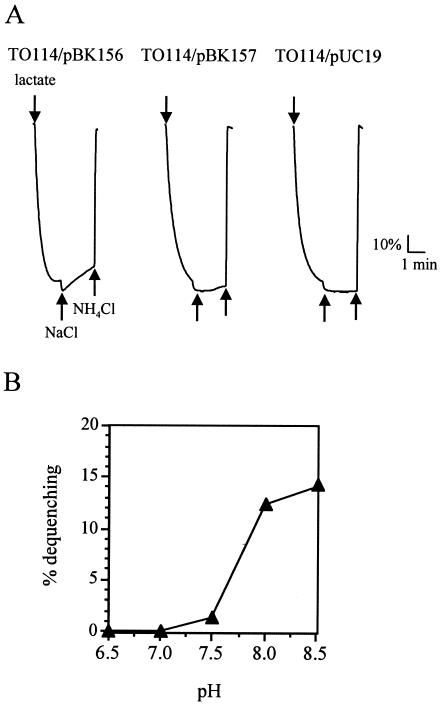
Next, we measured Na+/H+ antiport activity using everted membrane vesicles from the TO114 derivatives. We detected Na+/H+ antiport activity with membrane vesicles of TO114 with pBK156 but not in those of TO114 with pUC19 or pBK157 (Fig. 3A). The antiport activity is pH dependent, with a pH profile similar to that of NhaA of E. coli (20): the activity was higher at pH 8.5 (14%) and pH 8.0 (12.3%) than at pH 7.5 (1.3%) and undetectable at pH 7.0 and 6.5 (Fig. 3B). This pH profile was different from that of the Mnh system, which is reported to be maximized at pH 7.0 to 7.5 (9). These results show that the cluster PA1054 to PA1059 was responsible for Na+/H+ antiport activity.
FIG. 3.
Na+/H+ antiport activities measured by the fluorescence quenching method. (A) Everted membrane vesicles were prepared from cells of E. coli TO114 with pBK156, pBK157, or pUC19 grown in LBK medium. The first arrow indicates the addition of 10 mM Tris-lactate to initiate fluorescence quenching due to the inward movement of H+ by respiration. The second and third arrows indicate the additions of 10 mM NaCl and 25 mM NH4Cl, respectively. (B) The pH profile of the antiport activity conferred by the sha homologue cluster. Each value is shown as the mean of three replicates.
Disruption of PA1054: Na+-sensitive growth and impaired Na+ efflux.
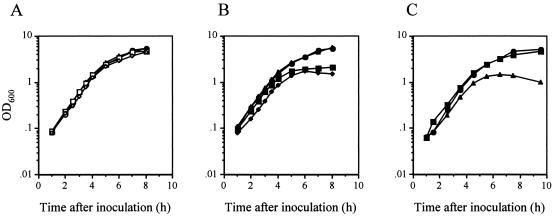
We disrupted PA1054, the first and largest gene in the cluster and homologous to shaA, and determined the effect on Na+ tolerance in growth and Na+ efflux. The growth of a PA1054-disrupted mutant (TR18) was impaired with an increase in the concentration of NaCl up to 0.3 M, though the wild type was not affected (Fig. 4A and B). The growth defect was apparent in the stationary phase rather than the exponential phase of growth. In the presence of 0.3 M NaCl, the OD600 and CFU of TR18 were significantly lower than those of the wild type at 24 h after inoculation, suggesting that lysis of cells occurred (Table 2). We also disrupted a single-gene type Na+/H+ antiporter, NhaP, to compare the effect on Na+ sensitivity in growth with the TR18 mutant. An nhaP-disrupted mutant (HK3; nhaP::Cmr) showed Na+ sensitivity in growth but more resistance to NaCl than TR18. The HK3 mutant showed a growth curve similar to that of PAO1 in the presence of 0.3 M NaCl (Fig. 4C). The OD600 and CFU of HK3 were 5.72 and 1.9 × 109 colonies per ml at 24 h after inoculation, respectively, which were equivalent to those of PAO1 (Table 2). These results showed that the Sha homologue system is more responsible for Na+ tolerance in P. aeruginosa than NhaP.
