Abstract
We have recently described a multicomponent cascade that regulates type III secretion in Bordetella. This cascade includes a group of proteins, BtrU, BtrW, and BtrV, that contain an array of domains that define partner-switching complexes previously characterized in gram-positive bacteria. BtrU contains a PP2C-like serine phosphatase domain, BtrW contains a serine kinase/anti-sigma factor motif, and BtrV includes an anti-sigma factor antagonist domain. On the basis of genetic studies and sequence similarity with the RsbU-RsbW-RsbV and SpoIIE-SpoIIAB-SpoIIAA partner switchers of Bacillus subtilis, a series of interactions between Bordetella orthologs have been proposed. Bacterial two-hybrid analysis, tagged protein pull-downs, and in vitro phosphorylation assays were used to characterize interactions between BtrW and BtrV. In addition, BtrV mutants predicted to mimic a constitutively phosphorylated form of BtrV or to be nonphosphorylatable and BtrW mutants defective in serine kinase activity or the ability to bind BtrV were constructed and analyzed. Our results demonstrate that (i) BtrW and BtrV interact with each other, (ii) BtrW phosphorylates BtrV at serine S55, (iii) the conserved serine residue S55 of BtrV plays a key role in BtrV-BtrW interactions, and (iv) the ability of BtrW to phosphorylate BtrV and disrupt BtrV-BtrW binding is essential for the type III secretion process.
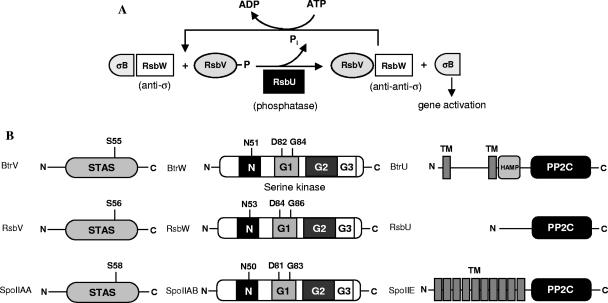
Studies of the regulation of σB activity in the general stress response pathway and σF activity in the sporulation pathway of Bacillus subtilis have led to the discovery of a new paradigm in bacterial signaling termed “partner switching” (1, 38). Canonical partner-switching regulatory modules include a phosphatase, a protein kinase/anti-sigma factor, and an antagonist protein/anti-anti-sigma factor. The RsbU-RsbW-RsbV module that controls σB activity in B. subtilis was among the first partner-switching units for which the interactions and functions of the components were deciphered (12, 21, 38). As summarized in Fig. 1A, σB activity is negatively regulated by the anti-sigma factor RsbW, which binds σB and prevents its interaction with core RNA polymerase. The anti-anti-sigma factor RsbV binds RsbW and disrupts the σB-RsbW complex, releasing σB to interact with RNA polymerase and initiate transcription of σB-dependent genes. In turn, RsbW acts as a protein kinase and phosphorylates RsbV at a conserved serine residue. Phosphorylated RsbV is unable to bind RsbW, which is then free to interact with σB. The serine phosphatase RsbU dephosphorylates RsbV∼P, restoring its ability to bind RsbW (18). This network of protein-protein interactions and reversible protein phosphorylation reactions, termed “partner switching” by Alper et al. in 1994 (1), has since been described in many gram-positive bacteria, including Bacillus anthracis (16), Staphylococcus aureus (27, 29), Staphylococcus epidermidis (22), Listeria monocytogenes (6), and Mycobacterium tuberculosis (3).
FIG. 1.
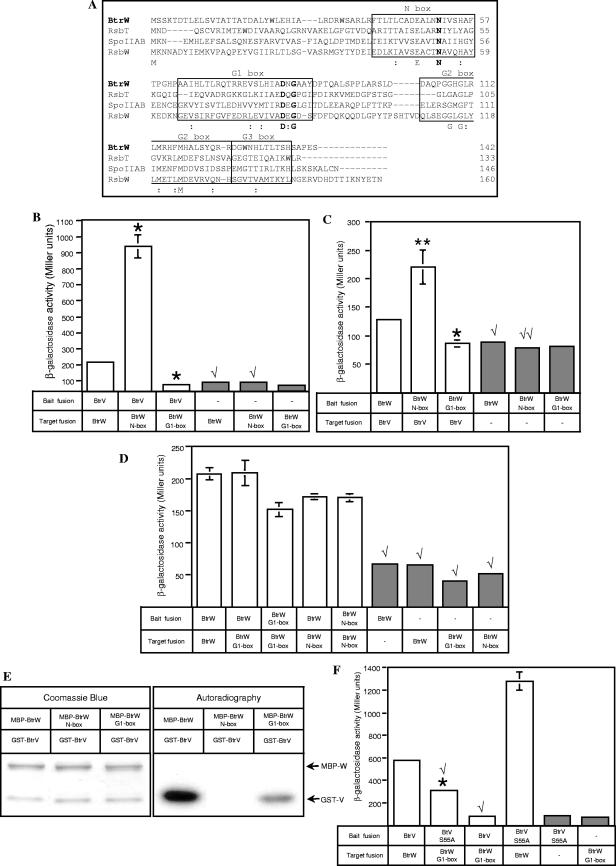
Partner switchers. (A) The B. subtilis partner-switching cycle regulates σB activity in the general stress response pathway (see the text). (B) Predicted protein structures of Bordetella BtrV, BtrW, and BtrU compared with B. subtilis orthologs. The conserved residues of BtrV and BtrW that were mutated and analyzed in this paper are shown. BtrV contains an anti-σ factor antagonist domain (STAS). BtrW contains an HPK-like serine kinase domain that forms a Bergerat fold with four motifs, called N, G1, G2, and G3 boxes, participating in binding to nucleotide ligands. BtrU contains two putative transmembrane domains separated by a stretch of mainly hydrophilic residues, a HAMP domain, and a PP2C-like serine phosphatase domain.
Recently, it was shown that partner-switching orthologs are involved in the regulation of type III secretion (TTS) in the gram-negative respiratory pathogen Bordetella (24). The Bordetella type III secretion system (TTSS) is encoded by the bsc locus and is preferentially expressed in virulent, Bvg+-phase cells (40). It has been most extensively studied in Bordetella bronchiseptica (24, 32, 33, 39, 40), the broad-host-range evolutionary progenitor of Bordetella pertussis and Bordetella parapertussis (35, 36). In vivo, the Bordetella TTSS contributes to persistent tracheal colonization in rats and mice and modulation of the host immune response (39, 40). In vitro, the TTSS causes nonapoptotic cytotoxicity in epithelial and phagocytic cells, activation of the MAP kinases ERK1 and ERK2, and aberrant localization of NF-κB (32, 33, 39). The Bordetella TTSS regulatory locus (btr) contains four genes, btrS, btrU, btrW, and btrV. btrS encodes an ortholog of extracytoplasmic-function sigma factors, and its product activates the transcription of genes encoding components of the TTS apparatus and secreted factors, as well as btrU, btrW, and btrV (24). The btrU gene product bears sequence similarity to the B. subtilis serine phosphatases RsbU and SpoIIE (Fig. 1B) (13, 31, 38). BtrW is orthologous to the RsbW and SpoIIAB anti-sigma factors (4, 26), and it contains an HPK (histidine protein kinase)-like serine kinase domain (17). Finally, the btrV gene product bears sequence similarity to the anti-anti-sigma factors RsbV and SpoIIAA (1, 12) and contains an anti-sigma factor antagonist (STAS) domain (2) (Fig. 1B).
Characterization of deletion mutations in btrS, btrU, btrW, and btrV suggested a novel mechanism for the regulation of type III secretion in Bordetella (24). In response to appropriate environmental signals, the master regulator BvgAS activates expression of btrS. BtrS then activates the transcription of genes encoding components of the TTS apparatus, secreted factors, and TTS regulatory genes. Mutational and ectopic expression studies demonstrated that BtrS is both necessary and sufficient for the transcription of genes included in the bsc locus. Mutational analysis of the partner switcher orthologs revealed a surprising phenotype. Although the transcription of TTS genes was unaffected, deletions in btrU, btrW, or btrV abrogated secretion of polypeptides through the TTS apparatus (24). Analogous to the partner-switching module summarized in Fig. 1A, a requirement for BtrV and BtrU would be predicted if BtrW plays an antagonistic role in the TTS process. The observation that deletion of btrW also eliminated TTS was unexpected, since elimination of an antagonist should result in constitutive activity of the system. These and other data suggested that the Bordetella partner-switching orthologs function with additional levels of complexity in comparison with their gram-positive counterparts.
