Abstract
Methylglyoxal (MG) is a toxic metabolite known to accumulate in various cell types. We detected in vivo conversion of MG to acetol in MG-accumulating Escherichia coli cells by use of 1H nuclear magnetic resonance (1H-NMR) spectroscopy. A search for homologs of the mammalian aldo-keto reductases (AKRs), which are known to exhibit activity to MG, revealed nine open reading frames from the E. coli genome. Based on both sequence similarities and preliminary characterization with 1H-NMR for crude extracts of the corresponding mutant strains, we chose five genes, yafB, yqhE, yeaE, yghZ, and yajO, for further study. Quantitative assessment of the metabolites produced in vitro from the crude extracts of these mutants and biochemical study with purified AKRs indicated that the yafB, yqhE, yeaE, and yghZ genes are involved in the conversion of MG to acetol in the presence of NADPH. When we assessed their in vivo catalytic activities by creating double mutants, all of these genes except for yqhE exhibited further sensitivities to MG in a glyoxalase-deficient strain. The results imply that the glutathione-independent detoxification of MG can occur through multiple pathways, consisting of yafB, yqhE, yeaE, and yghZ genes, leading to the generation of acetol.
Methylglyoxal (MG) is a widely occurring ketoaldehyde that is accumulated under physiological conditions with uncontrolled carbohydrate metabolism (1, 11, 20). MG synthesis is mediated by enzymes, including methylglyoxal synthase, cytochrome P450, and amine oxidase, which are involved in glycolytic bypass, acetone metabolism, and amino acid breakdown, respectively (8, 18). In eukaryotic cells, MG is also generated by nonenzymatic fragmentation of dihydroxyacetone phosphate or glyceraldehyde 3-phophate (28). MG is a highly toxic electrophile and reacts with cellular macromolecules, including DNA and proteins (16, 18).
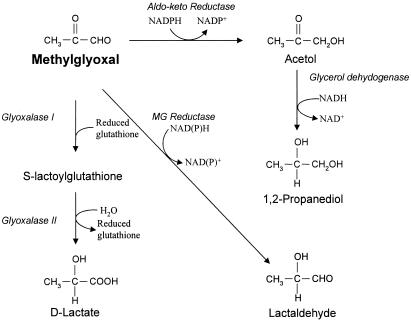
There are various ways that the cellular degradation of MG occurs (Fig. 1). The glyoxalase system, consisting of glyoxalase I and II, converts MG into d-lactate in the presence of glutathione (30). The conversion of MG into lactaldehyde by the MG reductase was also suggested (26, 29). The enzymes, presumably aldose and aldehyde reductases, mediating the reduction of MG to acetol and d-lactaldehyde have been reported for Escherichia coli, yeast (Saccharomyces cerevisiae), plants, and mammals (16, 25, 31). The mammalian aldo-keto reductase (AKR) family AKR1, AKR1A1 (EC 1.1.1.2), and AKR1B1 (EC 1.1.1.21) and the family AKR7, AKR7A2, and AKR7A5 convert methylglyoxal to acetol in the presence of NADPH (14, 15, 27, 31, 32). The E. coli YghZ protein, belonging to the AKR14 family, was recently characterized as an enzyme involved in MG reduction and was also shown to enhance resistance to MG when overproduced (12).
FIG. 1.
Metabolic pathways for methylglyoxal. MG can be converted to d-lactate, lactaldehyde, and acetol by glyoxalase, MG reductase, and aldo-keto reductase, respectively (15, 17). MG reductase and aldehyde reductase of E. coli were as reported previously (25, 29), with the identities of the corresponding genes unknown.
Aldo-keto reductases encompass a large superfamily of NADPH-dependent oxidoreductases that reduce various aldehydes and ketones (17). They all share a common (α/β)8-barrel motif characteristic of triose phosphate isomerase. Most AKRs are monomeric, with the exception of the dimeric mammalian AKR7 family enzymes (21) and yeast xylose reductases (19). The physiological functions of these enzymes are largely unknown, due to the broad spectrum of substrate specificities. Although a number of mammalian and eukaryotic AKRs have been characterized, only a small subset of bacterial AKRs has had their substrate specificities reported (10). E. coli YafB and YqhE have been characterized as 2,5-diketo-gluconate (2,5-DKG) reductases, while YghZ was shown to reduce aldehydes and ketones, including MG.
In this study, we observed the metabolic production of acetol in methylglyoxal-accumulating E. coli cells and demonstrated that four E. coli AKRs, YafB, YqhE, YeaE, and YghZ, are involved in the production of acetol from MG. These enzymes were purified and shown to catalyze NADPH-dependent MG reduction to acetol. In addition, strains lacking the corresponding genes, except for yqhE, exhibited increased MG susceptibilities in vivo. MG detoxification through multiple pathways appears to be due to the broad substrate specificities of the AKRs.
MATERIALS AND METHODS
Bacterial strains and plasmids.
All strains used are derivatives of E. coli K-12. MG1655 was used as a wild-type strain for gene disruption and for AKR gene amplification. The CK281 [leu thr his thi rpsL lacY xyl ara tonA tsx Δrbs(D-R)::spc] strain harboring pJK5 (RbsD-overproducing plasmid derived from pBR322) and pJK10 (containing rbsACBK, derived from pACYC184) was used for nuclear magnetic resonance (NMR) analysis of metabolites from the intact cell. This CK281/pJK5/pJK10 strain exhibits an enhanced production of mutarotase, which results in methylglyoxal accumulation (20). Disruptions of the yqhE and yajO genes were created using a previously described method for chromosomal gene inactivation (9). Other AKR mutants were obtained from the E. coli Genome Project (University of Wisconsin, Madison) and transferred to CK281 and MG1655 using P1. To construct AKR- and glyoxalase-deficient strains, we introduced the gloA::kan allele (MJF388) (23) into each AKR mutant. To transfer gloA::kan, the nearby mlc::Tn10 marker was used and verified by PCR. The BL21(DE3) strain was used (Novagen) for the overexpression and purification of proteins.
Sample preparation for NMR analysis.
