Abstract
The first step in the transfer of the Bacteroides conjugative transposon CTnDOT is excision of the integrated element from the chromosome to form a circular transfer intermediate. Excision occurs only after the bacteria are exposed to tetracycline. Previously, four excision genes were identified. One was the integrase gene intDOT, which appeared to be expressed constitutively. Three other genes essential for excision (orf2c, orf2d, and exc) were found located in a cluster 13 kbp downstream of intDOT. By using uidA fusions and real-time reverse transcriptase PCR, we demonstrate here that the excision genes orf2c, orf2d, and exc are part of an operon that also contains open reading frame orf3, previously shown not to be essential for excision. We also show that operon expression is regulated at the transcriptional level in response to tetracycline. The transcript start site for the operon has been localized. Three CTnDOT regulatory genes are thought to be involved in tetracycline regulation of excision, rteA, rteB, and rteC. By placing rteC under the control of a heterologous promoter, we found that RteC alone was sufficient for induction of the orf2c operon. If, however, the rteC gene was under the control of its own promoter, it was not able to induce orf2c operon expression unless rteA and rteB were present. Thus, RteA and RteB participate in excision by stimulating transcription of rteC. Using electrophoretic mobility shift analysis, we found that a purified His6-tagged form of RteC bound DNA upstream of the −33 region of the promoter. Changing the sequence in the region between bp −50 and −70 reduced the expression of the orf2c operon in vivo. Taken together, our results support the hypothesis that RteC acts as a DNA-binding protein that binds upstream of the orf2c promoter and is responsible for tetracycline-regulated transcriptional regulation of the orf2c operon.
Conjugative transposons (CTns) related to the Bacteroides CTn CTnDOT have been found in a number of Bacteorides species (22, 26). Members of CTnDOT family appear to be contributing significantly to transfer of antibiotic resistance genes among Bacteroides species (22, 26, 40). Transfer of CTnDOT occurs in three steps: excision from the chromosome to form a double-stranded circular intermediate, conjugative transfer of the circular intermediate to the recipient, and integration of the transferred circular form into the recipient's chromosome (40).
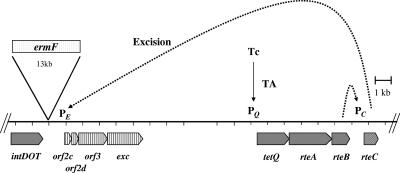
Excision is stimulated by tetracycline (7, 8, 30). In fact, no excision is detected unless the cells carrying CTnDOT are first exposed to tetracycline (8, 30). A previous study identified four genes that were essential for excision (8, 30). One was the integrase gene intDOT, which is located at one end of the CTn. The other genes were located in a cluster 13 kbp downstream of intDOT (Fig. 1). Single-crossover disruptions and deletions in three of these genes, orf2c, orf2d, and exc, abolished excision. A fourth gene located in this cluster, orf3, could be deleted without affecting excision. We report here that genes in the orf2c cluster are organized in an operon and are regulated at the transcriptional level.
FIG. 1.
Model for the regulation of the excision of CTnDOT. The genes important for the excision of CTnDOT are shown. The 13-kbp ermF region present in CTnDOT is indicated by the bar labeled ermF. The dashed lines indicate the hypothetical regulatory steps that are proven in this study. The intDOT gene, which is required both for integration and excision, is expressed constitutively, but expression of the orf2c-2d-orf3-exc operon is regulated. Growth of the cells in tetracycline stimulates the production of TetQ and RteA-RteB, a process that is regulated by translational attenuation (TA) (37), shown by the solid arrow. RteB activates the transcription of rteC, and the RteC protein then activates the transcription of the orf2c operon. IntDOT plus products from the orf2c operon interact to cause the excision of the CTn.
Expression of intDOT appeared to be constitutive because integration occurs with equal frequency in the absence and presence of tetracycline (7, 30, 35). Thus, the genes responsible for regulated excision were presumably in the orf2c gene cluster, but nothing was known about the regulation of these genes. Previous studies had identified three genes that might control the genes responsible for regulated excision of CTnDOT (28, 29). These genes were rteA, rteB, and rteC (Fig. 1). RteA and RteB had been identified tentatively as regulatory proteins because they had significant amino acid similarity to members of known two component regulatory systems, with RteA being the sensor component and RteB being the transcriptional regulator (Fig. 1) (29). By contrast, the amino acid sequence of RteC (accession no. AAA22922) did not exhibit significant similarity to any proteins in the databases. In particular, RteC did not have the helix-turn-helix motif that is found in many DNA binding proteins. The reason for thinking that rteC might be a regulatory gene was that a disruption in rteC abolished excision and transfer of CTnDOT.
Although RteA and RteB resemble regulatory proteins at the amino acid sequence level, the tetQ-rteA-rteB operon is not controlled by transcriptional activation and regulation of this operon does not require either RteA or RteB. Rather, exposure of cells to tetracycline brings into play a translational attenuation mechanism involving a leader region at the 5′ end of the operon. Presumably the rate of ribosome movement along the mRNA, which is influenced by tetracycline, is responsible for the tetracycline-induced increase in production of TetQ, RteA, and RteB (37). Since RteA and RteB do not control the expression of the tetQ-rteA-rteB operon at the transcriptional level, their function might be to control the expression of the downstream gene, rteC. In this report, we provide the first evidence that RteC is responsible for controlling the expression of genes in the orf2c operon and that expression of rteC itself is controlled by RteA and RteB.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used in this study are listed in Table 1. Bacteroides sp. strain BT4001ΩQABC contains a single copy of the central regulatory region of CTnDOT, tetQ-rteA-rteB-rteC, in the chromosome of BT4001 (39). BT4001ΩQAB has a single copy of tetQ-rteA-rteB without rteC (39). Bacteroides strains were grown in chopped meat (Remel) and then transferred to TYG (Trypticase-yeast extract-glucose) medium containing tetracycline (1 μg/ml; induced) or no tetracycline (uninduced) (12, 37). Cells were grown overnight. Previous experience has shown that cells in late exponential phase or in stationary phase exhibited the highest excision levels. Antibiotic concentrations (in micrograms per milliliter) were as follows: ampicillin, 100; cefoxitin, 20; chloramphenicol, 10; erythromycin, 10; gentamicin, 200; tetracycline, 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant phenotypea | Description (reference) |
|---|---|---|
| E. coli | ||
| DH5α MCR | recA | Gibco BRL |
| HB101 (RP1) | recA Strr | HB101 containing IncPα plasmid RP1 (25) |
| BL21 (DE3) | F−gal dcm (DE3) | E. coli with the bacteriophage T7 promoter-based expression system that carries the lambda DE3 lysogen (Invitrogen) (31) |
| B. thetaiotamicron 5482A | ||
| BT4001 | Rifr | Spontaneous rifampin mutant of B. thetaiotamicron 5482A (24) |
| BT4007 | Rifr Tcr Emr | B. thetaiotaomicron 4001 that contains wild-type CTnDOT |
| BT4001ΩQAB | Rifr Tcr | B. thetaiotamicron BT4001 that carries the CTnDOT region comprising tetQ, rteA, and rteB inserted into the chromosome via an NBU 2 minielement (40) |
