Abstract
Chlorocatechol 1,2-dioxygenase (CCD) is the first-step enzyme of the chlorocatechol ortho-cleavage pathway, which plays a central role in the degradation of various chloroaromatic compounds. Two CCDs, CbnA from the 3-chlorobenzoate-degrader Ralstonia eutropha NH9 and TcbC from the 1,2,4-trichlorobenzene-degrader Pseudomonas sp. strain P51, are highly homologous, having only 12 different amino acid residues out of identical lengths of 251 amino acids. But CbnA and TcbC are different in substrate specificities against dichlorocatechols, favoring 3,5-dichlorocatechol (3,5-DC) and 3,4-dichlorocatechol (3,4-DC), respectively. A study of chimeric mutants constructed from the two CCDs indicated that the N-terminal parts of the enzymes were responsible for the difference in the substrate specificities. Site-directed mutagenesis studies further identified the amino acid in position 48 (Leu in CbnA and Val in TcbC) as critical in differentiating the substrate specificities of the enzymes, which agreed well with molecular modeling of the two enzymes. Mutagenesis studies also demonstrated that Ile-73 of CbnA and Ala-52 of TcbC were important for their high levels of activity towards 3,5-DC and 3,4-DC, respectively. The importance of Ile-73 for 3,5-DC specificity determination was also shown with other CCDs such as TfdC from Burkholderia sp. NK8 and TfdC from Alcaligenes sp. CSV90 (identical to TfdC from R. eutropha JMP134), which convert 3,5-DC preferentially. Together with amino acid sequence comparisons indicating high conservation of Leu-48 and Ile-73 among CCDs, these results suggested that TcbC of strain P51 had diverged from other CCDs to be adapted to conversion of 3,4-DC.
Chlorocatechols are the central metabolites in the bacterial degradation processes of many chlorinated aromatic compounds (reviewed in references 50 and 60). The degradation processes via chlorocatechols have been studied intensively with simple chlorinated aromatics such as chlorobenzoates (7, 10, 18, 23, 25, 26), chlorobenzenes (21, 47, 51, 53, 54, 56, 57, 62), and 2,4-dichlorophenoxyacetic acid (2,4-D) (4, 9, 14). In the degradation of chlorocatechols, the chlorocatechol ortho-cleavage pathway (modified ortho-cleavage pathway) plays a central role, converting chlorocatechols into 3-oxoadipate (reviewed in references 24, 42, and 50), although several examples of degradation of chlorocatechols via the meta-cleavage pathway have been documented (1, 34).
The genes encoding enzymes of the chlorocatechol ortho-cleavage pathway have been found as a cluster and mostly on large plasmids, which are often transmissible (3, 6, 7, 8, 32, 39, 62, 67; for a review, see reference 38). Some of the chlorocatechol gene clusters have been found to be carried on transposons (30, 40, 45, 63) or a phage-like element (49, 55). Recent reports on the entire sequencing of two 2,4-D degradative plasmids, pJP4 and pEST4011, illustrated their genetic features of basic plasmid functions such as replication, maintenance, and transfer, shared with other broad-host-range IncP1 plasmids (59, 63). These findings have revealed some of the reasons underlying the wide distribution of the gene clusters of the chlorocatechol ortho-cleavage pathway.
In the first step of the chlorocatechol ortho-cleavage pathway, chlorocatechol 1,2-dioxygenase (CCD) cleaves the aromatic ring, converting chlorocatechols into chloro-cis,cis-muconates, and thus has been a subject of intensive study from an enzymatic viewpoint. The CCDs of several strains have been characterized, and some of them have been purified (2, 5, 10, 27, 31, 33, 35, 43, 44, 46, 61). The early enzymatic studies of the CCD from the 3-chlorobenzoate (3-CB)-degradative Pseudomonas sp. strain B13 revealed that the CCD is adapted for the conversion of chlorocatechols such as 3-chlorocatechol, unlike catechol 1,2-dioxygenase, which cannot process chlorocatechols efficiently (10, 11). Several studies have revealed a tendency such that CCDs responsible for the conversion of 3,5-dichlorocatechol (3,5-DC) produced as an intermediate of 2,4-D degradation are adapted for the preferential conversion of 3,5-DC (2, 43). CCDs engaged in degradative pathways for other compounds could show different substrate specificities towards congeners of chlorocatechols: one example is TcbC from the 1,2,4-trichlorobenzene-degrading Pseudomonas sp. strain P51, which favors 3,4-dichlorocatechol (3,4-DC) (61). Recently, the crystal structure of the 4-chlorocatechol 1,2-dioxygenase from Rhodococcus opacus strain 1CP has been solved (15, 17), and another CCD from the strain 1CP has been crystallized (16). The crystal structure of the evolutionarily related catechol 1,2-dioxygenase with its substrate has been also solved (65), and a catalytic mechanism for intradiol cleavage enzymes has been proposed (48, 64). While the crystal structure studies provide information of the enzymes including the amino acid residues that interact with substrates, there has been no report of the actual effect of relevant amino acids on the substrate specificities towards different congeners of chlorocatechols.
During the course of our previous study of the function of a chlorocatechol gene cluster including the comparison of the activities of several CCDs, we found that the substrate specificity of CbnA, a CCD from a 3-CB-degrading bacterium, Ralstonia eutropha NH9 (recently renamed Wautersia eutropha and further renamed Cupriavidus necator) (40), was different from that of TcbC from strain P51 despite their high similarity (95% identity at amino acid level); the former prefers 3,5-DC while the latter prefers 3,4-DC among dichlorocatechols (31, 61). Therefore, we conducted a comparative analysis of the two enzymes using chimeric and site-directed mutants. Our analysis revealed the importance of the amino acids in positions 48, 52, and 73 in the determination of the different substrate specificities of the two CCDs. The relevance of position 73 in the substrate specificity for 3,5-DC was further confirmed by using two 3,5-DC-type CCDs, TfdC from Burkholderia sp. NK8 (31) and TfdC from the 2,4-D-degrading Alcaligenes sp. CSV90 (identical to TfdC from R. eutropha JMP134) (3, 41). Our present study demonstrates that the CCDs of gram-negative bacteria have the three amino acid positions involved in the interaction with substrates in common with ClcA of R. opacus 1CP and CatA of Acinetobacter sp. ADP1 despite the distinct evolution of the (chloro)catechol 1,2-dioxygenases of gram-negative bacteria and gram-positive bacteria (12, 13).
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and chemicals.
Bacterial strains and plasmids used in this study are listed in Table 1. Burkholderia sp. NK8 and Ralstonia eutropha NH9 were grown in BSM medium (39) supplemented with 5 mM 3-CB at 30°C. Alcaligenes sp. CSV90 was grown in BSM containing 2 mM 2,4-D at 30°C. Plasmid-containing Escherichia coli strains were grown at 30°C or 37°C in Luria-Bertani (LB) medium containing ampicillin (Ap; 50 μg/ml) (52). 3-CB was purchased from Wako Pure Chemical Industries (Osaka, Japan). 2,4-D and 3-chlorocatechol were purchased from Tokyo Chemical Industry (Tokyo, Japan). Dichlorocatechols were purchased from Helix Biotech (New Westminster, Canada).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant propertiesa | Reference or source |
|---|---|---|
| Strains | ||
| Burkholderia sp. strain NK8 | 3-CB+ 4-CB+ | 18 |
| Ralstonia eutropha NH9 | 3-CB+ | 39 |
| Pseudomonas sp. strain P51 | 1,2-DCB+ 1,2,4-TCB+ | 62 |
| Alcaligenes sp. strain CSV90 | 2,4-D+ | 2 |
| Escherichia coli DH5α | Δ(lacZYA-argF)U169 hsdR17 relA1 supE44 endA1 recA1 thi gyrA96 φ80dlacZΔM15 | 22 |
| Plasmids | ||
| pUC18 | LacZ+ Apr | 68 |
| pUC9A | cbnA including SD sequence inserted in pUC18; Apr | 31 |
| pUC51 | tcbC including SD sequence inserted in pUC18; Apr | This study |
| pUC90 | tfdCCSV90 including SD sequence inserted in pUC18; Apr | This study |
| pUC8A | tfdCNK8 including SD sequence inserted in pUC18; Apr | 31 |
3-CB, 3-chlorobenzoate; 4-CB, 4-chlorobenzoate; 1,2-DCB, 1,2-dichlorobenzene; 1,2,4-TCB, 1,2,4-trichlorobenzene; 2,4-D, 2,4-dichlorophenoxyacetic acid; Apr, ampicillin resistance; SD, Shine-Dalgarno sequence.
DNA preparation and in vitro manipulation.
