Abstract
The yhcH gene is part of the nan operon in bacteria that encodes proteins involved in sialic acid catabolism. Determination of the crystal structure of YhcH from Haemophilus influenzae was undertaken as part of a structural genomics effort in order to assist with the functional assignment of the protein. The structure was determined at 2.2-Å resolution by multiple-wavelength anomalous diffraction. The protein fold is a variation of the double-stranded β-helix. Two antiparallel β-sheets form a funnel opened at one side, where a putative active site contains a copper ion coordinated to the side chains of two histidine and two carboxylic acid residues. A comparison to other proteins with a similar fold and analysis of the genomic context suggested that YhcH may be a sugar isomerase involved in processing of exogenous sialic acid.
Sialic acids (Sias) comprise a family of about 40 derivatives of the nine-carbon sugar neuraminic acid (56). The most widespread form of Sia is N-acetylneuraminic acid (Neu5Ac), which has an acetylated amino group at C-5. The hydroxylated form of Neu5Ac, N-glycolylneuraminic acid, is also common in many animal species (29, 39). Sias are usually located at the end of a glycan chain in vertebrate glycoconjugates and are involved in molecular and cellular recognition. Thus, the immune system can distinguish between self and nonself structures according to their Sia patterns. Sias as components of the capsular polysaccharide in pathogenic bacteria not only mask the organisms from the immune system of the host but also provide the means of host cell recognition.
Sia catabolism in bacteria involves cleavage of cell surface glycoconjugates by sialidases, transport of free Sia molecules through the membrane, and degradation of the molecules to N-acetylmannosamine and pyruvate through the action of Neu5Ac aldolase (lyase) (59). N-Acetylmannosamine is then phosphorylated by a specific kinase and isomerized to N-acetylglucosamine 6-phosphate, which enters the amino sugar metabolic pathways. Sia thus can serve as the sole carbon or nitrogen source in bacteria and as a source of amino sugars for cell wall synthesis (45).
In many bacteria the genes involved in Sia catabolism form an operon (59, 60). In Escherichia coli the operon includes the nanATEK-yhcH genes coding for the aldolase, the transporter, the epimerase, the kinase, and a protein with an unknown function, respectively. Expression of the operon is controlled by a repressor protein encoded by the upstream gene nanR (31). Since the discovery of Sia catabolism in E. coli (60), the pathway has also been described in Clostridium perfringens (61) and Haemophilus influenzae (58). The complete bacterial genomic DNA sequences revealed that nan systems are present in diverse species, including gamma-proteobacteria, clostridia, streptococci, staphylococci, and fusobacteria (59).
In this study, we focused on the uncharacterized protein encoded by the yhcH gene of H. influenzae. This protein emerged as a target in a structural genomics project aimed at the functional assignment of proteins through determination of their three-dimensional structures (17). It is highly expressed in H. influenzae and E. coli cells growing in rich medium (33, 34). The YhcH homologs are present in gram-negative and gram-positive bacteria but not in archaea or eukaryotes. No functional information for this protein family has been available, other than that one of its members, E. coli EbgC, showed up as a subunit of an experimentally evolved beta-galactosidase (18). Although this observation did not provide any direct clue to the protein function in vivo, it has been used for functional assignment in the Swiss-Prot and COG databases (5, 52). In contrast, the Pfam database (6) lists YhcH homologs as members of the Domain of Unknown Function (DUF386) protein family.
The YhcH protein was cloned and expressed, and the crystal structure was determined at 2.2-Å resolution. Analysis of the structure and of the genome context suggested that YhcH may function as a copper-dependent sugar isomerase. A possible role in Sia catabolism may involve the processing of exogenous glycolated neuraminic acid.
MATERIALS AND METHODS
Cloning, expression, and purification.
The YhcH (HI0227) gene was cloned by PCR, using genomic H. influenzae DNA as a template, into the pET15b vector (Novagen) and was expressed in E. coli strain B834(DE3). The cells were grown in LB medium supplemented with 100 μg/ml ampicillin until the A600 reached 0.8, when expression was induced with 1 mM isopropyl-β-d-thiogalactoside. The cells were harvested after 4 h, suspended in 20 mM Tris-HCl, pH 8.4, and lysed in a French press at 8,000 lb/in2. The soluble extract was applied to a Poros HQ50 anion-exchange column equilibrated with the same buffer. The flowthrough was collected, examined by polyacrylamide gel electrophoresis in the presence of sodium dodecyl sulfate, dialyzed against 20 mM 4-morpholinolineethanesulfonic acid (MES), pH 5.5, and applied to an HS20 cation-exchange column. The protein was eluted in an NaCl gradient, concentrated to 12 mg/ml, and dialyzed against 50 mM Tris-HCl, pH 7.5, 0.1 mM dithiothreitol, 0.1 mM EDTA. The yield was 65 mg from 3.4 g cells obtained from a 3-liter culture. The molecular mass measured by matrix-assisted laser-desorption ionization spectrometry (17,663 Da) was consistent with the molecular mass calculated from the amino acid sequence (17,670 Da).
Equilibrium sedimentation.
The oligomeric state of the protein was investigated by equilibrium sedimentation ultracentrifugation. The data were collected at 25°C and 4°C in 50 mM Tris-HCl, pH 7.5, 0.1 mM dithiothreitol, 0.1 mM EDTA buffer at a range of concentrations (0.1 to 2.2 mg/ml) and rotor speeds. The data were fitted to an ideal single-species model. No attempt to model a mixture of oligomeric states was made.
Crystallization and structure determination.
YhcH crystals were grown by the vapor diffusion hanging drop method at room temperature from 0.1 M HEPES, pH 7.5, 25% polyethylene glycol 4000, 1 M sodium acetate. These crystals belong to space group P21 with the following unit cell parameters: a = 41.9 Å, b = 153.9 Å, c = 53.8 Å, and β = 112.9°. There are four polypeptide chains in the asymmetric unit with a solvent content of 45%. For X-ray data collection, the crystals were flash-frozen in liquid propane in the crystallization solution.