| BT4001ΩQABC | Rifr Tcr | B. thetaiotamicron BT4001 that carries the CTnDOT region comprising tetQ, rteA, rteB, and rteC inserted into the chromosome via an NBU1 minielement (40) |
| BT4104ΩRDB1 | Thy− Tpr Tcr Emr ΩrteA | Chromosomal disruption of rteA in BT4104 (16) |
| Plasmids | ||
| pMJF2 | Apr (Emr) | A cloning vector to create a uidA fusion, also an E. coli-Bacteroides shuttle vector (11) |
| pLYL05 | Apr (Cefr) | An E. coli-Bacteroides shuttle vector containing cfxA (27, 34) |
| pLYL02 | Apr (Emr) | Same as pMJF2 with the uidA gene in opposite orientation to facilitate cloning (this report) |
| pC-COW | Apr Tcr Cmr (Cmr) | An E. coli-Bacteroides shuttle vector with IS4351-cat and Bacteroides plasmid pB8-51 that is compatible with pMJF2-based vectors (37) |
| pCR2.1 | Apr | A 3.9-kb cloning vector for PCR products (Invitrogen) |
| pYS32 | Apr (Emr) | A 2.9-kb HindIII fragment of pYS33 that contains uidA fused to the 1.0-kb promoter region of the orf2c operon cloned into pC-COW (33) |
| pGFK1 | Apr (Emr) | 1.2-kb BspEI-XmnI fragment of region of CTnDOT cloned into pLY02 to generate the fusion of uidA to the 188-bp N-terminal coding sequence of orf3 and its 1.0-kb upstream sequence (this study) |
| pGFK10 | Apr (Emr) | 2.7-kb SphI-SmaI PCR product containing 300 bp upstream of orf2c start codon cloned into the SphI-SmaI sites of pMJF2 (this study) |
| pGFK34 | Apr (Emr) | 0.3-kb SphI-SmaI PCR product containing 300 bp upstream of orf2c start codon cloned into the SphI-SmaI sites of pMJF2 (this study) |
| pGFK35 | Apr (Emr) | Plasmid containing a mutated sequence (AAAC) in the putative −7 region from pGFK34 and cloned into the SphI-SmaI site of pMJF2 (this study) |
| pGFK36 | Apr (Emr) | A plasmid containing a mutated sequence (TTTC) in the putative −7 region from pGFK34 and cloned into the SphI-SmaI site of pMJF2 (this study) |
| pGFK38 | Apr (Emr) | A plasmid containing a mutated sequence (TATG) in the putative −7 region from pGFK34 and cloned into the SphI-SmaI site of pMJF2 (this study) |
| pGFK43 | Apr (Cmr) | A plasmid containing a 20-bp mutated sequence between bp-70 and -50 upstream of the orf2c transcriptional start site from pGFK34 and cloned into the SphI-SmaI site of pMJF2 (this study) |
| PGFK63 | Apr (Emr) | 0.3-kb SphI-SmaI PCR product containing 300 bp upstream of orf2c start codon cloned into the SphI-SmaI site of pMJF2 (this study) |
| pGFK65 (pPc-rteC) | Apr (Cefr) | A clone containing rteC with its own promoter cloned into pLYL05 (this study) |
| pGFK59.3 (pPQ) | Apr (Cefr) | 0.2-kb SphI-SstI PCR product containing the tetQ promoter with an NcoI site cloned into SphI-SstI pLYL05 (this study) |
| pGFK67 (pPQ-rteC) | Apr (Cefr) | 1.2-kb NcoI-SstI PCR product containing the rteC coding region plus the tetQ promoter region cloned into the NcoI-SstI site on pGFK59.3 to produce an in-frame fusion of rteC to the tetQ promoter (this study) |
| pGFK69 (pPQ-rteC; His6 tagged) | Apr (Cefr) | 1.2-kb NcoI-SmaI PCR product containing a His6 tag on the C-terminal end of RteC cloned into the NcoI-SmaI site of pGFK59.3 (this study) |
| pET27b | Knr | A plasmid for the overexpression of His6-tagged proteins in E. coli (Novagen) |
| pGFK90.1 | Knr | 0.7-kb NcoI-XhoI rteC coding region cloned into the NcoI-XhoI site of pET27b (this study) |
Phenotypes in parentheses are expressed only in Bacteroides, and phenotypes outside parentheses are expressed in E. coli. Abbreviations: Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Emr, erythromycin resistance; Rifr, rifampin resistance; Tcr, tetracycline resistance; Tpr, trimethoprim resistance, Knr, kanamycin resistance; Thy−, thymidine auxotroph.
Construction of transcriptional and translational fusions.
To generate transcriptional fusions, two different restriction fragments were fused to the uidA reporter gene (11, 13, 21). pGFK1 was constructed with a 1.3-kb BspEI-XmnI fragment, which starts upstream of orf2c and continues into orf3, cloned into the uidA vector pLYL02 (this laboratory). pYS32 was constructed with a 1.2-kb HindIII-XmnI fragment cloned into pLYL02. These clones were confirmed to be transcriptional fusions by DNA sequence analysis. To generate the translation fusions, promoter region fragments were amplified by PCR. Genomic DNA from Bacteroides thetaiotaomicron BT4007, a strain that contains CTnDOT, was used as a template, and the Pfu (Stratagene) polymerase was used for high-fidelity amplification. The forward primer contained a SphI restriction enzyme site, and the reverse primer contained a SmaI site so that the cloned DNA created an in-frame fusion with the methionine codon of the gene to which the fusion was made. The PCR products were cloned into pCR2.1 (Invitrogen). After a colony with the correct insert was obtained, plasmid DNA was isolated and digested with SmaI and SphI to check the sizes of the various fragments.
The SphI-SmaI fragment from each pCR2.1 clone was then isolated and ligated into the SphI-SmaI site of the reporter vector pMJF2 (11). pMJF2 was used instead of pLYL02 to generate these fusions because of ease of cloning. pMJF2, like pLYL02, contains a promoterless uidA gene, but the orientation of the gene with respect to the multiple cloning region was reversed (Table 1). Each construct was sequenced, and the plasmids were transformed into Escherichia coli MCR. They were transferred into Bacteroides recipients by conjugation. DNA sequencing was performed by the University of Illinois Biotechnology Genetic Engineering Facility with an Applied Biosystems model 373A, version 2.0.1A, automated dye terminator.
Triparental matings.
Transcriptional fusion and translational fusion clones were transferred into Bacteroides strains by triparental matings (25). The two donors were E. coli DH5α MCR, which contained either a transcriptional or a translational fusion clone, and HB101, which contained the IncP plasmid RP1. BT4001 Bacteroides strains were recipients. In some cases the recipient strain BT4001 did not contain any sequences from CTnDOT. In other cases, the recipient contained tetQ-rteA-rteB only (BT4001ΩQAB) or tetQ-rteA-rteB and rteC (BT4001ΩQABC). These last two strains were constructed to provide a single copy of the regulatory genes stably integrated in the host chromosome (39). Matings were done aerobically on nitrocellulose filters as previously described (25). For pMJF2-based clones, transconjugants were selected on TYG plates containing erythromycin (10 μg/ml) and gentamicin (200 μg/ml). In some experiments, plasmid pC-COW (37) or pLYL05 (27, 34) was used to introduce rteC into the recipients. For pC-COW-based clones, the transconjugants were selected on TYG agar plates containing chloramphenicol (20 μg/ml) and gentamicin (200 μg/ml). For pLYL05-based clones, the transconjugants were selected on plates containing cefoxitin (20 μg/ml) and gentamicin (200 μg/ml) (27).
Cloning of rteC under the control of its own promoter and under the control of a heterologous promoter.
To construct a plasmid carrying rteC under the control of its own promoter, a fragment of approximately 2 kb which contained the rteC gene plus 900 bp of upstream sequence and 500 bp of downstream sequence was amplified by PCR. BT4007 genomic DNA was used as a template, and the Pfu polymerase was used for high-fidelity amplification. The forward primer contained a PstI restriction enzyme site, and the reverse primer contained an SstI site. The PCR products were first cloned into pCR2.1. After a colony with the correct insert was obtained, plasmid DNA was isolated and digested with PstI and SstI. The isolated fragment was cloned into the PstI-SstI sites of pLYL05 to produce pPC-rteC, which contains rteC behind its own promoter.
