Intelligence operations worldwide monitor individuals directly or indirectly linked to terrorist networks for their communication activities, collectively described as “chatter.” Considerable effort and resources are devoted to assessing intelligence chatter. A major challenge is to distinguish innocuous, everyday discourse from those missives that act to coordinate more nefarious activities. Microorganisms also chatter, and our ability to understand the routes, mechanisms, and purposes of these communications may have profound effects on human health, industry, agriculture, and the environment. Bacteria communicate and coordinate their activities through chemical signals that either diffuse through the extracellular environment or remain cell associated. As with the global surveillance of human chatter, research activity focused on microbial communication mechanisms has intensified over recent years. The study of bacterial communication systems is an extremely active area of microbiology and has generated a significant paradigm shift in the way we perceive and examine microbial populations.
Given the intense interest in microbial signaling systems, the American Society for Microbiology (ASM) sponsored its second conference on Cell-Cell Communication in Bacteria from 23 to 27 July 2004 (CCCB-04) in Banff, Canada. The Banff conference continued the trend initiated at the 2001 conference (in Snowbird, Utah) of outstanding presentations on bacterial signaling but also considerably expanded the scope of signaling systems covered, with 230 scientists in attendance delivering 48 oral presentations and 137 posters. Over the 3 1/2-day conference, oral presentations were given in seven thematically organized sessions in the mornings and evenings, and posters were viewed on each afternoon. The scientific discourse at this conference was highly stimulating, with work ranging from structure-function analysis of signaling mechanisms to the ecology of signaling in natural environments. We attempted in this review to select specific findings that embody recent experimental and conceptual advances in the area of microbial signaling.
SIGNALING BASICS: LANGUAGES AND LOGIC
Social and antisocial behavior in bacteria.
Bacteria exude a wide range of compounds into their external environment and also elaborate molecules that extend from their cell surfaces. Members of a growing list of diffusible molecules and cell-associated externalized structures function as signals (Table 1). Some of the first recognized diffusible signaling mechanisms were described as “autoinduction,” reflecting the observation that the bacteria themselves were the source of the signal (70). Some bacterial signals may function as simple autoinduction circuits, perhaps utilizing the stability of the signal as an indirect measure of relevant environmental parameters, such as pH or flow. Most bacteria, however, are tuned to higher concentrations of the self-produced signals than that which individual cells can attain (21, 22). The term quorum sensing collectively describes these systems, in that inducing levels of signal require a minimum bacterial population density referred to as a quorum. Although quorum sensing has predominantly been studied for single species, there is mounting evidence for multispecies quorum sensing (84, 88). In some cases, bacteria unable to synthesize their own signal are capable of responding to signals of other bacteria, eavesdropping on their competitors, or synchronizing with collaborators. There are also examples of interference, in which microbes and host organisms release signaling inhibitors or actively degrade bacterial signal molecules (107). Finally, there are multiple ways by which bacteria can communicate through cell contact-dependent mechanisms in which signals elaborated on the cell surface interact to stimulate concerted behaviors (46).
TABLE 1.
Microbial signaling systems
| Signal(s) | Microbe(s) | Synthesis | Precursor(s)a | Receptor(s) | Regulated function(s) | Reference |
|---|---|---|---|---|---|---|
| Acyl homoserine lactones (AHLs) | Proteobacteria | LuxI-type enzymes | SAM, acyl-ACP | LuxR-type proteins | Diverse processes | 21 |
| AinS-type enzymes | SAM, acyl-ACP, acyl-CoA | Two-component kinase | ||||
| Linear oligopeptides | Gram-positive organisms | Genetically encoded (e.g., B. subtilis) | Secreted prepeptide | Two-component systems, phosphorelays | Diverse processes | 77 |
| Cyclized oligopeptides | Gram-positive organisms | Genetically encoded (e.g., S. aureus) | Secreted post-syn mod. prepeptide | Two-component systems, phosphorelays | Virulence genes | 72 |
| γ-Butyrolactones (GBLs) | Streptomyces spp. | AfsA-type GBL synthase | Acyl-ACP, glycerol | ArpA-type repressor, GBL-binding protein | Secondary metabolism; antibiotics and sporulation | 37 |
| Furanosyl diester (+/− boron); AI-2 | Diverse taxa (no α-Proteobacteria) | LuxS AI-2 synthase | Methionine salvage (DPD) | Vibrio spp. LuxP/LuxQ/LuxO phosphorelay; unknown for other systems | Luminescence and diverse processes | 80 |
| cis-11-Methyl-2-dodecenoic acid (DSF) | Xanthomonas spp., perhaps others | RpfB, RpfF | acyl-CoA? | RpfC, RpfH | Virulence and pigmentation | 100 |
| 4-Hydroxy-2-alkyl quinolines (PQS, HAQs) | Pseudomonads | pqsABCDE, pqsH | Anthranilic acid | ? | Global regulation, virulence | 13 |
| Palmitic acid methyl esters (PAME) | Ralstonia solanacearum | PhcB | SAM (?), fatty acids | PhcR regulator | Virulence, exopolysaccharides | 18 |
| Putrescine | Proteus mirabilis | SpeA, SpeBb | Ornithine, arginine | ? | Swarming motility | 87 |
| A-signal (amino acids) | M. xanthus | AsgAB-dependent protease | Cell surface | SasS/SasR/SasN proteins | Early development, aggregation | 46 |
| C-signalc | M. xanthus | Translation product | CsgA protein | FruA, unknown HPK (csgA) | Coordinated motility, fruiting body development | 46 |
| Cyclic dipeptides | Many taxa | ? | Amino acids? | LuxR-type proteins? | Secondary metabolism | 35 |
DPD is 4,5-dihydroxy-2,3-pentanedione. CoA is coenzyme A.
Based on E. coli biosynthetic pathway.
C-signal is a cell surface-associated protein (17-kDa processed form of CsgA). HPK, histidine protein kinase; secreted post-syn mod. prepeptide, secreted post-synthetically modified prepeptide.
It has been recently suggested that in some cases, quorum sensing might be a side effect of cells monitoring their diffusion environment instead of communicating (78). These two ideas are far from mutually exclusive. Limits on diffusion and signal accumulation due to increasing population density are certainly related components of the multicellular processes that are often described as quorum sensing. It is, however, true that outside of a few examples, direct evidence for dedicated roles in monitoring population density is limited. Those of us working on microbial signaling should keep alternate explanations in mind when considering the function and benefit of these mechanisms for the microbes that utilize them.
MECHANISMS OF CELL-CELL COMMUNICATION
Chemical and functional diversity of diffusible signals.
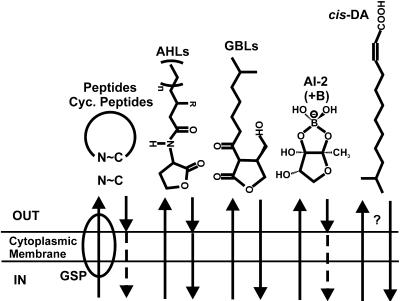
Collectively, bacteria produce a wide range of potential signal molecules regulating a diversity of functions (Table 1). A striking trend at the Banff conference was the diversity of different signals under active investigation. Acylated homoserine lactones (AHLs) in the Proteobacteria and peptide-based signals in gram-positive bacteria are well described and continue to be intensively investigated (Fig. 1 and Table 1). Likewise, the gamma-butyrolactones (GBLs) of filamentous Streptomyces spp., the A-signaling amino acids of Myxococcus xanthus, and autoinducer 2 (AI-2) systems remain the focus of a great deal of work and were well represented. In addition to these well-studied mechanisms, several novel signals and potential signals were also described (Table 1).
FIG. 1.
Examples of bacterial signal diversity. Several different signal types are depicted. N∼C indicates either linear or cyclized peptides, AHLs are acylated homoserine lactones, GBLs are γ-butyrolactones, AI-2 is the furanosyl borate diester, and cis-DA is cis-11-methyl-2-dodecenoic acid. Peptides may be externalized by the general secretion pathway (GSP) or through other more specific mechanisms. Arrows indicate transit of signals across the bacterial envelope. Arrowheads that contact the envelope indicate signals that are bound by membrane-associated receptors. Dashed arrows indicate active transport into cells. Peptides and AI-2 may be perceived externally but can also transported into the cell.
Pseudomonads, for example, synthesize not only AHL-based signals but also a series of 4-hydroxy-2-alkyl quinoline (HAQ) compounds, such as 2-heptyl-3-hydroxy-4-(1H)-quinolone, originally defined as PQS, the Pseudomonas quinolone signal (13, 75). Although some HAQ compounds are recognized antibiotics (e.g., phenazines) and cytochrome inhibitors, several groups are studying the HAQs as signal molecules, as well as their role in pathogenesis and their integration with AHL signaling in the pseudomonads (B. A. Iglewski, CCCB-04, abstr. S3:2; P. Williams et al., CCCB-04, abstr. S5:2). It is clear that some of the HAQs function as intercellular signals (or “messengers”; see below), expanding the breadth of communication in the pseudomonads.
Cell signaling mechanisms control virulence factor and pigment synthesis in the plant pathogen Xanthomonas campestris (3, 76). Until recently, the signal molecules were described as diffusible signaling factor (DSF) and diffusible factor, with limited chemical characterization. Findings from the group of Lian-Hui Zhang identified DSF as cis-11-methyl-2-dodecenoic acid (Fig. 1), and bioassays suggest that this compound or analogues are synthesized by diverse bacteria (100; L.-H. Zhang, CCCB-04, abstr. S3:3). DSF is structurally similar to farnesoic acid, a signaling molecule in the pathogenic yeast Candida albicans (38). DSF and farnesoic acid are cross-functional and may allow cross-kingdom signaling.
The signal originally defined as autoinducer 2 (AI-2) in Vibrio harveyi is a furanosyl diester compound containing a single boron atom (10). Structural analyses of the Salmonella enterica serovar Typhimurium AI-2 with its binding protein LsrB presented at the Banff conference by S. T. Miller reveal a different signal molecule, (2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran, lacking the boron of the V. harveyi AI-2 (65; S. T. Miller et al., CCCB-04, poster abstr. 38B). AI-2 and this molecule are derived from the same methionine salvage pathway intermediate (80). Indirect functional assays suggest that S. enterica serovar Typhimurium may require the boron-free form, while V. harveyi responds best when boron is present. These findings reveal chemical diversity for AI-2-type signals that may influence the degree of cross talk afforded through these signals.