Plasmid DNA was isolated using the QIAprep Spin Miniprep Kit as specified by the manufacturer's directions (QIAGEN, Hilden, Germany). Restriction endonuclease digestions, ligations, and detection of colonies carrying an insert were performed according to standard procedures (52). DNA fragments were purified from agarose gel slices using the GeneClean II system (Qbiogene, Inc., Carlsbad, CA). Transformation of E. coli DH5α was achieved by the meothod of Inoue et al. (29).
Construction of hybrid CCDs.
The DNA fragments encoding CbnA, TcbC, TfdC of Burkholderia sp. NK8 (TfdCNK8), and TfdC of Alcaligenes sp. CSV90 (TfdCCSV90) were amplified by PCR with primers against the start codon including the Shine-Dalgarno sequence and stop codon of the genes. The N-terminal primer contained an EcoRI site (5′-TTTTGAATTCCGACAAAGGAGACCGGGATGAACGAACG-3′ for cbnA and tcbC, 5′-TTTTGAATTCGACGGAGGCAAAGTGAACAAAAG-3′ for tfdCNK8, and 5′-TTTTGAATTCGGAACGGAGGCGAGGTGAACAAGCG-3′ for tfdCCSV90; the EcoRI site in the sequences above is underlined, and the start codon is shown in boldface type). The C-terminal primers contained a PstI site (5′-TTTTCTGCAGTCATGCGTGCTCCCGGGTCGTTCG-3′ for cbnA, 5′-TTTTCTGCAGTCATACGTGCTCCCGCGCGCGCTCG-3′ for tcbC, and 5′-TTTTCTGCAGTCACGGGTTTGCCCCCGCCTGCGCAC-3′ for tfdCNK8, and 5′-TTTTCTGCAGTCATGTGTTGGCTCCCGAGTGCTCGAGGGCGCG-3′ for tfdCCSV90; the PstI site is underlined, and the stop codon is shown in boldface type). The EcoRI-PstI fragments containing the cbnA, tcbC, tfdCNK8, and tfdCCSV90 genes were cloned into the EcoRI-PstI site of pUC18 to get pUC9A, pUC51C, pUC8A, and pUC90, respectively.
Hybrid CCD genes were composed of pUC9A and pUC51C by using fragment exchanges. Conserved BamHI, ApaI, and NspV restriction sites were located in the nucleotide sequences of the genes cbnA and tcbC. The three endonucleases were used together with EcoRI and PstI to digest pUC9A and pUC51C to obtain the EcoRI-BamHI, BamHI-ApaI, ApaI-NspV, and NspV-PstI fragments; the amino acid sequences which they encode were defined as regions 1, 2, 3, and 4, respectively. Using the four fragments of cbnA and tcbC, hybrid plasmids were constructed from one or two fragment exchanges. Site-directed mutagenesis was performed by PCR using primers that contained an StyI or a BamHI site. The primers used are listed in Table 2. All the mutants were verified by DNA sequencing.
TABLE 2.
Primers used for site-directed mutagenesis
| Construct | Mutation | Primera |
|---|---|---|
| MC12 | T70R | 5′-ACGCGCGGATCCAGCGCCCGCGATCC-3′ |
| MC35 | L48V | 5′-GGCCAAGGAAATGGCTGTCCTGCTGG-3′ |
| MC3n | I73M | 5′-ACGCGCGGATCCACGCCCGCGATGCAGGGCCCGTA-3′ |
| MC4 | V52A | 5′-GGCCAAGGAAATGGCTCTCCTGCTGGACGCGTTCTTCAACC-3′ |
| MC6 | I73L | 5′-GCGGATCCACGCCCGCGCTCCAGGGCCCGTATTCC-3′ |
| MC61 | I73G | 5′-GCGGATCCACGCCCGCGGGCCAGGGCCCGTATTCC-3′ |
| MC62 | I73V | 5′-GCGGATCCACGCCCGCGGTCCAGGGCCCGTATTCC-3′ |
| MC63 | I73F | 5′-GCGGATCCACGCCCGCGTTCCAGGGCCCGTATTCC-3′ |
| MC7 | L48I | 5′-GGCCAAGGAAATGGCTATCCTGCTGGAC-3′ |
| MC9 | M46V | 5′-GGCCAAGGAAGTGGCTCTCCTGCTGGAC-3′ |
| MT61 | A52G | 5′-GGCCAAGGAAGTGGCTGTCCTGCTGGACGGGTTCTTCAACCACACGATC-3′ |
| MT62 | A52L | 5′-GGCCAAGGAAGTGGCTGTCCTGCTGGACCTGTTCTTCAACCACACGATC-3′ |
| MT63 | A52F | 5′-GGCCAAGGAAGTGGCTGTCCTGCTGGACTTCTTCTTCAACCACACGATC-3′ |
| MT7 | V46M | 5′-GGCCAAGGAAATGGCTGTCCTGCTGGAC |
| I73MNK8 | I73M | 5-ACGCCAGCGATGGAGGGACC-3 |
| I73MCSV90 | I73M | 5-TTCAACGCCTGCTATGGAGGGAC-3 |
The underlined bases indicate the endonuclease restriction sites used for construction of plasmids. GGATCC, BamHI, CCAAGG, StyI. Base changes are shown in boldface.
Expression of CCD in E. coli.
Protein expression of CCDs was performed in E. coli DH5α as described previously (31). One milliliter of the E. coli cells precultured in 5 ml of LB plus Ap at 37°C for 10 h was used to inoculate 50 ml of LB medium containing Ap and 0.3 mM isopropyl β-d-thiogalactopyranoside (IPTG). The cultures were grown at 30°C for 16 h. Cells harvested by centrifugation were used to prepare a crude cell extract for the CCD assay.
Assay of CCD activity.
CCD activity was measured by the disappearance of chlorocatechol as described previously (31). Cell extracts were prepared by sonication, and cell debris was removed by centrifugation for 15 min at 15,000 rpm at 4°C. The concentration of chlorocatechols in the enzyme reaction mixture was analyzed by high-performance liquid chromatography with a C18 reversed-phase column (Wako, Tokyo). A solvent of 10 mM phosphoric acid:acetonitrile (1:1 [vol/vol]) was applied at a flow rate of 1 ml/min. The activities were measured in triplicate. Enzyme activities were expressed as the number of nanomoles of substrate per minute per milligram of protein. Protein concentrations were determined by the Bradford protein assay from Bio-Rad (Hercules, CA) using bovine serum albumin as a standard.
Western blot analysis of the mutant proteins with no CCD activity.
To check the production of the chimeric and site-directed mutated proteins with no CCD activity, Western blot analysis was performed using whole-cell protein samples from induced DH5α strains expressing the mutant genes. The cell lysates were prepared by sonication. Proteins (each, 10 mg) were separated by sodium dodecyl sulfate-12.5% polyacrylamide gel electrophoresis and transferred onto a Clear Blot Membrane-P (ATTO, Tokyo, Japan) by electroblotting. A synthetic peptide (WHSTPDGKYSGFHD) that was designed on the basis of the amino acid sequences of TfdCNK8 and CbnA was used by Sawady Technology (Tokyo, Japan) for the preparation of rabbit antiserum. The membranes were probed with rabbit antiserum and a secondary horseradish peroxidase-conjugated anti-rabbit immunoglobulin antibody. Immunoreactive bands were detected by using the ECL Western blot reagent (Amersham Biosciences, Piscataway, NJ).
Purification of CbnA and TcbC.
For enzyme kinetics experiments, E. coli cells containing the recombinant CCDs were grown at 30°C for 15 h in LB medium containing 50 μg/ml ampicillin and 0.3 mM IPTG. Cells were harvested by centrifugation (10,000 × g; 5 min at 4°C). The cell pellets were washed with 33 mM Tris-HCl, pH 8.0, and stored at −80°C. The cells were resuspended in 50 mM Tris-HCl, pH 8.0, and disrupted by sonication (20 s, repeated a total of 10 times). The crude cell extract was centrifuged at 16,000 × g for 30 min and filtered through a 0.22-μm membrane. The supernatant was loaded onto the HiPrep 16/10 DEAE FF column (Amersham Biosciences) equilibrated with 50 mM Tris-HCl, pH 8.0, and protein was eluted with a linear gradient of 0 to 700 mM NaCl in this buffer. The brownish fractions containing the desired activity were combined, concentrated, and desalted on an Amicon Centriprep YM-10 centrifugal filter device (Millipore, Bedford, MA). The concentrated enzyme solution was further purified by gel filtration carried out with a HiLoad 16/60 Superdex 200 pg column (Amersham Biosciences), equilibrated with the above-described Tris-HCl buffer containing 0.1 M NaCl; the same buffer was used as the eluent. All the purification steps were performed at 10°C.
Enzyme kinetics.