The structure was solved by the two-wavelength anomalous diffraction method (MAD) using a mercury derivative. Crystals were soaked in 2 mM KHgSCN overnight and appeared to be nonisomorphous compared to the native crystals (R-merge = 44.3%) with unit cell deviations of up to 3% (a = 43.1 Å, b = 152.3 Å, c = 53.4 Å, β = 113.6°). The 2.6-Å diffraction data for the derivative and the 2.2-Å data for the native crystal (Table 1) were collected on the IMCA-CAT beamline at the Advanced Photon Source (Argonne, IL) equipped with a MAR charge-coupled device detector. The following programs were used: HKL2000 (43) for data processing, SnB (37), MLPHARE (42), and DM (13) for phasing, O (30) for model building, and REFMAC (40) for refinement. The atomic model was built into the MAD-phased electron density and refined against the Hg derivative data. It was used for further refinement against the native data at 2.2-Å resolution. No noncrystallographic symmetry restraints were applied to the four independent protein molecules. The refinement statistics are shown in Table 1. Water molecules were added at the (Fo-Fc) electron density peaks using a cutoff level of 3σ. The same native crystal was used for an X-ray fluorescence absorption experiment at the copper edge (wavelength, 1.3804 Å). A complete data set was collected at the peak wavelength (1.3766 Å) and used for anomalous Fourier calculations. Programs from the CCP4 suite (12) were used for crystallographic calculations, CLUSTALW (53) and ESPRIPT (26) were used for sequence alignment, and MOLSCRIPT (35) and RASTER3D (36) were used for ribbon diagrams.
TABLE 1.
X-ray data and refinement statistics
| Parameter | Hg peaka | Hg edgea | Native |
|---|---|---|---|
| Wavelength (Å) | 1.0053 | 1.0087 | 1.1000 |
| Resolution range (Å) | 20-2.6 (2.65-2.6)b | 20-2.7 (2.76-2.7) | 20-2.2 (2.24-2.20) |
| Unique reflections | 36,092 (1,951) | 32,330 (2,079) | 29,577 (1,033) |
| Completeness (%) | 98.6 (97.2) | 98.4 (95.1) | 93.0 (64.4) |
| Redundancy | 1.8 (1.8) | 1.8 (1.8) | 7.2 (2.3) |
| Rsym (Σ|I-〈I〉|)/ΣI) | 0.036 (0.142) | 0.035 (0.112) | 0.055 (0.090) |
| 〈I/σ〉 | 19.9 (4.6) | 21.3 (6.2) | 29.8 (6.2) |
| Fraction of refls with I>3σ (%) | 84.6 (57.3) | 88.8 (64.9) | 86.5 (71.4) |
| Rcryst (Σ‖F0|-|Fc‖)/Σ|F0|) | 0.185 | 0.176 | |
| Rfree (5% data) | 0.247 | 0.246 | |
| No. of protein atoms | 4,508 | 4,758 | |
| No. of water molecules | 78 | 154 | |
| RMSD in bonds (Å) | 0.027 | 0.017 | |
| RMSD in angles (°) | 2.8 | 1.9 | |
| Mean B-factor | |||
| Molecule A (Å2) | 47.7 | 47.1 | |
| Molecule B (Å2) | 52.1 | 52.0 | |
| Molecule C (Å2) | 53.4 | 52.2 | |
| Molecule D (Å2) | 56.3 | 55.3 | |
| B-factor from Wilson plot (Å2) | 35.5 | 35.6 |
Anomalous pairs not merged.
The data in parentheses are the data for the highest-resolution shell.
Protein structure accession codes.
The atomic coordinates and experimental data for the native protein and the mercury derivative have been deposited in the Protein Data Bank under accession codes 1S4C and 1JOP, respectively.
RESULTS AND DISCUSSION
Molecular structure.
The MAD-phased electron density map allowed unambiguous modeling of the entire protein molecule, including the N- and C-terminal residues. The electron density is weak for residues 53 to 60 and 135 to 141, which were traced in only one of the four crystallographically independent molecules. The atomic B-factors are relatively high even for well-defined regions (around 30 Å2), which probably reflects the degree of disorder of the crystal rather than the flexibility of the protein molecule. This contention is supported by the comparison of the protein monomers present in the asymmetric part of the unit cell. The structures of these monomers are essentially identical despite different crystal environments. The monomers can be superimposed with a root mean square deviation of 0.22 to 0.31 Å for all Cα atoms. The maximum deviations do not exceed 1.3 Å. Clearly, the structure is not influenced by crystal contacts.
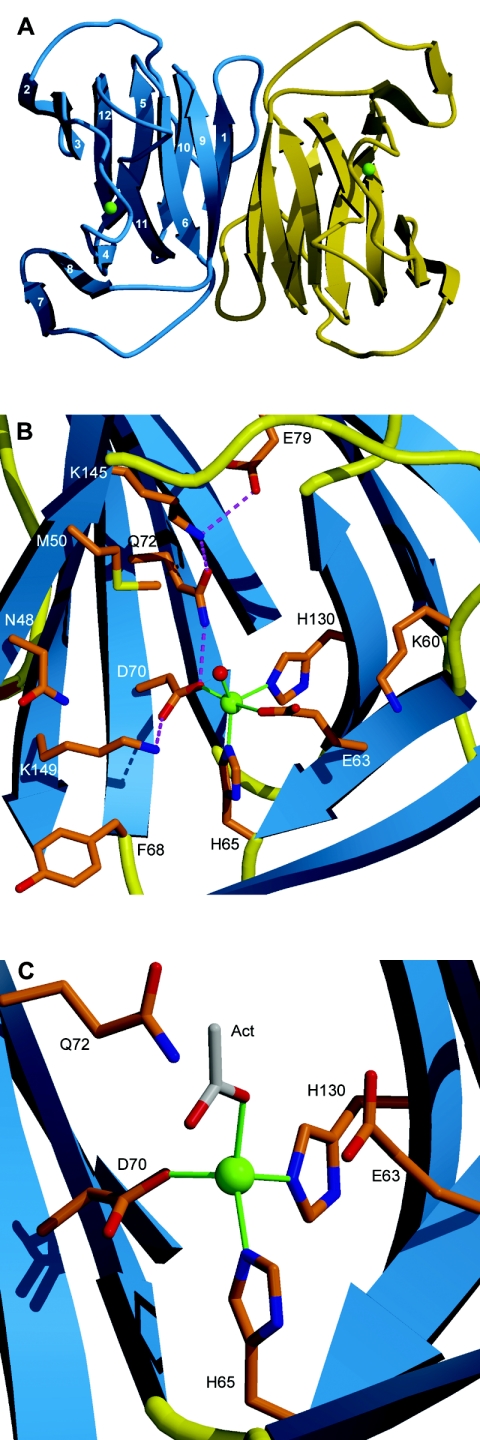
The YhcH structure is composed of two antiparallel β-sheets consisting of six β-strands each (Fig. 1A). The β-sheets form a sandwich of a jelly roll type. One of the β-sheets is twisted by almost 180°, as measured between the β-strands at the opposite ends of the sheet. This leaves the β-sandwich open at one end, so that the overall shape of the β-structure resembles a funnel.