FIG. 4.
Growth of P. aeruginosa. (A) PAO1 (wild type). (B) TR18 (PA1054::Smr). Cells were grown in LBK medium containing 0 M (circles), 0.1 M (triangles), 0.2 M (squares), and 0.3 M (diamonds) of NaCl. (C) The growth curves of PAO1 (circles), TR18 (triangles), and HK3 (nhaP::Cmr) (squares) in the LBK medium containing 0.3 M NaCl.
TABLE 2.
Effect of NaCl on the growth of P. aeruginosaa
| NaCl concn (M) | PAO1 (sha+)
|
TR18 (PA1054::Smr)
|
Ratiob | ||
|---|---|---|---|---|---|
| OD600 | CFU | OD600 | CFU | ||
| 0 | 3.98 | 1.5 × 109 | 3.96 | 1.2 × 109 | 0.81 |
| 0.1 | 5.08 | 4.4 × 109 | 4.80 | 4.2 × 109 | 0.95 |
| 0.2 | 5.92 | 4.0 × 109 | 4.18 | 2.8 × 109 | 0.70 |
| 0.3 | 5.96 | 4.0 × 109 | 0.35 | 1.8 × 106 | 4.5 × 10−4 |
Cells were grown in LB medium containing the indicated concentration of NaCl, and growth was examined at 24 h after inoculation.
The ratio of the number of CFU of TR18 to that of PAO1 in the same condition.
KCl did not have an inhibitory effect on the growth of the TR18 mutant and the wild type, although it improved Na+ sensitivity in the growth of TR18 when present with NaCl (data not shown). A similar observation was reported with E. coli mutant EP432, which lacks NhaA and NhaB Na+/H+ antiporters (7). We also examined the effect of pH on the growth of PAO1 and TR18 on buffered agar plates containing 10 g/liter of tryptone, 5 g/liter of yeast extract, and 70 mM bis-Tris propane, adjusted to pH 7, 8, or 9. P. aeruginosa grew at up to pH 9, although growth was less at pH 9 than at pH 7 or 8, and no difference was observed between PAO1 and TR18 at all pHs tested (data not shown). No substantial phenotypic variation in alginate production and antibiotic resistance was observed with the TR18 mutant.
Since PA1054 is necessary for Na+/H+ antiport activity (Fig. 3), the TR18 mutant was expected to show a decreased capacity of secondary Na+ efflux. We examined 22Na+ efflux from right-side-out membrane vesicles of PAO1, TR18, and HK3 upon energization of a transmembrane proton gradient (ΔpH), a component of proton motive force (Fig. 5). Imposition of ΔpH (inside alkaline) enhanced 22Na+ efflux in the membrane vesicles of PAO1, while the Na+ efflux was weakly accelerated in those of HK3 and more weakly accelerated in those of TR18. Taken together with these results, we concluded that the Sha homologue system is responsible for a major Na+ excretion capacity that is important for Na+ tolerance of P. aeruginosa.
FIG. 5.
22Na+ efflux from right-side-out membrane vesicles prepared from P. aeruginosa PAO1 (wild type) (circles), TR18 (PA1054::Smr) (triangles) and HK3 (nhaP::Cmr) (squares). Efflux was initiated by dilution of the membrane vesicles into buffers at pH 6 containing 100 mM potassium acetate (open symbols) or 100 mM potassium gluconate (closed symbols). Acetate diffusion generates a theoretical 2.9 units of ΔpH (inward direction). Each value is shown as the mean of three replicates.
The virulence of the PA1054-disrupted mutant is significantly reduced in mouse infection models.