The values of apparent kinetic constants (km, Vmax, and kcat) were obtained from [S]/v versus [S] plots. For this, the activities were assayed spectrophotometrically at 25°C with an air-saturated buffer of 50 mM Tris-HCl, pH 8.0. The reaction was initiated by the addition of the enzyme solution, and absorption at 260 nm was monitored with a spectrophotometer. Vmax was expressed as the number of units per milligram of protein; 1 U was defined as the amount of enzyme that transformed 1 μmol of substrate per min. The extinction coefficients for chlorocatechols were taken from Dorn and Knackmuss (11).
Theoretical models of CbnA and TcbC.
Theoretical models of CbnA and TcbC were prepared by using the Swiss model repository based on the crystal structures of 4-chlorocatechol 1,2-dioxygenase from R. opacus 1CP (ClcA1CP) (Protein Database identification no. 1s9a) and catechol 1,2-dioxygenase from Acinetobacter sp. strain ADP1 (CatAADP1) (Protein Database identification no. 1dlm and 1dlq). The obtained models were modified manually; the substrate was added to the active site on the basis of the earlier solved crystal structures of the complex of intradiol cleavage enzymes with catechol-type substrates (Protein Database identification no. 1dlt and 3pca); the side chains around the bound substrate were also modified. The obtained models were regularized using a routine in the program Quanta (Accelrys Software Inc., San Diego, CA).
RESULTS
Different substrate specificities of CbnA and TcbC. CbnA and TcbC are highly conserved at the amino acid sequence level, differing from each other at only 12 residues out of identical lengths of 251 residues (Table 3 and Fig. 1). Despite their high identity, the two enzymes possess very different substrate specificities against 3,4-DC and 3,5-DC. CbnA had a high relative activity against 3,5-DC (80.8%) and low activity against 3,4-DC (4.2%) when compared with its activity against 3-chlorocatechol (100%) (Table 3). On the contrary, TcbC had a high relative activity against 3,4-DC (98.8%) but a low activity against 3,5-DC (6.1%) compared to its activity against 3-chlorocatechol (Table 3). The differences should be derived from (some of) the 12 different residues.
TABLE 3.
Relative activities of chlorocatechol 1,2-dioxygenases constructed from cbnA and tcbC by using fragment exchange
| Method | Construct | Amino acid residuesa | Relative activityb
|
||
|---|---|---|---|---|---|
| 3-CCc | 3,4-DC | 3,5-DC | |||
| Fragment exchange | CbnA | M46L48V52-T70I73-F133L141V199-A229A239P247A251 | 100 (172.9 ± 6.3) | 4.2 ± 0 | 80.8 ± 1.6 |
| C123T4 | MLV-TI-FLV-TDAV | 100 (212.5 ± 6.8) | 6.6 ± 0.1 | 86.9 ± 3.9 | |
| C12T34 | MLV-TI-IMA-TDAV | 100 (268.7 ± 36.8) | 5.9 ± 0.9 | 87.3 ± 3.2 | |
| C1T234 | MLV-RM-IMA-TDAV | 100 (378.6 ± 9.8) | 1.8 ± 0.1 | 17.1 ± 0.4 | |
| C14T23 | MLV-RM-IMA-AAPA | 100 (137.7 ± 7.6) | 5.1 ± 1.3 | 8.7 ± 0.5 | |
| C134T2 | MLV-RM-FLV-AAPA | 100 (78.9 ± 9.3) | 9.1 ± 0.4 | 20.9 ± 1.1 | |
| T1C234 | VVA-TI-FLV-AAPA | 0 | 0 | 0 | |
| TcbC | V46V48A52-R70M73-I133M141A199-T229D239A247V251 | 100 (278.2 ± 6.4) | 98.8 ± 5.4 | 6.1 ± 0.9 | |
| T123C4 | VVA-RM-IMA-AAPA | 0 | 0 | 0 | |
| T12C34 | VVA-RM-FLV-AAPA | 0 | 0 | 0 | |
| T1C234 | VVA-TI-FLV-AAPA | 0 | 0 | 0 | |
| MT22 | VVA-RI-FLV-AAPA | 100 (54.3 ± 1.6) | 72.4 ± 2.2 | 22.3 ± 4.8 | |
| T14C23 | VVA-TI-FLV-TDAV | 100 (247.5 ± 13.0) | 57.7 ± 4.3 | 15.2 ± 2.9 | |
| T124C3 | VVA-RM-FLV-TDAV | 100 (154.3 ± 8.5) | 100 ± 5.2 | 7.9 ± 0.3 | |
| Site-directed | CbnA | M46L48V52-T70I73-F133L141V199-A229A239P247A251 | 100 (172.9 ± 6.3) | 4.2 ± 0 | 80.8 ± 1.6 |
| mutagenesis | MC9 | VLV-TI-FLV-AAPA | 100 (245.5 ± 9.0) | 5.9 ± 0.1 | 109 ± 1.7 |
| MC35 | MVV-TI-FLV-AAPA | 0 | 0 | 0 | |
| MC31 | MVV-TI-FLV-TDAV | 100 (205.8 ± 1.4) | 34.3 ± 0.4 | 13.3 ± 0.1 | |
| C123T4 | MLV-TI-FLV-TDAV | 100 (212.5 ± 6.8) | 6.6 ± 0.1 | 86.9 ± 3.9 | |
| MC4 | MLA-TI-FLV-AAPA | 100 (72.1 ± 1.7) | 23.8 ± 0.9 | 61.8 ± 1.5 | |
| MC41 | MLA-TI-FLV-TDAV | 100 (215.6 ± 1.6) | 25.8 ± 2.3 | 78.7 ± 1.3 | |
| MC12 | MLV-RI-FLV-AAPA | 100 (473.3 ± 2.1) | 7.1 ± 0.9 | 71.8 ± 1.8 | |
| MC3n | MLV-TM-FLV-AAPA | 100 (40.5 ± 2.4) | 12.3 ± 1.0 | 16.1 ± 1.9 | |
| TcbC | V46V48A52-R70M73-I133M141A199-T229D239A247V251 | 100 (278.2 ± 6.4) | 98.8 ± 5.4 | 6.1 ± 0.9 | |
| MT7 | MVA-RM-IMA-TDAV | 100 (91.4 ± 3.4) | 105 ± 0.5 | 4.4 ± 1.6 | |
| MT4 | MLA-RM-IMA-TDAV | 100 (192.3 ± 5.3) | 31.1 ± 0.3 | 13.5 ± 0.2 | |
| MT3 | MVV-RM-IMA-TDAV | 100 (146.7 ± 4.0) | 40.4 ± 1.6 | 6.9 ± 1.7 | |
| MT5 | VLV-RM-IMA-TDAV | 100 (241.4 ± 8.5) | 6.6 ± 0.1 | 12.3 ± 0.3 | |
| MT62 | VVL-RM-IMA-TDAV | 100 (51.6 ± 2.6) | 402 ± 7.5 | 1 ± 0.7 | |
The amino acid residues shown are the 12 different residues between CbnA and TcbC. The residues from CbnA are in italics; the residues from TcbC are underlined. The numbers show position of residues in the enzymes. The hyphens express the boundaries of the four restriction fragments 1, 2, 3, and 4, delineated by BamHI, ApaI and NspV (from left to right).
The activity against 3-CC was set as 100% for each construct. Values indicate means ± standard deviation of three replicates.
The values in parentheses are specific activities expressed as nanomoles per minute per milligram of protein. 3-CC, 3-chlorocatechol; 3,4-DC, 3,4-dichlorocatechol; 3,5-DC, 3,5-dichlorocatechol.
FIG. 1. .
Alignment of (chloro)catechol 1,2-dioxygenases. Unless stated otherwise below, the amino acid sequences used for the alignment are those of chlorocatechol 1,2-dioxygenase: CatA ADP1, catechol 1,2-dioxygenase from Acinetobacter sp. strain ADP1 (DDBJ accession number AF009224); CbnA NH9, Ralstonia eutropha NH9 (accession number AB019032); TcbC P51, Pseudomonas sp. strain P51 (accession number M57629); TfdC CSV90, 3,5-dichlorocatechol 1,2-dioxygenase from Alcaligenes sp. CSV90 (accession number D16356), identical to TfdC of R. eutropha JMP134 (3, 41); TfdC NK8, Burkholderia sp. NK8 (accession number AB050198); and ClcA 1CP, 4-chlorocatechol 1,2-dioxygenase from Rhodococcus opacus 1CP (accession number AF003948). The alignment is according to the study of ClcA of R. opacus 1CP (17). The conserved residues in all six enzymes are shown in boldface. The 12 residues differing between CbnA and TcbC are expressed in boldface italics, and numbers of the positions are indicated in italics above the alignment. The four regions delineated by restriction sites in cbnA and tcbC are indicated by the dotted arrows above the alignment. The residues which are considered to interact with substrate in CatA of ADP1 and ClcA of 1CP are labeled with # above the alignment and with * below the alignment, respectively. The four conserved ligands for iron are labeled with δ.
Substrate specificity of chimeric CCDs constructed from CbnA and TcbC.