FIG. 1.
Crystal structure of YhcH. (A) Ribbon diagram of the dimeric molecule of YhcH. β-Strands are represented by arrows and are numbered consecutively. Cu ions are indicated by green spheres. (B) Putative active site in subunit B of YhcH as viewed from the left with respect to panel A. (C) Cu coordination in subunit A. The acetate molecule is gray. Amino acids are labeled with their one-letter abbreviations. Cu coordination bonds are indicated by green lines, and the hydrogen bond network is indicated by dashed magenta lines.
The YhcH fold is a variation of the double-stranded β-helix that was given the name cupin (15). Cupins have two histidine-containing motifs, originally designated GX5HXHX4EX6G and GX5PXGX2HX3N, that correspond to strands β4/β5 and β10/β11 of YhcH. In many cupin proteins these conserved histidines bind the active site metal. Accumulation of new sequences during the past few years has revealed that the motifs are much less conserved than was first suggested (32). Histidine residues in the first motif may be replaced by glutamine, glutamate, or aspartate residues. Along with the sequence variability is the spectrum of metal ions employed by these proteins. Fe2+ has been found most frequently, although Mn2+, Zn2+, Co2+, Ni2+, and Cu2+ have also been detected. Some proteins, such as the seed storage 7S proteins, do not contain any metal at all despite the conservation of the three-dimensional structure (32).
Cupins represent one of the most functionally diverse protein superfamilies (16, 32). 2-Oxoglutarate and Fe2+-dependent dioxygenases constitute perhaps the largest group both in terms of the range of species and in terms of the substrates. Bacterial antibiotic synthases are the best-characterized members of this group. Another group of cupins, according to the SCOP database (41), includes germin-like seed storage proteins (62, 63), oxalate decarboxylase (3), phosphoglucose and phosphomannose isomerases (7, 11, 51), dTDP-4-dehydrorhamnose 3,5-epimerase RmlC (10, 14, 24), and dioxygenases acting on homogentisate (54), acireductone (46), and quercetin (21). These are the proteins that exhibit the greatest structural similarity to YhcH, as revealed by a DALI (27) search. Quercetin 2,3-dioxygenase (QDO) is ranked first, with the Z-score of 6.4. The r.m.s. deviation between the superimposed structures is 1.9 Å for 85 common Cα atoms when the “catalytic” N-terminal domain of QDO is used. Curiously, the inactive metal-free C-terminal domain of QDO fits YhcH better, with an r.m.s. deviation of 1.6 Å for the same 85 Cα atoms. The largest deviations between the structures occur in the two loops that are partially disordered in YhcH.
Oligomeric structure.
Cupins typically form dimers that may further assemble into hexamers. The dimer consists either of two separate polypeptide chains or of topologically identical domains within a single polypeptide. A common theme in dimer formation is the incorporation of an N-terminal segment of one subunit in the β-sheet of the other subunit. Such an arrangement yields a symmetrical dimer with an extensive interface. The active site of the cupin protein is located in the crevice between the β-sheets. In the dimer, both active sites remain accessible, although in some proteins (e.g., RmlC) the N-terminal protrusion from the other subunit forms part of the substrate binding site (24). There have been no reports on the cooperativity of substrate binding in these oligomeric enzymes.
YhcH also exists in a dimeric form according to the equilibrium sedimentation data collected at 25°C and 4°C at pH 7.5. The crystal structure reveals two tightly associated dimers in the asymmetric part of the unit cell. The solvent-accessible area buried upon dimerization is over 2,000 Å2, which is one-quarter of the total surface area of the monomer. However, the association of monomers in the YhcH dimer differs from that in the typical cupin dimer. The interface is formed by β-strands β1 and β9 at the narrow end of the β-funnel and their symmetry-related equivalents in the other molecule (Fig. 1A). The twofold molecular symmetry yields a continuous β-sandwich spanning the dimer. Besides the main chain hydrogen bonds between the β-strands, there are a few other contacts that include residues of the loop following β1. YhcH dimerization leaves the putative active sites accessible from the opposite ends of the dimer.
Metal binding.
The YhcH molecule contains a cation binding site at the opening of the β-funnel. The ion in the native crystal was identified as copper by using X-ray fluorescence spectroscopy. Scanning the crystal in the appropriate X-ray energy range revealed an absorption edge at 8,982 eV (1.3804 Å), which corresponds to the value for copper. The anomalous signal from the data collected at a peak wavelength of 1.3766 Å confirmed the presence of the Cu ion in the structure. Since copper was not added to the protein during purification and crystallization, this result suggests that copper is the physiological metal for YhcH.