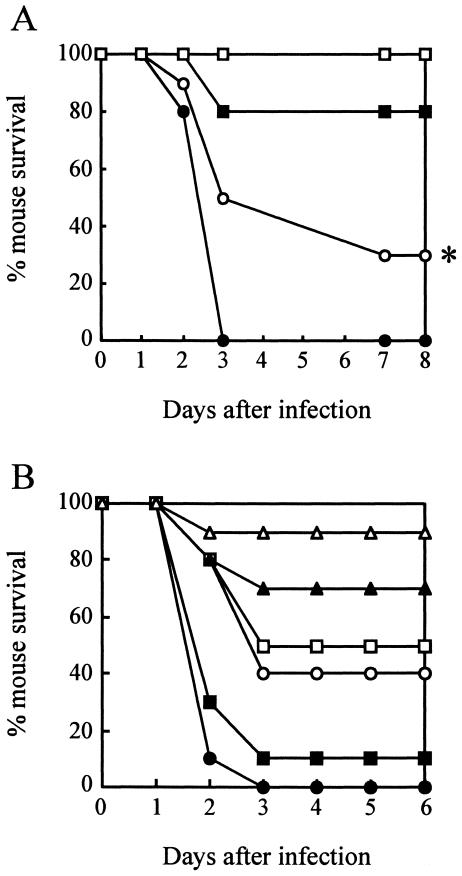
P. aeruginosa is known to cause chronic pulmonary, systemic, and urinary-tract infections (1). The virulence of the wild-type PAO1 and the TR18 mutant was examined in three kinds of mouse infection models. The TR18 mutant showed significantly less virulence compared to PAO1, as shown by the survival curves in Fig. 6. In the systemic infection model (Fig. 6A), with a relatively high dose of bacterial cells (5 × 105 CFU per mouse), the mice infected with TR18 had 50% survival at 3 days after infection, whereas mice infected with PAO1 had 100% mortality (P < 0.05). With a relatively low dose of cells (5 × 104 CFU per mouse), 20% mortality was observed with PAO1, but TR18 showed no mortality. In the pulmonary infection model 3 days post infection, the higher (1 × 105 CFU per mouse) and middle (3 × 104 CFU per mouse) doses of cells resulted in 100% and 90% mortality, respectively, with PAO1, whereas mortality was 60% and 50% with TR18 (Fig. 6B).
FIG. 6.
Virulence of P. aeruginosa PAO1 and TR18 in mouse infection models. (A) In the systemic infection model, mice were intravenously challenged with 5 × 104 CFU (squares) or 5 × 105 CFU (circles) per mouse. (B) In the pulmonary infection model, mice were challenged intranasally with 1 × 104 CFU (triangles), 3 × 104 CFU (squares), or 1 × 105 (circles) CFU per mouse. Survival rate was monitored post infection. Closed symbols, PAO1; open symbols, TR18. The asterisk indicates significant difference from the PAO1 value by Student's t test or an Aspin-Welch test (P < 0.05).
We next examined the number of viable bacteria in the infected organs of surviving mice. In the pulmonary infection model (Fig. 7), although the number of viable bacteria in the lung showed almost no change between PAO1 and TR18 at 1 day after infection, the number of bacteria significantly decreased with TR18 compared to PAO1 at 3 days after inoculation. In a urinary tract infection model (Table 3), the number of bacteria in the kidney was much less with TR18 than with PAO1 at 3 and 7 days after infection. Similar results were obtained in the bladder (Table 3). From these results, we conclude that disruption of PA1054 resulted in the attenuation of virulence of P. aeruginosa in mouse infections, probably due to impaired colonization of bacteria in the host.
FIG. 7.
Viability of P. aeruginosa PAO1 and TR18 in mouse lungs in the pulmonary infection model. Surviving mice were sacrificed at 1 and 3 days after infection, and bacterial titers in the lungs were measured as described in Materials and Methods. Results for low doses (1 × 104 CFU per mouse) and high doses (3 × 104 CFU per mouse) are shown. Closed bars, PAO1; open bars, TR18. Each value is shown as the mean and standard deviation for two to five replicates. The asterisk indicates significant difference from the PAO1 value by Student's t test or an Aspin-Welch test (P < 0.01).