Several putative signal molecules were also described, including cyclic dipeptides that regulate pathogenesis in Vibrio vulnificus (D. Park et al., CCCB-04, abstr. S1:6) and putrescine in control of Proteus mirabilis cellular differentiation during swarming (87; G. Sturgill et al., CCCB-04, poster abstr. 100). In these and other cases, it remains unclear whether these compounds are dedicated to signaling functions.
Only a relatively small percentage of microorganisms have been cultivated in the laboratory. It is a virtual certainty that uncultivated microbes also employ intercellular signaling mechanisms, but it is unclear which currencies of communication they use. Metagenomics is one strategy for addressing this issue (29). Fragmented total environmental DNA samples are fused to cloning vectors en masse to generate clone libraries that are subsequently shotgun sequenced or introduced into an appropriate host to screen for functions of interest. Jo Handelsman's group is using metagenomics to identify novel factors involved either positively or negatively in signal exchange by soil microorganisms (J. Handelsman, presented at CCCB-04). A large-insert metagenomic library from soil was introduced into Escherichia coli cells carrying the gfp reporter gene, under the control of the AHL-responsive protein LuxR from Vibrio fischeri. E. coli does not synthesize AHLs, and activation of LuxR or interference with AHL-dependent activation must be directed by the introduced genes. Isolation of transformants that alter LuxR activity using fluorescence-activated cell sorting allowed identification of several inserts that directed increased or decreased lux-gfp expression. Sequence analysis of five derivatives with increased transcription revealed that only one carried a luxI-like gene, whereas the other four encode novel proteins that may produce AHLs or related compounds. These and other studies suggest that we are only scratching the surface of signal diversity from the natural environment.
Signal synthesis and release.
Proteobacteria synthesize AHLs from S-adenosyl methionine (SAM), the source for the homoserine moiety, which is linked to acyl chains, usually donated by an acylated acyl carrier protein (acyl-ACP) (Fig. 2) (see reference 20). There are at least two different classes of enzymes that can catalyze AHL synthesis, but the most common of these are homologous to the LuxI protein from Vibrio fischeri. Each LuxI-type protein exhibits a preference for acyl chains of a certain length and oxidation state, but the determinants of chain length specificity are not well understood. Solveig Sjöblom from Tapio Palva's group described the comparison of two closely related LuxI-type proteins, ExpISCC1 and ExpISCC3193 from the strains Erwinia carotovora SCC1 and SCC3193, respectively (S. Sjöblom et al., CCCB-04, abstr. S6:7). Although highly similar in amino acid sequence, ExpISCC1 directs synthesis of 3-oxo-hexanoyl homoserine lactone (3-oxo-C6-HSL), whereas ExpISCC3193 catalyzes synthesis of the eight-carbon derivative (3-oxo-C8-HSL). Swapping of specific residues between the two proteins switches the AHL chain length that is specified. Simultaneous alteration of two residues in ExpISCC1 (F69L and M127T) converted its AHL product to 3-oxo-C8-HSL, although three additional changes were also required for normal levels of synthesis. Structural information on LuxI-type proteins is beginning to complement genetic and biochemical analyses. Mair Churchill's group was the first to report the structure of an AHL synthase, EsaI from Pantoea stewartii, and presented new findings on the structure of LasI from Pseudomonas aeruginosa (25, 102; M. Churchill et al., CCCB-04, abstr. S5:3). LasI catalyzes synthesis of 3-oxo-dodecanoyl HSL (3-oxo-C12-HSL), while EsaI directs 3-oxo-C6-HSL synthesis. Although the two proteins share only weak sequence conservation, their structures are highly similar, and each adopts a fold similar to GCN5 acyltransferases, enzymes that recognize the phosphopantetheine moiety that forms the thioester linkage between acyl chains and ACP (25, 102). The regions of these proteins thought to dictate acyl chain interactions have different structures, with a restricted pocket on EsaI and a tunnel in LasI, perhaps correlating to the relative sizes of their substrates. Churchill speculated that AHL synthases which synthesize long-chain AHLs derive chain length specificity from the acyl chain itself plus differential interactions with the ACP, whereas AHL synthases that specify shorter-chain AHLs can directly interact only with the ACP, due to limited accessibility of the short chains.
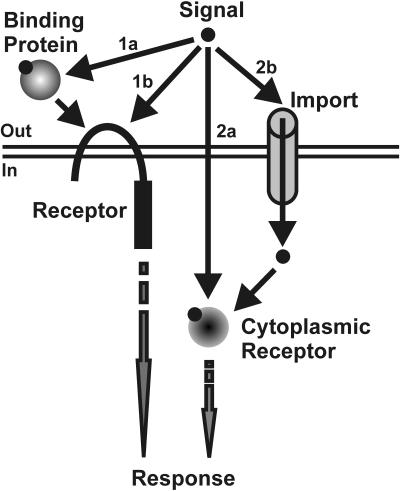
FIG. 2.
Mechanisms of signal perception. Generalized signal is depicted as a filled circle. Pathways 1a and 1b involve external perception of the signal either through an external binding protein intermediate that presents the signal to the transmembrane receptor (1a) or through direct interaction of the signal with a transmembrane receptor (1b). Receipt of the external signal is transduced to target processes within the cell. In pathways 2a and 2b, the signal is internalized through passive diffusion (2a) or through specific import mechanisms (2b) and interacts with an intracellular receptor, which in turn modulates cellular processes.
Oligopeptide signals produced by gram-positive microbes are often proteolytically liberated from secreted, genetically encoded proteins (Table 1) (53). An excellent example of such a signal molecule is the pentapeptide (ERGMT) called competence-stimulating factor (CSF) from Bacillus subtilis which controls initiation of the genetic competence pathway and influences sporulation (77). CSF is derived from the C terminus of the 40-amino-acid PhrC protein, a pre-pro-CSF (86). Export of pre-pro-CSF via the general secretion pathway and signal peptidase cleavage releases a 15-amino-acid pro-CSF that is further cleaved extracellularly to generate the pentapeptide. Beth Lazazzera presented recent work on analysis of pro-CSF cleavage (B. Lazazzera, CCCB-04, abstr. S7:1). Mutations in pro-CSF residues adjacent to the site for proteolytic cleavage block normal processing. As might be predicted, CSF cleavage activity is associated with the cell wall fraction. CSF accumulates extracellularly and is imported back into the cell via an oligopeptide permease(s), where it subsequently targets several different cytoplasmic receptor proteins (54).
Gary Dunny presented recent findings on the synthesis of oligopeptide mating pheromones that regulate plasmid conjugal transfer for Enterococcus faecalis (G. Dunny, CCCB-04, abstr. S1:1). Strains that do not carry the conjugal plasmid pCF10 produce a heptapeptide (LVTLVFV), to which conjugal donors respond by activating mating pair formation and subsequent conjugation (2). The short peptide signals are derived from putative lipoprotein substrates by cleavage through an endopeptidase called EEP (1). A major question in this system has been how donor cells prevent self-activation by endogenously produced pheromone. The pCF10 plasmid encodes a membrane protein called PrgY that may play a role in exit of the pheromone from cells (7). Dunny hypothesized that one mechanism to prevent donors from autoactivating might be PrgY-mediated expulsion of the pheromone following EEP-mediated processing.
Signal perception.
The mechanisms by which bacteria sense and respond to intercellular signals in their environments are of two basic types (Fig. 2). In the first, the signal is recognized extracytoplasmically by a specific sensor protein, which transmits the information to an intracellular regulator. Systems dependent on transmembrane sensor kinases often fall into this category. In some cases, an extracellular binding protein binds the signal, and this complex then interacts with the sensor kinase (Fig. 2, pathway 1a). For the second general mechanism, the signal transits across the cell membrane either by diffusion or by specific transport (Fig. 2, pathways 2a and b). Inside the cell, it interacts with and directly modulates the function of a cytoplasmic target protein, often a DNA binding protein. An interesting variation on this mechanism occurs when a diffusible molecule (described as a “messenger”) is chemically converted to an active signal only after it enters into the target cell (13).
Gram-positive bacteria utilize one or the other of these mechanisms. In several well-studied organisms, including Streptococcus pneumoniae and Staphylococcus aureus, oligopeptide signals are recognized by transmembrane sensor kinases that control the phosphorylation of cognate response regulators to alter gene transcription (Table 1) (68, 72). In other gram-positive bacteria, such as Bacillus subtilis, oligopeptide signals are imported through ABC transporters (54, 57). The internalized peptides then directly bind a target regulatory protein, such as a phosphatase enzyme, modifying its activity and subsequently altering the phosphorylation state of a key two-component response regulator(s).
Bacteria that utilize AHLs usually perceive the signals via cytoplasmic proteins. AHLs transit across the cellular envelope via diffusion, although they may be assisted by transporters (21). The target for the AHL is usually a member of the LuxR family of transcriptional activators. LuxR and several of its homologs, including TraR and LasR, directly bind to their cognate AHLs (30, 82, 109). LuxR-type proteins bind to DNA as multimers, usually dimers, and thereby regulate transcription (103).
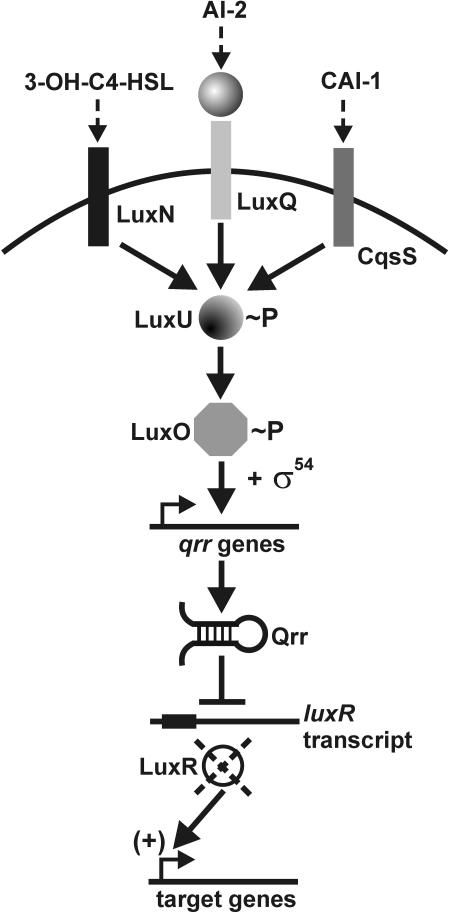
In contrast to most AHL-based signaling systems, Vibrio harveyi perceives an AHL signal (3-hydroxy-butanoyl-HSL) using a transmembrane sensor kinase, LuxN, that is localized to the cytoplasmic membrane (Fig. 3). It is not yet clear whether the AHL is sensed externally or through interactions with cytoplasmic portions of LuxN. AI-2 (furanosyl borate diester) is also a V. harveyi signal molecule. Extracellular AI-2 is bound by a periplasmic binding protein (LuxP), and the AI-2/LuxP complex is thought to interact with periplasmic portions of the LuxQ sensor kinase (Fig. 3). The mechanisms of signal perception for some of the more recently characterized signals and potential signals, such as cis-11-methyl-2-dodecenoic acid (DSF), are not well understood.