Central to the partner-switching paradigm are interactions between proteins containing HPK-like serine kinase domains and their STAS domain-containing partners. We therefore examined the interactions between wild-type and mutant forms of BtrW and BtrV using a variety of in vivo and in vitro assays. Our results suggest a requirement for BtrW-mediated phosphorylation of BtrV and dissociation of the BtrV-BtrW complex for the activation of TTS in Bordetella.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
Bordetella bronchiseptica strains RB50, RB53, RB54, WD3, ΔbtrW, and ΔbtrV have been previously described (8, 24, 40). All strains were cultured in Stainer-Scholte liquid medium or on Bordet-Gengou agar (Becton Dickinson Microbiology Systems, San Jose, CA) containing 7.5% defibrinated sheep blood (Mission Laboratories, Rosemead, CA). Antibiotics were used at the following concentrations: streptomycin, 40 μg ml−1, and chloramphenicol, 50 μg ml−1.
The Escherichia coli strains used were XL1Blue and XL1Blue MRF′ laqIq bla lacZ Kanr (Stratagene, La Jolla, CA), TB1 and ER2508 (New England Biolabs, Beverly, MA), and BL21 (Amersham Biosciences, Piscataway, NJ). E. coli was cultured in Luria-Bertani broth, on Luria-Bertani agar, in rich medium (1% tryptone, 0.5% yeast extract, 0.5% NaCl, 2% glucose), or in 2× TYG medium (1.6% tryptone, 1% yeast extract, 0.5% NaCl, 4% glucose). Antibiotics were used at the following concentrations: carbenicillin, 250 μg ml−1; ampicillin, 100 μg ml−1; chloramphenicol, 34 μg ml−1; kanamycin, 40 μg ml−1; and tetracycline, 15 μg ml−1.
Site-directed mutagenesis.
The minimal open reading frames of btrV and btrW plus an additional 40 bp upstream of the respective start codons were amplified from the wild-type B. bronchiseptica RB50 genomic templates by PCR and ligated into pUC19 (btrV) or pMTL20 (btrW) high-copy-number vectors. Mutants were constructed using the QuikChange Site-directed Mutagenesis kit (Stratagene) according to the manufacturer's protocol. The following primers were used to introduce the desired mutations: for btrV(S55A), 5′-GGCTTGACTACATTTCCGCCGCAGGGCTGCGCGTG-3′; for btrV(S55D), 5′-GGCTTGACTACATTTCCGACGCAGGGCTGCGCGTG −3′; for btrW(N51A), 5′-CGAGGCGCTGAACGCCATCGTGTCCCATGC-3′; for btrW(G82AD84A), 5′-GGTCGTAGGCGGCGGCGTTGGCGGCAATGTGCAGG-3′ (and their com- plementary strands). Constructs were confirmed by DNA sequencing. The mutated genes were used for the construction of vectors for bacterial two-hybrid analysis, protein purification, and introduction of the mutated genes into the Bordetella chromosome.
Bacterial two-hybrid analysis.
Primers BtrV_Out_F (5′-TTAGCGGCCGCAAGAACGCAGAGGATTCGT-3′) and BtrV_Out_R (5′-TATCTCGAGCTAGCGGGGCAAGGC-3′) were used to amplify the wild-type btrV gene from RB50, a wild- type strain of B. bronchiseptica. Primers BtrW_Out_F (5′-TTAGCGGCCGCATCAAGCAAAACC-3′) and BtrW_Out_R (5′-TATCTCGAGTCAGGATTCAGGCGC-3′) were used to amplify the wild-type btrW gene from RB50. The same set of primers was used to amplify btrV(S55A), btrV(S55D), btrW(N51A), and btrW(D82AG84A) from the constructs produced as a result of the site-directed mutagenesis. The amplified genes were ligated into the BacterioMatch (Stratagene) two-hybrid vector pBT, creating fusions with the C terminus of the λcI protein, and vector pTRG, creating fusions with the amino-terminal domain of the RNA polymerase α subunit. Clones were confirmed by sequence analysis and cotransformed into the BacterioMatch reporter strain (XL1Blue MRF′ laqIq bla lacZ Kanr). To assay for β-galactosidase (β-Gal) activity, overnight cultures were subcultured to an optical density at 600 nm (OD600) of 0.07 in LB medium containing tetracycline (15 μg ml−1) and chloramphenicol (34 μg ml−1). The cultures were incubated at 30°C until the OD600 reached 0.4, at which point isopropyl β-d-thiogalactoside (IPTG) was added to a final concentration of 10 μM. After 2 hours of IPTG induction, the cells were harvested and permeabilized with sodium dodecyl sulfate (SDS) and CHCl3, and β-galactosidase activity was measured using the method described by Miller (25).
Purification of wild-type and mutant proteins.
The wild-type and mutant btrV genes were amplified from RB50 and from the site-directed mutagenesis products, respectively, and subcloned into the pGEX-2T vector (Amersham Biosciences) to generate glutathione S-transferase (GST) N-terminal fusions. E. coli strain BL21 was transformed with the constructed plasmids, and overexpression of the GST fusion proteins was induced by the addition of 0.6 mM IPTG when the cultures reached an OD600 of 0.5. After being harvested, the cells were lysed with a French press and the collected supernatants were batch bound to glutathione-Sepharose 4B resin (Amersham Biosciences). The bound proteins were eluted with 10 mM glutathione, and the fractions containing GST fusion proteins were identified by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Coomassie blue staining and verified by immunoblot analysis using anti-GST (B-14) monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA). As a negative control for the protein-protein interaction and phosphorylation assays, E. coli strain BL21 was transformed with unmodified pGEX-2T and used for overexpression and purification of the GST tag as described above.
The wild-type and mutant btrW genes were amplified from RB50 and from the site-directed mutagenesis products, respectively, and subcloned into the pMAL-c2X vector (New England Biolabs) to generate E. coli maltose-binding protein (MBP) N-terminal fusions. E. coli strain ER2508 was transformed with the constructed vectors and induced with 0.8 mM IPTG for overexpression of the MBP fusion proteins. After induction, the harvested cells were lysed with a French press and the collected supernatants were batch bound to amylose resin (New England Biolabs). The bound proteins were eluted with 2.5, 5, 10, and 20 mM maltose, and the fractions containing MBP fusion proteins were identified by 10% SDS-PAGE followed by Coomassie blue staining and verified by immunoblot analysis using anti-MBP rabbit serum (New England Biolabs). As a negative control for the protein-protein interaction and phosphorylation assays, E. coli strain TB1 was transformed with unmodified pMAL-c2X, and this clone was used for the overexpression and purification of MBP-β-Gal α fragment fusion protein as described above.
Protein pull-down assays. (i) GST pull-down.
GST-BtrV, GST tag alone, MBP-BtrW, and MBP-β-Gal α fragment (2.5 μg) were added to 20 μl of 50% glutathione-Sepharose in the combinations shown in Fig. 2C and allowed to bind for 45 min at 30°C. The bound complexes were washed four times with 1 ml of phosphate-buffered saline (140 mM NaCl, 2.7 mM KCl, 10.1 mM Na2HPO4, and 1.8 mM KH2PO4, pH 7.3). The proteins were eluted with 20 mM glutathione, separated on 10% SDS-PAGE, and transferred onto Immobilon-P polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA). Monoclonal anti-GST antibody and anti-MBP rabbit serum were used in the immunoblot analysis to identify eluted proteins.
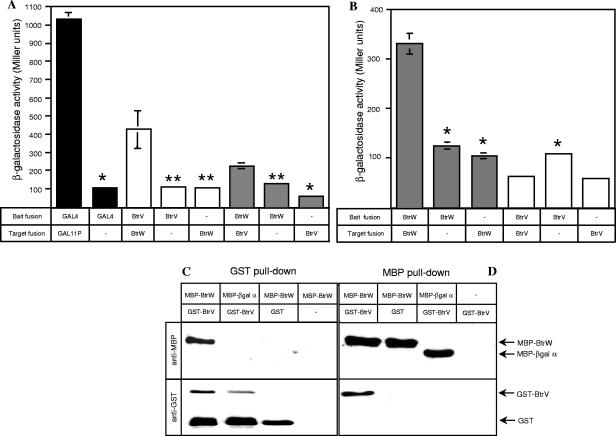
FIG. 2.