For the analysis of metabolites from intact cells, supernatants were taken from cells with excess or lack of mutarotase as described previously (20). The cells were cultured to an optical density at 600 nm (OD600) of 1.0 in an M9 medium containing 0.4% glycerol with the appropriate antibiotics to which 0.2% d-ribose was added, and they were incubated for a further 3 h. The cells were removed by centrifugation at 15,000 × g for 30 min, and the resulting supernatants were stored at −20°C until measurement. To detect the MG reduction activity of cell extracts from each AKR mutant, cells were cultured overnight in an LB medium. The cells were then harvested by centrifugation, washed twice with 100 mM potassium phosphate (pH 7.0), resuspended in the same buffer, and disrupted by use of a sonicator. Cell debris was removed by centrifugation at 15,000 × g for 30 min, and the resulting supernatants were dialyzed three times, each for 5 h, against 100 mM potassium phosphate (pH 7.0). The supernatants were then stored at −20°C until measurement.
NMR analyses of metabolites.
A Bruker AVANCE-400 NMR spectrometer equipped with a temperature controller was used for NMR experiments with a 5-mm NMR tube. The sample was kept at 28°C during measurement. The 1H-NMR measurement was carried out for quantitative analysis using a 300 pulse with a long relaxation delay. The duration of acquisition was approximately 5 min for each NMR spectrum. All measurements were carried out in 600 μl of solution with 10% D2O as a locking substance. For the characterization of metabolites from the enzymatic reactions of MG with crude extracts, the NMR measurement was carried out with crude extracts (0.6 mg), MG (3 mM), coenzymes (1 mM NADH, 1 mM NADPH), buffer (100 mM potassium phosphate, pH 7.0), and D2O. The NMR data were collected 12 h after mixing. For the reactions with the purified AKR proteins, 1H-NMR measurement was taken after 10 min of mixing with the purified protein (10 μg), MG (3 mM), coenzyme (1 mM NADPH), buffer (100 mM potassium phosphate, pH 7.0), and D2O.
Semiquantitative reverse transcription-PCR (RT-PCR).
To examine changes in the expressions of the five AKR genes upon MG addition, MG1655 cells were cultured to an OD600 of 0.4 in LB and divided into two separate flasks, one with and one without 0.2 mM MG. The samples were further incubated to an OD600 of 0.8, from which total RNAs were isolated using an RNeasy extraction kit (QIAGEN) according to the manufacturer's instructions. After the treatment of the extracted RNA (1 μg) with DNase I, single-stranded cDNA was synthesized with random hexamers in a 20-μl reaction volume using the SuperScript synthesis system (Invitrogen). PCR was then carried out with the cDNA by use of the same primers for cloning AKR genes. The ompA primers (5′-TCAGGGCGTTCAACTGACCG-3′ and 5′-GCCTGCGGCTGAGTTACAAC-3′) were used for an internal control. The PCR was performed with Taq DNA polymerase at 94°C for 3 min, followed by 23 cycles of reaction at 95°C for 30 s, 50°C for 40 s, and 72°C for 120 s, with a final elongation step at 72°C for 10 min. The PCR products were analyzed by agarose gel electrophoresis after normalization of template loading with an amount of ompA. The band intensities were compared using TINA (Raytest).
Identification of acetol by GC/MS.
For the identification of acetol, vacuum distillation was carried out to purify the compound from the culture medium. Initially, the culture medium was fractionally distilled under a vacuum, and the distillates were collected stepwise at the temperature ranges of 70 to 80°C, 80 to 85°C, and 85 to 90°C. After a preanalysis of the distillates, the distillations were repeated approximately 20 times at 85 to 90°C in order to concentrate the sample from 500 ml of the culture medium. A Shimadzu QP 5050A gas chromatography (GC)/ mass spectroscopy (MS) instrument was used to identify the final sample. After GC separation using a DB-WAX capillary column at an oven temperature of 300°C, the metabolite was separated, and the peaks obtained were analyzed with a mass spectrometer. An electron ionization with 70 eV energy was applied to acquire the mass spectrum of the sample.
Cloning of E. coli AKRs by PCR.
The genomic DNA of E. coli MG1655 was used as the template for the amplification of AKRs. The PCR primer pairs used were 5′-GGGCATATGGCTATCCCTGCATTTGGTTTAGG-3′ and 5′-GGGCTCGAGTTAATCCCATTCAGGAGCCAGAC-3′ for yafB, 5′-GGGCATATGGCTAATCCAACCGTTATTAAG-3′ and 5′-GGGCTCGAGTTAGCCGCCGAACTGGTCAGG-3′ for yqhE, 5′-GGGCATATGCAACAAAAAATGATTCAATTT-3′ and 5′-GGGCTCGAGTCACACCATATCCAGCGCAGT-3′ for yeaE, 5′-GGGCATATGGTCTGGTTAGCGAATCCC-3′ and 5′-GGGCTCGAGTCATTTATCGGAAGACGCCTG-3′ for yghZ, and 5′-GGGCATATGCAATACAACCCCTTAGGA-3′ and 5′-GGGCTCGAGTTTAAATCCTACGACAGGATGCG-3′ for yajO (the NdeI and XhoI sites are underlined). The primers contained NdeI and XhoI restriction sites at the 5′ and 3′ ends, respectively. The PCR products were digested with NdeI and XhoI and introduced into the NdeI and XhoI restriction sites of pET21b (Novagen). The YajO protein was expressed with an N-terminal His tag. Since all of the PCR products except for YajO harbor stop codons, all proteins were expressed in their native forms without His tags. The constructs were verified by DNA sequencing and named pET-YafB, pET-YqhE, pET-YeaE, pET-YghZ, and pET-YajO-His.
Purification of AKRs.