FIG. 3.
A model for three-way signal convergence in Vibrio harveyi. The three recognized signals are indicated prior to association with their cognate transmembrane receptors. In the absence of signal, receptors initiate the LuxU-LuxO phosphorelay, resulting in elevated levels of the Qrr regulatory RNAs, which block LuxR translation, thus preventing activation of lux genes and other target functions. Signal perception reverses phosphate flow, limiting qrr expression and allowing LuxR activation of target genes (see model in reference 32 for details).
Signal-dependent transcriptional control.
An active area of current research focuses on the regulatory proteins that transduce the response during cell-cell communication, typically transcription factors that bind to DNA. Identifying the targets of regulation, the mechanisms by which they are controlled, and the key features of transcriptional control are important research directions for a number of labs.
The TraR protein from Agrobacterium tumefaciens is arguably the best understood LuxR-type protein at a structural and biochemical level. It remains the only LuxR-type protein for which structural information is available (96, 108). TraR forms a dimer in the presence of 3-oxo-C8-HSL (one AHL molecule/protomer). The AHL-associated dimer is stabilized against proteolysis, while the apoprotein is proteolytically unstable (109). The TraR dimer is active for binding to specific DNA target sequences, typically located immediately upstream of target promoters. The TraR structure reveals that the AHL is completely enveloped within the N-terminal AHL binding domain of the protein, while the C-terminal region comprises the DNA binding motif. The inaccessibility of the AHL molecules within the mature dimer is consistent with a model in which TraR binds the AHL during protein folding but in which the apoprotein is unable to bind AHL once it is fully synthesized (110). Steve Winans reported recent work to identify TraR residues required for transcriptional activation (104; C. White and S. Winans, CCCB-04, abstr. S1:2). Extensive mutagenesis of C-terminal residues exposed to solvent in the TraR crystal structure identified mutants that retain the ability to bind DNA but are deficient in activating transcription, presumably due to inappropriate RNA polymerase contacts. Earlier studies from the V. fischeri LuxR protein had identified a similar region that is required to activate transcription but dispensable for DNA binding (17).
Biochemical analysis of the P. aeruginosa LasR protein by Martin Schuster in the Greenberg lab suggests that it shares many of the attributes found for TraR (M. Schuster et al., CCCB-04, poster abstr. 126). LasR forms dimers with each protomer bound extremely tightly to a single 3-oxo-C12-HSL molecule. LasR activates the expression of many different promoters in P. aeruginosa (81, 99). In vitro DNA binding assays with LasR and several different regulated promoters revealed that LasR can recognize a number of different promoter architectures (82). Although some of these promoters have easily identified las box DNA binding sites, several lack recognizable las boxes entirely but are strongly activated and bound by LasR. Schuster's analysis reveals surprisingly loose sequence conservation between LasR-regulated promoters, perhaps reflecting its role as a global regulator of diverse target genes.
Although many LuxR-type regulators are AHL-responsive transcriptional activators, several function as repressors. One such repressor is EsaR, found in Pantoea stewartii, a pathogen of maize (5). EsaR controls exopolysaccharide production in response to 3-oxo-C6-HSL synthesized by EsaI. Prior genetic studies suggested that EsaR functions as a negative regulator that is induced by its AHL. Susanne Beck von Bodman reported that EsaR formed a functional homodimer that could repress transcription of its own gene (esaR) in the absence of AHL (66; S. Beck von Bodman et al., CCCB-04, abstr. S4:4). The mechanism by which EsaR repressed exopolysaccharide synthesis was not known. As described by Beck von Bodman, EsaR represses expression of rcsA, encoding an activator of cps genes, required for exopolysaccharide production (6, 66a). The AHL signal inactivates EsaR, releasing it from its DNA binding site, elevating rcsA expression, in turn activating cps expression, and resulting in increased exopolysaccharide synthesis. Other LuxR homologs apparently function as repressors, and P. stewartii should continue to provide important insights into AHL-dependent derepression.
Streptomyces coelicolor is an actinomycete that produces a number of antibiotics during morphological differentiation. Cell-cell signaling involves the GBLs described above (Table 1 and Fig. 1), which bind to transcription factors that influence gene expression. One of these GBLs, SCB1, is the inducing ligand for the ScbR DNA binding transcriptional regulator (90). ScbR, in addition to autorepression, activates expression of the divergently transcribed scbA gene, likely to encode a GBL synthase. Despite clear evidence for regulation of antibiotic production, the target genes under direct control of ScbR-GBL were not known. E. Takano described microarray analysis of the ScbR regulon (E. Takano et al., CCCB-04, abstr. S4:5). The expression of greater than 20 genes was influenced by ScbR, but the promoter for only one of these, a presumptive polyketide biosynthesis regulator, kasO, was bound by ScbR in vitro (91). The regulatory network under GBL control (and linked through KasO) remains to be fully characterized.
REGULATORY NETWORKS: EXPANDING THE REACH OF COMMUNICATION
Although the fundamental mechanisms for sensing and responding to signal molecules can be relatively simple, the potential complexity of the signaling networks they control is impressive. Many bacteria that inhabit complex environments experience multiple signals, some quite similar to their own, and thus they must discriminate between true signals and background noise. In addition, many bacteria recognize and respond to multiple chemically distinct signals. These signals may be integrated to amplify expression of a particular gene(s) or independently control transcription of discrete sets of genes; thus, the regulatory cascades involved in the response can be staggeringly complex.
Densely overlapping signal response pathways in P. aeruginosa.
The opportunistic pathogen P. aeruginosa utilizes quorum sensing to regulate factors necessary for virulence (85). P. aeruginosa utilizes at least three different externalized signals, two of which are AHLs and one which is a hydroxy alkyl quinoline (HAQ) called PQS (Table 1). The complexity of the P. aeruginosa signaling network was the focus of Barbara Iglewski's presentation (B. H. Iglewski, CCCB-04, abstr. S5:2). Microarray analysis of gene expression patterns from the Iglewski group and others suggests that the LuxR-type protein LasR (responsive to 3-oxo-C12-HSL synthesized by LasI) sits at the top of a complex regulatory hierarchy, controlling the expression of a large number of genes, including rhlR, encoding a second LuxR-type protein (81, 99). RhlR (responsive to C4-HSL synthesized by RhlI) also controls many genes, including a subset of the same genes as LasR. Iglewski noted that LasR and RhlR in aggregate regulate upwards of 600 genes, including 31 predicted regulators. These downstream regulators considerably expand the impact of the initial cell-cell signals and contribute to the intricate response to quorum sensing observed for P. aeruginosa.
Multiple signaling pathways and regulatory RNA in Vibrio species.
Vibrio harveyi, a bioluminescent enteric bacterial species from fish, recognizes at least three distinct extracellular signals: (i) the AHL 3-hydroxy-butanoyl HSL, (ii) AI-2 (a furanosyl borate diester), and (iii) the CAI-1 signal, as yet chemically uncharacterized (33; J. Henke and B. Bassler, CCCB-04, poster abstr. 96). Although these three parallel pathways may seem redundant, it is clear from work in Bonnie Bassler's lab that integration of these signals produces a so-called “coincidence detection system” to allow modulation of the response of this organism to intraspecies and interspecies cues (67). Independent sensor kinase proteins respond to each cognate signal, converging through the LuxU phosphotransferase protein to control the phosphorylation state of the response regulator protein LuxO (Fig. 3). AHL, AI-2, and CAI-1 signals apparently reverse the flow of phosphate away from LuxO, leading to activation of target functions, including bioluminescence. LuxO was genetically defined as a repressor of lux gene expression but was also recognized as a member of the NtrC subfamily of response regulators and as such was predicted to interact with σ54 to activate the transcription of target genes (4, 58). Promoter sequences recognized by σ54-containing polymerases are highly conserved and are absent upstream of both the lux operon and the V. harveyi luxR gene encoding an activator of lux genes (not a V. fischeri LuxR-type AHL regulator) (89). It was therefore hypothesized that there is a regulatory intermediate(s) activated by LuxO, and Kenny Mok presented findings from the Bassler lab on identification of such intermediates (K. Mok et al., CCCB-04, abstr. S4:1). Mok and colleagues isolated a luminescent colony from a transposon mutant library of a nonluminescent V. harveyi derivative (harboring the constitutively active LuxO D47E, mimicking LuxO∼P). This mutant was disrupted in the V. harveyi hfq gene, which encodes an RNA chaperone known to promote the function of small regulatory RNA (sRNA) molecules in other bacteria (56). Hfq affected the stability of the V. harveyi luxR mRNA transcript and also the transcript encoding HapR, its homolog in Vibrio cholerae. The genome sequences of several Vibrio species with LuxO homologs (the V. harveyi genome has not been sequenced) were analyzed for the presence of sRNA genes, and four candidates were identified in V. cholerae. Each sRNA gene was subsequently shown to be controlled through LuxO and σ54, and these were designated qrr (quorum-regulated RNA) genes. Each Qrr has a region complementary to the ribosome binding site of the hapR and luxR transcripts and is likely to occlude translational initiation. Surprisingly, all four qrr genes must be mutated to mimic the constitutively bioluminescent phenotype of a luxO mutant. Mok speculated that the Qrr regulators allow for exquisite fine tuning of target gene expression, perhaps with each qrr gene differentially regulated by particular environmental conditions. The complex regulatory circuitry underlying quorum sensing control in these Vibrio species provides for multiple external signals and multiple physiological inputs through the phosphotransfer pathway and the Qrr regulatory RNAs.
Signal interference.