BtrW interacts with BtrV and can form homodimers. (A and B) Characterization of interactions between wild-type BtrW and BtrV using a bacterial two-hybrid system. The BacterioMatch two-hybrid reporter strains containing bait and target vectors that were empty or carried the indicated fusions of λcI to Gal4, BtrV, or BtrW (bait fusions) or fusions of the RNA polymerase α subunit to Gal11P, BtrV, or BtrW (target fusions) were assayed for β-galactosidase activity. The values are the averages of three independent samples. The unpaired t test was used to compare differences in activities of the negative controls versus the samples carrying both bait and target fusions for each fusion arrangement; *, P < 0.005; **, P < 0.05. In some cases, the error bars (standard errors) are too small to be seen. (A) BtrW interacts with BtrV. The black bars represent positive and negative controls for proper functioning of the bacterial two-hybrid system. The white bars represent samples in which BtrW was the bait and BtrV was the target, while the gray bars represent the reciprocal arrangement. (B) BtrW can form homodimers (gray bars), but BtrV does not (white bars). (C and D) GST and MBP pull-down assays confirm the BtrW-BtrV interaction. (C) Glutathione-Sepharose was incubated with affinity-purified GST-BtrV and MBP-BtrW. For controls, glutathione-Sepharose was mixed with GST-BtrV and MBP-β-Gal α, GST tag and MBP-BtrW, or MBP-BtrW alone. Separated by 10% SDS-PAGE, protein complexes were probed with anti-MBP or anti-GST antibody. A band corresponding to MBP-BtrW is seen only in a sample containing GST-BtrV. (D) Amylose slurry was mixed with affinity-purified MBP-BtrW and GST-BtrV. To test the specificity of binding, amylose slurry was also mixed with purified MBP-BtrW and GST tag, MBP-β-Gal α and GST-BtrV, or GST-BtrV alone. The samples were separated by 10% SDS-PAGE and probed with anti-MBP or anti-GST antibody. A band corresponding to GST-BtrV was detected only in the sample containing MBP-BtrW.
(ii) MBP pull-down.
GST-BtrV, GST tag alone, MBP-BtrW, and MBP-β-Gal α fragment protein fusions (2.5 μg each) were added to 35 μl of 50% amylose slurry in the combinations depicted in Fig. 2D. The samples were incubated for 2 h at 4°C and washed four times with 1 ml of column buffer (20 mM Tris-HCl, pH 7.4, 200 mM NaCl, 1 mM EDTA). The bound complexes were eluted with 10 mM maltose, separated on 10% SDS-PAGE, and transferred onto a PVDF membrane. Western blot analysis was conducted using monoclonal anti-GST antibody and anti-MBP rabbit serum.
In vitro phosphorylation assay.
Purified GST-BtrV (wild type or mutant; 1 μg) was preincubated for 10 min at 30°C in phosphorylation buffer (50 mM Tris-Cl, pH 7.5, 50 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol [DTT], 0.1 mM EDTA) containing 111.7 μM of [γ-32P]ATP (specific activity, 40 to 100 Ci/mmol). The reactions were initiated by addition of 1 μg of purified MBP-BtrW (wild type or mutants), proceeded for 15 min at 30°C, and were terminated by addition of 3× sample buffer (240 mM Tris-Cl, pH 6.8, 30% glycerol, 75 mM EDTA, 6% SDS). After being heated to 85°C for 5 min, proteins were separated by 10% SDS-PAGE. The gel was exposed to film at −80°C for at least 6 h, followed by Coomassie blue staining.
Micro-liquid chromatography with electrospray-ionization tandem mass spectrometry (μLC-MSMS).
Purified GST-BtrV (2 μg) was preincubated for 15 min at 30°C in phosphorylation buffer (50 mM Tris-Cl, 50 mM KCl, 5 mM MgCl2, 1 mM DTT, 0.1 mM EDTA, pH 7.5) containing 100 μM ATP (total reaction volume, 30 μl), after which 2 μg of purified MBP-BtrW was added, and the reaction proceeded for 30 min at 30°C. The phosphorylation reaction was stopped by addition of 0.2 μg of trypsin in 0.5 M NH4HCO3. The trypsin digestion proceeded for 3 h at 37°C, and then the digestion mixture was vacuum dried.
For μLC-MSMS, the sample was dissolved in 70% acetic acid (5 μl) and injected immediately. Micro-liquid chromatography was performed at 1.5 μl/min using buffers containing 0.1% formic acid (A, water, and B, acetonitrile) and a 200-μm polymeric reverse-phase column (PLRP/S; 5 μm; 300 Å; Polymer Laboratories, Amherst, MA). The column was equilibrated (5% B, 20 min) prior to sample injection, initiating a linear gradient (40% B, 30 min; 80% B, 50 min). The column eluent was delivered to a metal needle electrospray-ionization source (Proxeon) operated at 3.6 kV. The ion trap mass spectrometer (LCQ-DECA; Thermo Finnigan, San Jose, CA) was programmed to perform collision-activated dissociation tandem mass spectrometry on the doubly charged ion (M + 2H+) corresponding to the phosphorylated form of the BtrV tryptic peptide LDYISSAGLR (L50-R59), which contains both S54 and S55 residues (588 atomic mass units; 0.3 isolation width).
Construction of Bordetella mutants.
btrV(S55A), btrV(S55D), btrW(N51A), and btrW(D82AG84A) mutant genes were PCR amplified from the vectors obtained during site-directed mutageneis and cloned into the allelic-exchange vector pRE112 (15). The mutated alleles were introduced into the wild-type B. bronchiseptica chromosome by allelic exchange as previously described (24). The introduced alleles were PCR amplified and confirmed by sequencing.
Immunoblot analysis.
Wild-type and mutant B. bronchiseptica strains were grown overnight under Bvg+ conditions, and the cell cultures obtained were separated into the pellet and supernatant fractions. Proteins from the supernatant fractions were first precipitated with 10% trichloroacetic acid for 6 h on ice. OD equivalents of B. bronchiseptica whole-cell lysates and supernatants were electrophoresed on 13% SDS-polyacrylamide gels and transferred to PVDF membranes (Millipore). The blots were incubated with either mouse anti-BopD polyclonal antibody (1:5,000 dilution) or rat anti-Bsp22 polyclonal antibody (1:3,000 dilution). Both anti-BopD and anti-Bsp22 polyclonal antibodies were made by Covance Research Products Inc., Denver, PA, against purified GST-Bsp22 and GST-BopD fusions (39). Antigen-antibody complexes were detected with horseradish peroxidase-conjugated anti-mouse or anti-rat immunoglobulin (Amersham Biosciences) at a 1:5,000 dilution and visualized by chemiluminescence using SuperSignal West Pico Chemiluminescent Substrate (Pierce, Rockford, IL).
RESULTS
BtrW interacts with BtrV.
We tested the ability of BtrW to interact with BtrV using a bacterial two-hybrid system developed by Hochschild and coworkers (11, 19). In this system, bait polypeptides are fused to the C terminus of bacteriophage λcI protein and target sequences are expressed as C-terminal fusions to the amino-terminal domain of the RNA polymerase α subunit. If the bait and target interact, transcription of a reporter operon containing bla and lacZ is activated. The strength of the bait-target interaction is proportional to the levels of antibiotic resistance and β-galactosidase activity (11). To test for interactions between BtrW and BtrV, λcI and the N-terminal domain of the RNA polymerase α subunit were fused to full-length BtrW and BtrV, respectively. The interactions were also tested with BtrW and BtrV fused in the reciprocal order. Recombinant vectors were cotransformed into a reporter strain, and multiple individual colonies from each transformation were assayed for β-galactosidase activity following induction with 10 μM IPTG for 2 hours. As shown in Fig. 2A, the β-galactosidase activity of the reporter strain containing both Gal4 and Gal11P fusions (positive control) was greater than 1,000 Miller units, whereas the β-galactosidase activity of the negative control strain carrying the Gal4 fusion only was 10 times lower and represented the background level of activity. In reporter strains cotransformed with bait and target vectors carrying BtrW and BtrV fusions in either combination, β-galactosidase activities were significantly higher than those observed with negative control strains in which either the bait or the target vectors were empty (Fig. 2A). These data indicate that BtrW is capable of interacting with BtrV in a bacterial two-hybrid system.