The pET-AKR plasmids were transformed into the BL21(DE3) strain, and the transformed cells were grown in an LB medium to an OD600 at 0.5. The proteins were expressed by an addition of 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h. The cells were harvested by centrifugation, washed twice with 20 mM Tris-Cl (pH 8.3), resuspended in the same buffer with 0.1 mM phenylmethylsulfonyl fluoride, and disrupted by a sonicator. Cell debris was removed by centrifugation at 15,000 × g for 30 min, and the resulting supernatant was applied to a DEAE-Sepharose column (Amersham Biosciences) equilibrated with 20 mM Tris-Cl (pH 8.3). The proteins were eluted with a linear gradient of 0 to 0.5 M NaCl in the same buffer. The AKR proteins were eluted from within a range of between 0.15 and 0.2 M NaCl concentration. The fractions that contained AKRs were pooled and dialyzed against a 20 mM Tris-Cl (pH 7.0) buffer and applied to an Affi-Gel Blue column (Bio-Rad) equilibrated with 20 mM Tris-Cl (pH 7.0). The proteins were eluted with a linear gradient of 0 to 1 M NaCl in the same buffer. The AKR proteins were eluted from within a range between 0.3 and 0.5 M NaCl concentration. The fractions that contained AKRs were combined and concentrated with an ultrafiltration cell (Amicon, Beverly, MA) to 2 ml, and applied onto a Superdex 200 (Amersham Biosciences) gel filtration column (1.6 by 70 cm) equilibrated with a 50 mM potassium phosphate buffer (pH 7.0). The fractions containing AKRs were pooled and stored at −70°C.
The histidine-tagged YajO protein was purified as described by the manufacturer's instructions (Novagen). pET-YajO-His was transformed into E. coli BL21(DE3) and cultured in LB with ampicillin (100 μg/ml) at 30°C to the mid-exponential phase (OD600 = 0.5). IPTG (0.5 mM) was added, and then the cells were incubated for 3 h. The cells were harvested and resuspended in a binding buffer (20 mM Tris-Cl, pH 7.9, 5 mM imidazole, 500 mM NaCl). After disruption by sonication, cell debris was eliminated by centrifugation at 15,000 × g for 15 min, and the protein was purified by standard procedures with His-Bind resin (Novagen). The concentration and purity (over 95%) was determined using the Bradford reagent (6) and by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (22).
Enzyme assays.
Enzyme activities of the purified E. coli AKRs were measured at 25°C using a Beckman Coulter DU800 spectrophotometer by monitoring the initial rate at 340 nm with oxidation of NADPH/NADH or reduction of NADP+. The standard assay for reducing aldehyde or ketone was carried out in 1 ml of 100 mM potassium phosphate buffer (pH 7.0) with 0.1 mM NADPH as a coenzyme. The aldehyde and ketone substrates were obtained from Sigma-Aldrich and Wako (Osaka, Japan), and 2,5-diketo-d-gluconate was from J. G. Pan (Korea Research Institute of Bioscience and Biotechnology, Taejon, Korea). For measuring specific activities, concentrations of substrates were 0.1, 1, and 10 mM. In all reactions, the nonenzymatic rates were subtracted from the observed initial reaction rates. The apparent Km and kcat values were determined by measuring the initial rate over a range of substrate concentrations. The kinetic parameters were determined with Sigmaplot (SPSS Inc., Chicago, IL) by fitting to the Michaelis-Menten equation.
MG susceptibilities of strains with AKR deletions.
Cell viability after being exposed to MG was determined for strains with AKR deletions. Cells with AKR deletions were grown overnight at 37°C in an M9 medium supplemented with 0.4% (wt/vol) glycerol, and aliquots of 3 ml were washed with the M9 medium, resuspended in 5 ml of the M9 medium containing 0.4% (wt/vol) glycerol, and grown to an early exponential phase (OD600 = 0.4). The cells were diluted 10-fold into a fresh M9 medium with 0.4% (wt/vol) glycerol, to which 0.3 mM MG was added. After the MG treatment, samples were taken at various times during incubation and plated onto LB agar plates. After 20 h of incubation at 37°C, colonies were counted to measure rates of survival.
RESULTS
Detection of acetol from methylglyoxal-producing E. coli cells.
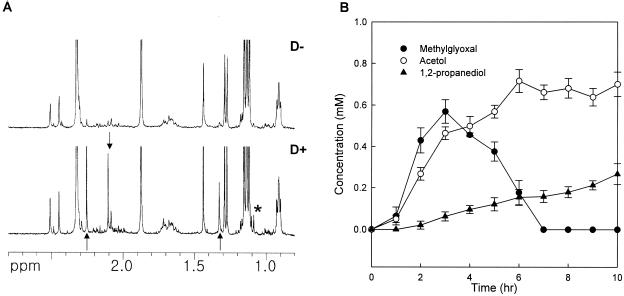
Previously, we observed that E. coli cells with enhanced mutarotase activity were killed by growth on ribose due to an accumulation of MG (20). The 1H-NMR spectroscopy analysis of these cells revealed that MG was converted to an unknown compound (Fig. 2A). The peak was further purified by vacuum distillation at 85 to 90°C, from which two fragment peaks corresponding to [CH3CO]+ and [CH2OH]+ were identified from the GC/MS spectrum. Based on these data, we characterized the unknown compound as acetol, which has not previously been reported as a fermentation product of prokaryotic cells. Commercially available acetol exhibits NMR and GC/mass spectra the same as those of the unknown compound (data not shown). The NMR spectrum of MG is seen as two peaks at 2.30 and 1.39 ppm and originating from —CH3 groups of two conformers with partial double bonds between the two carbonyls of the molecule. Commercially available MG exhibited the same NMR spectrum. On the other hand, acetol generates a single peak (2.16 ppm) for —CH3, because CH3CO rotates freely around the bond between CH3CO— and —CH2OH. The production of MG rises sharply, while it disappears almost completely after several hours (Fig. 2B). However, the level of acetol reaches a steady state, although a small amount of acetol was converted to 1,2-propanediol (1,2-PDO) (Fig. 2B). The presence of native reductases mediating acetol conversion to 1,2-PDO, including glycerol dehydrogenase, which indeed enhanced this activity when it was overproduced, has been proposed (4). The conversion of acetol to 1,2-PDO appears to be slow, and the compound was detected by 1H-NMR only in the declining stage of MG production (Fig. 2B).
FIG. 2.