Both cbnA and tcbC can be divided into four regions by using the restriction sites that are unique and conserved in the two genes. The regions of cbnA were named C1, C2, C3, and C4, starting from the N terminus. The corresponding regions of tcbC were named T1, T2, T3, and T4.
To determine which region(s) of cbnA was responsible for the specific activity pattern of the enzyme (higher activity against 3,5-DC than against 3,4-DC), a region(s) of cbnA was replaced by the corresponding region(s) of tcbC (Table 3). The activity pattern of C123T4 and C12T34, which consisted of at least two N-terminal regions of CbnA and the other C-terminal regions of TcbC, was almost the same as that of CbnA, i.e., high relative activities against 3,5-DC (ca. 87%) and low activities against 3,4-DC (ca. 6%) (Table 3). It was thus suggested that the difference in the activity pattern for the dichlorocatechols between CbnA and TcbC was not due to the differences in the amino acid residues in regions 3 and 4.
On the contrary, when region 2 of CbnA was replaced by that of TcbC, the relative activity against 3,5-DC decreased fourfold or more (C1T234, C14T23, and C134T2). It was thus shown that C2 of CbnA contributed to the higher level of activity of CbnA against 3,5-DC.
The activities of chimeric enzymes consisting of the N terminus of TcbC and the C terminus of CbnA are shown in Table 3. Three constructs (T123C4, T12C34, and T1C234) were inactive. Replacement of C4 of T12C34 and T1C234 by T4, generating T124C3 and T14C23, respectively, restored the activity. It was thus suggested that the interaction between the amino acids unique to TcbC in T1 and T4 was necessary to maintain the enzyme activity of TcbC. MT22, in which Thr-70 of T1C234 was replaced by Arg, not only regained activity but also exhibited a specificity pattern similar to that of TcbC, suggesting a suppression effect of Arg at this position in T1C234.
The fact that T14C23 exhibited higher specificity towards 3,4-DC, in contrast to C123T4, which favors 3,5-DC (Table 3 and above), suggested that region 1 of the enzymes was critical for the difference in the specificities towards the two dichlorocatechols.
While T124C3 retained high activity against 3,4-DC (100%), T14C23 exhibited decreased activity against 3,4-DC (58%), suggesting that T2 contributed to the activity pattern of TcbC to some extent. The decrease, however, was not so significant compared to that between the corresponding chimeric constructs of CbnA, where replacement of C2 by T2 decreased the relative activity against 3,5-DC to one-fourth or less (above).
The substrate specificity pattern of T124C3 was very similar to that of TcbC, indicating that amino acids unique to T3 were not relevant to the specificity pattern of TcbC.
Together, the above results strongly suggested that region 1 was critical for the difference in the specificity pattern between CbnA and TcbC. It was also suggested that region 2 of the two enzymes contributed, to some extent, to the higher relative activity towards the preferred dichlorocatechol of the respective enzymes.
Site-directed mutation.
To establish which amino acid residue(s) determines the difference between CbnA and TcbC in relative activity against the dichlorocatechols, we generated point mutants by replacing an amino acid in a relevant position in one CCD by that in a corresponding position of the other CCD or by another amino acid and analyzed their activities.
Amino acids in regions 1 and 2 of CbnA were replaced one by one (Table 3). Replacement of Met-46 of CbnA by Val, the amino acid at position 46 in TcbC, did not change the substrate specificity pattern (MC9), suggesting that the amino acid in position 46 was not important to differentiate the substrate specificity.
When Leu-48 of CbnA was replaced by Val, the amino acid at the corresponding position of TcbC, the resultant construct MC35 did not show any activity. The activity of MC35, however, was restored by the replacement of region 4 of CbnA by that of TcbC (MC31). These results suggested that, among the amino acids unique to T1, it was Val-48 that had some interaction with the amino acid(s) unique to T4, which rendered the enzyme active. From a comparison of the specificity patterns of MC31 and C123T4, which were different only at position 48, it was concluded that Leu-48 was critical for the specificity of CbnA favoring 3,5-DC.
When Val-52 of both CbnA and C123T4 was replaced by Ala, generating MC4 and MC41, respectively, the higher activity for 3,5-DC was retained while the relative activity for 3,4-DC was increased. It was, therefore, suggested that Val-52 was not critical for the high activity against 3,5-DC, while Ala-52 contributed to the 3,4-DC activity to some extent.
In region 2 of CbnA, the replacement of Thr-70 by Arg did not affect the substrate specificity pattern of CbnA (MC12) (Table 3). On the contrary, the replacement of Ile-73 by Met resulted in the enzyme having low relative specificity against 3,5-DC (MC3n; 16.1%). Replacement of Ile-73 of CbnA by other amino acids (Leu, Gly, Val, or Phe) resulted in inactive enzymes (data not shown). It was thus indicated that Ile-73 was important for the high specificity of CbnA for 3,5-DC.
The amino acids in region 1 of TcbC were replaced by the amino acids of the corresponding positions in region 1 of CbnA (Table 3). When Val-46 of TcbC was replaced by Met (MT7), the substrate specificity pattern of the enzyme was not changed. Thus, Val-46 was not critical in the determination of substrate specificity against 3,4-DC of TcbC.
When Val-48 and Ala-52 of MT7 was replaced by Leu and Val, respectively (resulting in MT4 and MT3, respectively), the relative activities against 3,4-DC decreased two- to threefold (31.1% and 40.4%, respectively). When positions 48 and 52 of TcbC were replaced simultaneously (resulting in MT5), the relative activity against 3,4-DC decreased considerably (6.6%). Similar results for specificity were obtained with C1T234 (Table 3). These results indicated that Val-48 and Ala-52 were involved in the high specificity against 3,4-DC of TcbC.
When Ala-52 of TcbC was replaced by Leu, the resulting construct MT62 exhibited a markedly increased relative activity for 3,4-DC (402%). On the contrary, when Ala-52 of TcbC was replaced by Gly or Phe, the resulting constructs did not show any activity (data not shown), suggesting the critical importance of the amino acid in position 52 for the activity and substrate specificity of TcbC.
In summary, it was indicated that Leu-48 and Ile-73 of CbnA were critical for its high specificity against 3,5-DC, while Val-48 and Ala-52 of TcbC were critical for its high specificity against 3,4-DC. On the other hand, Met-73 did not seem to be critical for the 3,4-DC specificity of TcbC, considering that MT22 and T14C23 retained relatively high specificity against 3,4-DC (Table 3).
Confirmation of production of the mutant proteins with no CCD activity.
To check if the null activity of some of the chimeric and site-directed mutant proteins in the above experiments was due to the failure of production of the proteins, Western blot analysis was performed using whole-cell protein samples from induced DH5α strains expressing the mutant CCD genes. Use of the antibody specific for the synthetic 14-residue peptides of CbnA demonstrated that the CCD mutant constructs described above that showed no CCD activity yielded full-length proteins (data not shown).
Involvement of Ile-73 in the high relative activities against 3,5-DC of CCDs from other strains.
Ile-73 is conserved in many CCDs including CbnA; TfdC from a chlorobenzoate-degrading strain, Burkholderia sp. NK8 (TfdCNK8) (31); and TfdC from a 2,4-D-degrading strain, Alcaligenes sp. CSV90 (TfdCCSV90) (Fig. 1) (3). These TfdCs have been shown in previous studies to have very high relative activities against 3,5-DC (2, 31). In the present study, Ile-73 of CbnA was shown to be critically important in the determination of its high substrate specificity against 3,5-DC (MC3n) (Table 3). When replacement of Ile-73 of CbnA by several amino acids was performed, only replacement by Met resulted in active enzyme (above). Thus, Ile-73 of TfdCNK8 and TfdCCSV90 were replaced by Met to test the specificity. The relative activities of the resultant mutant TfdCs against 3,5-DC decreased from 171% to 58% (for TfdCNK8) and from 151% to 52% (for TfdCCSV90) (Table 4). This result indicated that the Ile-73 of TfdCs was also involved in the determination of substrate specificity against 3,5-DC. Our study suggested a universal importance of Ile-73 in the determination of the substrate specificity of CCDs against 3,5-DC.
TABLE 4.
Relative activities of wild-type and mutant (I73M) enzymes of TfdCs from Burkholderia sp. NK8 and from Alcaligenes sp. CSV90a
| Strain | Enzyme | Activity against:
|
||
|---|---|---|---|---|
| 3-CC | 3,4-DC | 3,5-DC | ||
| NK8 | TfdC | 100 (182.7 ± 3.5) | 5 ± 1.3 | 171 ± 5.3 |
| TfdCI73M | 100 (146.6 ± 5.2) | 17 ± 4.5 | 58 ± 0.7 | |
| CSV90 | TfdC | 100 (620.2 ± 19.1) | 9 ± 0.2 | 151 ± 0.6 |
| TfdCI73M | 100 (565.3 ± 13.3) | 18 ± 0.5 | 52 ± 1.4 | |
The activity against 3-CC was set as 100%. The specific activities for 3-CC are expressed in parentheses as nanomoles per minute per milligram of protein. 3-CC, 3-chlorocatechol; 3,4-DC, 3,4-dichlorocatechol; 3,5-DC, 3,5-dichlorocatechol.