Bacterial signaling pathways can be disrupted in a variety of ways. A growing area of research is the study of natural signal interference processes and the design of deliberate interference strategies. Signaling can be blocked at the levels of synthesis, stability, and perception of the signal. Several natural products interfere with perception of signal molecules, including the halogenated furanone compounds that inhibit AHL-based quorum sensing and inhibitory oligopeptides that block the function of closely related oligopeptide signaling systems (23, 43). There are also several well-documented examples of signal degradation mechanisms. AHL degradation can be catalyzed by AHL acylases that cleave the acyl chain to produce homoserine lactone and fatty acids and by AHL lactonases that cleave the lactone ring (107). AHL degradation activity was initially discovered in species of Bacillus and Variovorax paradoxus but is relatively widespread among diverse bacteria (16, 55). Although this phenomenon has been described as quorum quenching, it remains unclear whether the AHL degradation is coincidental or specifically targeted to AHLs. Recent findings have demonstrated that AHLs may not in fact be the preferred substrate for these systems (8). Perhaps more importantly, it was not clear that the amount of in situ AHL-degradative activity was sufficient to impact AHL signaling. Jared Leadbetter's group has analyzed diverse soil samples for AHL inactivation using radiolabeled AHL substrates. Most soils tested contained significant degradative potential, often inactivating the AHLs with no lag time, suggesting that this process is active in these environments (J. R. Leadbetter, CCCB-04, abstr. S3:5). Based on the rates of AHL mineralization, optimal conditions for degradation, and the effective concentrations produced by typical AHL-synthesizing bacteria, Leadbetter concluded that the amount of AHL degradation activity in these soils could readily interfere with endogenous AHL signaling.
Lian-Hui Zhang's group has focused considerable effort on harnessing AHL degradation as a biocontrol strategy and has reported several striking examples of plant protection (15, 107). Current efforts to engineer transgenic mammalian cell lines that express AHL lactonase activity are under way, and early results suggest that these cells manifest resistance to AHL-producing pathogens (L.-H. Zhang, CCCB-04, abstr. S3:3). Peter Greenberg also reported on AHL lactonase activity in certain human epithelial cell lines (11), and thus the ability to degrade AHLs may be a normal component of host resistance (P. Greenberg et al., CCCB-04, abstr. S5:1).
In the gram-positive pathogen S. aureus, virulence is regulated via the cyclic thioester octapeptide autoinducing peptide (AIP) and a complex oligopeptide-based signaling mechanism (71). Inducing levels of AIP are detected by the AgrC/A two-component system, which drives elevated expression of a regulatory RNA (RNA III) from the P3 promoter, which in turn activates virulence functions. Prior work suggested that the presence of the commensal microbe Lactobacillus reuteri reduced the virulence of S. aureus in certain model systems. Results presented by Jennifer Laughton from J. K. McCormick's group have traced the probiotic mechanism of L. reuteri to inhibition of the P3 promoter and hence functions under its control (J. Laughton et al., CCCB-04, abstr. S3:4). An uncharacterized component of L. reuteri culture supernatants effectively inhibits activation of P3, demonstrating the potential efficacy of signal interference by competing bacteria in ameliorating virulence.
COMMUNICATION DURING MICROBIAL DEVELOPMENT
Cellular differentiation is observed in a wide range of different microbes. The generation of new cell types, such as environmentally resistant spores and metabolically dedicated nitrogen-fixing cells, is a well-studied phenomenon. In these systems, differentiation is often a group decision, influenced by the population structure and positioning of cells relative to one another.
Horizontal gene transfer during B. subtilis development.
The Phr (pheromone) peptides are small signal molecules secreted by B. subtilis, initially discovered as regulators of the sporulation process (77). Phr signals are processed from precursor proteins secreted across the cytoplasmic membrane to mature forms with five or more amino acids. The peptides function to inhibit Rap phosphatases, which in turn modulate phosphorelay circuits controlling a range of B. subtilis physiology. The Phr peptides are genetically encoded in the same operons as the Rap phosphatases, and the B. subtilis genome contains seven distinct Rap/Phr cassettes. Jennifer Auchtung from Alan Grossman's group described her recent findings regarding the RapI/PhrI cassette, encoded within an integrative and conjugal element (ICE) designated ICEBsuI (J. Auchtung et al., CCCB-04, abstr. S2:4). Microarray analyses suggested that RapI stimulates expression of ICEBsuI genes. Mating experiments reveal that ICEBsuI excises and conjugates to several different species of Bacillus, including B. subtilis and B. anthracis, and this process is inhibited by PhrI. An intriguing model emerges from this work in which cells harboring ICEBsuI produce the PhrI pheromone and thus inhibit conjugal transfer but potential recipient cells lacking ICEBsuI do not produce PhrI and therefore do not inhibit conjugal transfer. Remarkably, and in an interesting contrast to the recognized recipient-produced mating pheromones of E. faecalis, ICEBsuI transfer in B. subtilis is stimulated by the inability of recipient cells to produce a signal molecule. Several of the rap phr cassettes in B. subtilis are carried on plasmids, and several others are carried on ICEs, suggesting significant potential for their horizontal transfer.
Oligopeptide control of heterocyst differentiation.
The filamentous cyanobacterium Anabaena sp. PCC7120 regulates the pattern and frequency of vegetative cell differentiation into nitrogen-fixing heterocysts in part via a peptide signal molecule called PatS (Table 1) (106). PatS is derived from a 13- to 17-amino-acid precursor oligopeptide, of which the C-terminal five amino acids (RGSGR) can function to inhibit heterocyst formation. PatS is released from developing heterocysts and is thought to diffuse laterally along the filament, inhibiting heterocyst development in adjacent cells (24). PatS mutants develop heterocysts in nonregular intervals along the filament, and ectopic expression of patS can inhibit heterocyst formation. Jim Golden presented time lapse microscopy demonstrating that expression of the patS gene occurs in a stochastic fashion and that cells which initially express high levels of patS differentiate into heterocysts, inhibiting proximal cells in the filament (J. W. Golden, CCCB-04, abstr. S2:1). A protein called HetR, recently verified to be a DNA binding protein inhibited by PatS, has long been recognized as a regulator of heterocyst development, and HetR mutants form heterocysts (39). Findings from the Golden lab, in which specific HetR mutants are not inhibited by PatS, provide genetic evidence for the regulatory link between PatS and HetR (48).
Communication during coordinated motility.
A wide range of microbes substantially alter their mode of motility and often their cellular morphology on solid surfaces (31). Salmonella enterica serovar Typhimurium differentiates from a swimming cell into longer swarmer cells with dense flagella. Mike Surette proposed that the swim-to-swarmer transition represents more than simply a change in motility strategy but rather an adaptation to survival in the polymicrobial communities of the mammalian gut (50; M. G. Surette, CCCB-04, abstr. S2:3). Surette and colleagues harvested cells from different regions of a swarming Salmonella population, comparing the proteome of actively swimming cells to swarming cells, and observed a large number of proteins with 5- to 20-fold changes. The spectrum of proteome alterations suggests that Salmonella significantly changes its cellular metabolism in swarmer cells, consistent with increased resistance to a variety of antimicrobial agents. Swarmer cells activate genes in the AI-2 biosynthetic pathway and appear to synthesize high levels of AI-2. Despite this, swarmer cells respond poorly to the signal. Surette speculates AI-2 signaling and swarmer cell differentiation prepare Salmonella for competition with the resident gut microbiota.
Identification of the Streptomyces coelicolor SapB peptide: signal or surfactant?
Species of Streptomyces are known to produce diffusible signals (Table 1). The transition from vegetative mycelial growth to formation of aerial hyphae, and eventual sporulation, is a complex developmental process (9). A diffusible factor called SapB was identified in S. coelicolor and reported to rescue several developmental (bld) mutants blocked for aerial hyphae formation and sporulation (105). SapB is a hydrophobic peptide surfactant, exogenous addition of which allows elaboration of aerial hyphae in the bld mutants but does not promote spore formation (95). Joanne Willey described the culmination of years of work from several labs to determine that SapB is a highly modified lantibiotic-type peptide (J. M. Willey, CCCB-04, abstr. S2:2; see also reference 51). A key to successful structural determination was the correlation of SapB production with a cluster of S. coelicolor genes, designated ramCSAB (rapid aerial mycelium formation) (62 and 73) by Justin Nodwell and colleagues. The ramS gene encodes a 42-amino-acid protein, a likely SapB precursor, but does not match the mass or known short sequences of SapB. RamC provided a clue, with carboxy-terminal similarity to enzymes involved in production of posttranslationally modified peptide antibiotics called lantibiotics (79). Lantibiotics are cleaved from short precursor proteins that are dehydrated at Ser/Thr residues to generate didehydroalanine/didehydrobutyrine, which form intrapeptide thioester linkages with Cys residues. Willey, Nodwell, and colleagues pieced together additional information, including sequences and mass measurements, concluding that SapB is derived from the 21 carboxy-terminal amino acid residues in RamS, with two lanthionine bridges (Ser3 to Cys10 and Ser13 to Cys20) forming two loops (51). Modeling of the SapB structure predicts extensive exposed hydrophobic domains, consistent with the surfactant properties of SapB. RamC is likely to be required for modification of the RamS peptide prior to or during export (J. R. Nodwell, CCCB-04, abstr. S2:6). During the extension of aerial hyphae, RamC is localized at filament tips, while the vegetative mycelia show more uniform expression. Although similar to lantibiotics, SapB exhibits no antimicrobial properties and rather functions as a surfactant. SapB is a bacterially produced compound that seems to occupy the conceptual interface between a developmental morphogen, a structural lubricant, and a signal molecule.
PHYSICAL INTERACTIONS OF BACTERIA
Bacterial cell-cell signaling can occur through physical interaction with a neighboring cell or through the receipt of a diffusible signal molecule. In many cases, such as biofilm formation or fruiting body formation, these interactions allow bacteria to function cooperatively and form complex structures.
Contact-dependent signaling in Myxococcus xanthus.
The soil bacterium M. xanthus undergoes a complex developmental pathway, involving several different diffusible and cell contact-dependent signals, to form fruiting bodies and sporulate (46). Motility plays an important role in this process and is a target for signaling. Two motility mechanisms are simultaneously employed by M. xanthus to move across surfaces; social (S) motility involves multiple cells that move via the action of type IV pili, and adventurous (A) motility propels cells along surfaces by extrusion of extracellular slime through “nozzles” (45). The balance between these two distinct forms of motility, their regulation by environmental signals, and their role in M. xanthus development are active areas of investigation. Trish Hartzell and Hera Vlamakis presented findings linking specific regulatory pathways, the MglA Ras-type GTPase and one of eight Che-type clusters from M. xanthus, respectively, to the control of motility (T. Hartzell et al., CCCB-04, abstr. S2:5; H. Vlamakis et al., CCCB-04, abstr. S2:7). It remains unclear how these motility controls are integrated with cell-cell communication in M. xanthus.