The coordination of the Cu ion is different in the crystallographically independent molecules. In molecule B four residues (Glu63, His65, Asp70, and His130) and a water molecule are involved in the metal coordination (Fig. 1B). Both carboxylate groups are monodentate ligands so that the geometry can be described as a distorted square pyramid. The bond lengths are in the range from 1.9 to 2.2 Å for all ligands except Glu63, which is 2.7 Å from Cu. In molecule A the electron density at the solvent position is great enough to accommodate a four-atom molecule. The ion was modeled as an acetate ion because it was present at a high concentration in the crystallization solution. The Cu-O distances for the acetate are 2.1 and 2.9 Å. On the other hand, Glu63 in molecule A is farther away from the metal, so that the Cu geometry is close to tetrahedral (Fig. 1C). In molecules C and D, the electron density is not so well defined. It was modeled with the solvent position occupied by water and Glu63 oriented away from the Cu ion. The observed flexibility of Glu63 may have functional importance, as discussed below. It should be noted, however, that the difference in Cu coordination may reflect the effect of partial chelation by EDTA during protein purification. The structural differences between the four subunits are primarily restricted to the coordination sphere of the metal. There are no significant differences in the rest of the protein structure.
In proteins, copper has been observed in one of the two oxidation states, Cu+ or Cu2+ (28). While Cu+ is preferably complexed by cysteine and methionine residues, Cu2+ is ligated mostly by histidine, hydroxyl groups of serine, threonine, or tyrosine residues, and water. From this point of view, the likely species of the metal in YhcH is Cu2+.
Most cupins contain metal ions bound at the site observed in YhcH. Typically, two or three amino acid ligands (one or two histidines and a carboxylic acid) are located in a short stretch of the sequence that matches strands β4 and β5. Another ligand, which is invariably a histidine, may be separated by up to 150 residues in the sequence but spatially comes from a β-strand next to β4 (β11 in YhcH). The wide diversity of metal ions and their coordination geometries in the cupins contribute to the variety of reactions catalyzed by these enzymes. Interestingly, QDO (21) is the only Cu-dependent enzyme in this structural superfamily.
Comparison of the metal-binding sites in QDO and YhcH revealed remarkable similarity between the two proteins. First, the geometry of the site is the same. The amino acid ligands of Cu2+ in QDO, His66, His68, Glu73, and His112, match the YhcH ligands Glu63, His65, Asp70, and His130, respectively. These ligands are associated with the same secondary structural elements in both proteins. Second, QDO is the only known protein with carboxylate ligation of a Cu ion. Therefore, YhcH is possibly the first example of double carboxylate ligation. Third, the alternate conformations of the glutamate ligand have been observed in both structures. In apo-QDO, the metal is predominantly bound in a tetrahedral geometry by three histidines and a water molecule (21). In complexes with substrates and substrate analogs, Cu2+ is pentacoordinated with Glu73 bound to both the metal and the substrate (50). In YhcH different coordination states are observed in one crystal. In the apo form represented by molecule B, Cu2+ is bound by all four protein groups and a water molecule. When an acetate ion replaces a water ligand, Glu63 leaves the coordination sphere of Cu2+. Thus, in both proteins the metal coordination is sensitive to the presence of an exogenous molecule, and the glutamate ligand follows this rearrangement, albeit in opposite ways.
Amino acid sequence analysis.
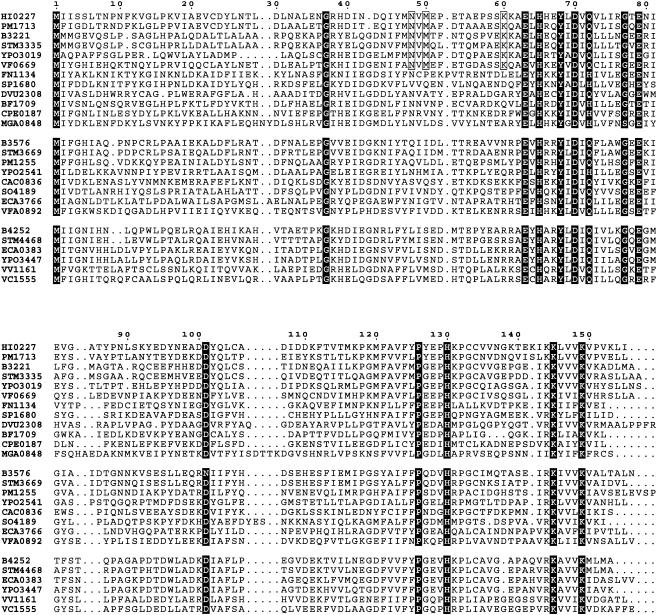
A BLAST (2) search in combination with a PROSITE (4) search using Cu-coordinating residues as a template identified over 40 YhcH homologs. These homologs are widely represented in gamma-proteobacteria as well as in streptococci, clostridia, and Mollicutes. No homologs have been found in archaea and eukaryotes. The levels of amino acid identity in the family range from 88% (between Salmonella enterica serovar Typhi STY4129 and Klebsiella oxytoca YiaL) to 18% (between S. enterica serovar Typhi STY4129 and Mycoplasma pulmonis MYPU6600). Three groups of highly conserved residues can be identified from the sequence alignment (Fig. 2). One group includes Glu63, His65, Asp70, and His130 involved in Cu2+ coordination. Another group includes residues that are likely important for the stability of the three-dimensional structure. These residues are Gly37 preceding β2, Gly77 in the loop between β5 and β6, Asp101 H bonded to the amino groups of the β4-β5 loop, and Pro126 in the β10-β11 loop. All of them are located in loops providing the necessary conformational flexibility (glycine) or rigidity (proline) at the sharp turns of the polypeptide chain. Asp101 stabilizes the reverse turn between β4 and β5 through hydrogen bonds to the main chain amino groups. The proper fold of this fragment is particularly important as it supports the conformation of the metal binding site.
FIG. 2.
Sequence alignment of the YhcH/YiaL/YjgK protein family. Residues conserved in the entire family are indicated by white letters on a black background. Residues of the “specificity triad” are enclosed in boxes. The numbering corresponds to that of YhcH from H. influenzae (HI0227). Sequences are labeled with the gene tags in the GenBank database (http://www.ncbi.nlm.nih.gov/GenBank). Abbreviations of organism names: PM, Pasteurella multocida; B, Escherichia coli; STM, Salmonella enterica serovar Typhimurium; YPO, Yersinia pestis; VF, Vibrio fischeri; FN, Fusobacterium nucleatum; SP, Streptococcus pneumoniae; DVU, Desulfovibrio vulgaris; BF, Bacteroides fragilis; CPE, Clostridium perfringens; MGA, Mycoplasma gallisepticum; CAC, Clostridium acetobutylicum; SO, Shewanella oneidensis; ECA, Erwinia carotovora; VV, Vibrio vulnificus; VC, Vibrio cholerae.