To construct a copy of the rteC clone with a heterologous promoter, the tetQ promoter (PQ) was cloned upstream of the rteC coding region. The PQ promoter fragment contained approximately 230 bp upstream of the start codon of the PQ region. It was amplified by PCR to generate SphI restriction enzyme sites at one end of the amplicons and NcoI plus SstI restriction enzyme sites at the other end. The PCR amplicon was digested with SphI and SstI and then cloned into the SphI-SstI sites of pLYL05 to generate pGFK59. pGFK59 was digested with NcoI and SstI. A 1.2-kb DNA fragment containing the 670-bp rteC coding region and 500 bp of downstream sequence was amplified to generate an NcoI site that overlapped the start codon of rteC and an SstI site at the 3′ end. This fragment was digested with NcoI and SstI and then ligated into the NcoI-SstI sites of pGFK59 to produce intact RteC behind PQ (pGFK67).
Construction and testing of a His6-tagged rteC gene.
To generate a His6-tagged rteC clone, with the His6 tag at the C terminus of the RteC protein, the rteC coding region was amplified with the forward primer containing an NcoI site and the reverse primer containing the His6 tag and a SmaI site. To test whether the His6-tagged form of the protein was active in vivo, the PCR amplicon was digested with NcoI and SmaI and inserted into NcoI-SmaI sites of pGFK59 to generate pGFK69. All three of the rteC constructs were mobilized into BT4001, with or without ΩQAB. The abilities of these clones to induce the expression of an orf2c-uidA fusion clone were measured.
GUS assays.
The uidA reporter gene on pMJF2 and pLYL02 encodes an E. coli β-glucuronidase (GUS). GUS assays were done as described by Feldhaus et al. (11). One unit was defined as 0.01 A415 U per min at 37°C. Protein concentrations were determined by the method of Lowry et al. (11). All GUS activities reported in this study are the averages of activities measured in phosphate buffer from at least three different transconjugants.
Site-directed mutagenesis.
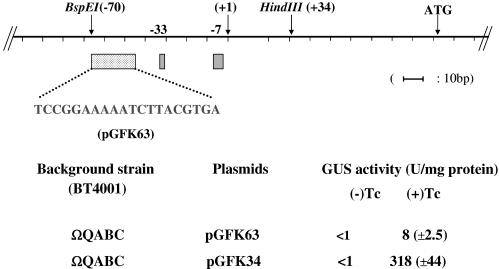
To determine if the putative −7 promoter sequence was in fact important for promoter activity, various clones in which single and multiple mutations had been made within the conserved GAnnTTTG motif (4) were constructed and fused with uidA. Using a QuikChange site-directed mutagenesis kit (Stratagene), mutations were created in the GAnnTTTG of the −7 motif. The mutations included GAnnAAAC (pGFK35), GAnnTTTC (pGFK36), and GAnnTATG (pGFK38). All mutations were confirmed by DNA sequence analysis. Clones were introduced into BT4001ΩABC. To determine if DNA upstream of the promoter consensus region centered on bp −33 was important, 20 base pairs between bp −50 and −70 were mutated (pGFK63).
RT-PCR analysis.
To determine whether orf2c, orf2d, orf3, and exc are part of the same operon, reverse transcriptase PCR (RT-PCR) analysis was done in which the primers amplified a segment of the mRNA extended from the 3′ end of one gene into the 5′ end of the next gene. To prepare the RNA samples, BT4007, which contains a single copy of CTnDOT in the chromosome, was grown either with or without tetracycline (10 ml). After cells were collected by centrifugation, 1 ml of TRIzol (Invitrogen) was used to extract total RNA from the samples (37). The samples were further extracted with phenol to remove proteins. Then, 2 volumes of absolute alcohol were added to each tube to precipitate RNA. After centrifugation at 13,000 rpm for 15 min, the RNA pellets were washed with 70% alcohol to remove salts. The pellets were dried at room temperature and dissolved in an RNA suspension solution (Ambion). After the optical density at 260 nm was measured to estimate total nucleic acid concentration, the samples were diluted to a concentration of 10 μg/20 μl, followed with DNase treatment to eliminate DNA in the samples. RT-PCR products were visualized on 2% agarose gels.
Real time RT-PCR analysis.
To determine more quantitatively whether expression of the orf2c operon is regulated at the transcriptional level, real-time RT-PCR was performed on RNA obtained from BT4007. From the purified RNA, cDNA was generated using 1 μg of RNA in a total volume of 20 μl plus random hexamers [d(N)6; New England Biolabs (NEB)] as primers and the Moloney murine leukemia virus reverse transcriptase (NEB). Bacteroides σ70 was used for the internal standard.
Real-time PCR was done using an iQcycler (Bio-Rad). Expression of the Bacteroides σ70 gene was used as an internal standard, and SYBR Green Supermix was used as a signal reporter. Reactions were done in a 96-well microtiter PCR plate using 1 μl of cDNA and (final concentrations) 0.4 μM sense and antisense primers for amplifying σ70, rteC, and exc; 3 μM MgCl2; and 1× iQ SYBR Green Supermix (Bio-Rad). Cycling conditions were as follows: denaturation (95°C for 3 min), amplification and quantification (95°C for 30 s, 50.1°C for 30 s, and 72°C for 30 s, with a single fluorescence measurement at both 53.7°C and 72°C for 30-s segments) repeated 40 times, a melting curve program (50 to 95°C with a heating rate of 0.1°C/s and continuous fluorescence measurement), and a cooling step to 50°C. For rteC and exc, the annealing temperatures were 53.7°C and 55.4°C, respectively. Each sample was tested in triplicate, and each experiment was repeated four times.
Data were analyzed using the iQcycler analysis software (Bio-Rad). Relative quantitation, which determines the changes in steady-state mRNA levels of a gene across multiple samples and provides a result relative to the levels of an internal control RNA, was used (36). The results were expressed as the difference (N) in the number of target gene copies relative to the number of σ70 gene copies and were determined from N = 2ΔΔCt = 2 (ΔCt target − ΔCt σ70 RNA), where ΔΔCt is ΔCt induced − ΔCt uninduced and ΔCt is the difference in threshold cycles for target and reference (3, 23).
RT-PCR was also used to assess the expression of rteC and the role of rteA and rteB in this regulation. RT-PCR analysis of rteC messages was done as described for the orf2c operon. Real-time RT-PCR analysis of rteC messages was done using RNA from cells containing CTnDOT that had been exposed or not exposed to tetracycline, as described for the orf2c operon. RT-PCR was also used to assess the effects of single-crossover disruptions in tetQ and rteA on the expression of rteC.
Primer extension.
Primer extension analysis was performed using the Promega primer extension system. The oligonucleotide primer 5′ TCC GTC AAT GAC CGA AAT ACG GAA CTT TCC A 3′ was complementary to nucleotides 17 to 48 of the orf2c gene coding region. The primer (10 pmol) was labeled with [γ-32P]dATP (3,000 Ci/mmol, 10 mCi/ml; Perkin-Elmer) (19, 20). Total RNA (40 μg) from cells induced or not induced by tetracycline (1 μg/ml) was precipitated with radioisotope-labeled primers. The pellet was air dried, suspended in primer extension buffer, and then incubated at 58°C for 1 h. After annealing primers to the mRNA, avian myeloblastosis virus reverse transcriptase (Promega) was added and the mixture was incubated at 42°C for 40 min. The extended labeled product was electrophoresed on an 8% polyacrylamide gel containing urea. A DNA sequencing ladder was prepared with a template encompassing the transcriptional start site region, using the same radiolabeled primer for the primer extension reaction. DNA sequencing was done by a sequence version 2.0 DNA sequencing kit (U.S. Biochemicals).