Studies of the B. subtilis RsbV and RsbW partner switchers suggest that the RsbV-RsbW interaction involves two RsbV monomers and one RsbW dimer (9). The availability of both bait and target vectors containing BtrW and BtrV fusions allowed us to test whether BtrW and BtrV were able to form homodimers. As shown in Fig. 2B, the β-galactosidase activity of the reporter strain cotransformed with bait and target vectors both carrying BtrW fusions was significantly higher than that of the negative controls. The level of β-galactosidase activity of the reporter strain containing both bait and target vectors carrying BtrV fusions did not differ from background. These data suggest that BtrW is able to bind to itself. We therefore propose that, similar to the B. subtilis RsbV-RsbW complex (9), BtrV-BtrW exists as a complex containing multimerized BtrW.
To provide an independent assessment of their interaction, BtrW and BtrV were cloned and overexpressed as MBP-BtrW and GST-BtrV fusions. Affinity-purified fusion proteins were used in GST and MBP pull-down assays. As shown in Fig. 2C, MBP-BtrW was specifically bound by GST-BtrV but not by GST or glutathione slurry alone. Likewise, GST-BtrV was not bound by the amylose resin or MBP-β-Gal α fragment fusion but specifically associated with MBP-BtrW (Fig. 2D). These results confirm the interaction between BtrW and BtrV identified in the bacterial two-hybrid system.
BtrW phosphorylates BtrV.
Based on the presence of a putative HPK-like serine kinase domain (17), we hypothesized that BtrW phosphorylates its binding partner, BtrV. To test whether BtrW possesses kinase activity, affinity-purified GST-BtrV and MBP-BtrW were incubated alone or together in kinase buffer containing [γ-32P]ATP (see Materials and Methods). After the reaction was terminated, the proteins were separated by SDS-PAGE. As shown in Fig. 3A, 32P-labeled GST-BtrV was detected when GST-BtrV was incubated with MBP-BtrW and [γ-32P]ATP, but no incorporation of 32P was detected when MBP-BtrW was omitted from the reaction. This indicates that BtrW phosphorylates BtrV in vitro. Incubation of MBP-BtrW alone with [γ-32P]ATP did not result in 32P incorporation, indicating that BtrW does not autophosphorylate under these conditions.
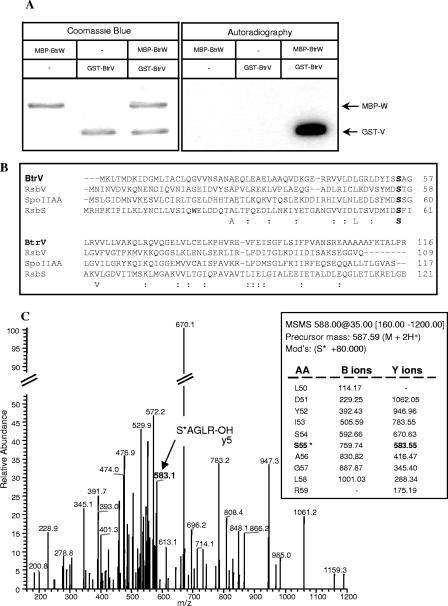
FIG. 3.
BtrW phosphorylates BtrV in vitro. (A) Affinity-purified MBP-BtrW, GST-BtrV, or a mixture of both was incubated with [γ-32P]ATP as described in Materials and Methods. After proteins from each sample were separated by SDS-PAGE, a 32P-labeled band was detected by autoradiography (right). Coomassie blue staining (left) indicated that this band corresponded to GST-BtrV. (B) Multiple sequence alignment between BtrV and its B. subtilis orthologs. The DNA-derived amino acid sequence of BtrV (GenBank database accession number NP_888191) was aligned with the sequences of B. subtilis RsbV, RsbS, and SpoIIAA (GenBank accession numbers P17903, P42410, and P10727, respectively) using CLUSTAL_X (34). Identical residues are shown below the alignment in one-letter amino acid code, and “:” indicates the positions of strongly conserved residues. The conserved serine residues that align with serine 55 of BtrV are shown in boldface. (C) μLC-MSMS of the singly phosphorylated form of the peptide spanning amino acids 50 to 59 of BtrV. The presence of an ion of m/z 583.1 corresponding to y5 indicates that S55 rather than S54 is a major site of phosphorylation.
Sequence alignment of BtrV with its B. subtilis orthologs RsbV, RsbS, and SpoIIAA indicates that S55 of BtrV aligns with S58 of SpoIIAA, S56 of RsbV, and S59 of RsbS and is the only conserved serine residue shared among all four proteins (Fig. 1B and 3B). Genetic and biochemical analyses of B. subtilis orthologs have shown that these conserved serine residues are sites of phosphorylation by cognate serine protein kinases (10, 28, 38). Thus, we hypothesized that S55 of BtrV is the target of phosphorylation by BtrW. The possibility that the adjacent serine (S54) is exclusively or additionally phosphorylated, however, could not be eliminated. Tandem mass spectrometry was therefore used to identify the site(s) of phosphorylation in BtrV. Affinity-purified GST-BtrV and MBP-BtrW fusion proteins were incubated with 100 μM ATP, tryptically digested, and analyzed by mass spectrometry as described in Materials and Methods. Good sequence coverage was obtained, and as shown in Fig. 3C, the presence of y5 ion SAGLR-OH at m/z 583.1 indicates that S55 is the predominant site of phosphorylation. The results of the in vitro phosphorylation assay, together with tandem mass spectrometry data, indicate that BtrW phosphorylates BtrV primarily at the conserved serine residue S55.
BtrV S55 plays a key role in BtrV-BtrW interactions.
We next constructed BtrV mutants in which S55 was replaced with a neutral residue (S55A) or a negatively charged amino acid (S55D) that could potentially mimic the phosphorylated state (10, 38). By analogy with anti-anti-sigma factors described in B. subtilis (Fig. 1B), we predicted that BtrV(S55A) would interact stably with BtrW while BtrV(S55D) would be deficient in binding (10, 23, 38). As expected, the level of β-galactosidase activity expressed by the two-hybrid reporter strain containing λcI-BtrV(S55A) and RNA polymerase α-BtrW fusions was considerably higher than that of the reporter strain carrying wild-type λcI-BtrV and RNA polymerase α-BtrW (Fig. 4A). This suggests that BtrW interacts with BtrV(S55A) more stably than with wild-type BtrV. Surprisingly, a significantly higher level of β-galactosidase activity (compared to that of the reporter strain with wild-type BtrV and BtrW fusions) was also detected in the strain containing λcI-BtrV(S55D) and RNA polymerase α-BtrW fusions. These data indicate that BtrV(S55D) is able to bind BtrW and that it does so with greater stability than wild-type BtrV. Even though the strong interaction between BtrW and BtrV(S55D) was unexpected, these results highlight the critical role of the S55 residue in the BtrV-BtrW interaction. It is clear that Bordetella partner switcher orthologs do not entirely follow the patterns deciphered in their Bacillus counterparts, since SpoIIAA(S58D) and RsbV(S56D) mutant proteins display reduced affinity for SopIIAB and RsbW, respectively (23, 38).
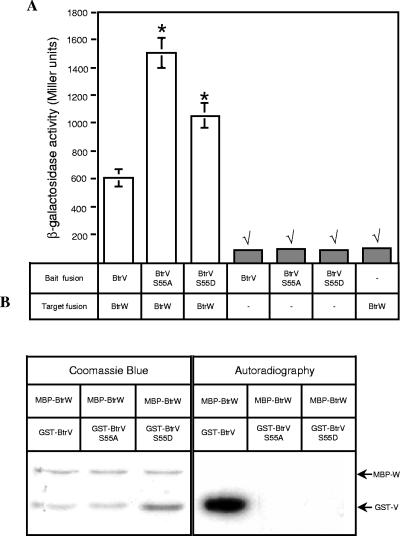
FIG. 4.