TABLE 3.
Number of viable cells in the kidney and bladder organs in a urinary tract infection model
| No. of CFU infected | Strain | Log CFU/kidney at indicated daya
|
Log CFU/bladder at indicated daya
|
||||
|---|---|---|---|---|---|---|---|
| 1 | 3 | 7 | 1 | 3 | 7 | ||
| 1.6 × 103 | PAO1 | 2.91 ± 0.17 | 4.53 ± 0.64 | 6.46 ± 0.74 | 2.06 ± 0.09 | 2.63 ± 0.33 | 3.55 ± 0.55 |
| TR18 | 3.25 ± 0.47 | 2.98 ± 0.45 | 2.67 ± 0.52b | 2.57 ± 0.18c | 1.97 ± 0.34 | 2.24 ± 0.30 | |
| 1.6 × 104 | PAO1 | 4.24 ± 0.33 | 5.73 ± 0.35 | 6.42 ± 0.76 | 3.21 ± 0.17 | 3.04 ± 0.45 | 3.67 ± 0.60 |
| TR18 | 4.03 ± 0.50 | 3.70 ± 0.70c | 3.50 ± 0.47c | 2.90 ± 0.21 | 2.21 ± 0.14 | 1.90 ± 0.37c | |
Values are shown as the means and standard deviations for five replicates.
Significantly different from PAO1 value (P < 0.01) by Student's t test or Aspin-Welch test.
P < 0.05.
DISCUSSION
In this study we showed that the sha homologue genes of P. aeruginosa (PA1054 to PA1059) are responsible for Na+/H+ antiport activity and Na+ tolerance. This is the third Na+/H+ antiporter to be reported in P. aeruginosa. Two kinds of Na+/H+ antiporters from P. aeruginosa have been reported, i.e., NhaP and NhaB. NhaP is rather specific to Na+ and is considered to be a major Na+ extrusion system (17, 34). In contrast, NhaB is likely to be the major Li+ extrusion system in P. aeruginosa (17). Database searching suggests that P. aeruginosa PAO1 does not have a homologue of NhaA, which is a principal Na+/H+ antiporter that confers high Na+ resistance in E. coli (19, 21). A single mutation in PA1054 resulted in more severe Na+ sensitivity in the growth of P. aeruginosa than the nhaP mutation (Fig. 4). The PA1054-to-PA1059 cluster conferred higher Na+ resistance to a Na+/H+ antiporter-deficient E. coli (at 0.6 M and higher NaCl concentrations; Fig. 2) than NhaP (up to 0.4 M NaCl) (17). We thus consider that the PA1054-to-PA1059 cluster rather than NhaP is responsible for the major capacity for Na+ extrusion in P. aeruginosa. The PA1054-to-PA1059 cluster is not likely to possess a major capacity for pH homeostasis, since disruption of PA1054 did not affect tolerance to alkaline pH (described above). We propose that the genes PA1054 and PA1055 to PA1059 be named sha (sodium hydrogen antiporter) genes shaAB and shaCDEFG, respectively.
The loss of Sha function resulted in the attenuation of virulence as well as reduced colonization in mice, clearly indicating that the Sha system has a role in the pathogenicity of P. aeruginosa. It can be considered that the reduced virulence and colonization of the TR18 mutant is a result of a decreased adaptive ability to the Na+ environment inside the host, in similarity to the case of pha mutants of S. meliloti (24). Given that Sha homologues are distributed among animal and plant pathogens, including Pseudomonas, Vibrio, Brucella, Xanthomonas, Agrobacterium, Staphylococcus, and Listeria spp., the Na+ tolerance conferred by Sha may be a potential virulence factor. Since Sha proteins, except ShaF, which slightly shows a similarity to bile salt transporters (11, 18), are not related to eukaryotic Na+/H+ antiporters and other proteins, they could be potential targets of inhibitors to suppress their pathogenicity.