Kinetics of CbnA and TcbC.
The two-step purification procedure yielded CbnA and TcbC of about 80 to 90% purity for each enzyme (data not shown). Using the partially purified proteins, it was shown that the Km values (in micromoles) with 3,4-DC and 3,5-DC for CbnA were 0.58 and 0.38, respectively and those for TcbC were 0.67 and 0.83, respectively. Vmax values (in units per milligram of protein) with 3,4-DC and 3,5-DC for CbnA were 0.07 and 1.6, respectively, and 2.68 and 0.24 for TcbC, respectively. These results suggested that the turnover rates were critical for the substrate specificities of CbnA and TcbC. The details of enzyme kinetics remain to be elucidated.
Theoretical molecular models.
Molecular models of CbnA and TcbC were prepared on the basis of the crystal structures of ClcA1CP (Protein Database identification no. 1s9a) (17) and CatAADP1 (Protein Database identification no. 1dlm and 1dlq) (65). Because the amino acid sequence identities between the target and model proteins are about 30 to 40%, the obtained models seem to be reliable in terms of predicting the location of amino acid residues.
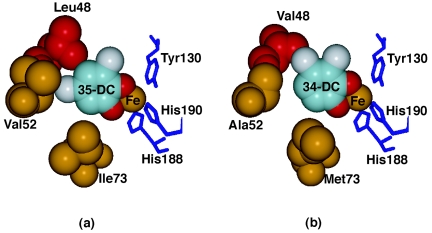
The active site of the CCDs is in the hydrophobic cavity around the Fe ion (17, 65). 3,5-DC and 3,4-DC could be modeled into the active site of CbnA and TcbC without close contacts to the protein atoms, respectively (Fig. 2). In the models, the Fe(III) ion coordinates two Nɛ atoms of histidines (His188 and His190), one Oη atom of tyrosine (Tyr130), and two hydroxyl groups of the substrate. Arg185 and Pro76 sandwich the substrate and seem to be important for fixing the substrate in the active site. Residues in positions 48, 52, and 73, which affect the substrate specificity of CCDs, are located around the bound substrate and close to the C4, C5, and C6 atoms, respectively (Fig. 2).
FIG. 2.
Molecular modeling of CbnA and TcbC with substrates. (a) CbnA and 3,5-dichlorocatechol; (b) TcbC and 3,4-dichlorocatechol. Molecular models of CbnA and TcbC were prepared on the basis of the crystal structures of ClcA from Rhodococcus opacus 1CP (Protein Database identification no. 1s9a) (17) and CatA from Acinetobacter sp. ADP1 (Protein Database identification no. 1dlm and 1dlq) (65).
DISCUSSION
In this study, amino acid residues responsible for the difference in the substrate specificities against the dichlorocatechols between CbnA and TcbC have been identified. For the specific activity of CbnA, high activity against 3,5-DC and low activity against 3,4-DC, Leu-48, and Ile-73 were critical. For the high activity against 3,4-DC and low activity against 3,5-DC of TcbC, Val-48 and Ala-52 were important.
The definitive importance of the amino acid residue in position 48 to the substrate specificity pattern was clear by comparing, for example, the high 3,5-DC specificity of C123T4 and high 3,4-DC specificity of MC31 (Table 3). Furthermore, the amino acid residue in position 48 seemed to have a greater effect on the specificity pattern than that in position 73 (below).
While the importance of Ile-73 for the 3,5-DC specific activity of CbnA was apparent by comparing CbnA and MC3n (Table 3), the contribution of Met-73 to the 3,4-DC specificity of TcbC did not seem to be so significant as outlined below. The mutants, MT22, T14C23, and MC31 (Table 3), which contained Val-48 and Ile-73, exhibited higher levels of activity towards 3,4-DC than 3,5-DC. This was also supported by the result that MC3n, which was different from CbnA only in having Met at position 73, did not exhibit increased activity for 3,4-DC.
In the mutants considered above (MT22, T14C23, MC31, and MC3n), in which the combination of the amino acids in positions 48 and 73 was different from the combination of those in the wild-type CbnA and TcbC, the preferred dichlorocatechol seemed to be dictated by the residue in position 48. This suggested a hierarchical relationship in the protein structure whereby the amino acid in position 48 could have a greater effect on the determination of the preferred dichlorocatechol than the amino acid in position 73. Although MT4 was an exception to this tendency, with Leu-48 and Met-73 and showing higher specificity toward 3,4-DC (31.1%) than toward 3,5-DC (13.5%), all the other mutants in this study agreed with this watershed rule. In this regard, it is noteworthy that we could not obtain a chimeric or site-directed mutant CCD that exhibited high activity with both 3,4-DC and 3,5-DC. This might indicate a constraint of the structure of the CCDs, especially regarding the substrate-binding pocket.
It was apparent that Ala-52 contributed to the activity pattern of TcbC from the results for MT7, MT3, and MT5 (Table 3). Comparison of MC4 with CbnA and MC41 with C123T4, where the replacement of Val-52 by Ala in the CbnA-type enzymes increased the relative 3,4-DC activity about four to fivefold, also revealed the contribution of Ala-52 to the 3,4-DC specificity. At the same time, MC4 and MC41 retained high activity with 3,5-DC, denying the possibility that Val-52 of CbnA was critical for its high 3,5-DC specificity.
In our kinetic experiments, the difference between CbnA and TcbC was more obvious in the values of Vmax against 3,4-DC and 3,5-DC than in the values of Km with these substrates. A more detailed quantitative analysis would reveal the role of the relevant amino acid residues in the enzymatic function of CbnA and TcbC.
A comparison among the site-directed mutants mutated in region 1 of CbnA (MC9, active; MC35, inactive; MC4, active) and MC31 (active) revealed that Val-48 required interaction with the amino acid(s) unique to TcbC region 4 for the enzyme to be active. However, although the chimeric constructs T12C34 and T1C234 did not have enzyme activity, the construct MT22 which retained Arg-70 of TcbC in place of Thr-70 in T1C234 exhibited CCD activity and TcbC-type specificity. This result suggested a suppressive effect of the combination of Arg-70 and Ile-73, resulting in the active TcbC-region 1-type enzyme regardless of the interaction with the amino acid(s) unique to TcbC region 4.
The above results of the amino acid positions involved in the difference of the substrate specificities between CbnA and TcbC agree well with the information on the crystal structures of ClcA1CP and CatAADP1 (which is evolutionarily related to the CCDs) (17, 65). The overall structures of ClcA1CP and CatAADP1 are similar in that they are dimers of two identical subunits containing N-terminal helices and C-terminal strands, each subunit containing a substrate-binding cavity and that the linker domain consisting of five N-terminal helices from each subunit forms the molecular dimer axis. Several differences between the crystal structures of the two enzymes are shown, for example, in the shapes of catalytic cavity resulting from shifts of relevant amino acids (17).
The amino acid residues of ClcA1CP and CatAADP1 which are considered to interact with the substrate, are shown in Fig. 1 (17, 65). The alignment in Fig. 1 shows that amino acid positions 48, 52, and 73 of CbnA and TcbC correspond to positions 49, 53, and 74, respectively, of ClcA1CP, which are considered to interact with the substrate in ClcA1CP. Leu-49 and Ala-53 of ClcA1CP are located in helix 3 of the protein, and Leu-49 of ClcA1CP is one of the amino acids that compose the active site entrance of the enzyme (17). The amino acid positions 48 and 73 of CbnA and TcbC correspond to positions 73 and 105, respectively, of CatAADP1, which are considered to interact with the substrate in CatAADP1 (Fig. 1) (65). Amino acid sequences of CbnA and TcbC show that they are more homologous to ClcA1CP than to CatAADP1; the numbers of identical amino acids at the corresponding position in the alignment are 97 between CbnA and ClcA1CP and 77 between CbnA and CatAADP1 (39% and 31% in the length of 251 amino acids of CbnA, respectively). Besides this overall identity, higher similarity of CbnA/TcbC to ClcA1CP than to CatAADP1 is shown in the regions that correspond to the helix 3 of ClcA1CP, which provides Leu-49, Asp-52, and Ala-53 to the active-site cavity of ClcA1CP. The amino acid alignment thus confirms that the residues of CbnA and TcbC that affect the substrate specificity are located at the positions corresponding to the positions of the residues interacting with substrates in the crystal structures of ClcA1CP and CatAADP1. The results that substitution of position 46 of CbnA or TcbC did not influence the substrate specificities are consistent with findings that the corresponding position of ClcA1CP (position 47) is outside the catalytic site in the structure of ClcA1CP.