Serine 55 of BtrV plays a key role in BtrV-BtrW interactions. (A) Bacterial two-hybrid analysis of the interactions between BtrV S55 mutants and BtrW. The β-galactosidase activities of the reporter strains containing either empty or recombinant vectors carrying wild-type BtrV, BtrV(S55A), or BtrV(S55D) as bait fusions and BtrW as a target fusion were measured. The unpaired t test was used to compare differences in the activities of the strains carrying BtrV(S55A)-BtrW and BtrV(S55D)-BtrW pairs versus the strain carrying the wild-type BtrV-BtrW pair; *, P < 0.005. In addition, the unpaired t test was used to compare differences between the β-galactosidase activities of the negative controls (gray bars) versus the reporter strains containing fusions of BtrV mutants and BtrW (white bars); √, P < 0.005. In some cases, the error bars (standard errors) are too small to be seen. (B) In vitro serine kinase assay to compare GST-BtrV wild type, GST-BtrV(S55A), and GST-BtrV(S55D) as phosphorylation substrates. The affinity-purified GST fusions of wild-type and mutant forms of BtrV were tested for the ability to be phosphorylated by MBP-BtrW as described in Materials and Methods. Coomassie blue staining (left) indicated that the radiolabeled band on the autoradiogram (right) corresponded to GST-BtrV. Neither GST-BtrV(S55A) nor GST-BtrV(S55D) was phosphorylated by MBP-BtrW.
We also examined phosphorylation of GST-BtrV(S55A) and GST-BtrV(S55D) by MBP-BtrW. As expected, MBP-BtrW failed to phosphorylate either mutant protein (Fig. 4B), thus supporting the mass spectrometry data showing that BtrW phosphorylates BtrV at S55.
An N-box mutation in BtrW eliminates kinase activity, and G1-box mutations weaken binding to BtrV.
The crystal structure of SpoIIAB from B. stearothermophilus, which is closely related to B. stublilis, revealed that the HPK-like serine kinase domain forms an ATP-binding Bergerat fold shared by members of the GHKL (gyrase, Hsp90, histidine kinase, and MutL) superfamily (5, 14). The Bergerat fold contains four conserved motifs called N, G1, G2, and G3 boxes that participate in binding nucleotide ligands (14). Sequence alignment of BtrW and its B. subtilis orthologs RsbW, RsbT, and SpoIIAB indicates the presence of several conserved residues within the N, G1, and G2 boxes (Fig. 1B and 5A). Site-directed mutagenesis was used to replace the invariant asparagine at position 51 in the N box with alanine (N51A). Crystal structure analysis of GHKL superfamily proteins indicates that this absolutely conserved N-box asparagine chelates a catalytic Mg2+ ion, which in turn connects the ATP phosphoryl groups to the nucleotide binding pocket (14). Therefore, N51 of BtrW is predicted to be essential for ATP binding and serine kinase activity. In addition, a double alanine replacement of conserved aspartate and glycine residues within the G1 box (D82AG84A) was constructed. For the GHKL superfamily of proteins, it is proposed that the conserved G1-box aspartate interacts directly with ATP whereas the conserved glycine forms one of the hinges required for flexibility of the ATP lid, which covers the nucleotide binding pocket (14). We therefore predicted that both D82 and G84 would also be important for the serine kinase activity of BtrW.
FIG.5.
BtrW N-box mutant binds but cannot phosphorylate BtrV, whereas BtrW G1-box mutant preserves its serine kinase activity despite its low affinity for BtrV. (A) Multiple sequence alignment between B. bronchiseptica and B. subtilis anti-sigma factors/serine kinase orthologs. CLUSTAL_X was used to align the DNA-derived amino acid sequence of BtrW (GenBank accession number NP_888190) with the sequences of B. subtilis RsbW, RsbT, and SpoIIAB (GenBank accession numbers P17904, P42411, and P10728, respectively). The conserved N, G1, G2, and G3 boxes of the Bergerat fold are outlined. The sequences of the conserved motifs were identified based on the alignment of the B. bronchiseptica BtrW and B. subtilis RsbW, RsbT, and SpoIIAB sequences with B. stearothermophilus SpoIIAB (GenBank accession number 1L0OB) (5). Identical residues are shown below the alignment in one-letter amino acid code, and “:” indicates the positions of strongly conserved residues. The conserved arginine residue corresponding to N51 of BtrW within the N box and the conserved aspartate and glycine residues corresponding to D82 and D84, respectively, of BtrW within the G1 box are shown in boldface. (B and C) Bacterial two-hybrid analysis of interactions between wild-type BtrV and BtrW N- and G1-box mutants. The BacterioMatch reporter strain was cotransformed with bait and target vectors carrying fusions of wild-type BtrV and wild-type BtrW, the BtrW N-box mutant, or the BtrW G1-box mutant. For the negative controls, the reporter strain was cotransformed with one vector carrying fusions to wild-type or mutant BtrW and another empty vector. The unpaired t test was used to compare differences between the activities of strains carrying BtrV-BtrW(N51A) and BtrV-BtrW(D82G84) pairs versus that of the strain with the wild-type BtrV-BtrW pair. *, P < 0.005; **, P < 0.05. The unpaired t test was also used to compare differences between the activity of the negative controls carrying a single fusion of wild-type or mutant BtrW (gray bars) versus that of the reporter strains carrying that BtrW fusion together with the BtrV fusion (white bars). √, P < 0.005; √√, P < 0.05. Note the differences in scales of β-galactosidase activity for panels B and C. In some cases, the error bars (standard errors) are too small to be seen. (B) β-Galactosidase activities of the reporter strains cotransformed with bait vectors carrying the BtrV fusions and target vectors carrying BtrW fusions. (C) β-Galactosidase activities of the reporter strains containing the reciprocal arrangement of BtrV and BtrW fusions compared to panel B. (D) Test for the ability of BtrW N-box and G1-box mutants to form homodimers. The reporter strain was cotransformed with bait and target plasmids carrying fusions of wild-type BtrW or BtrW N-box or BtrW G1-box mutants, and the β-galactosidase activities of the transformed strains were measured. The unpaired t tests to compare differences between the activity of the reporter strains carrying BtrW mutant-BtrW wild-type or double-BtrW mutant pairs versus the activity of the strain carrying a double-wild-type BtrW pair indicate that the activity levels are comparable. The unpaired t test was used to compare differences between the activities of the negative controls (gray bars) versus the strains carrying fusions on both bait and target vectors (white bars); √, P < 0.005. In some cases, the error bars are too small to be seen. (E) In vitro phosphorylation assay to assess serine kinase activities of MBP-BtrW N-box and MBP-BtrW G1-box mutants. Affinity-purified MBP-BtrW(N51A) and MBP-BtrW(D82AG84A) proteins were tested for the ability to phosphorylate GST-BtrV upon incubation with [γ-32P]ATP. The 32P-labeled bands were detected by autoradiography (right). The same amounts of the purified fusion proteins as were used in the kinase reactions were mixed and separated by SDS-PAGE, followed by Coomassie blue staining (left). The Coomassie blue staining indicated that 32P-labeled bands corresponded to GST-BtrV. The MBP-BtrW G1-box mutant can phosphorylate GST-BtrV, albeit more weakly than the MBP-BtrW wild type. (F) Assay for the ability of BtrV(S55A) to interact stably with the BtrW G1-box mutant in the bacterial two-hybrid system. β-Galactosidase activity was measured in the reporter strain cotransformed with bait vectors carrying the wild type or the S55A mutant of BtrV and the target vector carrying the wild type or the G1-box mutant of BtrW. Negative controls are in gray. The unpaired t test was used to compare the differences between the activity of the strain containing the BtrV(S55A)-BtrW G1-box mutant pair versus the strain containing the BtrV wild type-BtrW G1-box mutant pair; *, P < 0.005. In addition, an unpaired t test was conducted to evaluate the differences between the activities of the strains used in the test described above versus the reporter strain with wild-type BtrV and BtrW fusions; √, P < 0.005. In some cases, the error bars are too small to be seen.
The bacterial two-hybrid system was used to assess interactions between BtrW mutants and wild-type BtrV (Fig. 5B and 5C). The β-galactosidase activity of the reporter strains carrying the BtrW N-box mutant and wild-type BtrV was significantly higher than that of the reporter strains carrying wild-type BtrW and BtrV. These data suggest that BtrV binds BtrW(N51A) with significantly greater affinity than wild-type BtrW. The detection of a stable BtrV-BtrW(N51A) complex in the context of a bacterial two-hybrid system was reminiscent of the stable complexes between the nonphosphorylatable BtrV mutants and wild-type BtrW (Fig. 4A) and suggested that the BtrW N-box mutant might be defective primarily in its ability to phosphorylate BtrV. Conversely, the reporter strains carrying the BtrW G1-box mutant and wild-type BtrV produced levels of β-galactosidase activity similar to those of the negative controls, suggesting that the BtrW(D82AG84A) mutant is unable to form a stable complex with BtrV.