Detection of acetol from methylglyoxal-producing E. coli cells. (A) 1H-NMR spectra of culture media accumulating MG from ribose. The upper and lower NMR spectra were obtained from culture media of CK281/pBR322/pJK10 (D−) and CK281/pJK5/pJK10 (D+). The samples were taken after 3 h of ribose addition. The strain producing excess mutarotase (RbsD protein) was previously shown to accumulate methylglyoxal (20). The arrow pointing downward indicates acetol, the arrows pointing upward indicate methylglyoxal, and the asterisk indicates 1,2-propanediol. (B) After 0.2% d-ribose was added, samples were taken at the indicated times. Amounts of the metabolites shown in the key were determined by 1H-NMR spectroscopy. The error bars show standard deviations of three replicates.
Search for E. coli enzymes involved in acetol production.
As in mammals with considerable numbers of aldo-keto reductase enzymes, E. coli has nine open reading frames with amino acid similarities to these enzymes (http://www.sanger.ac.uk/Software/Pfam/). Among these, yqhE, yafB, and yeaE share similarities with both AKR1B1 (36%, 32%, and 30% amino acid identities with BLAST, respectively) and AKR1A1 (36%, 29%, and 28%), which have been demonstrated to catalyze NADPH-dependent conversions of MG to acetol. The yajO and yghZ genes share similarities (27% and 31% identities) with AKR7A5 members that were also shown to be involved in MG reduction. The purified E. coli YghZ protein was demonstrated to have MG reduction properties, with the identity of the product unknown, and its overproduction conferred slightly enhanced protection to MG (12). These five E. coli AKRs have the conserved catalytic tetrad of amino acids (Tyr, His, Asp, and Lys) found in most AKRs, as do the other four E. coli homologs: Tas, YdjG, YdbC, and YdhF.
In order to test whether these E. coli AKRs confer activity to MG, we obtained deletions of these genes by PCR-based gene inactivation (see Materials and Methods for the procedures for yqhE and yajO) or received mutants from the E. coli genome project (University of Wisconsin, Madison). The initial assessment of the genes for their MG-reducing capabilities was made with the strain that accumulates MG upon the addition of ribose. Unexpectedly, the levels of metabolites released from the cells varied greatly among different mutants (data not shown). Although levels of MG or acetol in the AKR mutants are generally higher or lower, respectively, than those in the wild type (not shown), analysis of other fermentation products, such as acetate, lactate, and pyruvate, revealed a fairly complicated nature of cell metabolism, perhaps due to the permeability of metabolites, the synthesis/utilization of metabolites, and the intracellular availability of reduced pyridine nucleotides.
MG-reducing activity in crude extracts of the mutant cells.
In order to exclude the possibility of physiological complication in intact cells, we prepared crude extracts (0.6 mg) from the AKR mutants of E. coli and incubated them with 3 mM MG in both the presence and the absence of NADPH (1 mM) and NADH (1 mM), which were analyzed by 1H-NMR spectroscopy. Without NADPH, acetol was undetected. In this condition, the major product from MG was lactate, which accumulated up to about 2 mM after several hours of saturation (data not shown). The remaining metabolic products included acetol, 1,2-PDO, and pyruvate, whose concentrations were in the range of 0.1 to 0.4 mM. These levels varied in different AKR mutants. For example, accumulations of acetol in yafB, yqhE and yghZ mutants after a couple of hours of MG addition were about 75% of that in the wild type (ca. 0.2 mM), whereas other mutants produced levels of acetol similar to that in the wild type.
Since prolonged incubation with MG promotes further conversion of these metabolites into 1,2-PDO, we compared the combined amount of acetol and 1,2-PDO in the mutants with that in the wild type. After 12 h of incubation, yafB, yqhE, and yghZ mutant cell extracts with NADPH and MG exhibited 36%, 27%, and 23% decreases in the amount of acetol plus 1,2-PDO, respectively, compared to that in the CK281 control (Table 1). This indicates that YafB, YqhE, and YghZ significantly contribute to cellular MG detoxification, generating acetol. For unknown reasons, the level of acetol plus 1,2-PDO was higher in the yajO mutant than in the wild type. Based on these results, we have chosen five genes (yafB, yqhE, yghZ, yeaE, and yajO) that show greater similarities to the mammalian AKRs as well as significant decreases in the production of acetol and 1,2,-PDO for further study.
TABLE 1.
Methylglyoxal reductions in E. coli AKR mutants
| Strain | Concentration (mM) of acetol plus 1,2-PDOb |
|---|---|
| Wild typea | 0.44 (1.00) |
| ΔyafB | 0.28 (0.64) |
| ΔyqhE | 0.32 (0.73) |
| ΔyeaE | 0.41 (0.93) |
| ΔyghZ | 0.34 (0.77) |
| ΔyajO | 0.61 (1.39) |
| ΔydhF | 0.45 (1.02) |
| ΔydjG | 0.41 (0.93) |
| ΔydbC | 0.50 (1.14) |
| ΔTAS | 0.44 (1.00) |
CK281/pJK5/pJK10 was used. AKR deletion mutants were constructed in the same background by P1.
Concentrations of acetol plus 1,2-propanediol were determined by 1H-NMR spectroscopy. The NMR measurements were carried out with crude extracts (0.6 mg), MG (3 mM), 1 mM NADPH, 100 mM potassium phosphate, pH 7.0, and D2O. The NMR data were collected after 12 hours of mixing. The values in parenthesis represent mutant-to-wild-type ratios.
Reduction of methylglyoxal to acetol by purified E. coli AKRs.