To further consider the relationship of the relevant amino acids of CbnA and TcbC with the dichlorocatechols, a molecular modeling study was conducted based on the structures of ClcA1CP and CatAADP1. Although it is difficult to predict the precise conformations of these residues when recognizing the substrate, the model structures not only confirm that residues 48, 52, and 73 are important to the substrate specificity of CbnA and TcbC, but also suggests possible structural necessities. In CbnA, the side chain of Leu-48 seems to be located just above the C4 atom (Fig. 2a). A substrate with a chloride atom at the C4 position is unlikely to bind to the substrate-binding pocket, due to the close contact between the chloride atom and the side chain of Leu-48. The upside-down-binding mode is also unlikely due to the close contact between the chloride atom on C5 and the side chain of Leu-48. On the other hand, residue 48 of TcbC is Val, which seems to generate sufficient space above the C4 atom, thus accommodating the chloride atom in the space (Fig. 2b). Our model thus confirms that residue 48 seems to be critical to the substrate specificity of CbnA and TcbC. In the mutant MT4, which had Leu-48 and showed higher specificity toward 3,4-DC than toward 3,5-DC, the amino acids in positions 52 and 73 were the same as in TcbC (Ala and Met, respectively). The combination of these amino acids forming the substrate-binding pocket of MT4 might have altered the conformation of the substrate-binding pocket to better accommodate 3,4-DC. The other two residues, Val52/Ala52 and Ile73/Met73, are also located close to the C5 and C6 atoms, respectively (Fig. 2). Although these residues are likely to interact with the substrate, it is difficult to predict the role of these residues in recognizing the substrates. To reveal the mechanism of the substrate specificity fully, detailed conformational changes upon substrate binding should be analyzed, which is hard to achieve by the theoretical approach with the model structure. Crystal structure analyses of CbnA and TcbC in complexes with substrates are required to unveil the details of functional roles of these residues in recognition of dichlorocatechols.
Intradiol dioxygenases for catechol and chlorocatechols fall into three groups based on their amino acid sequences: catechol 1,2-dioxygenases of gram-negative bacteria, CCDs of gram-negative bacteria, and both catechol 1,2-dioxygenases and CCDs of gram-positive bacteria (12, 13, 37). Leu-48 and Ile-73, the determinants of the high specificity for 3,5-DC in CbnA, or Leu and Ile at respective corresponding positions, are conserved in most of the members of the three groups reported thus far. Among these are CCDs, the substrate specificities of which have been documented: ClcA from Pseudomonas putida AC866 (5, 19) and Pseudomonas sp. B13 (10, 11), ClcA1CP (13, 17, 33), and the two TfdCs (2, 3, 31, 41, 43). These CCDs also show comparable or higher levels of activity against 3,5-DC than that against 3-chlorocatechol. The exceptions to the Leu-48 and Ile-73 rule are TcbC; TetC (Val-73) of Pseudomonas chlororaphis RW71 (46), which is highly homologous to CbnA; and DccAI and DccAII of Sphingomonas herbicidovorans MH (Leu-73) (36). However, TetC, with Leu in position 48, exhibits threefold-higher catalytic activity for 3,5-DC than for 3,4-DC (46). The difference in the residues in the position 73 between the highly similar CCDs CbnA and TetC suggests that they could have slightly different substrate specificities. DccAI and DccAII have Leu-48 and show a preference for 3,5-DC rather than for monochlorocatechols (36), in agreement with our results indicating the importance of Leu-48 for 3,5-DC specificity.
The primary importance of position 48 in the determination of substrate specificity for CbnA and TcbC has been demonstrated by the chimeric and site-directed mutagenesis study and agrees well with the molecular models described above. Therefore, the importance of the residue in position 73 was further tested with TfdCNK8 and TfdCCSV90. TfdCCSV90 has an amino acid sequence identical to that of TfdC of R. eutropha JMP134 (3, 20, 41). The replacement of Ile-73 by Met in TfdCNK8 and TfdCCSV90 resulted in a marked loss of 3,5-DC specificity. On the contrary, activity against 3,4-DC of TfdCNK8 (Met-73) and TfdCCSV90 (Met-73) did not increase notably, supporting the lesser importance of the residue in position 73 to 3,4-DC specificity.
The almost perfect conservation of Leu-48 and Ile-73 (or at the respective corresponding positions) among CCDs and catechol 1,2-dioxygenases throughout both the gram-negative and gram-positive bacteria suggests that an ancestor of both enzymes had these two residues at the corresponding positions. If this is so, the ancestral CCD might have high specificity for 3,5-DC inherently.
On the other hand, position 52 or its corresponding position is occupied by several different amino acids, depending on the phylogenetic location of the CCD or the catechol 1,2-dioxygenase. In the CCDs from gram-negative bacteria and catechol 1,2-dioxygenases from gram-positive bacteria, the amino acid at position 52 or the corresponding position is Val, except in TcbC (Ala-52). However, the amino acid at the corresponding position of catechol 1,2-dioxygenases of gram-negative bacteria is His or Lys (18, 58) and that of ClcA from R. opacus 1CP is Ala (13).
The above comparison and the enzymatic data of this study cast light upon the uniqueness of TcbC in terms of its specificity toward 3,4-DC and the amino acids occupying positions 48, 52, and 73, strongly suggesting the adaptation of TcbC for conversion of 3,4-DC produced by the upstream enzymes TcbA and TcbB in strain P51 (61, 66).
The two gene clusters cbnRABorfXCD and tcbRCDorfXEF are highly homologous. When identity between the corresponding genes of the two clusters is compared at the amino acid sequence level, it is the lowest between CbnA and TcbC (95%) (40). Among the chlorocatechol ortho-cleavage gene clusters reported thus far, two chlorocatechol gene clusters (the tetRCDorfXEF gene cluster of chlorobenzene-degrading P. chlororaphis RW71 and the tfdRCDorfIEF gene cluster of 2,4-D-degrading Delftia acidovorans P4a) constitute a highly homologous subgroup together with the cbn and tcb clusters with identical lengths of the corresponding genes, and the two clusters are more homologous to the cbn cluster than to the tcb cluster (28, 46). The N-terminal regions of CbnA, TetC, and TcbC differ only in five positions (46, 48, 52, 70, and 73). TfdC of strain P4a is different from CbnA only at position 47 (Asp in TfdC of P4a and Ala in CbnA). Among these, positions 48, 52, and 73 have been shown to be important for the differentiation of substrate specificity between CbnA and TcbC. In addition, at least one of the amino acids in region 4 unique to TcbC should be responsible for maintaining the enzyme activity by cooperating with Val-48 of TcbC. Since CbnA and TcbC are different only at 12 amino acid residues, a significant portion of the amino acid substitutions between the two enzymes plays some role in differentiating the activity pattern. Considering that the difference in the amino acid sequence encoded by the cbn and tcb gene clusters is the highest between CbnA and TcbC and that the differences between other corresponding genes are very low, this may indicate the critical importance of the substrate specificity of CCD in the adaptation of the chlorocatechol ortho-cleavage pathway to the substrate.
Acknowledgments
This work was supported by the Program for the Promotion of Basic Research Activity for Innovative Bioscience (PROBRAIN) and by a grant-in-aid (Hazardous Chemicals) from the Ministry of Agriculture, Forestry, and Fisheries of Japan (HC-05-2324-1).