Quite unusually, the position of Met1 is conserved in the alignment, indicating the importance of the length of the N-terminal β-strand. Strand β1 is part of the dimer interface, and the exact position of the N terminus may influence the dimerization of the protein.
There are three other strictly conserved residues (Glu79, Lys145, and Lys149) that are located close to the metal binding site and may therefore be functionally important. Together with Gln72 they form a network of H-bonded side chains that connects the Cu2+-bound carboxylate of Asp70 with the solvent-inaccessible carboxylate of Glu79 located deep in the active site cavity (Fig. 1B). Gln72 is replaced by a histidine in some members of the family, while it retains the ability to be part of the network. The buried position of Glu79 surrounded by hydrophobic residues implies its basic character and suggests that the network may function as a relay system.
Some bacteria possess several genes coding for the YhcH homologs. E. coli, for instance, has three such paralogs (YhcH, YiaL, and YjgK), and the levels of sequence identity between them are around 30%. Each of these three proteins belongs to a separate subfamily, the members of which are characterized by higher levels of sequence similarity to each other than to the proteins belonging to the other subfamilies. Thus, the entire family is usually referred to as YhcH/YiaL/YjgK. The three subfamilies must have the same fold but may differ in substrate specificity or regulation.
The YhcH crystal structure indicates three residues that may define the substrate specificity of the group of proteins from proteobacteria. Asn48, Met50, and Lys60 are located at the rim of the active site entrance (Fig. 1B) and may directly interact with a substrate bound close to the Cu ion. Their conservation in proteobacteria (Fig. 2) suggests a common substrate for this group of proteins (e.g., an amino sugar with a particular substituent). The same positions in the YjgK subfamily are occupied by Leu, Ser, and Arg, which are also highly conserved in the sequences. The lack of a conservation pattern in the YiaL subfamily may reflect broader substrate specificity among the members of this subfamily.
Genome context.
In bacteria, metabolism of Sia can proceed by either of two routes; the molecule can be catabolized to GlcNAc and eventually enter glycolysis, or it can be used for sialylation of the surface lipopolysaccharide (57, 58). Besides these routes, pathogenic bacteria have developed a pathway for Sia biosynthesis from GlcNAc that includes GlcNAc phosphorylation and epimerization (siaA, neuC, or nnaA) and consecutive synthesis of Neu5Ac (siaC, neuB, or nnaB) and CMP-Neu5Ac (siaB, neuA, or nnaC), which is incorporated into the polysaccharide by a specific transferase (neuS or siaD) (19, 20, 23). The corresponding genes are part of an operon that is present in pathogens such as Campylobacter jejuni, Neisseria meningitidis, Fusobacterium nucleatum, and E. coli K1. However, the operon is missing from nonpathogenic strains of E. coli K-12 and H. influenzae KW20, suggesting that the gene products of the nan and nna operons have nonoverlapping functions despite the similar reactions catalyzed by the enzymes.
Analysis of the genome context that relies on characteristics such as conserved gene neighborhoods, phylogenetic patterns, and coexpression in microarray experiments may provide certain clues to the function of a “hypothetical” protein (22, 44). Complete genome sequences are available for all members of the YhcH/YjgK/YiaL family. Although the H. influenzae HI0227 gene itself does not belong to any apparent gene string, its homologs in many other organisms are part of the nan operon that encodes the enzymes of the Sia degradation pathway (33, 59). The three-dimensional structure of the protein suggests an isomerase (epimerase) function, as it is typical for cupins. However, an epimerase, which catalyzes the interconvertion of N-acetylmannosamine 6-phosphate and GlcNAc-6-phosphate, is encoded by the nanE gene. As an epimerase, YhcH may have different substrate specificities depending on the tolerance of the nanT and nanA gene products involved in the first steps of Sia uptake. Neu5Ac aldolase (NanA) is specific to Neu5Ac as the most ubiquitous Sia in host organisms. However, the original study using the E. coli deletion strains indicated that the nature of the C-5 amino substituent in Sia does not affect transport or degradation (60). The aldolases from C. perfringens and E. coli are capable of cleaving a range of neuraminic acid derivatives with different substituents at C-5, including formyl, succinyl, and glycolyl neuraminic acids (1, 48). Regarding the NanT permease, it has been established that many bacteria, both gram negative and gram positive, exhibit an active proton symporter-type mechanism (59). Since it is highly specific for Sias, the NanT transporter can bind a range of neuraminic acid derivatives. For instance, inhibition studies with Pasteurella hemolytica revealed that N-glycolylneuraminic acid, Neu5Ac methyl ester, and 2,3-dihydro-2-deoxy-Neu5Ac may be taken up by a common transport system (49).
The cupin superfamily includes dioxygenases, isomerases/epimerases, and sugar binding proteins lacking any enzymatic activity (16, 32). Given that the YhcH protein is encoded in the nan operons of several strictly anaerobic bacteria, such as C. perfringens and F. nucleatum (33, 59), it is very unlikely that it could function as a dioxygenase. We suggest that YhcH may be an epimerase specific to neuraminic acid derivatives other than Neu5Ac, so that its activity would be complementary to NanE. This would allow utilization by YhcH-encoding bacterial pathogens of alternatively substituted neuraminic acids, such as those found in blood (9). One possible candidate for the YhcH substrate is a hydroxylated form of Neu5Ac, N-glycolylneuraminic acid. This molecule is one of the two major Sias on the surfaces of most primate cell types (29, 39).