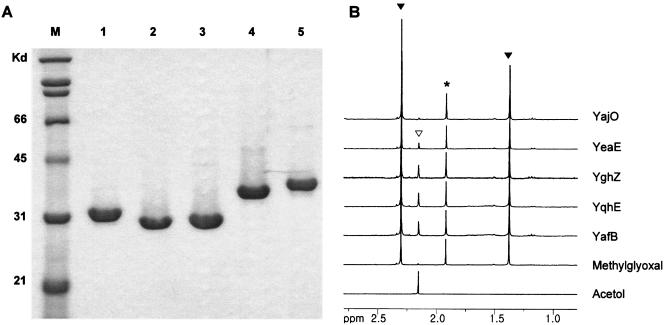
We cloned the E. coli AKR genes into the pET21b vector, from which the enzymes were purified as described in Materials and Methods. The purified proteins visualized by SDS-polyacrylamide gel electrophoresis were over 95% homogeneous (Fig. 3A). The apparent molecular masses of YafB, YqhE, YeaE, YghZ, and YajO-His6 proteins were in good agreement with the calculated ones of 29.4, 31.1, 31.0, 38.8, and 37.2 kDa, respectively.
FIG. 3.
Production of acetol from methylglyoxal by purified E. coli AKRs. (A) Native as well as His-tagged AKRs were purified as described in Materials and Methods and were visualized by 12% SDS-PAGE. Lanes: M, molecular mass marker; 1, YafB; 2, YqhE; 3, YeaE; 4, YghZ; 5, YajO-His6. (B) Production of acetol by the AKRs after reactions with 10 μg purified proteins, 3 mM MG, and 1 mM NADPH in 100 mM potassium phosphate buffer (pH 7.0) was detected by 1H-NMR spectroscopy. For the standard, NMR spectra of commercially available acetol and MG were also shown (▾, MG; ▿, acetol; *, contaminated acetate from the MG reagent). In these reactions, 1, 2-PDO was undetected.
The purified YafB, YqhE, YeaE, and YghZ proteins convert MG to acetol in the presence of NADPH, as determined by NMR spectroscopy, whereas the activity of YajO protein was negligible (Fig. 3B). The results are consistent with those obtained from the crude extracts. Since we were unable to detect lactaldehyde peaks on the NMR spectra after 10-h incubations of YafB, YqhE, YeaE, and YghZ with MG and NADPH, these enzymes appear to be specific for the acetol, but not for the lactaldehyde, production of E. coli. In a previous experiment (12), YghZ was shown to reduce MG, although the identity of the reaction product was undetermined.
For the purified enzymes, the apparent kinetic constants for MG were determined by measuring the initial rate of coenzyme oxidation (Table 2). As in the NMR experiments, the YafB, YqhE, YeaE, and YghZ proteins showed significant MG-reducing activities (over 1,800 nmol/min/mg with 1 mM MG). The enzyme activity was negligible with NADH (<3% of the NADPH-dependent activity). The reverse reaction, from acetol to MG, was not seen under our experimental conditions (data not shown). The Km values of MG for YafB, YqhE, and YeaE are 2.46, 2.05, and 2.09 mM, respectively, which are lower than that of YghZ (3.24 mM [13]). According to previous data, MG can be accumulated to concentrations of up to 1 to 1.4 mM in a culture medium (11, 20). Therefore, it appears that such Km values are physiologically relevant. The kcat/Km values for YafB, YqhE, YeaE, and YghZ (12) are 7.13 × 105, 8.09 × 105, 8.2 × 104, and 4.68 × 104 min−1 M−1, respectively (Table 2).
TABLE 2.
Apparent Michaelis constants for methylglyoxal
| Enzyme (reference) | Apparent Michaelis constants for MGa
|
||
|---|---|---|---|
| Km (mM) | kcat (min−1) | kcat/Km (min−1 M−1) | |
| YafB | 2.46 ± 0.12 | 1,749 ± 31 | 7.13 × 105 |
| YqhE | 2.05 ± 0.10 | 1,657 ± 29 | 8.09 × 105 |
| YeaE | 2.09 ± 0.06 | 171 ± 3 | 0.82 × 105 |
| YghZ | 6.91 ± 0.06 | 661 ± 3 | 0.96 × 105 |
| YghZ (13) | 3.24 ± 0.24 | 151 ± 5 | 4.68 × 104 |
The assays were carried out in triplicate. The Km and kcat values were obtained by measuring initial reaction velocities over a range of substrate concentrations. The numbers represent the means ± standard errors of the means.
Substrate spectrum of E. coli AKR enzymes.
One feature of the AKRs is their broad spectrum of substrate specificities. To determine whether these five AKRs have aldehyde- and ketone-reducing activities, we tested various aldehydes and ketones as substrates (Table 3). YafB and YqhE are similar in their substrate specificities, favoring aromatic aldehydes (4-nitrobenzaldehyde, 3-nitrobenzaldehyde, and benzaldehyde), MG, and phenylglyoxal, while catalyses of 2-carboxybenzaldehyde, xylose, and glucuronic acid are less efficient. YeaE also has a similar substrate specificity. YafB and YqhE display considerable homology with both AKR1A and AKR1B members of mammalian enzymes, although their substrate specificities are closer to that of the AKR1A family. The only differences lie in the specificities toward 1,2-naphthoquinone and glucuronic acid, which are known as good substrates for AKR1A1 (27). YajO actively converts 2-carboxybenzaldehyde, as in the case of the AKR7A family, although specificities toward other substrates are not seen (14, 27). Although YajO was annotated as a putative xylose reductase in GenBank, its xylose reduction activity was detected only marginally. In summary, the AKRs examined in this study appear to have unique substrate specificities, showing some similarity to those of the known AKR family.
TABLE 3.