REFERENCES
- 1.Arensdorf, J. J., and D. D. Focht. 1994. Formation of chlorocatechol meta cleavage products by a pseudomonad during metabolism of monochlorobiphenyls. Appl. Environ. Microbiol. 60:2884-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhat, M. A., T. Ishida, K. Horiike, C. S. Vaidyanathan, and M. Nozaki. 1993. Purification of 3,5-dichlorocatechol 1,2-dioxygenase, a nonheme iron dioxygenase and a key enzyme in the biodegradation of a herbicide, 2,4-dichlorophenoxyacetic acid (2,4-D), from Pseudomonas cepacia CSV90. Arch. Biochem. Biophys. 300:738-746. [DOI] [PubMed] [Google Scholar]
- 3.Bhat, M. A., M. Tsuda, K. Horiike, M. Nozaki, C. S. Vaidyanathan, and T. Nakazawa. 1994. Identification and characterization of a new plasmid carrying genes for degradation of 2,4-dichlorophenoxyacetate from Pseudomonas cepacia CSV90. Appl. Environ. Microbiol. 60:307-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bollag, J.-M., C. S. Helling, and M. Alexander. 1968. 2,4-D metabolism. Enzymatic hydroxylation of chlorinated phenols. J. Agr. Food Chem. 16:826-828. [Google Scholar]
- 5.Broderick, J. B., and T. V. O'Halloran. 1991. Overproduction, purification, and characterization of chlorocatechol dioxygenase, a non-heme iron dioxygenase with broad substrate tolerance. Biochemistry 30:7349-7358. [DOI] [PubMed] [Google Scholar]
- 6.Chatterjee, D. K., and A. M. Chakrabarty. 1983. Genetic homology between independently isolated chlorobenzoate-degradative plasmids. J. Bacteriol. 153:532-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatterjee, D. K., S. T. Kellogg, S. Hamada, and A. M. Chakrabarty. 1981. Plasmid specifying total degradation of 3-chlorobenzoate by a modified ortho pathway. J. Bacteriol. 146:639-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Don, R. H., and J. M. Pemberton. 1981. Properties of six pesticide degradation plasmids isolated from Alcaligenes paradoxus and Alcaligenes eutrophus. J. Bacteriol. 145:681-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Don, R. H., A. J. Weightman, H.-J. Knackmuss, and K. N. Timmis. 1985. Transposon mutagenesis and cloning analysis of the pathways for degradation of 2,4-dichlorophenoxyacetic acid and 3-chlorobenzoate in Alcaligenes eutrophus JMP134(pJP4). J. Bacteriol. 161:85-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorn, E., and H.-J. Knackmuss. 1978. Chemical structure and biodegradability of halogenated aromatic compounds. Two catechol 1,2-dioxygenases from a 3-chlorobenzoate-grown pseudomonad. Biochem. J. 174:73-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorn, E., and H.-J. Knackmuss. 1978. Chemical structure and biodegradability of halogenated aromatic compounds. Substituent effects on 1,2-dioxygenation of catechol. Biochem. J. 174:85-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eulberg, D., L. A. Golovleva, and M. Schlömann. 1997. Characterization of catechol catabolic genes from Rhodococcus erythropolis 1CP. J. Bacteriol. 179:370-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eulberg, D., E. M. Kourbatova, L. A. Golovleva, and M. Schlömann. 1998. Evolutionary relationship between chlorocatechol catabolic enzymes from Rhodococcus opacus 1CP and their counterparts in proteobacteria: sequence divergence and functional convergence. J. Bacteriol. 180:1082-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans, W. C., B. S. W. Smith, H. N. Fernley, and J. I. Davies. 1971. Bacterial metabolism of 2,4-dichlorophenoxyacetate. Biochem. J. 122:543-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferraroni, M., M. Y. Ruiz Tarifa, F. Briganti, A. Scozzafava, S. Mangani, I. P. Solyanikova, M. P. Kolomytseva, and L. Golovleva. 2002. 4-Chlorocatechol 1,2-dioxygenase from the chlorophenol-utilizing gram-positive Rhodococcus opacus 1CP: crystallization and preliminary crystallographic analysis. Acta Crystallogr. D Biol. Crystallogr. 58:1074-1076. [DOI] [PubMed] [Google Scholar]
- 16.Ferraroni, M., M. Y. Ruiz Tarifa, A. Scozzafava, I. P. Solyanikova, M. P. Kolomytseva, L. Golovleva, and F. Briganti. 2003. Preliminary crystallographic analysis of 3-chlorocatechol 1,2-dioxygenase of a new modified ortho-pathway from the gram-positive Rhodococcus opacus 1CP grown on 2-chlorophenol. Acta Crystallogr. D Biol. Crystallogr. 59:188-190. [DOI] [PubMed] [Google Scholar]
- 17.Ferraroni, M., I. P. Solyanikova, M. P. Kolomytseva, A. Scozzafava, L. Golovleva, and F. Briganti. 2004. Crystal structure of 4-chlorocatechol 1,2-dioxygenase from the chlorophenol-utilizing gram-positive Rhodococcus opacus 1CP. J. Biol. Chem. 279:27646-27655. [DOI] [PubMed] [Google Scholar]
- 18.Francisco, P. B., Jr., N. Ogawa, K. Suzuki, and K. Miyashita. 2001. The chlorobenzoate dioxygenase genes of Burkholderia sp. strain NK8 involved in the catabolism of chlorobenzoates. Microbiology 147:121-133. [DOI] [PubMed] [Google Scholar]
- 19.Frantz, B., and A. M. Chakrabarty. 1987. Organization and nucleotide sequence determination of a gene cluster involved in 3-chlorocatechol degradation. Proc. Natl. Acad. Sci. USA 84:4460-4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghosal, D., and I.-S. You. 1988. Nucleotide homology and organization of chlorocatechol oxidation genes of plasmids pJP4 and pAC27. Mol. Gen. Genet. 211:113-120. [DOI] [PubMed] [Google Scholar]
- 21.Haigler, B. E., S. F. Nishino, and J. C. Spain. 1988. Degradation of 1,2-dichlorobenzene by a Pseudomonas sp. Appl. Environ. Microbiol. 54:294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanahan, D., 1985. Techniques for transformation of E. coli, p. 109-135. In D. M. Glover (ed.), DNA cloning, vol. 1. IRL Press, Oxford, United Kingdom.
- 23.Hartmann, J., W. Reineke, and H.-J. Knackmuss. 1979. Metabolism of 3-chloro-, 4-chloro-, and 3,5-dichlorobenzoate by a pseudomonad. Appl. Environ. Microbiol. 37:421-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harwood, C. S., and R. E. Parales. 1996. The β-ketoadipate pathway and the biology of self-identity. Annu. Rev. Microbiol. 50:553-590. [DOI] [PubMed] [Google Scholar]
- 25.Hernandez, B. S., F. K. Higson, R. Kondrat, and D. D. Focht. 1991. Metabolism of and inhibition by chlorobenzoates in Pseudomonas putida P111. Appl. Environ. Microbiol. 57:3361-3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hickey, W. J., and D. D. Focht. 1990. Degradation of mono-, di-, and trihalogenated benzoic acids by Pseudomonas aeruginosa JB2. Appl. Environ. Microbiol. 56:3842-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hinteregger, C., M. Loidl, and F. Streichsbier. 1992. Characterization of isofunctional ring-cleaving enzymes in aniline and 3-chloroaniline degradation by Pseudomonas acidovorans CA28. FEMS Microbiol. Lett. 76:261-266. [DOI] [PubMed] [Google Scholar]
- 28.Hoffmann, D., S. Kleinsteuber, R. H. Müller, and W. Babel. 2003. A transposon encoding the complete 2,4-dichlorophenoxyacetic acid degradation pathway in the alkalitolerant strain Delftia acidovorans P4a. Microbiology 149:2545-2556. [DOI] [PubMed] [Google Scholar]
- 29.Inoue, H., H. Nojima, and H. Okayama. 1990. High efficiency transformation of Escherichia coli with plasmids. Gene 96:23-28. [DOI] [PubMed] [Google Scholar]
- 30.Leveau, J. H. J., and J. R. van der Meer. 1997. Genetic characterization of insertion sequence ISJP4 on plasmid pJP4 from Ralstonia eutropha JMP134. Gene 202:103-114. [DOI] [PubMed] [Google Scholar]
- 31.Liu, S., N. Ogawa, and K. Miyashita. 2001. The chlorocatechol degradative genes, tfdT-CDEF, of Burkholderia sp. strain NK8 are involved in chlorobenzoate degradation and induced by chlorobenzoates and chlorocatechols. Gene 268:207-214. [DOI] [PubMed] [Google Scholar]
- 32.Mäe, A. A., R. O. Marits, N. R. Ausmees, V. M. Kôiv, and A. L. Heinaru. 1993. Characterization of a new 2,4-dichlorophenoxyacetic acid degrading plasmid pEST4011: physical map and localization of catabolic genes. J. Gen. Microbiol. 139:3165-3170. [Google Scholar]
- 33.Maltseva, O. V., I. P. Solyanikova, and L. A. Golovleva. 1994. Chlorocatechol 1,2-dioxygenase from Rhodococcus erythropolis 1CP. Kinetic and immunochemical comparison with analogous enzymes from gram-negative strains. Eur. J. Biochem. 226:1053-1061. [DOI] [PubMed] [Google Scholar]
- 34.Mars, A. E., J. Kingma, S. R. Kaschabek, W. Reineke, and D. B. Janssen. 1999. Conversion of 3-chlorocatechol by various catechol 2,3-dioxygenases and sequence analysis of the chlorocatechol dioxygenase region of Pseudomonas putida GJ31. J. Bacteriol. 181:1309-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miguez, C. B., C. W. Greer, and J. M. Ingram. 1993. Purification and properties of chlorocatechol 1,2-dioxygenase from Alcaligenes denitrificans BRI 6011. Can. J. Microbiol. 39:1-5. [Google Scholar]
- 36.Müller, T. A., S. M. Byrde, C. Werlen, J. R. van der Meer, and H.-P. E. Kohler. 2004. Genetic analysis of phenoxyalkanoic acid degradation in Sphingomonas herbicidovorans MH. Appl. Environ. Microbiol. 70:6066-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murakami, S., N. Kodama, R. Shinke, and K. Aoki. 1997. Classification of catechol 1,2-dioxygenase family: sequence analysis of a gene for the catechol 1,2-dioxygenase showing high specificity for methylcatechols from gram+ aniline-assimilating Rhodococcus erythropolis AN-13. Gene 185:49-54. [DOI] [PubMed] [Google Scholar]
- 38.Ogawa, N., A. M. Chakrabarty, and O. Zaborina. 2004. Degradative plasmids, p. 341-376. In B. E. Funnell and G. J. Phillips (ed.), Plasmid biology. American Society for Microbiology, Washington, D.C.