M. xanthus develops fruiting bodies from simpler cell aggregates, in part, through a series of cell reversals (46). When two cells meet end to end, they can exchange a contact-dependent cue called C-signal (Table 1) (49). This signal involves the CsgA protein processed to a 17-kDa form, localized to the cell surface, and then recognized by presumptive C-signal receptors on colliding cells (59). Receipt of C-signaling results in an increase in gene expression of the actABCDE operon, which up-regulates csgA expression, thus elevating C-signal production. C-signal receipt also causes phosphorylation of the regulator FruA. This event influences the methylation/demethylation state of the Che-like proteins FrzCD, which in turn affects phosphorylation of FrzE. FrzE phosphorylation influences waves and streaming of aggregating M. xanthus cells. Finally, FruA phosphorylation also impacts sporulation. Increased C-signaling is required for each of the three developmental events (waves, streaming, and sporulation) (47). Dale Kaiser presented new data on the stages of fruiting body formation (D. Kaiser, CCCB-04, abstr. S4:2). Kaiser described the end stages of fruiting body formation as a three-dimensional cell traffic jam, such as might occur at the Place de la Concorde if cars could climb on top of each other. Computer simulations of fruiting body formation based on C-signaling rules for each developmental stage accurately predict many aspects of fruiting body formation (41). These findings suggest that the signaling and motility mechanisms established to date are sufficient to account for much of fruiting body formation.
Signaling and biofilm formation.
Bacteria that reside within biofilms, surface-adherent communities, have extensive opportunities to communicate and physically interact. In some cases, communication is required to properly assemble the biofilm (12, 40). In the gram-positive bacterium E. faecalis, oligopeptide-mediated signaling occurs through the FsrAB two-component system (69). Work from two different groups demonstrated that the FsrA-controlled gelA gene, encoding a zinc metalloprotease, is required for E. faecalis biofilm formation (28, 52). In addition to its role in biofilm formation, GelE functions in determining the length of the chains formed by E. faecalis cells, promoting degradation of misfolded surface proteins, and controlling the levels of the conjugation pheromone in culture supernatants (101). Marta Perego described the construction of strains with mutations in each of the 18 two-component systems of E. faecalis (27; M. Perego and L. Hancock, CCCB-04, abstr. S4:3). Of these, only fsr was required for biofilm formation, suggesting that other factors required for biofilm formation might be controlled via alternate regulatory networks.
Bacteria in nature are often in mixed communities. It is clear that interactions among multiple bacterial taxa are quite complex, and even the presence of two bacterial species can significantly complicate matters. Dingding An from Matt Parsek's lab described the results of competition experiments between P. aeruginosa and A. tumefaciens in dispersed liquid culture and in biofilms (D. An et al., CCCB-04, abstr. S7:6). In liquid culture, P. aeruginosa cells numerically dominated A. tumefaciens cells, and this was dependent on the AHL synthases LasI and RhlI. In biofilms formed on abiotic surfaces, P. aeruginosa also numerically dominated A. tumefaciens by developing into a thick blanket that covered the A. tumefaciens biofilm. Blanketing by P. aeruginosa was diminished when either type IV pili (pilA) or flagella (flgK) were defective, although these mutants were unaffected for competition in liquid culture. The Las and Rhl signaling mutants were only modestly reduced in blanketing efficiency, suggesting that the competitive mechanisms of P. aeruginosa were different in the two distinct environments. Supporting this idea, A. tumefaciens more effectively competed with P. aeruginosa during biofilm formation on plant roots.
HOST-MICROBE COMMUNICATION
In addition to mediating chatter among different bacterial species, signals are clearly exchanged with or intercepted by eukaryotic hosts. Signals originally thought to be dedicated to interbacterial communication can be recognized by eukaryotes, inducing specific responses. Zoospores of the marine alga Ulva intestinalis respond to the presence of AHL-producing derivatives of Vibrio anguillarum and synthetic AHLs, enhancing their colonization of surfaces (44; D. Wheeler et al., CCCB-04, abstr. S6:4). Plants also mount extensive responses to AHLs (64). Conversely, several different plants are known to produce quorum-sensing mimics (as yet uncharacterized), possibly to manipulate microbial rhizosphere populations (94). The marine alga Delisea pulchra produces halogenated furanones that disrupt AHL-based quorum sensing (23). Increasingly, it is apparent that metazoans have developed mechanisms by which they detect and in some cases manipulate microbial signaling pathways.
Signaling and signal interference with mammalian cells.
Several reports suggest that 3-oxo-C12-HSL, produced by P. aeruginosa, can stimulate cytokine production in mammalian cells (14, 36, 93). Manuba Horikawa reported that 3-oxo-C12-HSL also can induce apoptosis in macrophages and fibroblasts (92; M. Horikawa et al., CCCB-04, abstr. S6:6). It remains unclear whether these immune responses benefit or further harm the host cells (Fig. 4A). Similarly, the mammalian AHL lactonase activity reported by Peter Greenberg may be a component of innate defenses (P. Greenberg et al., CCCB-04, abstr. S5:1).
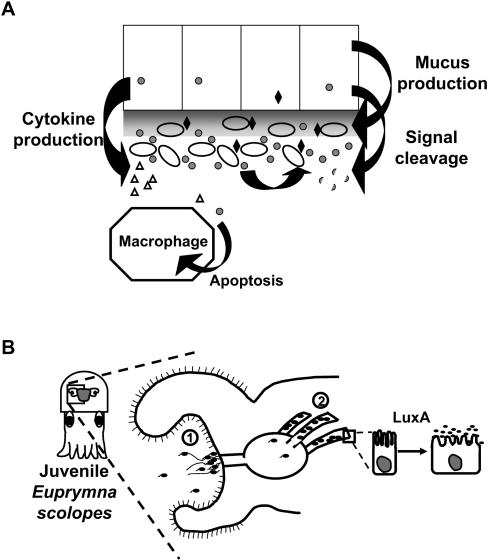
FIG. 4.
Diffusible signaling in bacteria-host associations. (A) Not only do bacteria recognize their own secreted signals (gray circles) to coordinate a response (black diamonds), but eukaryotic cells in the vicinity may also recognize these signals and produce a response. Some of the potential interactions are diagrammed. Various host cells may alter cytokine production (open triangles), mucus production (shaded gray), or other developmental events, such as apoptosis, in response to either the signal itself or the bacterial product of the signal transduction pathway. Hosts may also induce or constitutively synthesize signal-degrading enzymes. (B) Symbiotic Euprymna scolopes-V. fischeri interaction during light organ colonization. LuxA activity is dependent on AHL quorum sensing. The locations at which bacterial cell-cell signaling likely occurs are labeled as 1 and 2. Region 1 is the site of the initial attachment, or aggregation, by bacteria on the surface of the light organ. Region 2 is the site of colonization inside, where the bacteria multiply to high cell density and induce developmental changes, such as the AHL-dependent LuxA activity that is required for host epithelial cell swelling.
An environmental condition encountered by many bacteria, pathogens, and symbionts alike is that of mucus that lines and protects host surfaces. In the lungs of cystic fibrosis (CF) patients, P. aeruginosa stimulates mucus production (Fig. 4A). Sputum recovered from CF patients has been shown to contain AHL activity, and more recently, the HAQ signal PQS was shown to be enhanced in isolates from the CF lung (26, 83). Kelly Palmer from Marvin Whiteley's lab reported that CF sputum caused early induction of PQS-controlled genes as measured in microarrays (K. Palmer et al., CCCB-04, abstr. S6:5). Palmer also reported that coculturing P. aeruginosa with S. aureus resulted in the specific lysis of S. aureus and that growth in mucus caused that lysis to occur earlier. These data emphasize how host-produced factors can profoundly impact the timing, strength, and composition of an ensuing signaling cascade.
Signaling in host-associated vibrios.
AHL signaling was first discovered in Vibrio spp., and they continue to be intensively investigated, particularly for studying interactions that occur in the natural environment. Symbiosis of bioluminescent V. fischeri with the squid host Euprymna scolopes has provided a powerful model system (Fig. 4B). Successful symbiosis of V. fischeri with the squid requires LuxR, LuxI, and the lux genes they control, as well as the alternate AHL synthase AinS and the AI-2 synthase LuxS, which, like those of V. harveyi, signal through LuxO (60, 61). Furthermore, a nonluminescent mutant defective for the LuxRI-controlled luxA gene fails to induce normal host development (Fig. 4B) (97). In addition to these interbacterial and bacteria-host communications, symbiotic colonization by V. fischeri requires the hybrid sensor kinase RscS, which exhibits sequence similarity to LuxQ of V. harveyi (98). The rscS sensor kinase gene is not genetically linked to a response regulator gene. Karen Visick reported disruption of many of the 43 predicted response regulators encoded in the V. fischeri genome in an attempt to identify the cognate response regulator for RscS (E. A. Hussa et al., CCCB-04, abstr. S5:4). Disruption of one of these response regulator genes (VFA1026) causes a severe defect, similar to the rscS mutant, in the ability of V. fischeri to initiate symbiotic colonization. Further work will determine whether this regulator functions with RscS or as part of a separate pathway.
Nematode worms and their bacteria.
Xenorhabdus nematophila is a gram-negative bacterium that forms a symbiotic association with the nematode Steinernema carpocapsae. Together, the two organisms infect and kill insect larvae (19). The bacterium is not found free-living in the soil, and development of the infectious juvenile stage of the nematode life cycle requires uptake of the bacterium. Extensive communication between the host nematodes and the symbiotic bacteria is likely to underlie this process. In the lab, nematodes feed on X. nematophila (and other bacteria), leading to the question of how the switch occurs between using X. nematophila for nutrients and maintaining it as a symbiotic partner. Careful examination of initial infection by the Goodrich-Blair lab has revealed that one or two “founder” bacteria initiate symbiotic colonization (63; H. Goodrich-Blair, CCCB-04, abstr. S7:2). Growth and division, within a vesicle associated with the nematode intestine, is not linear but rather occurs as cycles of growth and possibly death of the microbe. The nematode contains an intravesicular space to which the symbiotic bacteria initially attach. Goodrich-Blair speculated that this intravesicular space could confer protection to the bacteria that will ultimately colonize the nematode. Genetic characterization of bacterial genes necessary for symbiosis should provide insights into these processes (34).