Substrate specificities of E. coli AKRs
| Substrate | Specific activity (nmol min−1 mg−1) of:a
|
||||
|---|---|---|---|---|---|
| YafB | YqhE | YeaE | YghZ | YajO-His6 | |
| 1,2-Naphthoquinone | 42.4 ± 9.5 | 127 ± 28 | 80.6 ± 4.4 | 32.4 ± 7.7 | 70.8 ± 13.1 |
| 2-Carboxybenzaldehyde | <10 | <10 | <10 | <10 | 22,900 ± 827 |
| 3-Nitrobenzaldehyde | 2,230 ± 130 | 2,500 ± 110 | 3,080 ± 170 | 53.7 ± 2.8 | <10 |
| 4-Nitrobenzaldehyde | 6,530 ± 280 | 10,300 ± 600 | 6,230 ± 140 | 3,680 ± 130 | 39.7 ± 5.0 |
| Benzaldehyde | 2,790 ± 40 | 3,880 ± 100 | 520 ± 30 | 593 ± 16 | 17.4 ± 1.2 |
| d,l-Glyceraldehyde | 1,360 ± 70 | 2,210 ± 10 | 77.7 ± 1.6 | 850 ± 37 | 23.6 ± 5.0 |
| d-Glucuronic acid | 15.3 ± 0.0 | 24.7 ± 0.9 | <10 | 18.6 ± 2.7 | <10 |
| 2,5-Diketo-d-gluconate | 40.1 ± 1.1 | 51.9 ± 1.5 | <10 | 11.9 ± 1.9 | <10 |
| Methylglyoxal | 14,900 ± 500 | 21,000 ± 400 | 1,890 ± 30 | 2,200 ± 20 | <10 |
| Phenylglyoxal | 39,100 ± 1,800 | 64,400 ± 1,500 | 2,710 ± 260 | 564 ± 57 | 22.4 ± 4.3 |
| d-Xylose | <10 | 94.9 ± 9.7 | <10 | 13.8 ± 1.9 | <10 |
The substrates 1,2-naphthoquinone, d-xylose, and d-glucuronic acid were used at 0.1, 10, and 10 mM concentrations, respectively. All other substrates were used at 1.0 mM concentrations. The values represent the means ± standard errors of the means.
MG-dependent induction and MG detoxification activities of E. coli AKRs in vivo.
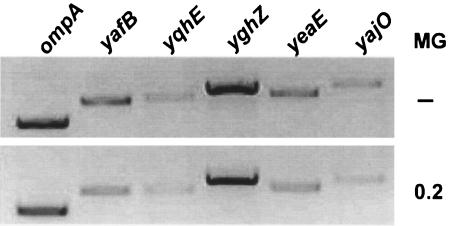
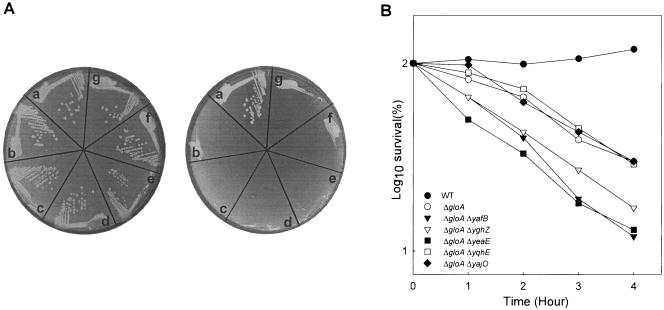
We tested whether MG affects expressions of AKR genes, as in yeast (3), by carrying out semiquantitative RT-PCR. As shown in Fig. 4, the total RNAs from the wild-type E. coli cells treated with and without 0.2 mM MG were isolated and compared. The results indicated that the expressions of the five AKR genes were not significantly altered upon MG addition, excluding the possibility of MG-dependent induction of the AKR genes. In order to assess the MG detoxification ability of AKRs, deletion strains were constructed to test their capabilities for tolerating MG (0.3 mM) in the medium. After incubation for designated times, viable cells were counted. Since none of the AKR single mutants exhibited differences in MG susceptibilities, we generated a double mutant lacking the gloA gene in addition to AKR genes. While the double mutants were able to form single colonies on the LB plate (Fig. 5A, left), the addition of 0.3 mM MG inhibited growth of the yafB, yghZ, and yeaE mutant cells (Fig. 5A, right). We also examined effect of MG (0.3 mM) in a liquid medium (Fig. 5B). After 4 h of incubation, the three AKR mutants exhibited increased sensitivities to MG, while the other mutants were indistinguishable from the gloA background. This suggests that yafB, yghZ, and yeaE play a crucial role in MG detoxification in vivo. However, the yqhE double mutant did not show any significant change in MG susceptibility relative to the gloA single mutant.
FIG. 4.
Expressions of the AKR genes upon MG addition. Total RNAs were isolated from the MG1655 cells both without (−) and with 0.2 mM MG, and an RT-PCR was carried out as described in Materials and Methods. The products were separated by agarose gel electrophoresis with the mRNA level of ompA as an internal control.
FIG. 5.
MG susceptibilities of the AKR and glyoxalase double mutants. Cell viability was determined in response to MG. (A) Cells were grown on LB plates without (left) and with (right) 0.3 mM MG. Segments: a, wild type (MG1655); b, ΔgloA; c, ΔgloA ΔyafB; d, ΔgloA ΔyghZ; e, ΔgloA ΔyeaE; f, ΔgloA ΔyajO; g, ΔgloA ΔyqhE. (B) Cells were grown overnight at 37°C in the M9 medium with 0.4% glycerol and resuspended in an M9 medium. After growing to an early exponential phase, cells were diluted 10-fold in fresh M9 medium containing 0.4% glycerol, to which 0.3 mM MG was added. Samples were taken at the indicated times, from which viable cells of the types indicated in the key were counted. WT, wild type.
DISCUSSION
Since methylglyoxal is a strong electrophile known to modify a variety of macromolecules, cells have developed various ways of detoxification (8). Acetol appears to be one of the detoxification products from MG. Other ways of removing MG include the glyoxalase and MG reductase systems, which generate d-lactate via a glutathione derivative or l-lactaldehyde. Previously, we demonstrated that unregulated production of MG occurs from enhanced ribose utilization, perhaps due to a rapid increase in the levels of the glycolysis intermediates (20). In such conditions, acetol accumulated in the culture medium, indicating that the acetol-producing metabolic pathway exists in E. coli. In this study, we investigated the gene(s) responsible for MG conversion into acetol in E. coli based on previous studies of enzymes with similar activities, i.e., the mammalian AKR family 1 and 7 members. A database search for sequence similarity revealed that the yqhE, yafB, and yeaE genes of E. coli share homologies to the mammalian AKR1 family, and yajO and yghZ genes share homologies with the AKR7 members, which have been the primary targets.