- 39.Ogawa, N., and K. Miyashita. 1995. Recombination of a 3-chlorobenzoate catabolic plasmid from Alcaligenes eutrophus NH9 mediated by direct repeat elements. Appl. Environ. Microbiol. 61:3788-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogawa, N., and K. Miyashita. 1999. The chlorocatechol-catabolic transposon Tn5707 of Alcaligenes eutrophus NH9, carrying a gene cluster highly homologous to that in the 1,2,4-trichlorobenzene-degrading bacterium Pseudomonas sp. strain P51, confers the ability to grow on 3-chlorobenzoate. Appl. Environ. Microbiol. 65:724-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perkins, E. J., M. P. Gordon, O. Caceres, and P. F. Lurquin. 1990. Organization and sequence analysis of the 2,4-dichlorophenol hydroxylase and dichlorocatechol oxidative operons of plasmid pJP4. J. Bacteriol. 172:2351-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pieper, D. H., and W. Reineke. 2004. Degradation of chloroaromatics by Pseudomona(d)s, p. 509-574. In J.-L. Ramos (ed.), Pseudomonas, vol. 3. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 43.Pieper, D. H., W. Reineke, K.-H. Engesser, and H.-J. Knackmuss. 1988. Metabolism of 2,4-dichlorophenoxyacetic acid, 4-chloro-2-methylphenoxyacetic acid and 2-methylphenoxyacetic acid by Alcaligenes eutrophus JMP134. Arch. Microbiol. 150:95-102. [Google Scholar]
- 44.Plumeier, I., D. Pérez-Pantoja, S. Heim, B. González, and D. H. Pieper. 2002. Importance of different tfd genes for degradation of chloroaromatics by Ralstonia eutropha JMP134. J. Bacteriol. 184:4054-4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poh, R. P.-C., A. R. W. Smith, and I. J. Bruce. 2002. Complete characterisation of Tn5530 from Burkholderia cepacia strain 2a (pIJB1) and studies of 2,4-dichlorophenoxyacetate uptake by the organism. Plasmid 48:1-12. [DOI] [PubMed] [Google Scholar]
- 46.Potrawfke, T., J. Armengaud, and R.-M. Wittich. 2001. Chlorocatechols substituted at positions 4 and 5 are substrates of the broad-spectrum chlorocatechol 1,2-dioxygenase of Pseudomonas chlororaphis RW71. J. Bacteriol. 183:997-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Potrawfke, T., K. N. Timmis, and R.-M. Wittich. 1998. Degradation of 1,2,3,4-tetrachlorobenzene by Pseudomonas chlororaphis RW71. Appl. Environ. Microbiol. 64:3798-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Que, L., Jr., and R. Y. Ho. 1996. Dioxygen activation by enzymes with mononuclear non-heme iron active sites. Chem. Rev. 96:2607-2624. [DOI] [PubMed] [Google Scholar]
- 49.Ravatn, R., S. Studer, A. J. B. Zehnder, and J. R. van der Meer. 1998. Int-B13, an unusual site-specific recombinase of the bacteriophage P4 integrase family, is responsible for chromosomal insertion of the 105-kilobase clc element of Pseudomonas sp. strain B13. J. Bacteriol. 180:5505-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reineke, W. 1998. Development of hybrid strains for the mineralization of chloroaromatics by patchwork assembly. Annu. Rev. Microbiol. 52:287-331. [DOI] [PubMed] [Google Scholar]
- 51.Reineke, W., and H.-J. Knackmuss. 1984. Microbial metabolism of haloaromatics: isolation and properties of a chlorobenzene-degrading bacterium. Appl. Environ. Microbiol. 47:395-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 53.Sander, P., R.-M. Wittich, P. Fortnagel, and W. Francke. 1991. Degradation of 1,2,4-trichloro- and 1,2,4,5-tetrachlorobenzene by Pseudomonas strains. Appl. Environ. Microbiol. 57:1430-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schraa, G., M. L. Boone, M. S. Jetten, A. R. W. van Neerven, P. J. Colberg, and A. J. B. Zehnder. 1986. Degradation of 1,4-dichlorobenzene by Alcaligenes sp. strain A175. Appl. Environ. Microbiol. 52:1374-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sentchilo, V., A. J. B. Zehnder, and J. R. van der Meer. 2003. Characterization of two alternative promoters for integrase expression in the clc genomic island of Pseudomonas sp. strain B13. Mol. Microbiol. 49:93-104. [DOI] [PubMed] [Google Scholar]
- 56.Spain, J. C., and S. F. Nishino. 1987. Degradation of 1,4-dichlorobenzene by a Pseudomonas sp. Appl. Environ. Microbiol. 53:1010-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spiess, E., C. Sommer, and H. Görisch. 1995. Degradation of 1,4-dichlorobenzene by Xanthobacter flavus 14p1. Appl. Environ. Microbiol. 61:3884-3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suzuki, K., A. Ichimura, N. Ogawa, A. Hasebe, and K. Miyashita. 2002. Differential expression of two catechol 1,2-dioxygenases in Burkholderia sp. strain TH2. J. Bacteriol. 184:5714-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trefault, N., R. De la Iglesia, A. M. Molina, M. Manzano, T. Ledger, D. Pérez-Pantoja, M. A. Sánchez, M. Stuardo, and B. González. 2004. Genetic organization of the catabolic plasmid pJP4 from Ralstonia eutropha JMP134 (pJP4) reveals mechanisms of adaptation to chloroaromatic pollutants and evolution of specialized chloroaromatic degradation pathways. Environ. Microbiol. 6:655-668. [DOI] [PubMed] [Google Scholar]
- 60.van der Meer, J. R., W. M. de Vos, S. Harayama, and A. J. B. Zehnder. 1992. Molecular mechanisms of genetic adaptation to xenobiotic compounds. Microbiol. Rev. 56:677-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van der Meer, J. R., R. I. L. Eggen, A. J. B. Zehnder, and W. M. de Vos. 1991. Sequence analysis of the Pseudomonas sp. strain P51 tcb gene cluster, which encodes metabolism of chlorinated catechols: evidence for specialization of catechol 1,2-dioxygenases for chlorinated substrates. J. Bacteriol. 173:2425-2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van der Meer, J. R., A. R. W. van Neerven, E. J. de Vries, W. M. de Vos, and A. J. B. Zehnder. 1991. Cloning and characterization of plasmid-encoded genes for the degradation of 1,2-dichloro-, 1,4-dichloro-, and 1,2,4-trichlorobenzene of Pseudomonas sp. strain P51. J. Bacteriol. 173:6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vedler, E., M. Vahter, and A. Heinaru. 2004. The completely sequenced plasmid pEST4011 contains a novel IncP1 backbone and a catabolic transposon harboring tfd genes for 2,4-dichlorophenoxyacetic acid degradation. J. Bacteriol. 186:7161-7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vetting, M. W., D. A. D'Argenio, L. N. Ornston, and D. H. Ohlendorf. 2000. Structure of Acinetobacter strain ADP1 protocatechuate 3, 4-dioxygenase at 2.2 Å resolution: implications for the mechanism of an intradiol dioxygenase. Biochemistry 39:7943-7955. [DOI] [PubMed] [Google Scholar]
- 65.Vetting, M. W., and D. H. Ohlendorf. 2000. The 1.8 Å crystal structure of catechol 1,2-dioxygenase reveals a novel hydrophobic helical zipper as a subunit linker. Structure 8:429-440. [DOI] [PubMed] [Google Scholar]
- 66.Werlen, C., H.-P. E. Kohler, and J. R. van der Meer. 1996. The broad substrate chlorobenzene dioxygenase and cis-chlorobenzene dihydrodiol dehydrogenase of Pseudomonas sp. strain P51 are linked evolutionarily to the enzymes for benzene and toluene degradation. J. Biol. Chem. 271:4009-4016. [DOI] [PubMed] [Google Scholar]
- 67.Xia, X.-S., S. Aathithan, K. Oswiecimska, A. R. W. Smith, and I. J. Bruce. 1998. A novel plasmid pIJB1 possessing a putative 2,4-dichlorophenoxyacetate degradative transposon Tn5530 in Burkholderia cepacia strain 2a. Plasmid 39:154-159. [DOI] [PubMed] [Google Scholar]
- 68.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]