Overexpression and purification of RteC.
The promoterless His6-rteC was amplified from BT4007. The forward primer contained an NcoI site at the ATG start codon, and the reverse primer added the His6 immediately before the stop codon followed by an XhoI site. Phusion DNA polymerase (MJ Research) was used to obtain high-fidelity PCR amplification. The cycle conditions were (i) 30 s at 98°C; (ii) 30 cycles of 30 s at 98°C, 1 min at 58°C, and 1 min at 72°C; and (iii) final extension of 10 min at 72°C. The 670-bp PCR product was cloned into pCR2.1 and then isolated as an NcoI-XhoI fragment which was cloned into the NcoI-XhoI sites of pET27b to generate pGFK90.1.
Escherichia coli BL21(DE3) was used as the host strain for pGFK90.1. The His6-tagged protein was purified following the protocol provided by the QIAexpressionis kit (QIAGEN). Cells were grown overnight at 37°C in 12.5 ml Luria-Bertani (LB) medium with kanamycin (50 μg/ml). The overnight cultures were used to inoculate 250 ml of LB medium containing kanamycin (50 μg/ml). The culture was incubated at 37°C. When the optical density at 600 nm reached 0.6, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 1 mM. The cells were grown at 37°C for 4 h with vigorous shaking. Cells were harvested by centrifugation at 4,000 × g for 20 min at 4°C. After the supernatant was discarded, the cells were kept at −80°C until use. Overexpression of His6-tagged RteC protein was confirmed by Western blotting with Pentra-His monoclonal antibody as a primary antibody (QIAGEN) and anti-mouse horseradish peroxidase raised from sheep (Amersham) as a secondary antibody. Purification of RteC was carried out at 4°C using a Ni-nitrilotriacetic acid agarose matrix of the QIAexpressionis kit (QIAGEN). The protein was eluted with 50 mM Tris-HCl (pH 8.0), 1 mM EDTA, 50 mM NaCl, 250 mM imidazole, and 10% glycerol. The purified RteC was diluted to 1 μg/μl and stored at −80°C.
To test the activity of the His6-tagged RteC in Bacteroides, the same His6-tagged RteC construct was cloned into pLYL05 behind the tetQ promoter (PQ). This vector was transferred to BT4001 containing pGFK43 (an orf2c-uidA fusion) to test for GUS activity as mentioned above.
EMSA.
To examine if RteC binds to the promoter region of the orf2c operon, electromobility shift assays (EMSA) were performed using the promoter region of orf2. The reaction mixture contained 32P-labeled target DNA in 50 mM Tris-HCl (pH 8.0), 50 mM NaCl, 1 mM EDTA, 2.5 mg/ml of bovine serum albumin, and 10% glycerol with 0.75 μg/μl herring sperm DNA. Different concentrations of RteC were added to the reaction mixture, followed by incubation at room temperature for 10 min. The samples were subjected to electrophoresis on a 5% native polyacrylamide gel in 0.5× Tris-borate-EDTA at room temperature. Gels were dried on filter paper in a vacuum drier and exposed to X-ray film for 18 h.
RESULTS
GUS activities of transcriptional and translational fusions.
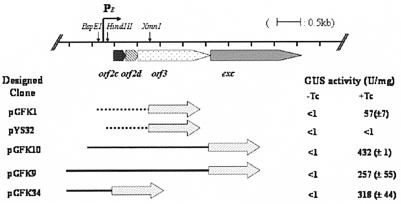
The results of GUS analysis of the various transcriptional and translational fusion clones constructed for this study are shown in Fig. 2. Cells containing pGFK1, a transcriptional fusion, exhibited an 80-fold increase in GUS activity when cells were exposed to tetracycline. Extracts from cells containing pYS32, which had a shorter upstream region, exhibited no GUS activity, even when exposed to tetracycline. Thus, sequences essential for promoter function are located between the HindIII and BspEI sites, 200 bp upstream of orf2c.
FIG. 2.
Fusion constructs and deletions in the promoter region of the orf2c excision gene cluster (PE) fused to uidA. At the top of the figure is a map of the orf2c operon and its promoter region (PE). The locations of important restriction sites are indicated. Below the map are various constructs containing the portions of the upstream region that were fused to uidA. The filled arrows show the location of the fusion point between the uidA gene and the promoter region segments. The constructs with dashed lines are transcriptional fusions and the constructs with solid lines are translational fusions to the uidA reporter gene. Each construct was transferred into BT4001ΩQABC, which contains the regulatory genes of CTnDOT integrated in the chromosome to measure the GUS activity. The GUS activities were determined in extracts from cells grown without (−Tc) or with (+Tc) tetracycline. The GUS activity is expressed as U/mg of protein. Each value is the average of at least two experiments done with three separate clones, and the calculated standard deviations are indicated in parentheses.
Translational fusions were also tested. pGFK9, which contains a 2.9-kb SphI-SmaI fragment spanning the 500-bp region, upstream of orf2c, exhibited approximately 640-fold-higher enzymatic activity in cells exposed to tetracycline than in its absence. Compared to the transcriptional fusions, the translational fusion increased the induced GUS activity by about sixfold. This increase in expression is probably due to the fact that these fusions have a native Bacteroides ribosome binding site rather than the region upstream of the E. coli uidA. Various amounts of DNA upstream of the start codon of orf2c were tested, but, except for the constructs that were deleted past the BspEI site, all of the fusions tested, including those shown in Fig. 2, had comparable activities. Combined results from these experiments indicate that a promoter is located upstream of the start codon of orf2c and that only about 200 bp is needed for the full expression. Also, orf2c, orf2d, orf3, and exc appear to be organized in an operon because fusions to them have similar expression patterns.
Direct test of whether orf2c, orf2d, orf3, and exc are in the same operon.
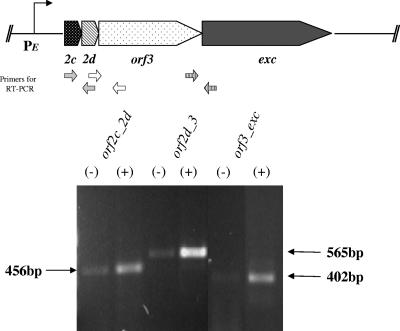
The results of the uidA fusion experiments suggested that orf2c, orf2d, orf3, and exc were regulated similarly and thus might share the same promoter. To test this hypothesis more directly, RT-PCR was performed to determine if there was, in fact, a single mRNA transcript. The results are shown in Fig. 3. The primers amplified transgene fragments in all cases, supporting the hypothesis that these genes are all in the same operon. Also, a tetracycline-associated increase in the level of message was seen in all cases.
FIG. 3.
RT-PCR analysis to determine whether the open reading frames in the orf2c gene cluster are part of an operon. The excision gene cluster is shown at the top. Abbreviations are as follows: 2c, orf2c; 2d, orf2d. The locations of the primers used for the RT-PCR analysis are shown below the map, with both primers in the same set given the same fill. The promoter region is indicated as PE. The mRNA was prepared from BT4007, which contains CTnDOT integrated in the chromosome, from cells grown in medium containing 1 μg/ml tetracycline (Tc) (+) or no Tc (−). Portions (5 μl) of the products of the RT-PCRs were electrophoresed on a 2% agarose gel. The sizes in bp of the products are indicated by arrows on either side of the gel. All products were of the expected sizes.