Recently, Potvin et al. (22) reported a systematic high-throughput screening of virulence factors and identified PA1054 as one of the mutated loci resulting in reduced virulence in a rat model of chronic respiratory infection. Our results were consistent with the results of Potvin et al. (22). It was also shown that biofilm formation decreases slightly for a mutant of PA1054 (22), but we could not find a difference in alginate production between PAO1 and TR18.
Several alternative sigma factors are known to be involved in the pathogenicity of P. aeruginosa. AlgU is an extracytoplasmic function-type sigma factor that transcribes genes of alginate biosynthesis, which is related to a mucoid phenotype that causes chronic pulmonary infections in patients with cystic fibrosis (5). σ54 encoded by rpoN is involved in motility, the production of pyocyanin, and pathogenicity in diverse hosts (8). RpoS is a global regulator that influences large number of genes in stationary phase and affects the production of several virulence factors, including alginate, exotoxin A, and pyocianin (28, 32). Since the loss of Sha function affects the activation of several alternative sigma factors in B. subtilis (13), we cannot exclude the possibility that the disruption of Sha may affect the induction of the above-named alternative sigma factors involved in pathogenicity, leading to a reduction of virulence.
While the transport mechanism of the Sha system is still not clear, the robust homeostatic capacity of Sha plays a critical role in various functions, such as infection, symbiosis, sporulation, and alkaliphily. We are interested in the molecular structure, assembly, and mechanism of the Sha system, which may support the robust homeostatic capacity of monovalent cations, including Na+ and H+. We are also interested in the effect of sha mutations on induction of alternative sigma factors in P. aeruginosa.
Acknowledgments
We thank H. Kobayashi (Chiba University, Japan) for providing E. coli TO114. We are grateful for R. H. Doi (University of California, Davis) for critical reading of the manuscript.
This work was partially supported by grants for the Bioarchitect Research Program and the Eco Molecular Science Research Program from RIKEN. This work was also supported by a Grant-in-Aid for Scientific Research for Young Scientists (B) from the Ministry of Education, Culture, Sports, Science and Technology of Japan to S.K.
REFERENCES
- 1.Bodey, G. P., R. Bolivar, V. Fainstein, and L. Jadeja. 1983. Infections caused by Pseudomonas aeruginosa. Rev. Infect. Dis. 5:279-313. [DOI] [PubMed] [Google Scholar]
- 2.Gemperli, A. C., P. Dimroth, and J. Steuber. 2002. The respiratory complex I (NDH I) from Klebsiella pneumoniae, a sodium pump. J. Biol. Chem. 277:33811-33817. [DOI] [PubMed] [Google Scholar]
- 3.Gemperli, A. C., P. Dimroth, and J. Steuber. 2003. Sodium ion cycling mediates energy coupling between complex I and ATP synthase. Proc. Natl. Acad. Sci. USA 100:839-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldberg, E. B., T. Arbel, J. Chen, R. Karpel, G. A. Mackie, S. Schuldiner, and E. Padan. 1987. Characterization of a Na+/H+ antiporter gene of Escherichia coli. Proc. Natl. Acad. Sci. USA 84:2615-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goven, J. R., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamamoto, T., M. Hashimoto, M. Hino, M. Kitada, Y. Seto, T. Kudo, and K. Horikoshi. 1994. Characterization of a gene responsible for the Na+/H+ antiporter system of alkaliphilic Bacillus species strain C-125. Mol. Microbiol. 14:939-946. [DOI] [PubMed] [Google Scholar]