It is possible that the failure to detect interactions between BtrV and the BtrW G1-box mutant in the bacterial two-hybrid analysis is due to gross disruption of the secondary or tertiary structure of BtrW by simultaneous alanine replacements of two closely located residues within the G1 box. To address this, we measured the abilities of BtrW mutants to interact with themselves. Two-hybrid analysis indicated that the BtrW G1-box mutant was able to bind both wild-type BtrW and another BtrW G1-box mutant equally well (Fig. 5D). Homotypic interactions by the BtrW N-box mutant were also examined, and as expected, there were no significant changes in the ability to bind wild-type BtrW or another BtrW N-box mutant. In addition, a biophysical characterization of MBP-BtrW, MBP-BtrW(N51A), and MBP-BtrW(D82AG84A) was conducted to assess whether the N51A or D82AG84A substitution measurably altered the structure of BtrW. Circular-dichroism spectroscopy in the far-UV region (190 to 250 nm) did not detect any significant changes in the secondary structure of either BtrW N-box or BtrW G1-box mutants in comparison to wild-type BtrW. Similarly, fluorescence emission spectroscopy did not show any major disruptions in the tertiary structure of BtrW mutants (data not shown). These data suggest that the D82AG84A mutation did not cause a global change in the structure of BtrW. Instead, this double alanine substitution most likely results in localized alterations in a region involved in binding BtrV.
We next analyzed BtrW N-box and G1-box mutant proteins in in vitro phosphorylation assays. As predicted, MBP-BtrW(N51A) failed to phosphorylate GST-BtrV (Fig. 5E), confirming that mutation of the conserved N51 residue within the N box disrupts the kinase activity of BtrW. To our surprise, the MBP-BtrW G1-box mutant was able to phosphorylate GST-BtrV, albeit to a lesser extent than the MBP-BtrW wild type, despite its inability to form stable interactions with BtrV in the bacterial two-hybrid assay. The level of β-galactosidase activity measured in the two-hybrid system reflects the ability of bait and target polypeptides to form stable complexes rather than simply the ability to interact. If a bait and a target bind to each other, but only for a short period of time, the two-hybrid system would reflect this by displaying a low level of activity. Therefore, it is possible that the BtrW G1-box mutant is able to transiently bind BtrV to allow phosphorylation, but the stability of the interaction is insufficient for detection in a two-hybrid system. We hypothesized that it may be possible to detect BtrW G1-box mutant binding by stabilizing the interaction with a nonphosphorylatable form of BtrV. Indeed, when the reporter strain was cotransformed with a bait vector carrying BtrV(S55A) and a target vector carrying BtrW(D82AG84A), the β-galactosidase activity was significantly higher than that of the reporter strain containing wild-type BtrV and BtrW G1-box mutant fusions (Fig. 5F). The ability to stabilize binding between the BtrW G1-box mutant and BtrV by providing a nonphosphorylatable form of BtrV suggests that the stability of the BtrV-BtrW complex depends both on the affinity of BtrW for BtrV and on the ability of BtrW to phosphorylate BtrV.
Effects of BtrV and BtrW mutations on TTS.
The BtrV and BtrW mutants described above can be classified into two groups: (i) mutants such as BtrV(S55A), BtrV(S55D), and BtrW(N51A), which form stable complexes with their binding partners but are defective in phosphorylation, and (ii) the BtrW(D82AG84A) mutant, which is predicted to form an unstable, transient complex with BtrV but possesses detectable kinase activity. The availability of these two classes of mutants provided an opportunity to investigate the properties of BtrV and BtrW that are required for the proper functioning of the Bordetella TTSS.
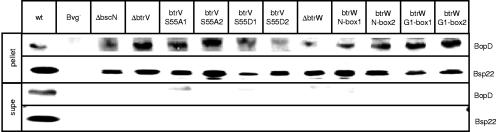
Allelic exchange was used to generate btrV(S55A), btrV(S55D), btrW(N51A), and btrW(D82AG84A) mutant derivatives of B. bronchiseptica strain RB50, and the effects on TTS were examined. Overnight cultures of wild-type RB50, the btrV and btrW mutants, and several control strains known to be deficient in TTS were separated into supernatant and pellet fractions and subjected to immunoblot analysis using antibodies directed against two type III secreted proteins, Bsp22 and BopD (Fig. 6). As shown previously, the wild-type strain expressed and secreted both Bsp22 and BopD, whereas the Bvg− phase-locked strain did not express either of these proteins (40). The ΔbscN strain, which lacks the ATPase required for TTS, expressed but did not secrete Bsp22 or BopD. For ΔbtrW and ΔbtrV mutants, the type III secreted proteins Bsp22 and BopD were detected in the pellet but not in the supernatant fractions. Although our initial study failed to detect Bsp22 or BopD in pellet fractions of a ΔbtrV strain (24), this was most likely due to protein instability in the absence of secretion as opposed to a block in translation of secreted polypeptides in the absence of BtrV.
FIG. 6.
B. bronchiseptica btrV(S55A), btrV(S55D), btrW(N51A), and btrW(D82AG84A) mutants are defective in type III secretion. Pellet and supernatant fractions of B. bronchiseptica wild-type and various mutant strains defective in type III secretion were separated by SDS-PAGE, transferred to PVDF membranes, and probed with polyclonal antibodies generated against Bsp22 and BopD.
As shown in Fig. 6, the btrV(S55A), btrV(S55D), btrW(N51A), and btrW(D82AG84A) mutants all had the same phenotype, namely, the inability to secrete Bsp22 and BopD. The failure of the btrV(S55A), btrV(S55D), and btrW(N51A) alleles to secrete Bsp22 and BopD suggests that the ability of BtrW to phosphorylate BtrV and/or disrupt BtrV-BtrW binding is required for the activation of TTS. Because TTS is inactive in the btrW(D82AG84A) mutant, we further propose that the D82AG84A substitutions eliminate interactions between BtrW and an alternative binding partner and/or decrease the overall level of BtrV∼P below a threshold required for activity. Our results support a model, described in detail below, in which phosphorylation and interactions with additional binding partners regulate TTS in Bordetella.
DISCUSSION
Early studies of the partner-switching mechanism focused on the general stress response and sporulation pathways of B. subtilis (1, 4, 21, 30, 38). More recently, it has become apparent that partner-switching modules are also involved in the regulation of virulence in several gram-positive pathogens, including S. aureus (20, 41), L. monocytogenes (6), and M. tuberculosis (3, 7). Since the σB-regulated stress response is highly conserved in gram-positive bacteria, these effects are likely due to a compromised ability to withstand environmental stresses encountered during infection. Interestingly, perhaps the most illustrative example of a role for partner switcher orthologs in the regulation of a system that is entirely dedicated to virulence can be found in the gram-negative respiratory pathogen B. bronchiseptica, which has adapted a variation of the partner-switching paradigm to control type III secretion.
Our previous studies demonstrated that nonpolar, in-frame deletions in btrU, btrW, or btrV eliminate TTS in Bordetella (24). In this report, we have demonstrated that BtrW binds and phosphorylates BtrV at the conserved serine residue S55. In addition, we have shown that BtrW is capable of homotypic interactions. These data suggest that the BtrV-BtrW complex may be similar to the B. subtilis RsbV-RsbW complex, in which RsbW forms a homodimer, with each RsbW monomer interacting with and phosphorylating one molecule of RsbV (9).
Characterization of BtrV and BtrW mutants carrying substitutions at conserved residues gave further insight into the biochemical properties of these partner switcher orthologs. By analogy to BtrV orthologs described in B. subtilis (23, 38), it was predicted that alanine substitution at S55 would prevent phosphorylation of BtrV by BtrW and stabilize the BtrW-BtrV(S55A) complex. On the other hand, mutation of S55 in BtrV to a negatively charged aspartate was predicted to mimic phosphorylated serine and prevent BtrW binding. To our surprise, the high levels of β-galactosidase activity produced by the reporter strains carrying BtrV(S55A)-BtrW and BtrV(S55D)-BtrW fusions indicated that BtrW formed stable complexes with both BtrV(S55A) and BtrV(S55D). The ability of Bordetella BtrV(S55D) to interact with BtrW distinguishes this mutant from the B. subtilis RsbV(S56D), RsbS(S59D), and SpoIIAA(S58D) orthologs, which do not bind their cognate anti-sigma factor/serine kinase partners (23, 38). Moreover, mutation of BtrV S55 to a negatively charged glutamic acid did not prevent BtrW binding (data not shown). We propose that the structures of Bordetella BtrW and/or BtrV differ from their B. subtilis orthologs such that replacement of the phosphorylatable serine in BtrV with negatively charged residues does not create sufficient electrostatic or steric clashes to disrupt interactions with BtrW. Instead, BtrW binds these mutant proteins with increased stability, presumably because phosphorylation does not occur. To our knowledge, this is the first report of a STAS domain-containing protein that preserves its ability to bind an anti-sigma factor upon mutation of the phosphorylated serine residue to a negatively charged amino acid.