Real-time RT-PCR analysis.
Real-time RT-PCR analysis allows mRNA quantification. Its detection limit is 10- to 100-fold better than those of other methods of quantification such as RNA protection assays and Northern hybridizations (18). Real-time RT-PCR compares the message being quantified to an internal standard gene that is constitutively expressed (5). No such internal standard had been identified previously for any Bacteroides species. The most commonly used internal standard, the 16S rRNA gene, proved to be unsuitable in BT4001 because the expression of the 16SrRNA genes was much higher than that of the orf2c or rteC gene. Accordingly, we tried three other reference genes that appear to be expressed constitutively in Bacteroides: σ70, malR (a regulatory gene that controls an α-glucosidase gene) (10), and thyA (a gene for thymidylate synthase). All three genes provided comparable and reproducible standard curves with high PCR efficiency (90 to 100%), with and without tetracycline induction. σ70 was chosen as the internal standard for this study, however, because results using this standard were the most reproducible. At first, we were concerned that σ70 expression might be growth phase regulated, but cells harvested at various growth phases, including the late-exponential-phase and stationary-phase cells used in this study, did not exhibit any variation in expression (data not shown).
The correlation of the standard curves for both the target gene and the reference gene fell between 0.998 and 1.000, and the PCR efficiency for each set of the experiment was between 90 and 105%. We performed each experiment with two different amounts of the cDNA, 5 ng and 1 ng of total transcripts per reaction. The melt curve analysis showed that neither primer dimers nor nonspecific products were formed. Using this method, we calculated the induction of the exc mRNA to be 84-fold for 5 ng total RNA and 96-fold for 1 ng total RNA (Table 2). Thus, the induction estimated from the transcriptional GUS fusion data (approximately 80-fold, from 0.7 U/mg protein in the absence of tetracycline to 57 U/mg protein in the presence of tetracycline) and the induction calculated using RT-PCR were comparable.
TABLE 2.
Real-time RT-PCR quantitation of tetracycline-induced transcription of exc and rteC
| Target genea | Amt of total DNA (ng) | Fold Tc inductionb (+/−SD) |
|---|---|---|
| exc | 5 | 83 (+/−1.2) |
| 1 | 96 (+/−4.3) | |
| rteC | 5 | 6 (+/−1.2) |
| 1 | 6 (+/−1.4) |
The internal standard, a constitutively expressed single-copy gene, was the σ70 gene. In our experience, expression of this gene is independent both of tetracycline stimulation and growth phase.
Triplicate runs were analyzed in four independent experiments. Induction (N) was calculated by the following formula: N = 2ΔΔCt = 2(ΔCt target − ΔCt σ70 RNA), where ΔΔCt is ΔCt target − ΔCt σ70 RNA and ΔCt is the difference in threshold cycles for the target and the σ70 reference.
Location of the transcriptional start site.
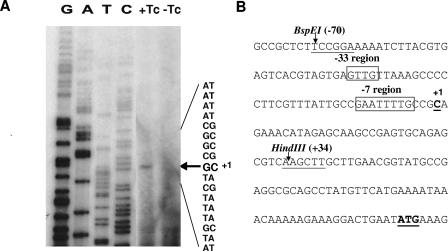
To localize the promoter region more precisely, primer extension analysis was used to determine the location of the 5′ end of the message. An oligonucleotide primer was used that was complementary to nucleotides 17 to 48 from the start codon of orf2c. The results shown in Fig. 4 indicated that the transcriptional start site was the C located 7 bp downstream of a putative −7 promoter region identified by comparison to the consensus sequence derived for Bacteroides (4).
FIG. 4.
Mapping of the transcriptional start site of the orf2c operon. The results of the primer extension analysis for the PE promoter are shown in lanes +Tc and-Tc in panel A. The cells containing CTnDOT were grown overnight with (+) or without (−) tetracycline (Tc), and 40 μg of total RNA was used for the primer extension analysis. The sequencing ladder is to the left of the primer extension lanes. On the right side, the sequences of both strands of the DNA are provided. The arrow indicates the transcriptional start site. In panel B the sequence of the entire promoter region is given, from the BspEI site to the ATG start of orf2c. The −7 and −33 consensus sequence motifs identified by comparison to the consensus Bacteroides promoter sequence motifs determined by Bayley et al. (4) are boxed and labeled above the line. The bp numbers are relative to the transcriptional start site (C), which is labeled +1. The HindIII site shown to be important in the cloning of a functional promoter region is shown at bp +34.
Bayley et al. (4), by comparing promoter regions from a number of Bacteroides genes, had previously suggested that Bacteroides consensus promoter regions are centered at −7 and −33 from the transcript start site of the genes for which this start site had been determined. They found that the −7 consensus sequence was essential for expression but that the −33 consensus was less important. Similar sequences were found at the expected distances from the orf2c operon transcription start site. To confirm that the predicted −7 region for orf2c was essential for expression, we constructed various clones in which single and multiple base pairs within the conserved −7 (GAnnTTTG) motif were mutated (4). These included GAnnAAAC (pGFK35), GAnnTTTC (pGFK36), and GAnnTATG (pGFK38). These clones were tested for GUS activity in the BT4001ΩABC strain. The GUS activity for each of these mutated sequences was reduced to basal level (<1 U/mg protein). This result and the location of the TTTG sequence relative to the transcription start site were consistent with the −7 region being an essential part of the promoter.
We suspected that RteC might be acting as an activator of orf2c operon expression because disruption of rteC abolished excision. As a test for the hypothesis that a region upstream of the −33 consensus sequence might be a binding site for an activator, possibly RteC, we did additional site-directed mutagenesis by changing multiple base pairs upstream of the putative −33 region. This region was identified on the basis of studies of other promoters that showed that the −33 region (or −35 region in E. coli) (17) could extend up to −50, especially when enhancer sequences called UP sequences are included. The size of the largest activator binding site is about 20 bp, so the 20 bp between −50 and −70 was changed. The changes were to complementary sequences rather than random sequences to preserve the %G+C composition. When all 20 bp (between −50 and −70) were mutated, the GUS activity in the absence of tetracycline stimulation was 0.3 U/mg protein and the tetracycline-induced level was reduced to 8 U/mg protein, a 27-fold reduction in the induced GUS specific activity compared to the positive control (pGFK34; Fig. 5). This result suggested that an activator binding site may be located in the −50 to −70 region.
FIG. 5.
Site-directed mutagenesis of a 20-bp region upstream of the −33 region of PE. A map indicating the location of the −33 and −7 consensus regions and the transcriptional start site (+1) is shown at the top. Two important restriction sites are indicated, as is the ATG start of orf2c. The sequence of the 20-bp region upstream of the −33 (bp −50 to bp −70) was altered by site-directed mutagenesis to a complementary sequence that preserved the spacing and G+C% composition of the region. The wild-type sequence cloned into pGFK34 was used as the positive control. pGFK63 contained the mutated sequence cloned into same vector. GUS activity was measured in extracts from cells containing each of the two vectors. The values given are for two separate assays done on three individual isolates. Standard deviations are given in parentheses.
Requirement for RteC.
The next question to be answered was whether RteC was indeed the activator protein. To confirm that RteC is essential for expression of the orf2c operon and that other regulatory proteins on CTnDOT were not involved, four plasmids carrying orf2c-uidA fusions (pGFK9, pGFK10, pGFK11, and pGFK34) were introduced separately into BT4001ΩQAB or BT4001ΩABC and GUS activity was measured. If RteC was required, only the clones in the latter strain should give tetracycline-induced GUS activity. The BT4001ΩQAB strain did not support induced expression of GUS from the plasmid unless a plasmid carrying wild-type rteC was added into this strain (Table 3, lines 1 and 3). These results showed that RteC is necessary for expression of the operon but did not indicate whether RteC, without RteA and RteB, could control operon expression.