In general, the physiological role of AKR is not clearly established, perhaps due to a broad spectrum of substrate specificities. From the results obtained with the AKR deletion mutants and the purified AKR proteins, we demonstrated that YafB, YqhE, YeaE, and YghZ participate in the detoxification of MG. The reason for the discrepancy between the in vivo and in vitro activities of YqhE is uncertain. One possibility is that the enzyme activity is suppressed in vivo. The cell extracts may eliminate other MG-detoxifying activity requiring a different coenzyme, although unknown regulatory interactions might be involved in vivo. In addition, since we examined the effect of the gene in a gloA mutant background abolishing major MG detoxification activity, the situation might be different from that for the wild type. It is unlikely that the yqhE gene is not expressed under our conditions, based on the result of RT-PCR (Fig. 4).
YafB and YqhE were initially reported as 2,5-diketo-d-gluconate reductases on the basis of their similarity to that of Corynebacterium spp. (13, 33). The YafB and YqhE proteins were shown to reduce 2,5-DKG, a key step in the microbial synthesis of vitamin C, although 2,5-DKG does not appear to be an endogenous substrate of the E. coli cell. The Km value (26 mM) of the Corynebacterium reductase for 2,5-DKG is too high for it to be a physiological substrate (24). The kcat/Km value of YqhE for 2,5-DKG was determined as 5.03 × 104 min−1 M−1 (Km = 3.1 ± 0.5 mM; kcat = 2.6 ± 0.2 s−1 [13]), which is lower than those of YafB, YqhE, YeaE, and YghZ for MG, as presented in this work. Although we do not exclude the possibility that other endogenous substrates exist, MG clearly serves as an efficient endogenous substrate for these AKRs.
It was previously reported that acetol is produced by an enzymatic reduction of MG in E. coli (25). However, it is unclear whether this enzyme is identical with one of the AKRs studied here, since those researchers were unable to identify gene(s) responsible for such an enzymatic activity. Furthermore, it is unclear if the reported enzyme activity was due to a single protein, since they were unable to purify the protein(s). Although they insisted that acetol was identified by UV absorption spectroscopy of the metabolite after the 2,4-dintrophenyl hydrazine treatment, this is hardly to be regarded as a proof. The molecular mass (100,000 ± 3,000 Da) characterized by gel permeation chromatography is also different from those of E. coli AKRs. However, it is possible that the enzyme is a multimeric form of an AKR. Considering the similarity of the substrate spectrum, the NADPH dependency, and the irreversibility of the reaction, the reported enzyme might be a single AKR or a mixture of AKRs characterized here. Since we tested the MG-reducing activity of all the AKR candidates of E. coli, it is unlikely that other AKRs exist.
The known AKR enzymes exhibit various aldehyde- and ketone-reducing activities. Similarly, the AKRs tested here are able to reduce aldehydes and ketones that are known as substrates of other AKRs. They show a unique spectrum of substrate specificities, although some similarities to known AKRs were noticed. For example, YajO efficiently reduces 2-carboxybenzaldehyde as in mammalian AKR7 enzymes but is incapable of reducing MG, known as a good substrate for AKR7 enzymes. Site-directed mutagenesis and deletion studies of mammalian AKRs revealed that the C-terminal loops are important in determining the substrate specificities of the enzymes (5). Although residues for the catalytic tetrad and coenzyme binding site are conserved in E. coli and mammalian AKRs, the lengths and amino acid compositions of the C-terminal loops vary greatly. This variation may explain the differences in substrate specificities.
Determining the physiological role of AKRs has been unsuccessful due to their broad substrate specificities and functional redundancies. While deletion of the yeast GRE3 gene, which encodes the aldose reductase converting MG to acetol, does not significantly decrease the MG-reducing activity or osmotic sensitivity relative to the wild type (2), the triple AKR deletion (i.e., deletions of YPR1, GRE3, and GCY1) results in enhanced sensitivity to heat shock stress as well as an alteration of global gene expression (7). Similarly, none of the single AKR deletions characterized here showed complete abolition of MG-reducing activity. However, by introducing the deletion of glyoxalase, we were able to demonstrate that three AKR genes contribute to MG detoxification in vivo. Further experiments are necessary to elucidate the roles of these enzymes in cellular stress situations with excess aldo and keto compounds, including MG.
Acknowledgments
This work was supported by the BK21, Glycomics, and 21C Frontier Microbial Genomics and Application Center Program, Ministry of Science & Technology, Republic of Korea, grant MG05-0202-2-0 to C. Park.
We thank F. R. Blattner, I. R. Booth, and J. G. Pan for providing strains and materials.