In Haemophilus ducreyi, the neu gene locus is part of a larger cluster that also includes rmlBACD genes responsible for the synthesis of l-rhamnose for incorporation into lipopolysaccharide (25). The rmlC gene product, dTDP-4-keto-6-deoxy-d-glucose 3,5-epimerase, catalyzes the third step of the pathway. This enzyme belongs to the cupin structural superfamily, although unlike most cupins, it is metal independent. Assuming that YhcH may also be involved in sugar processing, this structural similarity between YhcH and RmlC may be a case of protein fold accommodation for different but structurally similar substrates. Such cases, which often occur within a single pathway, have been observed for many functionally related proteins (55). For instance, in amino sugar metabolism, the gene products of neuA (Neu5Ac cytidylyltransferase) and glmU (GlcN-1P uridyltransferase) have a common fold (8, 38).
An alternative evolutionary path, adaptation of structurally unrelated proteins for the same biochemical activity, has also been documented. For sugar isomerization, there is the case of phosphoglucose isomerase (PGI), which is represented by two distinct protein families. In most organisms, the enzyme is a homodimer of 60- to 70-kDa subunits with an αβα sandwich topology. The general acid-base catalysis by PGI is metal independent (47). PGI of the second type has been found in some Euryarchaeota species. This type forms dimers of 21-kDa subunits with the cupin fold and catalyzes Glc-6P isomerization in a metal (presumably Fe2+)-dependent manner (7). If YhcH is a sugar epimerase, this PGI may represent the closest analog in terms of the reaction mechanism.
Acknowledgments
We are grateful to Eric R. Vimr for useful discussions on the possible function of YhcH.
This work was supported by National Institutes of Health grant P01-GM57890. The use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract W-31-109-Eng-38.
Certain commercial materials, instruments, and equipment are identified in this paper in order to specify the experimental procedure as completely as possible. In no case does such identification imply a recommendation or endorsement by the National Institute of Standards and Technology or the National Institutes of Health, nor does it imply that the materials, instruments, or equipment identified is necessarily the best available for the purpose.
REFERENCES
- 1.Aisaka, K., A. Igarashi, K. Yamaguchi, and T. Uwajima. 1991. Purification, crystallization and characterization of N-acetylneuraminate lyase from Escherichia coli. Biochem. J. 276:541-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anand, R., P. C. Dorrestein, C. Kinsland, T. P. Begley, and S. E. Ealick. 2002. Structure of oxalate decarboxylase from Bacillus subtilis at 1.75 Å resolution. Biochemistry 41:7659-7669. [DOI] [PubMed] [Google Scholar]
- 4.Bairoch, A. 1991. PROSITE: a dictionary of sites and patterns in proteins. Nucleic Acids Res. 19:2241-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bairoch, A., R. Apweiler, C. H. Wu, W. C. Barker, B. Boeckmann, S. Ferro, E. Gasteiger, H. Huang, R. Lopez, M. Magrane, M. J. Martin, D. A. Natale, C. O'Donovan, N. Redaschi, and L. S. Yeh. 2005. The Universal Protein Resource (UniProt). Nucleic Acids Res. 33:D154-D159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berrisford, J. M., J. Akerboom, A. P. Turnbull, D. de Geus, S. E. Sedelnikova, I. Staton, C. W. McLeod, C. H. Verhees, J. van der Oost, D. W. Rice, and P. J. Baker. 2003. Crystal structure of Pyrococcus furiosus phosphoglucose isomerase. Implications for substrate binding and catalysis. J. Biol. Chem. 278:33290-33297. [DOI] [PubMed] [Google Scholar]
- 8.Brown, K., F. Pompeo, S. Dixon, D. Mengin-Lecreulx, C. Cambillau, and Y. Bourne. 1999. Crystal structure of the bifunctional N-acetylglucosamine 1-phosphate uridyltransferase from Escherichia coli: a paradigm for the related pyrophosphorylase superfamily. EMBO J. 18:4096-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bulai, T., D. Bratosin, A. Pons, J. Montreuil, and J. P. Zanetta. 2003. Diversity of the human erythrocyte membrane sialic acids in relation with blood groups. FEBS Lett. 534:185-189. [DOI] [PubMed] [Google Scholar]
- 10.Christendat, D., V. Saridakis, A. Dharamsi, A. Bochkarev, E. F. Pai, C. H. Arrowsmith, and A. M. Edwards. 2000. Crystal structure of dTDP-4-keto-6-deoxy-d-hexulose 3,5-epimerase from Methanobacterium thermoautotrophicum complexed with dTDP. J. Biol. Chem. 275:24608-24612. [DOI] [PubMed] [Google Scholar]
- 11.Cleasby, A., A. Wonacott, T. Skarzynski, R. E. Hubbard, G. J. Davies, A. E. Proudfoot, A. R. Bernard, M. A. Payton, and T. N. Wells. 1996. The x-ray crystal structure of phosphomannose isomerase from Candida albicans at 1.7 Å resolution. Nat. Struct. Biol. 3:470-479. [DOI] [PubMed] [Google Scholar]
- 12.Collaborative Computational Project Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 13.Cowtan, K., and P. Main. 1998. Miscellaneous algorithms for density modification. Acta Crystallogr. Sect. D 54:487-493. [DOI] [PubMed] [Google Scholar]
- 14.Dong, C., L. L. Major, A. Allen, W. Blankenfeldt, D. Maskell, and J. H. Naismith. 2003. High-resolution structures of RmlC from Streptococcus suis in complex with substrate analogs locate the active site of this class of enzyme. Structure 11:715-723. [DOI] [PubMed] [Google Scholar]
- 15.Dunwell, J. M. 1998. Cupins: a new superfamily of functionally diverse proteins that include germins and plant storage proteins. Biotechnol. Genet. Eng. Rev. 15:1-32. [DOI] [PubMed] [Google Scholar]
- 16.Dunwell, J. M., A. Purvis, and S. Khuri. 2004. Cupins: the most functionally diverse protein superfamily? Phytochemistry 65:7-17. [DOI] [PubMed] [Google Scholar]
- 17.Eisenstein, E., G. L. Gilliland, O. Herzberg, J. Moult, J. Orban, R. J. Poljak, L. Banerjei, D. Richardson, and A. J. Howard. 2000. Biological function made crystal clear—annotation of hypothetical proteins via structural genomics. Curr. Opin. Biotechnol. 11:25-30. [DOI] [PubMed] [Google Scholar]