TABLE 3.
Effects of RteA/RteB and/or RteC on the GUS activity of the PE-uidA fusion
| BT4001a (pPE-uidA fusion) | + rteC plasmidb | GUS activityc (U/mg of protein)
|
|
|---|---|---|---|
| −Tc | +Tc | ||
| ΩQAB | 1.4 | 1 | |
| ΩQABC | 1 | 250 (+/−42) | |
| ΩQAB | pPc-rteC | <1 | 438 (+/−86) |
| ΩQAB | pPq-rteC | 447 (+/−38) | 517 (+/−70) |
| pPc-rteC | <1 | NAd | |
| pPq-rteC | 668 (+/−76) | NA | |
| pPq-rteC (His6 tagged) | 429 (+/−46) | NA | |
BT4001 strain containing the pPE-uidA fusion vector with and without a copy of tetQ-rteA-rteB (ΩQAB) or tetQ-rteA-rteB + rteC (ΩQABC) integrated in the chromosome.
BT4001 strain in column 1 with a plasmid carrying rteC controlled by its native promoter (pPc-rteC), rteC controlled by the tetQ promoter (pPq-rteC), or the His6-tagged form of rteC controlled by the Pq promoter.
See Materials and Methods for details. Values are the averages of two separate assays of four independent clones, uninduced (−Tc) or induced with 1 μg/ml tetracycline (+Tc).
NA, nonapplicable because there is no tetracycline resistance gene (tetQ) in the host strain.
The expression of rteC itself was first examined by real-time RT-PCR analysis. Initially, this analysis was done using RNA from BT4007, a derivative of BT4001 that contained CTnDOT. Results of this analysis indicated that expression of rteC was much less affected by tetracycline induction than expression of the orf2c genes (Table 2). The results in Table 2 showed that the induction of rteC was only sixfold, regardless of whether 1 ng or 5 ng of total RNA was the template.
The strain in which the RT-PCR analysis was done (BT4007) contained rteA and rteB as well as rteC. To determine whether RteC alone could support expression of the pGFK67 translational orf2c fusion, the rteC gene with its own promoter, pPC-rteC, was introduced into BT4001 (no copy of CTnDOT) along with the fusion clone. Given the relatively high level of uninduced expression of rteC, it seemed possible that a plasmid carrying this gene (copy number of 5 to 8 per cell) might allow expression of the orf2c fusion, but no GUS activity was detected (<1 U/mg protein; Table 3, line 5). Since this result could be explained if RteA and RteB are necessary for expression of rteC, we placed a heterologous promoter, the promoter of the tetQ operon (PQ), upstream of rteC (pGFK67). The fusion was a translational fusion in which the start codon of tetQ was fused with the start codon of rteC. This was done to ensure that a suitable ribosome binding site was available. The rteC gene fused to the tetQ operon promoter, provided in trans with the orf2c-uidA fusion plasmid, gave high levels of GUS activity, even in the absence of RteA and RteB and in the absence of tetracycline stimulation (Table 3, line 6). We had found previously that transcription controlled by the tetQ promoter was constitutive and that tetracycline control of the production of operon proteins was mediated by a translational attenuation mechanism (37).
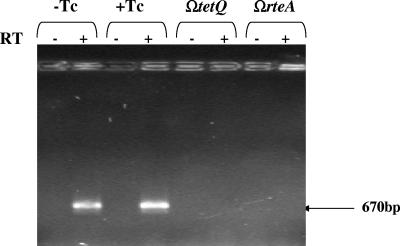
To confirm that RteA and RteB were needed for expression of the wild-type rteC gene, RT-PCR was used to detect rteC transcripts in a strain that had a single-crossover disruption in tetQ or rteA. Due to polarity, these mutant strains do not produce RteA or RteB. No rteC transcripts were detected (Fig. 6).
FIG. 6.
RT-PCR analysis of the effect of disruptions in rteA and rteB on transcription of rteC. Transcription of rteC from strain BT4007 was determined from RNA preparations made from cells grown with tetracycline (Tc) or without Tc (+Tc and-Tc, respectively) are shown in the first two sets of lanes. Reactions in which reverse transcriptase was added or not (+ or −, respectively) are also shown. Note that mRNA is detected both in cells grown with and without Tc because regulation of the tetQ-rteA-rteB operon, which controls expression of rteC, is at the level of translation rather than transcription (37). The third and fourth sets of lanes show the effect of insertions in tetQ (ΩtetQ) or rteA (ΩrteA) on mRNA from cells grown in the absence of Tc. Both of these insertions are polar on rteB, which is part of the tetQ-rteA-rteB operon. The expected location of the 670-bp rteC product is indicated by an arrow at the right.
RteC is a DNA binding protein.
To obtain purified RteC protein for use in EMSA, it is convenient to use a His-tagged form of the protein, but it is first important to make sure that this form of the protein is active in vivo. To determine whether a C-terminal His6-tagged RteC is active in vivo, we tested GUS production in a BT4001 strain that contained pGFK69, a plasmid that differed from pGFK7 in that it carried the His6-tagged version of rteC under the control of the tetQ promoter. The strain also contained pGFK43, which carried the orf2c-uidA fusion. The GUS activity in extracts from this strain was comparable to that in extracts from strains that had the nontagged rteC gene cloned in the same plasmid (Table 3, lines 6 and 7).
Since the His6-tagged form of the protein appeared to be active in vivo, we constructed an overexpression clone of the His6-tagged rteC gene in E. coli BL21. The preparation of overexpressed His6-RteC protein contained two proteins (data not shown). The predominant protein had the expected size for RteC (26 kDa). The larger, less abundant protein was 85 kDa in size. Western blotting of this partially purified preparation with antibodies that detected the His6 tag cross-reacted only with the 26-kDa band. We do not know the identity of the larger protein, but a protein of this size has been seen previously in other nickel column-purified preparations resulting from overexpression of a protein in E. coli. This preparation was used for EMSA of RteC binding to DNA upstream of orf2c.
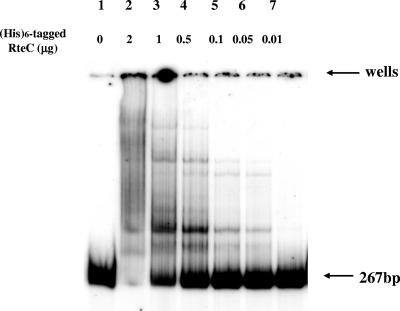
To determine if RteC binds to the promoter region of the orf2c operon, EMSA was performed with a DNA fragment of the upstream region of the orf2c promoter. It was 267 bp in size, containing DNA sequences between −143 and +124. The concentrations of purified His6-tagged RteC used for DNA binding assay were 2 μg/μl, 1 μg/μl, 0.5 μg/μl, 0.1 μg/μl, and 0.05 μg/μl. Results of this experiment are shown in Fig. 7. The DNA fragment exhibited altered migration when incubated with RteC. At the highest concentrations, all of the labeled DNA was shifted. More than one shifted band was observed, and some of the label was trapped in the wells. Reducing the amount of RteC increased the amount of unbound DNA. This, together with the fact that unlabeled nonspecific DNA was included in the EMSA analysis of the PE region, confirms that the binding of RteC to the orf2c promoter region DNA segment is specific.
FIG. 7.
Gel shift (EMSA) assay of the PE region using purified His6-tagged RteC protein. The tagged gene was shown to be active in vivo (Table 3, line 7). The DNA substrate used for EMSA was the 260-bp PE fragment that extended from bp +160 to −100 (ATG of rteC). The fragment was end labeled with 32P. The concentrations of the His6-tagged RteC added to the reaction mixtures in μg of protein are shown above each well. “0” indicates no RteC was added.