REFERENCES
- 1.Ackerman, R. S., N. R. Cozzarelli, and W. Epstein. 1974. Accumulation of toxic concentrations of methylglyoxal by wild-type Escherichia coli K-12. J. Bacteriol. 119:357-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguilera, J., and J. A. Prieto. 2001. The Saccharomyces cerevisiae aldose reductase is implied in the metabolism of methylglyoxal in response to stress conditions. Curr. Genet. 39:273-283. [DOI] [PubMed] [Google Scholar]
- 3.Aguilera, J., and J. A. Prieto. 2004. Yeast cells display a regulatory mechanism in response to methylglyoxal. FEMS Yeast Res. 4:633-641. [DOI] [PubMed] [Google Scholar]
- 4.Altaras, N. E., and D. C. Cameron. 1999. Metabolic engineering of a 1,2-propanediol pathway in Escherichia coli. Appl. Environ. Microbiol. 65:1180-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barski, O. A., K. H. Gabbay, and K. M. Bohren. 1996. The C-terminal loop of aldehyde reductase determines the substrate and inhibitor specificity. Biochemistry 35:14276-14280. [DOI] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Chang, Q., T. M. Harter, L. T. Rikimaru, and J. M. Petrash. 2003. Aldo-keto reductases as modulators of stress response. Chem.-Biol. Interact. 143:325-332. [DOI] [PubMed] [Google Scholar]
- 8.Cooper, R. A. 1984. Metabolism of methylglyoxal in microorganisms. Annu. Rev. Microbiol. 38:49-68. [DOI] [PubMed] [Google Scholar]
- 9.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellis, E. M. 2002. Microbial aldo-keto reductases. FEMS Microbiol. Lett. 216:123-131. [DOI] [PubMed] [Google Scholar]
- 11.Freedberg, W. B., W. S. Kistler, and E. C. Lin. 1971. Lethal synthesis of methylglyoxal by Escherichia coli during unregulated glycerol metabolism. J. Bacteriol. 108:137-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grant, A. W., G. Steel, H. Waugh, and E. M. Ellis. 2003. A novel aldo-keto reductase from Escherichia coli can increase resistance to methylglyoxal toxicity. FEMS Microbiol. Lett. 218:93-99. [DOI] [PubMed] [Google Scholar]
- 13.Habrych, M., S. Rodriguez, and J. D. Stewart. 2002. Purification and identification of an Escherichia coli beta-keto ester reductase as 2,5-diketo-d-gluconate reductase YqhE. Biotechnol. Prog. 18:257-261. [DOI] [PubMed] [Google Scholar]
- 14.Hinshelwood, A., G. McGarvie, and E. Ellis. 2002. Characterisation of a novel mouse liver aldo-keto reductase AKR7A5. FEBS Lett. 523:213-218. [DOI] [PubMed] [Google Scholar]
- 15.Hinshelwood, A., G. McGarvie, and E. M. Ellis. 2003. Substrate specificity of mouse aldo-keto reductase AKR7A5. Chem.-Biol. Interact. 143:263-269. [DOI] [PubMed] [Google Scholar]
- 16.Inoue, Y., and A. Kimura. 1995. Methylglyoxal and regulation of its metabolism in microorganisms. Adv. Microb. Physiol. 37:177-227. [DOI] [PubMed] [Google Scholar]
- 17.Jez, J. M., M. J. Bennett, B. P. Schlegel, M. Lewis, and T. M. Penning. 1997. Comparative anatomy of the aldo-keto reductase superfamily. Biochem. J. 326:625-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalapos, M. P. 1999. Methylglyoxal in living organisms: chemistry, biochemistry, toxicology and biological implications. Toxicol. Lett. 110:145-175. [DOI] [PubMed] [Google Scholar]
- 19.Kavanagh, K. L., M. Klimacek, B. Nidetzky, and D. K. Wilson. 2002. The structure of apo and holo forms of xylose reductase, a dimeric aldo-keto reductase from Candida tenuis. Biochemistry 41:8785-8795. [DOI] [PubMed] [Google Scholar]
- 20.Kim, I., E. Kim, S. Yoo, D. Shin, B. Min, J. Song, and C. Park. 2004. Ribose utilization with an excess of mutarotase causes cell death due to accumulation of methylglyoxal. J. Bacteriol. 186:7229-7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozma, E., E. Brown, E. M. Ellis, and A. J. Lapthorn. 2002. The crystal structure of rat liver AKR7A1. A dimeric member of the aldo-keto reductase superfamily. J. Biol. Chem. 277:16285-16293. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 23.MacLean, M. J., L. S. Ness, G. P. Ferguson, and I. R. Booth. 1998. The role of glyoxalase I in the detoxification of methylglyoxal and in the activation of the KefB K+ efflux system in Escherichia coli. Mol. Microbiol. 27:563-571. [DOI] [PubMed] [Google Scholar]
- 24.Miller, J. V., D. A. Estell, and R. A. Lazarus. 1987. Purification and characterization of 2,5-diketo-d-gluconate reductase from Corynebacterium sp. J. Biol. Chem. 262:9016-9020. [PubMed] [Google Scholar]
- 25.Misra, K., A. B. Banerjee, S. Ray, and M. Ray. 1996. Reduction of methylglyoxal in Escherichia coli K12 by an aldehyde reductase and alcohol dehydrogenase. Mol. Cell. Biochem. 156:117-124. [DOI] [PubMed] [Google Scholar]
- 26.Murata, K., Y. Fukuda, M. Simosaka, K. Watanabe, T. Saikusa, and A. Kimura. 1985. Metabolism of 2-oxoaldehyde in yeasts. Purification and characterization of NADPH-dependent methylglyoxal-reducing enzyme from Saccharomyces cerevisiae. Eur. J. Biochem. 151:631-636. [DOI] [PubMed] [Google Scholar]
- 27.O'Connor, T., L. S. Ireland, D. J. Harrison, and J. D. Hayes. 1999. Major differences exist in the function and tissue-specific expression of human aflatoxin B1 aldehyde reductase and the principal human aldo-keto reductase AKR1 family members. Biochem. J. 343:487-504. [PMC free article] [PubMed] [Google Scholar]
- 28.Phillips, S. A., and P. J. Thornalley. 1993. The formation of methylglyoxal from triose phosphates. Investigation using a specific assay for methylglyoxal. Eur. J. Biochem. 212:101-105. [DOI] [PubMed] [Google Scholar]
- 29.Saikusa, T., H. Rhee, K. Watanabe, K. Murata, and A. Kimura. 1987. Metabolism of 2-oxoaldehydes in bacteria: purification and characterization of methylglyoxal reductase from E. coli. Agric. Biol. Chem. 7:1893-1899. [Google Scholar]
- 30.Thornalley, P. J. 1990. The glyoxalase system: new developments towards functional characterization of a metabolic pathway fundamental to biological life. Biochem. J. 269:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vander Jagt, D. L., B. Robinson, K. K. Taylor, and L. A. Hunsaker. 1992. Reduction of trioses by NADPH-dependent aldo-keto reductases. Aldose reductase, methylglyoxal, and diabetic complications. J. Biol. Chem. 267:4364-4369. [PubMed] [Google Scholar]
- 32.Wermuth, B., J. D. Munch, and J. P. von Wartburg. 1977. Purification and properties of NADPH-dependent aldehyde reductase from human liver. J. Biol. Chem. 252:3821-3828. [PubMed] [Google Scholar]
- 33.Yum, D. Y., B. Y. Lee, and J. G. Pan. 1999. Identification of the yqhE and yafB genes encoding two 2,5-diketo-d-gluconate reductases in Escherichia coli. Appl. Environ. Microbiol. 65:3341-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]