- 18.Elliott, A. C., S. K., M. L. Sinnott, P. J. Smith, J. Bommuswamy, Z. Guo, B. G. Hall, and Y. Zhang. 1992. The catalytic consequences of experimental evolution. Studies on the subunit structure of the second (ebg) beta-galactosidase of Escherichia coli, and on catalysis by ebgab, an experimental evolvant containing two amino acid substitutions. Biochem. J. 282:155-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frosch, M., C. Weisgerber, and T. F. Meyer. 1989. Molecular characterization and expression in Escherichia coli of the gene complex encoding the polysaccharide capsule of Neisseria meningitidis group B. Proc. Natl. Acad. Sci. USA 86:1669-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frosch, M., U. Edwards, K. Bousset, B. Krausse, and C. Weisgerber. 1991. Evidence for a common molecular origin of the capsule gene loci in gram-negative bacteria expressing group II capsular polysaccharides. Mol. Microbiol. 5:1251-1263. [DOI] [PubMed] [Google Scholar]
- 21.Fusetti, F., K. H. Schroter, R. A. Steiner, P. I. van Noort, T. Pijning, H. J. Rozeboom, K. H. Kalk, M. R. Egmond, and B. W. Dijkstra. 2002. Crystal structure of the copper-containing quercetin 2,3-dioxygenase from Aspergillus japonicus. Structure 10:259-268. [DOI] [PubMed] [Google Scholar]
- 22.Galperin, M. Y., and E. V. Koonin. 2000. Who's your neighbor? New computational approaches for functional genomics. Nat. Biotechnol. 18:609-613. [DOI] [PubMed] [Google Scholar]
- 23.Ganguli, S., G. Zapata, T. Wallis, C. Reid, G. Boulnois, W. F. Vann, and I. S. Roberts. 1994. Molecular cloning and analysis of genes for sialic acid synthesis in Neisseria meningitidis group B and purification of the meningococcal CMP-NeuNAc synthetase enzyme. J. Bacteriol. 176:4583-4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giraud, M. F., G. A. Leonard, R. A. Field, C. Berlind, and J. H. Naismith. 2000. RmlC, the third enzyme of dTDP-l-rhamnose pathway, is a new class of epimerase. Nat. Struct. Biol. 7:398-402. [DOI] [PubMed] [Google Scholar]
- 25.Giraud, M. F., and J. H. Naismith. 2000. The rhamnose pathway. Curr. Opin. Struct. Biol. 10:687-696. [DOI] [PubMed] [Google Scholar]
- 26.Gouet, P., E. Courcelle, D. I. Stuart, and F. Metoz. 1999. ESPript: multiple sequence alignments in PostScript. Bioinformatics 15:305-308. [DOI] [PubMed] [Google Scholar]
- 27.Holm, L., and C. Sander. 1998. Touring protein fold space with Dali/FSSP. Nucleic Acids Res. 26:316-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holm, R. H., P. Kennepohl, and E. I. Solomon. 1996. Structural and functional aspects of metal sites in biology. Chem. Rev. 96:2239-2314. [DOI] [PubMed] [Google Scholar]
- 29.Howard, R. J., G. Reuter, J. W. Barnwell, and R. Schauer. 1986. Sialoglycoproteins and sialic acids of Plasmodium knowlesi schizont-infected erythrocytes and normal rhesus monkey erythrocytes. Parasitology 92:527-543. [DOI] [PubMed] [Google Scholar]
- 30.Jones, T. A., J. Y. Zou, S. W. Cowan, and M. Kjeldgaard. 1991. Improved methods for building models in electron density maps and the location of errors in these models. Acta Crystallogr. Sect. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 31.Kalivoda, K. A., S. M. Steenbergen, E. R. Vimr, and J. Plumbridge. 2003. Regulation of sialic acid catabolism by the DNA-binding protein NanR in Escherichia coli. J. Bacteriol. 185:4806-4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khuri, S., F. T. Bakker, and J. M. Dunwell. 2001. Phylogeny, function, and evolution of the cupins, a structurally conserved, functionally diverse superfamily of proteins. Mol. Biol. Evol. 18:593-605. [DOI] [PubMed] [Google Scholar]
- 33.Kolker, E., K. S. Makarova, S. Shabalina, A. F. Picone, S. Purvine, T. Holzman, T. Cherny, D. Armbruster, R. S. Munson, Jr., G. Kolesov, D. Frishman, and M. Y. Galperin. 2004. Identification and functional analysis of ‘hypothetical’ genes expressed in Haemophilus influenzae. Nucleic Acids Res. 32:2353-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kolker, E., S. Purvine, M. Y. Galperin, S. Stolyar, D. R. Goodlett, A. I. Nesvizhskii, A. Keller, T. Xie, J. K. Eng, E. Yi, L. Hood, A. F. Picone, T. Cherny, B. C. Tjaden, A. F. Siegel, T. J. Reilly, K. S. Makarova, B. O. Palsson, and A. L. Smith. 2003. Initial proteome analysis of model microorganism Haemophilus influenzae strain Rd KW20. J. Bacteriol. 185:4593-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kraulis, P. J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946-950. [Google Scholar]
- 36.Merritt, E. A., and D. J. Bacon. 1997. Raster3D—photorealistic molecular graphics. Methods Enzymol. 277:505-524. [DOI] [PubMed] [Google Scholar]
- 37.Miller, R., S. M. Gallo, H. G. Khalak, and C. M. Weeks. 1994. SnB: crystal structure determination via Shake-and-Bake. J. Appl. Crystallogr. 27:613-621. [Google Scholar]
- 38.Mosimann, S. C., M. Gilbert, D. Dombroswki, R. To, W. Wakarchuk, and N. C. Strynadka. 2001. Structure of a sialic acid-activating synthetase, CMP-acylneuraminate synthetase, in the presence and absence of CDP. J. Biol. Chem. 276:8190-8196. [DOI] [PubMed] [Google Scholar]
- 39.Muchmore, E. A., S. Diaz, and A. Varki. 1998. A structural difference between the cell surfaces of humans and the great apes. Am. J. Phys. Anthropol. 107:187-198. [DOI] [PubMed] [Google Scholar]
- 40.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by maximum-likelihood method. Acta Crystallogr. Sect. D 53:240-255. [DOI] [PubMed] [Google Scholar]
- 41.Murzin, A. G., S. E. Brenner, T. Hubbard, and C. Chothia. 1995. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247:536-540. [DOI] [PubMed] [Google Scholar]
- 42.Otwinowski, Z. 1991. Maximum likelihood refinement of heavy atom parameters, p. 80-88. In W. Wolf, P. R. Evans, and A. G. W. Leslie (ed.), Proceedings of the CCP4 Study Weekend. Daresbury Laboratory, Warrington, United Kingdom.