DISCUSSION
In Fig. 1, we show the hypothetical cascade of events that leads to expression of excision genes in the orf2c operon. Results reported here confirm that this cascade is correct. Previously, Wang et al. (37) had shown that the operon containing tetQ and the regulatory genes rteA and rteB was regulated in response to tetracycline by a translational attenuation mechanism, but neither RteA nor RteB had any role in this regulation. Since single-crossover insertions into rteA, rteB, and rteC all resulted in abolition of excision (8, 29), it seemed reasonable to expect that the RteC protein might regulate the expression of excision genes and that the RteA/RteB system might control the expression of rteC.
We first had to answer the question of whether the orf2c-orf2d-orf3-exc gene cluster was organized in a single operon and, if so, whether expression of that operon occurred at the transcriptional level. Results of our uidA fusion and real-time RT-PCR experiments support these hypotheses. We also localized the orf2c operon promoter region and determined that the transcription start site was at the expected distance from a consensus −7 promoter region. Bayley et al. have shown for a number of Bacteroides promoters that the consensus promoter sequences were at −7 and −33, with the −7 sequence being the most important (4). Since it is not yet clear that the genes on CTnDOT are of Bacteroides origin, we felt that it was important to confirm that the presumed −7 sequence was in fact essential for expression of the orf2c operon. The position of the orf2c transcript start site and the effect of mutagenizing the TTTG sequence, which is centered at −7 compared to the transcript start site, are both consistent with this being a site for RNA polymerase binding. It is interesting that a single mutation in this sequence was sufficient to stop expression whereas, in the promoters studied by Bayley et al. (4), more than a single mutation was needed.
The integrase gene intDOT appears to be expressed constitutively and in CTnDOT is separated by 13 kbp from the orf2c operon (7, 38). In a closely related CTn, CTnERL, the intERL gene is closer to the orf2c operon (9, 34). In other excising elements, such as phage lambda and the gram-positive conjugative transposon Tn916, the integrase (int) and the excisionase (xis) genes are adjacent to each other (2, 6, 7, 15, 32). Clearly, from the arrangement of genes in CTnDOT and CTnERL, proximity of integration and excision genes is not a requirement for efficient excision. In the case of phage lambda, expression of the int gene and expression of xis gene are controlled differently by a repressor mechanism so that only int is expressed during integration and both int and xis are expressed during excision (1, 14). The CTnDOT system appears to be a variation on this strategy, in which excision is controlled by increased expression of the genes whose products will cooperate with IntDOT to catalyze excision of CTDOT.
Our results demonstrate that the CTnDOT excision genes located in the orf2c operon are controlled by an activator protein, RteC, rather than a repressor. We considered the possibility that the orf2c operon might be regulated by a repressor. There are two lines of evidence that argue against this hypothesis. First, if RteC were a repressor, eliminating it (in the BT4001 ΩAB strain) should have resulted in tetracycline-independent expression of the orf2c operon. This was not the case; no expression of orf2c was detected in BT4001 ΩAB. Second, our mutagenesis experiments and EMSA experiments suggest that RteC is a DNA binding protein that binds upstream of the orf2c promoter, the usual site for activator binding.
The stimulatory effect of tetracycline on excision appears to be exerted indirectly through the tetQ operon gene products. More production of RteA and RteB, the presumed sensor and transcriptional activator proteins, results in more expression of the rteC gene, and the resulting increase in RteC protein concentration leads to activation of orf2c operon expression. What RteA is sensing is still a mystery, if in fact it is sensing anything in Bacteroides hosts. It is certainly not sensing tetracycline because the tetracycline effect on production of proteins encoded by the tetQ operon occurs independently of RteA and RteB (37).
A somewhat surprising finding was that the presence of the PQ-rteC plasmid did not result in regulated expression of the orf2c-uidA fusions. Although transcription of the tetQ operon message is constitutive, the production of proteins from genes in this operon is regulated, presumably due to the interaction of tetracycline with ribosomes, which stall on a leader peptide in the tetQ leader region and change the stem-loop structure of this region so as to make the ribosome binding site of the tetQ gene available (37). Since the mRNA sequence up to the start codon of rteC was replaced by the leader region and ribosome binding site of tetQ, this same type of translational attenuation should have been operational in the case of RteC production. A possible explanation of this apparent anomaly is that some production of proteins encoded in the tetQ operon occurs even when tetracycline is absent and that this basal level of protein production from a plasmid (estimated copy number of 5 to 8 per cell) is sufficient to trigger enough expression of rteC to provide maximal stimulation of the orf2c operon. A finding that supports this hypothesis is that real-time RT-PCR analysis shows that the level of rteC expression rises by only about sixfold after stimulation of cells with tetracycline. Insertions in tetQ and rteA eliminate the noninduced level of rteC transcript (Fig. 6). This shows that a low level of rteA/rteB is being made without tetracycline induction. Yet the small sixfold rise seems to be sufficient to activate orf2c operon expression. Thus, expression of rteC from the heterologous tetQ promoter may well have been sufficient, even in the absence of tetracycline, to fully activate expression of the orf2c operon. Whatever the explanation, it is clear from the results shown in Table 3 that RteC alone is sufficient for orf2c operon expression and that the contribution of RteA and RteB is to control the amount of RteC in the cell.
The picture of tetracycline regulation of excision of CTnDOT that is emerging from our results is that RteA and RteB act to stimulate expression of rteC. In turn, RteC acts as an activator to stimulate the expression of the genes in the orf2c operon. Finally, the products of genes in this region supplement the action of IntDOT to form the excision complex that allows the circular form of CTnDOT to form.
Acknowledgments
We thank Yuri Sutanto and Yanping Wang for their groundwork on this project and for providing valuable input and the clone pYS32.
The work reported here was supported by grants from the U.S. National Institutes of Health, AI/GM 22383 (A.A.S.) and GM28717 (J.F.G.).
REFERENCES
- 1.Abremski, K., and S. Gottesman. 1981. Site specific recombination: Xis-dependent excisive recombination of bacteriophage lambda. J. Mol. Biol. 153:67-78. [DOI] [PubMed] [Google Scholar]
- 2.Azaro, M. A., and A. Landy. 2002. λ integrase and the λ Int family, p. 118-148. In N. L. Craig, R. Cragie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, D.C.