LESSONS FROM METAZOAN SIGNALING
The Banff conference included several excellent presentations on eukaryotic multicellular signaling. John Carlson presented fascinating results on the ability of Drosophila melanogaster to smell and taste. The issue of how fruit flies distinguish different odors and tastes using the same receptor systems certainly has relevance to signal specificity and response for bacterial systems. While the anatomic and molecular details are clearly distinct (neuronal bundles and G protein-coupled receptors), there is unity at both the conceptual and mechanistic levels in which some odor and gustatory receptors act as broadly tuned systems while others are highly specialized. Similarly, it is clear that certain bacterial systems act to recognize a broad range of signals while others are exquisitely selective. Carol Manahan presented findings on the mechanisms by which the slime mold Dictyostelium discoideum coordinates development and motility during fruiting body formation in response to starvation, a process with facile similarity to myxobacterial fruiting body formation. In the slime mold, cyclic AMP acts to stimulate chemotaxis resulting in waves of cellular response. The slime mold cells coalesce to form a motile multicellular colony (a “slug”) that moves in concerted fashion. Manahan's work focuses on the coordination of motility via the action and subcellular localization of the second messenger phosphoinositol trisphosphate. While there is no exact microbial equivalent of the phosphoinositol trisphosphate system, cyclic diguanosine monophosphate (c-di-GMP) has been described as a possible microbial second messenger that can be linked to multicellular activity, such as quorum sensing and biofilm formation (42). Findings presented by Max Dow suggest that c-di-GMP signaling may be an important component in the DSF signaling pathway of X. campestris (M. Dow et al., CCCB-04, poster abstr. 4A).
Laurent Keller provided an insightful ecological analysis of signaling with his work on insect behavior as a backdrop. Keller pointed out that even among colonial insects in which the organization and cooperation between individuals are very high, significant conflicts of interest exist within the colony, such as the skewing of sex ratios in conflicts between queens and worker class ants (74). Similarly, Keller argued that obvious conflicts exist between different bacteria competing for common resources and that true cooperation will be rare. Behavioral ecologists often divide communication mechanisms into several different classes: (i) honest signals, (ii) cues, and (iii) chemical manipulation. Honest signals involve active participation of both the signal producer and the responder, while cues involve coincidental production of a signal by an individual, subsequently perceived by another individual. Cues benefit the responder but are neutral or even detrimental to the signal producer. Chemical manipulation is the production of signals to direct the behavior of another individual, often harming the responder. Keller suggested that all of these mechanisms of communication exist among bacteria. Honest signals are likely to be intraspecific, cues can be inter- and intraspecific, and manipulative chemicals are usually interspecific. As the field of microbial cell-cell communication begins to address the questions of why microbes produce signals and to what benefit, the concepts developed in fields such as insect behavior are an important foundation on which to build.
SUMMARY AND PERSPECTIVE
The 2004 ASM conference on Cell-Cell Communication in Bacteria provided outstanding examples of the diversity and volume of microbial chatter and our growing understanding of the mechanisms by which bacteria communicate. We have gained an in-depth understanding about several model systems, including AHL-based quorum sensing, oligopeptide signaling mechanisms, and development in M. xanthus. Additional signaling systems are being discovered at an impressive rate, and it is clear that chatter among microorganisms is extensive and pervasive. The interconnected regulatory networks derived from cell-cell communication have far-reaching consequences for the understanding and potential control of bacterial behavior. Antimicrobial strategies that focus on signaling are being realized in preliminary applied studies. It is also clear that nature has already developed such strategies by which bacteria and other organisms specifically detect and interfere with the signaling mechanisms of their competitors, such as with the degradation of AHL signals by lactonases or acylases.
As with other areas of the life sciences, technical advances in genomics, proteomics, structural biology, and microscopy are having a tremendous impact on studies of bacterial cell-cell communication systems. The number of studies defining genome-wide responses to signaling is increasing, and the resolution provided through structural analysis of proteins and RNAs involved in communication circuitry is already resulting in novel strategies for interfering with and manipulating bacterial behavior. The ability to analyze cell-cell communication at many different scales, from community-level analysis, through genomic and proteomic networks, to the atomic scale of three-dimensional protein structure, will profoundly influence the direction of this area of research in the future. An understanding of the ecological consequences and driving evolutionary forces that influence bacterial signaling systems is also crucial for integrating our observations at multiple scales into a picture of bacterial interactions with each other and their environment. The consolidation of current observations, the application of increasingly powerful new technologies, and the novel application of our growing understanding of cell-cell communication in bacteria will provide the momentum for years to come in this exciting area of microbiology.
Acknowledgments
We thank all of the participants of this stimulating and thought-provoking conference, with special credit to organizers Bonnie Bassler and Steve Winans. Many intriguing findings were reported. Regretably, only a portion of these could be included in this necessarily brief conference review. Research on cell-cell communication systems is supported in the authors' labs by the National Science Foundation (MCB 0223724 and 0238515 to C.F.) and the National Institutes of Health (GM59690 to K.L.V.).
REFERENCES
- 1.An, F. Y., M. C. Sulavik, and D. B. Clewell. 1999. Identification and characterization of a determinant (eep) on the Enterococcus faecalis chromosome that is involved in production of the peptide sex pheromone cAD1. J. Bacteriol. 181:5915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antiporta, M. H., and G. M. Dunny. 2002. ccfA, the genetic determinant for the cCF10 peptide pheromone in Enterococcus faecalis OG1RF. J. Bacteriol. 184:1155-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barber, C. E., J. L. Tang, J. X. Feng, M. Q. Pan, T. J. G. Wilson, H. Slater, J. M. Dow, P. Williams, and M. J. Daniels. 1997. A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Mol. Microbiol. 24:555-566. [DOI] [PubMed] [Google Scholar]
- 4.Bassler, B. L., M. Wright, and M. R. Silverman. 1994. Sequence and function of LuxO, a negative regulator of luminescence in Vibrio harveyi. Mol. Microbiol. 12:403-412. [DOI] [PubMed] [Google Scholar]
- 5.Beck von Bodman, S., G. T. Hayman, and S. K. Farrand. 1992. Opine catabolism and conjugal transfer of the nopaline Ti plasmid pTiC58 are coordinately regulated by a single repressor. Proc. Natl. Acad. Sci. USA 89:643-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernhard, F., K. Poetter, K. Geider, and D. L. Coplin. 1990. The rcsA gene from Erwinia amylovora: identification, nucleotide sequence, and regulation of exopolysaccharide biosynthesis. Mol. Plant-Microbe Interact. 3:429-437. [DOI] [PubMed] [Google Scholar]
- 7.Buttaro, B. A., M. H. Antiporta, and G. M. Dunny. 2000. Cell-associated pheromone peptide (cCF10) production and pheromone inhibition in Enterococcus faecalis. J. Bacteriol. 182:4926-4933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlier, A., R. Chevrot, Y. Dessaux, and D. Faure. 2004. The assimilation of gamma-butyrolactone in Agrobacterium tumefaciens C58 interferes with the accumulation of the N-acyl-homoserine lactone signal. Mol. Plant-Microbe Interact. 17:951-957. [DOI] [PubMed] [Google Scholar]
- 9.Champness, W. 2000. Actinomycete development, antibiotic production, and phylogeny: questions and challenges, p. 11-31. In Y. V. Brun and L. J. Shimkets (ed.), Prokaryotic development. ASM Press, Washington, D.C.
- 10.Chen, X., S. Schauder, N. Potier, A. Van Dorsselaer, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 11.Chun, C. K., E. A. Ozer, M. J. Welsh, J. Zabner, and E. P. Greenberg. 2004. Inactivation of a Pseudomonas aeruginosa quorum-sensing signal by human airway epithelia. Proc. Natl. Acad. Sci. USA 101:3587-3590. (First published 17 February 2004; 10.1073/pnas.0308750101.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 13.Deziel, E., F. Lepine, S. Milot, J. He, M. N. Mindrinos, R. G. Tompkins, and L. G. Rahme. 2004. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc. Natl. Acad. Sci. USA 101:1339-1344. (First published 22 January 2004; 10.1073/pnas.0307694100.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dimango, E., H. J. Zar, R. Bryan, and A. Prince. 1995. Diverse Pseudomonas aeruginosa gene products stimulate respiratory epithelial cells to produce interleukin-8. J. Clin. Investig. 5:2204-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong, Y.-H., L.-H. Wang, J.-L. Xu, H.-B. Zhang, X.-F. Zhang, and L.-H. Zhang. 2001. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 411:813-817. [DOI] [PubMed] [Google Scholar]
- 16.Dong, Y.-H., J.-L. Xu, X.-Z. Li, and L.-H. Zhang. 2000. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates virulence of Erwinia carotovora. Proc. Natl. Acad. Sci. USA 97:3526-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egland, K. A., and E. P. Greenberg. 2001. Quorum sensing in Vibrio fischeri: analysis of the LuxR DNA binding region by alanine-scanning mutagenesis. J. Bacteriol. 183:382-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flavier, A. B., M. A. Schell, and T. P. Denny. 1997. Identification of 3-hydroxypalmitic acid methyl ester as a novel autoregulator controlling virulence in Ralstonia solanacearum. Mol. Microbiol. 26:251-259. [DOI] [PubMed] [Google Scholar]
- 19.Forst, S., B. Dowds, N. Boemare, and E. Stackebrandt. 1997. Xenorhabdus and Photorhabdus spp.: bugs that kill bugs. Annu. Rev. Microbiol. 51:47-72. [DOI] [PubMed] [Google Scholar]
- 20.Fuqua, C., and A. Eberhard. 1999. Signal generation in autoinduction systems: synthesis of acylated homoserine lactones by LuxI-type proteins, p. 211-230. In G. M. Dunny and S. C. Winans (ed.), Cell-cell signaling in bacteria. ASM Press, Washington, D.C.