- 7.Harel-Bronstein, M., P. Dibrov, Y. Olami, E. Pinner, S. Schuldiner, and E. Padan. 1995. MH1, a second-site revertant of an Escherichia coli mutant lacking Na+/H+ antiporters (ΔnhaAΔnhaB), regains Na+ resistance and a capacity to excrete Na+ in a ΔμH+-independent fashion. J. Biol. Chem. 270:3816-3822. [DOI] [PubMed] [Google Scholar]
- 8.Hendrickson, E. L., J. Plotnikova, S. Mahajan-Miklos, L. G. Rahme, and F. M. Ausubel. 2001. Differential roles of the Pseudomonas aeruginosa PA14 rpoN gene in pathogenicity in plants, nematodes, insects, and mice. J. Bacteriol. 183:7126-7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiramatsu, T., K. Kodama, T. Kuroda, T. Mizushima, and T. Tsuchiya. 1998. A putative multisubunit Na+/H+ antiporter from Staphylococcus aureus. J. Bacteriol. 180:6642-6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ito, M., A. A. Guffanti, and T. A. Krulwich. 2001. Mrp-dependent Na+/H+ antiporters of Bacillus exhibit characteristics that are unanticipated for completely secondary active transporters. FEBS Lett. 496:117-120. [DOI] [PubMed] [Google Scholar]
- 11.Ito, M., A. A. Guffanti, B. Oudega, and T. A. Krulwich. 1999. mrp, a multigene, multifunctional locus in Bacillus subtilis with roles in resistance to cholate and to Na+ and in pH homeostasis. J. Bacteriol. 181:2394-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ito, M., A. A. Guffanti, W. Wang, and T. A. Krulwich. 2000. Effects of nonpolar mutations in each of the seven Bacillus subtilis mrp genes suggest complex interactions among the gene products in support of Na+ and alkali but cholate resistance. J. Bacteriol. 182:5663-5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kosono, S., K. Asai, Y. Sadaie, and T. Kudo. 2004. Altered gene expression in the transition phase by disruption of a Na+/H+ antiporter gene (shaA) in Bacillus subtilis. FEMS Microbiol. Lett. 232:93-99. [DOI] [PubMed] [Google Scholar]
- 14.Kosono, S., S. Morotomi, M. Kitada, and T. Kudo. 1999. Analyses of a Bacillus subtilis homologue of the Na+/H+ antiporter gene which is important for pH homeostasis of alkaliphilic Bacillus sp. C-125. Biochim. Biophys. Acta 1409:171-175. [DOI] [PubMed] [Google Scholar]
- 15.Kosono, S., Y. Ohashi, F. Kawamura, M. Kitada, and T. Kudo. 2000. Function of a principal Na+/H+ antiporter, ShaA, is required for initiation of sporulation in Bacillus subtilis. J. Bacteriol. 182:898-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krulwich, T. A., J. Cheng, and A. A. Guffanti. 1994. The role of monovalent cation/proton antiporters in Na+-resistance and pH homeostasis in Bacillus: an alkaliphile versus a neutralophile. J. Exp. Biol. 196:457-470. [DOI] [PubMed] [Google Scholar]
- 17.Kuroda, T., N. Fujita, J. Utsugi, M. Kuroda, T. Mizushima, and T. Tsuchiya. 2004. A major Li+ extrusion system NhaB of Pseudomonas aeruginosa: comparison with the major Na+ extrusion system. Microbiol. Immunol. 48:243-250. [DOI] [PubMed] [Google Scholar]
- 18.Mathiesen, C., and C. Haegerhaell. 2003. The ‘antiporter module’ of respiratory chain complex I includes the MrpC/NuoK subunit-a revision of the modular evolution scheme. FEBS Lett. 549:7-13. [DOI] [PubMed] [Google Scholar]
- 19.Padan, E., and S. Schuldiner. 1994. Molecular physiology of Na+/H+ antiporters, key transporters in circulation of Na+ and H+ in cells. Biochim. Biophys. Acta 1185:129-151. [DOI] [PubMed] [Google Scholar]