- 3.Ball, T. B., F. A. Plummer, and K. T. HayGlass. 2003. Improved mRNA quantitation in LightCycler RT-PCR. Int. Arch. Allergy Immunol. 130:82-86. [DOI] [PubMed] [Google Scholar]
- 4.Bayley, D. P., E. R. Rocha, and C. J. Smith. 2000. Analysis of cepA and other Bacteroides fragilis genes reveals a unique promoter structure. FEMS Microbiol. Lett. 193:149-154. [DOI] [PubMed] [Google Scholar]
- 5.Bustin, S. A. 2002. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J. Mol. Endocrinol. 29:23-39. [DOI] [PubMed] [Google Scholar]
- 6.Celli, J., and P. Trieu-Cuot. 1998. Circularization of Tn916 is required for expression of the transposon-encoded transfer functions: characterization of long tetracycline-inducible transcripts reading through the attachment site. Mol. Microbiol. 28:103-117. [DOI] [PubMed] [Google Scholar]
- 7.Cheng, Q., B. J. Paszkiet, N. B. Shoemaker, J. F. Gardner, and A. A. Salyers. 2000. Integration and excision of a Bacteroides conjugative transposon, CTnDOT. J. Bacteriol. 182:4035-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng, Q., Y. Sutanto, N. B. Shoemaker, J. F. Gardner, and A. A. Salyers. 2001. Identification of genes required for the excision of CTnDOT, a Bacteroides conjugative transposon. Mol. Microbiol. 41:625-632. [DOI] [PubMed] [Google Scholar]
- 9.Cheng, Q., N. Wesslund, N. B. Shoemaker, A. A. Salyers, and J. F. Gardner. 2002. Development of an in vitro integration assay for the Bacteroides conjugative transposon CTnDOT. J. Bacteriol. 184:4829-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho, K. H., D. Cho, G. R. Wang, and A. A. Salyers. 2001. New regulatory gene that contributes to control of Bacteroides thetaiotaomicron starch utilization genes. J. Bacteriol. 183:7198-7205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feldhaus, M. J., V. Hwa, Q. Cheng, and A. A. Salyers. 1991. Use of an Escherichia coli β-glucuronidase gene as a reporter gene for investigation of Bacteroides promoters. J. Bacteriol. 173:4540-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holdeman, L. V., and W. E. C. Moore. 1975. Anaerobe laboratory manual, 4th ed. Virginia Polytechnic Institute and State University, Blacksburg, Va.
- 13.Jefferson, R. A., S. M. Burgess, and D. Hirsh. 1986. β-Glucuronidase from Escherichia coli as a gene-fusion marker. Proc. Natl. Acad. Sci. USA 83:8447-8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kikuchi, Y., and H. A. Nash. 1979. Nicking-closing activity associated with bacteriophage lambda int gene product. Proc. Natl. Acad. Sci. USA 76:3760-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Landy, A. 1989. Dynamic, structural, and regulatory aspects of site-specific recombination. Annu. Rev. Biochem. 58:913-950. [DOI] [PubMed] [Google Scholar]
- 16.Li, L. Y., N. B. Shoemaker, and A. A. Salyers. 1995. Location and characteristics of the transfer region of a Bacteroides conjugative transposon and regulation of transfer genes. J. Bacteriol. 177:4992-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Neill, M. C. 1989. Escherichia coli promoters. I. Consensus as it relates to spacing class, specificity, repeat substructure, and three-dimensional organization. J. Biol. Chem. 264:5522-5530. [PubMed] [Google Scholar]
- 18.Pfaffl, M. W., and M. Hafeleit. 2001. Validities of mRNA quantificating using recombinant RNA and recombinant DNA eternal calibration curve in real time RT-PCR. Biotechnol. Lett. 23:275-282. [Google Scholar]
- 19.Rocha, E. R., and C. J. Smith. 1997. Regulation of Bacteriodes fragilis katB mRNA by oxidative stress and carbon limitation. J. Bacteriol. 179:7033-7039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryu, S., and S. Garges. 1994. Promoter switch in the Escherichia coli pts operon. J. Biol. Chem. 269:4767-4772. [PubMed] [Google Scholar]
- 21.Salyers, A. A., N. Shoemaker, A. Cooper, J. D'Elia, and J. A. Shipman. 1999. Genetic methods for Bacteroides species. Methods Microbiol. 29:229-249. [Google Scholar]
- 22.Salyers, A. A., N. B. Shoemaker, A. M. Stevens, and L. Y. Li. 1995. Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol. Rev. 59:579-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Savli, H., A. Karadenizli, F. Kolayli, S. Gundes, U. Ozbek, and H. Vahaboglu. 2003. Expression stability of six housekeeping genes: a proposal for resistance gene quantification studies of Pseudomonas aeruginosa by real-time quantitative RT-PCR. J. Med. Microbiol. 52:403-408. [DOI] [PubMed] [Google Scholar]
- 24.Shoemaker, N. B., C. Getty, E. P. Guthrie, and A. A. Salyers. 1986. Regions in Bacteroides plasmids pBFTM10 and pB8-51 that allow Escherichia coli-Bacteroides shuttle vectors to be mobilized by IncP plasmids and by a conjugative Bacteroides tetracycline resistance element. J. Bacteriol. 166:959-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shoemaker, N. B., and A. A. Salyers. 1987. Facilitated transfer of IncPβ R751 derivatives from the chromosome of Bacteroides uniformis to Escherichia coli recipients by a conjugative Bacteroides tetracycline resistance element. J. Bacteriol. 169:3160-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shoemaker, N. B., H. Vlamakis, K. Hayes, and A. A. Salyers. 2001. Evidence for extensive resistance gene transfer among Bacteroides spp. and between Bacteroides and other genera in the human colon. Appl. Environ. Microbiol. 67:561-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shoemaker, N. B., G. R. Wang, and A. A. Salyers. 2000. Multiple gene products and sequences required for excision of the mobilizable integrated Bacteroides element NBU1. J. Bacteriol. 182:928-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stevens, A. M., J. M. Sanders, N. B. Shoemaker, and A. A. Salyers. 1992. Genes involved in production of plasmidlike forms by a Bacteroides conjugal chromosomal element share amino acid homology with two-component regulatory systems. J. Bacteriol. 174:2935-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stevens, A. M., N. B. Shoemaker, L. Y. Li, and A. A. Salyers. 1993. Tetracycline regulation of genes on Bacteroides conjugative transposons. J. Bacteriol. 175:6134-6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stevens, A. M., N. B. Shoemaker, and A. A. Salyers. 1990. The region of a Bacteroides conjugal chromosomal tetracycline resistance element which is responsible for production of plasmidlike forms from unlinked chromosomal DNA might also be involved in transfer of the element. J. Bacteriol. 172:4271-4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 32.Su, Y. A., and D. B. Clewell. 1993. Characterization of the left 4 kb of conjugative transposon Tn916: determinants involved in excision. Plasmid 30:234-250. [DOI] [PubMed] [Google Scholar]
- 33.Sutanto, Y. 2003. Excision of conjugative transposon in Bacteroides thetaiotamicron. Ph.D. thesis, University of Illinois at Urbana-Champaign, Urbana-Champaign, Ill.
- 34.Sutanto, Y., J. M. DiChiara, N. B. Shoemaker, J. F. Gardner, and A. A. Salyers. 2004. Factors required in vitro for excision of the Bacteroides conjugative transposon, CTnDOT. Plasmid 52:119-130. [DOI] [PubMed] [Google Scholar]
- 35.Valentine, P. J., N. B. Shoemaker, and A. A. Salyers. 1988. Mobilization of Bacteroides plasmids by Bacteroides conjugal elements. J. Bacteriol. 170:1319-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, J., N. B. Shoemaker, G. R. Wang, and A. A. Salyers. 2000. Characterization of a Bacteroides mobilizable transposon, NBU2, which carries a functional lincomycin resistance gene. J. Bacteriol. 182:3559-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, Y., N. B. Shoemaker, and A. A. Salyers. 2004. Regulation of a Bacteroides operon that controls excision and transfer of the conjugative transposon CTnDOT. J. Bacteriol. 186:2548-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whittle, G., B. D. Hund, N. B. Shoemaker, and A. A. Salyers. 2001. Characterization of the 13-kb ermF region of Bacteroides conjugative transposon CTnDOT. Appl. Environ. Microbiol. 67:3488-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whittle, G., N. B. Shoemaker, and A. A. Salyers. 2002. Characterization of genes involved in modulation of conjugal transfer of the Bacteroides conjugative transposon CTnDOT. J. Bacteriol. 184:3839-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whittle, G., N. B. Shoemaker, and A. A. Salyers. 2002. The role of Bacteroides conjugative transposons in the dissemination of antibiotic resistance genes. Cell. Mol. Life Sci. 59:2044-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]