Mutagenesis of the conserved asparagine residue in the BtrW N box eliminated the ability to phosphorylate BtrV. In addition, the bacterial two-hybrid analysis indicated that the reporter strain carrying BtrW(N51A) and wild-type BtrV fusion pairs produced higher levels of β-galactosidase activity than the reporter strain carrying wild-type BtrW and BtrV fusions. These observations are consistent with those obtained with the BtrV(S55A) and BtrV(S55D) mutants. Together, they suggest that some level of phosphorylation occurs in the E. coli two-hybrid system and that phosphorylation reduces the stability of the BtrV-BtrW complex. The properties of BtrW(N51A) support the prediction that BtrW forms a Bergerat ATP-binding fold, similar to the one in SpoIIAB (5), with the conserved asparagine residue at position 51 playing an essential role in serine kinase activity.
In contrast to the expected behavior of the BtrW N-box mutant, the dual alanine replacements of the conserved aspartate and glycine residues in the G1 box of BtrW resulted in an unexpected phenotype. Based on similarity to GHKL superfamily proteins (14), it was predicted that mutation of both D82 and G84 would eliminate the kinase activity of BtrW. Surprisingly, BtrW(D82AG84A) retained detectable phosphorylation activity upon incubation with wild-type BtrV, despite the inability to form stable complexes with BtrV, as indicated by a negligible level of β-galactosidase activity in the two-hybrid analysis. The ability of the S55A mutation in BtrV to partially rescue the binding defect observed with the D82AG84A mutations in BtrW may explain this apparent inconsistency. We propose that BtrW(D82AG84A) interacts with BtrV at a level that is sufficient for detectable phosphorylation but is below the minimum threshold of stability required for detection in the two-hybrid system. Substitution of a nonphosphorylatable substrate, BtrV(S55A), decreases the dissociation rate of the BtrV(S55A)-BtrW(D82AG84A) complex to a level that allows detection by the two-hybrid analysis. Interestingly, alanine substitutions for the conserved aspartate and two surrounding residues in the G1 box of the B. subtilis ortholog RsbT also disrupted binding of the RsbT mutant with its antagonist RsbS (37). Although the conserved aspartate and glycine residues in the G1 box of BtrW play an important role in formation of the BtrV-BtrW complex, they do not seem to be involved in intramolecular interactions, since alanine substitutions did not affect binding to another molecule of BtrW, nor did they distort the secondary or tertiary structure of the mutant protein as determined by circular dichroism and fluorescence emission spectroscopy.
On the basis of results obtained with in-frame deletion mutations, we initially proposed that formation of a stable BtrV-BtrW complex is required for TTS in Bordetella and that phosphorylation plays a negative role by disrupting the BtrV-BtrW interaction (24). Although this model agrees with the observed lack of TTS in strains carrying ΔbtrW, ΔbtrV, or ΔbtrU alleles, it is inconsistent with our current observations. Most notably, although BtrV(S55A), BtrV(S55D), and BtrW(N51A) are incapable of undergoing phosphorylation and consequently form stable complexes, they fail to support TTS. Our cumulative results, which are summarized in Table 1, suggest a revised model for partner switcher-mediated regulation of Bordetella TTS. We propose that the ability of BtrW to phosphorylate BtrV is required for the activation of TTS. Upon phosphorylation, the BtrV-BtrW complex is expected to dissociate, releasing BtrW and BtrV∼P. Free BtrW may then associate with an alternative partner, which could be a part of the TTS machinery, a chaperone, or another regulatory molecule. We predict that this association is essential for TTS.
TABLE 1.
Compilation of data obtained from genetic and biochemical analyses of BtrW and BtrV mutantsa
| Strain or protein | Type III secretion | BtrV phosphorylation | BtrV-BtrW binding |
|---|---|---|---|
| Wild type | + | + | + |
| BtrV (S55A) | − | − | + |
| BtrV (S55D) | − | − | + |
| BtrW (N51A) | − | − | + |
| BtrW (D82AG84A) | − | + | − |
+, present; −, absent.
The suggestion that BtrW interacts with an alternative binding partner to activate TTS is supported by several lines of evidence. Deletion of btrW eliminates TTS, and all BtrW orthologs that have been sufficiently characterized have alternative binding partners in addition to their BtrV-like antagonists. Furthermore, the phenotype associated with the btrW(D82AG84A) allele is consistent with this hypothesis. In vitro analyses indicated that the BtrW G1-box mutant protein is defective in the ability to form a stable complex with BtrV. BtrW orthologs, such as SpoIIAB and RsbT, use overlapping sites for interacting with both of their alternative binding partners (17, 37), and it is therefore possible that the same domain of BtrW is involved in binding to both BtrV and its competing partner. If so, the G1-box mutation might not only weaken binding of BtrW to BtrV but also disrupt the interactions between BtrW and one or more alternative binding partners that are required for TTS.
Although the prediction that activation of Bordetella TTS requires the ability of BtrW to phosphorylate BtrV and disrupt the BtrV-BtrW complex is consistent with phenotypes obtained with the btrV(S55A), btrV(S55D), btrW(N51A), btrW(D82AG84A), and ΔbtrW alleles, it is also necessary to account for the observation that deletion of btrU or btrV eliminates TTS. In the model described above, both BtrU and BtrV are expected to negatively regulate TTS, since BtrU-mediated dephosphorylation of BtrV∼P would promote formation of a BtrV-BtrW complex, which presumably inactivates BtrW. To accommodate the fact that TTS is inactive in ΔbtrU and ΔbtrV mutants, we propose that BtrU and BtrV play positive roles in addition to their negative roles in partner switcher-mediated regulation of TTS. Although the exact nature of these additional functions remains to be determined, BtrU and BtrV could form part of the TTS apparatus and/or stabilize binding partners required for secretion.
It is apparent that the Bordetella BtrW, BtrV, and BtrU proteins participate in a complex regulatory cascade that represents a variation of the partner-switching paradigm. Analysis of the biochemical properties of BtrW and BtrV suggests that uncharacterized interactions with components of the TTS apparatus and other regulatory factors are likely to play a key role in governing secretion. The identification of additional proteins that associate with BtrW and perhaps BtrV∼P will be essential for understanding the regulatory mechanisms that control TTS.
Acknowledgments
We are grateful to our reviewers for their thoughtful and helpful comments. We thank Jérôme Zoidakis for technical assistance with circular dichroism and fluorescence spectroscopy.
N.A.K. was supported by Microbial Pathogenesis Training Grant T32AI07323-16, University of California, Los Angeles. This work was supported by NIH grants AI 38417 and AI 61598 to J.F.M.