- 20.Padan, E., T. Tzubery, K. Herz, L. Kozachkov, A. Rimon, and L. Galili. 2004. NhaA of Escherichia coli, as a model of a pH-regulated Na+/H+ antiporter. Biochim. Biophys. Acta 1658:2-13. [DOI] [PubMed] [Google Scholar]
- 21.Padan, E., M. Venturi, Y. Gerchman, and N. Dover. 2001. Na+/H+ antiporters. Biochim. Biophys. Acta 1505:144-157. [DOI] [PubMed] [Google Scholar]
- 22.Potvin, E., D. E. Lehoux, I. Kukavica-Ibrulj, K. L. Richard, F. Sanschagrin, G. W. Lau, and R. C. Levesque. 2003. In vivo functional genomics of Pseudomonas aeruginosa for high-throughput screening of new virulence factors and antibacterial targets. Environ. Microbiol. 5:1294-1308. [DOI] [PubMed] [Google Scholar]
- 23.Prentki, P., and H. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 24.Putnoky, P., A. Kereszt, T. Nakamura, G. Endre, E. Grosskopf, P. Kiss, and A. Kondorosi. 1998. The pha gene cluster of Rhizobium meliloti involved in pH adaptation and symbiosis encodes a novel type of K+ efflux system. Mol. Microbiol. 28:1091-1101. [DOI] [PubMed] [Google Scholar]
- 25.Rosen, B. P. 1986. Ion extrusion systems in Escherichia coli. Methods Enzymol. 25:328-336. [DOI] [PubMed] [Google Scholar]
- 26.Sakuma, T., N. Yamada, H. Saito, T. Kakegawa, and H. Kobayashi. 1998. pH dependence of the function of sodium ion extrusion systems in Escherichia coli. Biochim. Biophys. Acta 1363:231-237. [DOI] [PubMed] [Google Scholar]
- 27.Sasakawa, C., and M. Yoshikawa. 1987. A series of Tn5 variants with various drug-resistance markers and suicide vector for transposon mutagenesis. Gene 56:283-288. [DOI] [PubMed] [Google Scholar]
- 28.Schuster, M., A. C. Hawkins, C. S. Harwood, and E. P. Greenberg. 2004. The Pseudomonas aeruginosa RpoS regulon and its relationship to quorum sensing. Mol. Microbiol. 51:973-985. [DOI] [PubMed] [Google Scholar]
- 29.Steuber, J. 2003. The C-terminally truncated NuoL subunit (ND5 homologue) of the Na+-dependent complex I from Escherichia coli transports Na+. J. Biol. Chem. 278:26817-26822. [DOI] [PubMed] [Google Scholar]
- 30.Steuber, J., C. Schmid, M. Rufibach, and P. Dimroth. 2000. Na+ translocation by complex I (NADH:quinone oxidoreductase) of Escherichia coli. Mol. Microbiol. 35:428-434. [DOI] [PubMed] [Google Scholar]
- 31.Stolpe, S., and T. Friedrich. 2004. The Escherichia coli NADH:ubiquinone oxidoreductase (complex I) is a primary proton pump but may be capable of secondary sodium antiport. J. Biol. Chem. 279:18377-18383. [DOI] [PubMed] [Google Scholar]
- 32.Suh, S.-J., L. Silo-Suh, D. E. Woods, D. J. Hassett, S. E. H. West, and D. E. Ohman. 1999. Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J. Bacteriol. 181:3890-3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swartz, T. H., M. Ito, D. B. Hicks, M. Nuqui, A. A. Guffanti, and T. A. Krulwich. 2005. The Mrp Na+/H+ antiporter increases the activity of the malate:quinone oxidoreductase of an Escherichia coli respiratory mutant. J. Bacteriol. 187:388-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Utsugi, J., K. Inaba, T. Kuroda, M. Tsuda, and T. Tsuchiya. 1998. Cloning and sequencing of a novel Na+/H+ antiporter gene from Pseudomonas aeruginosa. Biochim. Biophys. Acta 1398:330-334. [DOI] [PubMed] [Google Scholar]
- 35.Yoshinaka, T., H. Takasu, R. Tomizawa, S. Kosono, and T. Kudo. 2003. A shaE deletion mutant showed lower Na+ sensitivity compared to other mutants in the Bacillus subtilis sodium/hydrogen antiporter (Sha) system. J. Biosci. Bioeng. 95:306-309. [DOI] [PubMed] [Google Scholar]