- 21.Fuqua, C., and E. P. Greenberg. 2002. Listening in on bacteria: acylhomoserine lactone signalling. Nat. Rev. Mol. Cell Biol. 3:685-695. [DOI] [PubMed] [Google Scholar]
- 22.Fuqua, W. C., S. C. Winans, and E. P. Greenberg. 1994. Quorum sensing in bacteria: the LuxR/LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Givskov, M., R. de Nys, M. Manefield, L. Gram, R. Maximilien, L. Eberl, S. Molin, P. D. Steinberg, and S. Kjelleberg. 1996. Eukaryotic interference with homoserine lactone-mediated prokaryotic signalling. J. Bacteriol. 178:6618-6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Golden, J. W., and H. S. Yoon. 2003. Heterocyst development in Anabaena. Curr. Opin. Microbiol. 6:557-563. [DOI] [PubMed] [Google Scholar]
- 25.Gould, T. A., H. P. Schweizer, and M. E. Churchill. 2004. Structure of the Pseudomonas aeruginosa acyl-homoserinelactone synthase LasI. Mol. Microbiol. 53:1135-1146. [DOI] [PubMed] [Google Scholar]
- 26.Guina, T., S. O. Purvine, E. C. Yi, J. Eng, D. R. Goodlett, R. Aebersold, and S. I. Miller. 2003. Quantitative proteomic analysis indicates increased synthesis of a quinolone by Pseudomonas aeruginosa isolates from cystic fibrosis airways. Proc. Natl. Acad. Sci. USA 100:2771-2776. (First published 24 February 2003; 10.1073/pnas.0435846100.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hancock, L., and M. Perego. 2002. Two-component signal transduction in Enterococcus faecalis. J. Bacteriol. 184:5819-5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hancock, L. E., and M. Perego. 2004. The Enterococcus faecalis fsr two-component system controls biofilm development through production of gelatinase. J. Bacteriol. 186:5629-5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Handelsman, J. 2004. Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 68:669-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanzelka, B. L., and E. P. Greenberg. 1995. Evidence that the N-terminal region of the Vibrio fischeri LuxR protein constitutes an autoinducer-binding domain. J. Bacteriol. 177:815-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harshey, R. M. 2003. Bacterial motility on a surface: many ways to a common goal. Annu. Rev. Microbiol. 57:249-273. [DOI] [PubMed] [Google Scholar]
- 32.Henke, J. M., and B. L. Bassler. 2004. Bacterial social engagements. Trends Cell Biol. 14:648-656. [DOI] [PubMed] [Google Scholar]
- 33.Henke, J. M., and B. L. Bassler. 2004. Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi. J. Bacteriol. 186:6902-6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heungens, K., C. E. Cowles, and H. Goodrich-Blair. 2002. Identification of Xenorhabdus nematophila genes required for mutualistic colonization of Steinernema carpocapsae nematodes. Mol. Microbiol. 45:1337-1353. [DOI] [PubMed] [Google Scholar]
- 35.Holden, M. T. , S. R. Chhabra, R. de Nys, P. Stead, N. J. Bainton, P. J. Hill, M. Manefield, N. Kumar, M. Labatte, D. England, S. Rice, M. Givskov, G. P. C. Salmond, G. S. A. B. Stewart, B. W. Bycroft, S. Kjelleberg, and P. Williams. 1999. Quorum-sensing crosstalk: isolation and chemical characterization of cyclic dipeptides from Pseudomonas aeruginosa and other Gram-negative bacteria. Mol. Microbiol. 33:1254-1266. [DOI] [PubMed] [Google Scholar]
- 36.Hooi, D. S., B. W. Bycroft, S. R. Chhabra, P. Williams, and D. I. Pritchard. 2004. Differential immune modulatory activity of Pseudomonas aeruginosa quorum-sensing signal molecules. Infect. Immun. 72:6463-6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horinouchi, S. 2002. A microbial hormone, A-factor, as a master switch for morphological differentiation and secondary metabolism in Streptomyces griseus. Front. Biosci. 7:d2045-d2057. [DOI] [PubMed] [Google Scholar]
- 38.Hornby, J. M., E. C. Jensen, A. D. Lisec, J. J. Tasto, B. Jahnke, R. Shoemaker, P. Dussault, and K. W. Nickerson. 2001. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl. Environ. Microbiol. 67:2982-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang, X., Y. Dong, and J. Zhao. 2004. HetR homodimer is a DNA-binding protein required for heterocyst differentiation, and the DNA-binding activity is inhibited by PatS. Proc. Natl. Acad. Sci. USA 101:4848-4853. (First published 29 March 2004; 10.1073/pnas.0400429101.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huber, B., K. Riedel, M. Hentzer, A. Heydorn, A. Gotschlich, M. Givskov, S. Molin, and L. Eberl. 2001. The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology 147:2517-2528. [DOI] [PubMed] [Google Scholar]
- 41.Igoshin, O. A., R. Welch, D. Kaiser, and G. Oster. 2004. Waves and aggregation patterns in myxobacteria. Proc. Natl. Acad. Sci. USA 101:4256-4261. (First published 12 March 2004; 10.1073/pnas.0400704101.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jenal, U. 2004. Cyclic di-guanosine-monophosphate comes of age: a novel secondary messenger involved in modulating cell surface structures in bacteria? Curr. Opin. Microbiol. 7:185-191. [DOI] [PubMed] [Google Scholar]
- 43.Ji, G., R. Beavis, and R. P. Novick. 1997. Bacterial interference caused by autoinducing peptide variants. Science 276:2027-2030. [DOI] [PubMed] [Google Scholar]
- 44.Joint, I., K. Tait, M. E. Callow, J. A. Callow, D. Milton, P. Williams, and M. Camara. 2002. Cell-to-cell communication across the prokaryote-eukaryote boundary. Science 298:1207. [DOI] [PubMed] [Google Scholar]
- 45.Kaiser, D. 2003. Coupling cell movement to multicellular development in myxobacteria. Nat. Rev. Microbiol. 1:45-54. [DOI] [PubMed] [Google Scholar]
- 46.Kaiser, D. 2004. Signaling in myxobacteria. Annu. Rev. Microbiol. 58:75-98. [DOI] [PubMed] [Google Scholar]
- 47.Kaiser, D., and R. Welch. 2004. Dynamics of fruiting body morphogenesis. J. Bacteriol. 186:919-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khudyakov, I. Y., and J. W. Golden. 2004. Different functions of HetR, a master regulator of heterocyst differentiation in Anabaena sp. PCC 7120, can be separated by mutation. Proc. Natl. Acad. Sci. USA 101:16040-16045. (First published 1 November 2004; 10.1073/pnas.0405572101.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim, S. K., and D. Kaiser. 1990. C-factor: a cell-cell signaling protein required for fruiting body morphogenesis of M. xanthus. Cell 61:19-26. [DOI] [PubMed] [Google Scholar]
- 50.Kim, W., and M. G. Surette. 2004. Metabolic differentiation in actively swarming Salmonella. Mol. Microbiol. 54:702-714. [DOI] [PubMed] [Google Scholar]
- 51.Kodani, S., M. E. Hudson, M. C. Durrant, M. J. Buttner, J. R. Nodwell, and J. M. Willey. 2004. The SapB morphogen is a lantibiotic-like peptide derived from the product of the developmental gene ramS in Streptomyces coelicolor. Proc. Natl. Acad. Sci. USA 101:11448-11453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kristich, C. J., Y. H. Li, D. G. Cvitkovitch, and G. M. Dunny. 2004. Esp-independent biofilm formation by Enterococcus faecalis. J. Bacteriol. 186:154-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lazazzera, B. A., and A. D. Grossman. 1998. The ins and outs of peptide signaling. Trends Microbiol. 6:288-294. [DOI] [PubMed] [Google Scholar]
- 54.Lazazzera, B. A., J. M. Solomon, and A. D. Grossman. 1997. An exported peptide functions intracellularly to contribute to cell density signaling in Bacillus subtilis. Cell 89:917-925. [DOI] [PubMed] [Google Scholar]
- 55.Leadbetter, J. R., and E. P. Greenberg. 2000. Metabolism of acyl-homoserine lactone quorum-sensing signals by Variovorax paradoxus. J. Bacteriol. 182:6921-6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lenz, D. H., K. C. Mok, B. N. Lilley, R. V. Kulkarni, N. S. Wingreen, and B. L. Bassler. 2004. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118:69-82. [DOI] [PubMed] [Google Scholar]
- 57.Leonard, B. A., A. Podbielski, P. J. Hedberg, and G. M. Dunny. 1996. Enterococcus faecalis pheromone binding protein, PrgZ, recruits a chromosomal oligopeptide permease system to import sex pheromone cCF10 for induction of conjugation. Proc. Natl. Acad. Sci. USA 93:260-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lilley, B. N., and B. L. Bassler. 2000. Regulation of quorum sensing in Vibrio harveyi by LuxO and sigma-54. Mol. Microbiol. 36:940-954. [DOI] [PubMed] [Google Scholar]
- 59.Lobedanz, S., and L. Søgaard-Andersen. 2003. Identification of the C-signal, a contact-dependent morphogen coordinating multiple developmental responses in Myxococcus xanthus. Genes Dev. 17:2151-2161. (First published 15 August 2003; 10.1101/gad.274203.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lupp, C., and E. G. Ruby. 2004. Vibrio fischeri LuxS and AinS: comparative study of two signal synthases. J. Bacteriol. 186:3873-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lupp, C., M. Urbanowski, E. P. Greenberg, and E. G. Ruby. 2003. The Vibrio fischeri quorum-sensing systems ain and lux sequentially induce luminescence gene expression and are important for persistence in the squid host. Mol. Microbiol. 50:319-331. [DOI] [PubMed] [Google Scholar]
- 62.Ma, H., and K. Kendall. 1994. Cloning and analysis of a gene cluster from Streptomyces coelicolor that causes accelerated aerial mycelium formation in Streptomyces lividans. J. Bacteriol. 176:3800-3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martens, E. C., K. Heungens, and H. Goodrich-Blair. 2003. Early colonization events in the mutualistic association between Steinernema carpocapsae nematodes and Xenorhabdus nematophila bacteria. J. Bacteriol. 185:3147-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mathesius, U., S. Mulders, M. Gao, M. Teplitski, G. Caetano-Anolles, B. G. Rolfe, and W. D. Bauer. 2003. Extensive and specific responses of a eukaryote to bacterial quorum-sensing signals. Proc. Natl. Acad. Sci. USA 100:1444-1449. (First published 2 January 2003; 10.1073/pnas.262672599.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miller, S. T., K. B. Xavier, S. R. Campagna, M. E. Taga, M. F. Semmelhack, B. L. Bassler, and F. M. Hughson. 2004. Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol. Cell 15:677-687. [DOI] [PubMed] [Google Scholar]
- 66.Minogue, T. D., M. Wehland-von Trebra, F. Bernhard, and S. Beck von Bodman. 2002. The autoregulatory role of EsaR, a quorum sensing regulator in Pantoea stewartii subsp. stewartii: evidence for a repressor function. Mol. Microbiol. 44:1625-1635. [DOI] [PubMed] [Google Scholar]
- 66a.Minogue, T. D., A. L. Carlier, M. D. Koutsoudis, and S. B. von Bodman. 2005. The cell density-dependent expression of stewartan exopolysaccharide in Pantoea stewartii spp. stewartii is a function of EsaR-mediated repression of the rcsA gene. Mol. Microbiol. 56:189-203. [DOI] [PubMed] [Google Scholar]
- 67.Mok, K. C., N. S. Wingreen, and B. L. Bassler. 2003. Vibrio harveyi quorum sensing: a coincidence detector for two autoinducers controls gene expression. EMBO J. 22:870-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morrison, D. A., and M. S. Lee. 2000. Regulation of competence for genetic transformation in Streptococcus pneumoniae: a link between quorum sensing and DNA processing genes. Res. Microbiol. 151:445-451. [DOI] [PubMed] [Google Scholar]
- 69.Nakayama, J., Y. Cao, T. Horii, S. Sakuda, A. D. Akkermans, W. M. de Vos, and H. Nagasawa. 2001. Gelatinase biosynthesis-activating pheromone: a peptide lactone that mediates a quorum sensing in Enterococcus faecalis. Mol. Microbiol. 41:145-154. [DOI] [PubMed] [Google Scholar]
- 70.Nealson, K. H. 1977. Autoinduction of bacterial luciferase: occurrence, mechanism, and significance. Arch. Microbiol. 112:73-79. [DOI] [PubMed] [Google Scholar]
- 71.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429-1449. [DOI] [PubMed] [Google Scholar]
- 72.Novick, R. P. 1999. Regulation of pathogenicity in Staphylococcus aureus by a peptide-based density-sensing system, p. 129-146. In G. M. Dunny and S. C. Winans (ed.), Cell-cell signaling in bacteria. ASM Press, Washington, D.C.