REFERENCES
- 1.Alper, S., L. Duncan, and R. Losick. 1994. An adenosine nucleotide switch controlling the activity of a cell type-specific transcription factor in B. subtilis. Cell 77:195-205. [DOI] [PubMed] [Google Scholar]
- 2.Aravind, L., and E. V. Koonin. 2000. The STAS domain—a link between anion transporters and anti-sigma-factor antagonists. Curr. Biol. 10:R53-R55. [DOI] [PubMed] [Google Scholar]
- 3.Beaucher, J., S. Rodrigue, P. E. Jacques, I. Smith, R. Brzezinski, and L. Gaudreau. 2002. Novel Mycobacterium tuberculosis anti-sigma factor antagonists control σF activity by distinct mechanisms. Mol. Microbiol. 45:1527-1540. [DOI] [PubMed] [Google Scholar]
- 4.Benson, A. K., and W. G. Haldenwang. 1993. Bacillus subtilis sigma B is regulated by a binding protein (RsbW) that blocks its association with core RNA polymerase. Proc. Natl. Acad. Sci. USA 90:2330-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell, E. A., S. Masuda, J. L. Sun, O. Muzzin, C. A. Olson, S. Wang, and S. A. Darst. 2002. Crystal structure of the Bacillus stearothermophilus anti-sigma factor SpoIIAB with the sporulation sigma factor σF. Cell 108:795-807. [DOI] [PubMed] [Google Scholar]
- 6.Chaturongakul, S., and K. J. Boor. 2004. RsbT and RsbV contribute to σB-dependent survival under environmental, energy, and intracellular stress conditions in Listeria monocytogenes. Appl. Environ. Microbiol. 70:5349-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, P., R. E. Ruiz, Q. Li, R. F. Silver, and W. R. Bishai. 2000. Construction and characterization of a Mycobacterium tuberculosis mutant lacking the alternate sigma factor gene, sigF. Infect. Immun. 68:5575-5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cotter, P. A., and J. F. Miller. 1994. BvgAS-mediated signal transduction: analysis of phase-locked regulatory mutants of Bordetella bronchiseptica in a rabbit model. Infect. Immun. 62:3381-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delumeau, O., R. J. Lewis, and M. D. Yudkin. 2002. Protein-protein interactions that regulate the energy stress activation of σB in Bacillus subtilis. J. Bacteriol. 184:5583-5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diederich, B., J. F. Wilkinson, T. Magnin, M. Najafi, J. Erringston, and M. D. Yudkin. 1994. Role of interactions between SpoIIAA and SpoIIAB in regulating cell-specific transcription factor sigma F of Bacillus subtilis. Genes Dev. 8:2653-2663. [DOI] [PubMed] [Google Scholar]
- 11.Dove, S. L., J. K. Joung, and A. Hochschild. 1997. Activation of prokaryotic transcription through arbitrary protein-protein contacts. Nature 386:627-630. [DOI] [PubMed] [Google Scholar]
- 12.Dufour, A., and W. G. Haldenwang. 1994. Interactions between a Bacillus subtilis anti-sigma factor (RsbW) and its antagonist (RsbV). J. Bacteriol. 176:1813-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duncan, L., S. Alper, F. Arigoni, R. Losick, and P. Stragier. 1995. Activation of cell-specific transcription by a serine phosphatase at the site of asymmetric division. Science 270:641-644. [DOI] [PubMed] [Google Scholar]
- 14.Dutta, R., and M. Inouye. 2000. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem. Sci. 25:24-28. [DOI] [PubMed] [Google Scholar]
- 15.Edwards, R. A., L. H. Keller, and D. M. Schifferli. 1998. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207:149-157. [DOI] [PubMed] [Google Scholar]
- 16.Fouet, A., O. Namy, and G. Lambert. 2000. Characterization of the operon encoding the alternative σB factor from Bacillus anthracis and its role in virulence. J. Bacteriol. 182:5036-5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garsin, D. A., D. M. Paskowitz, L. Duncan, and R. Losick. 1998. Evidence for common sites of contact between the antisigma factor SpoIIAB and its partners SpoIIAA and the developmental transcription factor σF in Bacillus subtilis. J. Mol. Biol. 284:557-568. [DOI] [PubMed] [Google Scholar]
- 18.Helmann, J. D. 1999. Anti-sigma factors. Curr. Opin. Microbiol. 2:135-141. [DOI] [PubMed] [Google Scholar]
- 19.Hu, J. C., M. G. Kornacker, and A. Hochschild. 2000. Escherichia coli one- and two-hybrid systems for the analysis and identification of protein-protein interactions. Methods 20:80-94. [DOI] [PubMed] [Google Scholar]
- 20.Jonsson, I. M., S. Arvidson, S. Foster, and A. Tarkowski. 2004. Sigma factor B and RsbU are required for virulence in Staphylococcus aureus-induced arthritis and sepsis. Infect. Immun. 72:6106-6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalman, S., M. L. Duncan, S. M. Thomas, and C. W. Price. 1990. Similar organization of the sigB and spoIIA operons encoding alternate sigma factors of Bacillus subtilis RNA polymerase. J. Bacteriol. 172:5575-5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knobloch, J. K., S. Jager, M. A. Horstkotte, H. Rohde, and D. Mack. 2004. RsbU-dependent regulation of Staphylococcus epidermidis biofilm formation is mediated via the alternative sigma factor σB by repression of the negative regulator gene icaR. Infect. Immun. 72:3838-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lord, M., T. Magnin, and M. D. Yudkin. 1996. Protein conformational change and nucleotide binding involved in regulation of σF in Bacillus subtilis. J. Bacteriol. 178:6730-6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mattoo, S., M. H. Yuk, L. L. Huang, and J. F. Miller. 2004. Regulation of type III secretion in Bordetella. Mol. Microbiol. 52:1201-1214. [DOI] [PubMed] [Google Scholar]
- 25.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Min, K. T., C. M. Hilditch, B. Diederich, J. Errington, and M. D. Yudkin. 1993. Sigma F, the first compartment-specific transcription factor of B. subtilis, is regulated by an anti-sigma factor that is also a protein kinase. Cell 74:735-742. [DOI] [PubMed] [Google Scholar]
- 27.Miyazaki, E., J. M. Chen, C. Ko, and W. R. Bishai. 1999. The Staphylococcus aureus rsbW (orf159) gene encodes an anti-sigma factor of SigB. J. Bacteriol. 181:2846-2851.10217777 [Google Scholar]
- 28.Najafi, S. M., A. C. Willis, and M. D. Yudkin. 1995. Site of phosphorylation of SpoIIAA, the anti-anti-sigma factor for sporulation-specific sigma F of Bacillus subtilis. J. Bacteriol. 177:2912-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palma, M., and A. L. Cheung. 2001. σB activity in Staphylococcus aureus is controlled by RsbU and an additional factor(s) during bacterial growth. Infect. Immun. 69:7858-7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmidt, R., P. Margolis, L. Duncan, R. Coppolecchia, C. P. Moran, Jr., and R. Losick. 1990. Control of developmental transcription factor sigma F by sporulation regulatory proteins SpoIIAA and SpoIIAB in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 87:9221-9225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schroeter, R., S. Schlisio, I. Lucet, M. Yudkin, and R. Borriss. 1999. The Bacillus subtilis regulator protein SpoIIE shares functional and structural similarities with eukaryotic protein phosphatases 2C. FEMS Microbiol. Lett. 174:117-123. [DOI] [PubMed] [Google Scholar]
- 32.Skinner, J. A., A. Reissinger, H. Shen, and M. H. Yuk. 2004. Bordetella type III secretion and adenylate cyclase toxin synergize to drive dendritic cells into a semimature state. J. Immunol. 173:1934-1940. [DOI] [PubMed] [Google Scholar]
- 33.Stockbauer, K. E., A. K. Foreman-Wykert, and J. F. Miller. 2003. Bordetella type III secretion induces caspase 1-independent necrosis. Cell Microbiol. 5:123-132. [DOI] [PubMed] [Google Scholar]
- 34.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van der Zee, A., F. Mooi, J. Van Embden, and J. Musser. 1997. Molecular evolution and host adaptation of Bordetella spp.: phylogenetic analysis using multilocus enzyme electrophoresis and typing with three insertion sequences. J. Bacteriol. 179:6609-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.von Wintzingerode, F., G. Gerlach, B. Schneider, and R. Gross. 2002. Phylogenetic relationships and virulence evolution in the genus Bordetella. Curr. Top. Microbiol. Immunol. 264:177-199. [PubMed] [Google Scholar]
- 37.Woodbury, R. L., T. Luo, L. Grant, and W. G. Haldenwang. 2004. Mutational analysis of RsbT, an activator of the Bacillus subtilis stress response transcription factor, σB. J. Bacteriol. 186:2789-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang, X., C. M. Kang, M. S. Brody, and C. W. Price. 1996. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev. 10:2265-2275. [DOI] [PubMed] [Google Scholar]
- 39.Yuk, M. H., E. T. Harvill, P. A. Cotter, and J. F. Miller. 2000. Modulation of host immune responses, induction of apoptosis and inhibition of NF-κB activation by the Bordetella type III secretion system. Mol. Microbiol. 35:991-1004. [DOI] [PubMed] [Google Scholar]
- 40.Yuk, M. H., E. T. Harvill, and J. F. Miller. 1998. The BvgAS virulence control system regulates type III secretion in Bordetella bronchiseptica. Mol. Microbiol. 28:945-959. [DOI] [PubMed] [Google Scholar]
- 41.Ziebandt, A. K., H. Weber, J. Rudolph, R. Schmid, D. Hoper, S. Engelmann, and M. Hecker. 2001. Extracellular proteins of Staphylococcus aureus and the role of SarA and sigma B. Proteomics 1:480-493. [DOI] [PubMed] [Google Scholar]