- 43.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 44.Overbeek, R., M. Fonstein, M. D'Souza, G. D. Pusch, and N. Maltsev. 1999. The use of gene clusters to infer functional coupling. Proc. Natl. Acad. Sci. USA 96:2896-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plumbridge, J., and E. Vimr. 1999. Convergent pathways for utilization of the amino sugars N-acetylglucosamine, N-acetylmannosamine, and N-acetylneuraminic acid by Escherichia coli. J. Bacteriol. 181:47-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pochapsky, T. C., S. S. Pochapsky, T. Ju, H. Mo, F. Al-Mjeni, and M. J. Maroney. 2002. Modeling and experiment yields the structure of acireductone dioxygenase from Klebsiella pneumoniae. Nat. Struct. Biol. 9:966-972. [DOI] [PubMed] [Google Scholar]
- 47.Read, J., J. Pearce, X. Li, H. Muirhead, J. Chirgwin, and C. Davies. 2001. The crystal structure of human phosphoglucose isomerase at 1.6 A resolution: implications for catalytic mechanism, cytokine activity and haemolytic anaemia. J. Mol. Biol. 309:447-463. [DOI] [PubMed] [Google Scholar]
- 48.Schauer, R., M. Wember, F. Wirtz-Peitz, and C. Ferreira do Amaral. 1971. Studies on the substrate specificity of acylneuraminate pyruvate-lyase. Hoppe-Seyler's Z. Physiol. Chem. 352:1073-1080. [DOI] [PubMed] [Google Scholar]
- 49.Solana, S., A. A. Reglero, H. Martinez-Blanco, B. Revilla-Nuin, I. G. Bravo, L. B. Rodriguez-Aparicio, and M. A. Ferrero. 2001. N-acetylneuraminic acid uptake in Pasteurella (Mannheimia) haemolytica A2 occurs by an inducible and specific transport system. FEBS Lett. 509:41-46. [DOI] [PubMed] [Google Scholar]
- 50.Steiner, R. A., K. H. Kalk, and B. W. Dijkstra. 2002. Anaerobic enzyme substrate structures provide insight into the reaction mechanism of the copper-dependent quercetin 2,3-dioxygenase. Proc. Natl. Acad. Sci. USA 99:16625-16630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swan, M. K., J. T. Solomons, C. C. Beeson, T. Hansen, P. Schonheit, and C. Davies. 2003. Structural evidence for a hydride transfer mechanism of catalysis in phosphoglucose isomerase from Pyrococcus furiosus. J. Biol. Chem. 278:47261-47268. [DOI] [PubMed] [Google Scholar]
- 52.Tatusov, R. L., N. D. Fedorova, J. D. Jackson, A. R. Jacobs, B. Kiryutin, E. V. Koonin, D. M. Krylov, R. Mazumder, S. L. Mekhedov, A. N. Nikolskaya, B. S. Rao, S. Smirnov, A. V. Sverdlov, S. Vasudevan, Y. I. Wolf, J. J. Yin, and D. A. Natale. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Titus, G. P., H. A. Mueller, J. Burgner, S. Rodriguez De Cordoba, M. A. Penalva, and D. E. Timm. 2000. Crystal structure of human homogentisate dioxygenase. Nat. Struct. Biol. 7:542-546. [DOI] [PubMed] [Google Scholar]
- 55.Todd, A. E., C. A. Orengo, and J. M. Thornton. 2001. Evolution of function in protein superfamilies, from a structural perspective. J. Mol. Biol. 307:1113-1143. [DOI] [PubMed] [Google Scholar]
- 56.Traving, C., and R. Schauer. 1998. Structure, function and metabolism of sialic acids. Cell. Mol. Life Sci. 54:1330-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vimr, E., and C. Lichtensteiger. 2002. To sialylate, or not to sialylate: that is the question. Trends Microbiol. 10:254-257. [DOI] [PubMed] [Google Scholar]
- 58.Vimr, E., C. Lichtensteiger, and S. Steenbergen. 2000. Sialic acid metabolism's dual function in Haemophilus influenzae. Mol. Microbiol. 36:1113-1123. [DOI] [PubMed] [Google Scholar]
- 59.Vimr, E. R., K. A. Kalivoda, E. L. Deszo, and S. M. Steenbergen. 2004. Diversity of microbial sialic acid metabolism. Microbiol. Mol. Biol. Rev. 68:132-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vimr, E. R., and F. A. Troy. 1985. Identification of an inducible catabolic system for sialic acids (nan) in Escherichia coli. J. Bacteriol. 164:845-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walters, D. M., V. L. Stirewalt, and S. B. Melville. 1999. Cloning, sequence, and transcriptional regulation of the operon encoding a putative N-acetylmannosamine-6-phosphate epimerase (nanE) and sialic acid lyase (nanA) in Clostridium perfringens. J. Bacteriol. 181:4526-4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Woo, E. J., J. M. Dunwell, P. W. Goodenough, A. C. Marvier, and R. W. Pickersgill. 2000. Germin is a manganese containing homohexamer with oxalate oxidase and superoxide dismutase activities. Nat. Struct. Biol. 7:1036-1040. [DOI] [PubMed] [Google Scholar]
- 63.Woo, E. J., J. Marshall, J. Bauly, J. G. Chen, M. Venis, R. M. Napier, and R. W. Pickersgill. 2002. Crystal structure of auxin-binding protein 1 in complex with auxin. EMBO J. 21:2877-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]