- 73.O'Connor, T. J., P. Kanellis, and J. R. Nodwell. 2002. The ramC gene is required for morphogenesis in Streptomyces coelicolor and expressed in a cell type-specific manner under the direct control of RamR. Mol. Microbiol. 45:45-57. [DOI] [PubMed] [Google Scholar]
- 74.Pearcy, M., S. Aron, C. Doums, and L. Keller. 2004. Conditional use of sex and parthenogenesis for worker and queen production in ants. Science 306:1780-1783. [DOI] [PubMed] [Google Scholar]
- 75.Pesci, E. C., J. B. J. Milbank, J. P. Pearson, S. McKnight, A. S. Kende, E. P. Greenberg, and B. H. Iglewski. 1999. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:11229-11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Poplawsky, A. R., and W. Chun. 1997. pigB determines a diffusible factor needed for extracellular polysaccharide slime and xanthomonadin production in Xanthomonas campestris pv. campestris. J. Bacteriol. 179:439-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pottathil, M., and B. A. Lazazzera. 2003. The extracellular Phr peptide-Rap phosphatase signaling circuit of Bacillus subtilis. Front. Biosci. 8:d32-d45. [DOI] [PubMed] [Google Scholar]
- 78.Redfield, R. J. 2002. Is quorum sensing a side effect of diffusion sensing? Trends Microbiol. 10:365-370. [DOI] [PubMed] [Google Scholar]
- 79.Sahl, H. G., and G. Bierbaum. 1998. Lantibiotics: biosynthesis and biological activities of uniquely modified peptides from gram-positive bacteria. Annu. Rev. Microbiol. 52:41-79. [DOI] [PubMed] [Google Scholar]
- 80.Schauder, S., K. Shokat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 81.Schuster, M., C. P. Lostroh, T. Ogi, and E. P. Greenberg. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185:2066-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schuster, M., M. L. Urbanowski, and E. P. Greenberg. 2004. Promoter specificity in Pseudomonas aeruginosa quorum sensing revealed by DNA binding of purified LasR. Proc. Natl. Acad. Sci. USA 101:15833-15839. (First published 25 October 2004; 10.1073/pnas.0407229101.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 84.Smith, J. N., and B. M. Ahmer. 2003. Detection of other microbial species by Salmonella: expression of the SdiA regulon. J. Bacteriol. 185:1357-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Smith, R. S., and B. H. Iglewski. 2003. P. aeruginosa quorum-sensing systems and virulence. Curr. Opin. Microbiol. 6:56-60. [DOI] [PubMed] [Google Scholar]
- 86.Solomon, J. M., B. A. Lazazzera, and A. D. Grossman. 1996. Purification and characterization of an extracellular peptide factor that affects two different developmental pathways in B. subtilis. Genes Dev. 10:2014-2024. [DOI] [PubMed] [Google Scholar]
- 87.Sturgill, G., and P. N. Rather. 2004. Evidence that putrescine acts as an extracellular signal required for swarming in Proteus mirabilis. Mol. Microbiol. 51:437-446. [DOI] [PubMed] [Google Scholar]
- 88.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Swartzman, E., M. Silverman, and E. A. Meighen. 1992. The luxR gene product of Vibrio harveyi is a transcriptional activator of the lux promoter. J. Bacteriol. 174:7490-7493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Takano, E., R. Chakraburtty, T. Nihira, Y. Yamada, and M. J. Bibb. 2001. A complex role for the gamma-butyrolactone SCB1 in regulating antibiotic production in Streptomyces coelicolor A3(2). Mol. Microbiol. 41:1015-1028. [DOI] [PubMed] [Google Scholar]
- 91.Takano, E., H. Kinoshita, V. Mersinias, G. Bucca, G. Hotchkiss, T. Nihira, C. P. Smith, M. Bibb, W. Wohlleben, and K. Chater. 2005. A bacterial hormone (the SCB1) directly controls the expression of a pathway-specific regulatory gene in the cryptic type I polyketide biosynthetic gene cluster of Streptomyces coelicolor. Mol. Microbiol. 56:465-479. [DOI] [PubMed] [Google Scholar]
- 92.Tateda, K., Y. Ishii, M. Horikawa, T. Matsumoto, S. Miyairi, J. C. Pechere, T. J. Standiford, M. Ishiguro, and K. Yamaguchi. 2003. The Pseudomonas aeruginosa autoinducer N-3-oxododecanoyl homoserine lactone accelerates apoptosis in macrophages and neutrophils. Infect. Immun. 71:5785-5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Telford, G., D. Wheeler, P. Williams, P. T. Tomkins, P. Appleby, H. Sewell, G. S. A. B. Stewart, B. W. Bycroft, and D. I. Pritchard. 1998. The Pseudomonas aeruginosa quorum-sensing signal molecule N-(3-oxododecanoyl)-l-homoserine lactone has immunomodulatory activity. Infect. Immun. 66:36-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Teplitski, M., J. B. Robinson, and W. D. Bauer. 2000. Plants secrete substances that mimic bacterial N-acyl homoserine lactone signal activities and affect population density-dependent behaviors in associated bacteria. Mol. Plant-Microbe Interact. 13:637-648. [DOI] [PubMed] [Google Scholar]
- 95.Tillotson, R. D., H. A. Wosten, M. Richter, and J. M. Willey. 1998. A surface active protein involved in aerial hyphae formation in the filamentous fungus Schizophillum commune restores the capacity of a bald mutant of the filamentous bacterium Streptomyces coelicolor to erect aerial structures. Mol. Microbiol. 30:595-602. [DOI] [PubMed] [Google Scholar]
- 96.Vannini, A., C. Volpari, C. Gargiola, E. Muraglia, R. Cortese, R. De Francesco, P. Nedderman, and S. Di Marco. 2002. The crystal structure of the quorum-sensing protein TraR bound to its autoinducer and target DNA. EMBO J. 21:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Visick, K. L., J. Foster, J. Doino, M. McFall-Ngai, and E. G. Ruby. 2000. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J. Bacteriol. 182:4578-4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Visick, K. L., and L. M. Skoufos. 2001. Two-component sensor required for normal symbiotic colonization of Euprymna scolopes by Vibrio fischeri. J. Bacteriol. 183:835-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wagner, V. E., D. Bushnell, L. Passador, A. I. Brooks, and B. H. Iglewski. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185:2080-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang, L. H., Y. He, Y. Gao, J. E. Wu, Y. H. Dong, C. He, S. X. Wang, L. X. Weng, J. L. Xu, L. Tay, R. X. Fang, and L. H. Zhang. 2004. A bacterial cell-cell communication signal with cross-kingdom structural analogues. Mol. Microbiol. 51:903-912. [DOI] [PubMed] [Google Scholar]
- 101.Waters, C. M., M. H. Antiporta, B. E. Murray, and G. M. Dunny. 2003. Role of the Enterococcus faecalis GelE protease in determination of cellular chain length, supernatant pheromone levels, and degradation of fibrin and misfolded surface proteins. J. Bacteriol. 185:3613-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Watson, W. T., T. D. Minogue, D. L. Val, S. Beck von Bodman, and M. E. A. Churchill. 2002. Structural basis and specificity of acyl-homoserine lactone signal production in bacterial quorum sensing. Mol. Cell 9:1-20. [DOI] [PubMed] [Google Scholar]
- 103.Welch, M., D. E. Todd, N. A. Whitehead, S. J. McGowan, B. W. Bycroft, and G. P. C. Salmond. 2000. N-Acyl homoserine lactone binding to the CarR receptor determines quorum-sensing specificity in Erwinia. EMBO J. 19:631-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.White, C. E., and S. C. Winans. 2005. Identification of amino acid residues of the Agrobacterium tumefaciens quorum-sensing regulator TraR that are critical for positive control of transcription. Mol. Microbiol. 55:1473-1486. [DOI] [PubMed] [Google Scholar]
- 105.Willey, J. M., R. Santamaria, J. Guijarro, M. Geistlich, and R. Losick. 1991. Extracellular complementation of a developmental mutation implicates a small sporulation protein in aerial mycelium formation by Streptomyces coelicolor. Cell 65:641-650. [DOI] [PubMed] [Google Scholar]
- 106.Yoon, H.-S., and J. W. Golden. 1998. Heterocyst pattern formation controlled by a diffusible peptide. Science 282:935-938. [DOI] [PubMed] [Google Scholar]
- 107.Zhang, L. H., and Y. H. Dong. 2004. Quorum sensing and signal interference: diverse implications. Mol. Microbiol. 53:1563-1571. [DOI] [PubMed] [Google Scholar]
- 108.Zhang, R. G., T. Pappas, J. L. Brace, P. C. Miller, T. Oulmassov, J. M. Molyneaux, J. C. Anderson, J. K. Bashkin, S. C. Winans, and A. Joachimiak. 2002. Structure of a bacterial quorum-sensing transcription factor complexed with autoinducer-type pheromone and DNA. Nature 417:971-974. [DOI] [PubMed] [Google Scholar]
- 109.Zhu, J., and S. C. Winans. 1999. Autoinducer binding by the quorum-sensing regulator TraR increases affinity for target promoters in vitro and decreases TraR turnover rates in whole cells. Proc. Natl. Acad. Sci. USA 96:4832-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhu, J., and S. C. Winans. 2001. The quorum-sensing transcriptional regulator TraR requires its cognate signaling ligand for protein folding, protease resistance, and dimerization. Proc. Natl. Acad. Sci. USA 98:1507-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]