Abstract
The alternative sigma factor σB is an important regulator of the stress response of Bacillus cereus. Here, the role of the regulatory proteins RsbV, RsbW, and RsbY in regulating σB activity in B. cereus is analyzed. Functional characterization of RsbV and RsbW showed that they act as an anti-sigma factor antagonist and an anti-sigma factor, respectively. RsbW can also act as a kinase on RsbV. These data are in line with earlier functional characterizations of RsbV and RsbW homologs in B. subtilis. The rsbY gene is unique to B. cereus and its closest relatives and is predicted to encode a protein with an N-terminal CheY domain and a C-terminal PP2C domain. In an rsbY deletion mutant, the σB response upon stress exposure was almost completely abolished, but the response could be restored by complementation with full-length rsbY. Expression analysis showed that rsbY is transcribed from both a σA-dependent promoter and a σB-dependent promoter. The central role of RsbY in regulating the activity of σB indicates that in B. cereus, the σB activation pathway is markedly different from that in other gram-positive bacteria.
The gram-positive rod-shaped bacterium Bacillus cereus is a frequent cause of food-borne disease, which has vomiting or diarrhea as its relatively mild main symptoms (22). B. cereus can also cause dangerous nongastrointestinal infections, including periodontitis, fulminant endophthalmitis, and meningitis in immunocompromised patients (6, 9, 12, 15). B. cereus is part of a group of bacteria which has been named the B. cereus group (reviewed in reference 13). This group also includes Bacillus thuringiensis, which is an insect pathogen and therefore is widely used as a biopesticide, and Bacillus anthracis, which can cause the disease anthrax. Recent complete-genome sequence studies confirmed the close genetic relationships between the organisms in the B. cereus group (21). In several gram-positive bacteria, such as Bacillus subtilis, Staphylococcus aureus, and Listeria monocytogenes, the alternative sigma factor σB plays an important role in redirecting gene expression under stress conditions (reviewed in references 10, 20, and 28). In B. cereus, σB is activated upon environmental stress and entry into stationary phase (5, 26). Phenotypic analysis of the sigB deletion mutant of B. cereus showed that σB is involved in the adaptive heat stress response (26).
Extensive studies in B. subtilis have addressed the topic of the regulation of σB activity (reviewed in reference 20). In nonstressed cells, σB is present in an inactive form by complexation with the anti-sigma factor RsbW. In this form, σB is unable to bind to RNA polymerase and thus cannot initiate the transcription of stress response genes. Under stress, an anti-sigma factor antagonist, RsbV, can bind to RsbW, thereby forming an RsbV-RsbW complex. This leads to the release of σB, which can then bind to RNA polymerase, leading to the transcription of σB-dependent genes. RsbW not only acts as an anti-sigma factor for σB but also is a kinase for RsbV, in which it phosphorylates a serine residue. The phosphorylated form of RsbV is unable to complex with RsbW and thus cannot release σB from its complex with RsbW. However, under stress conditions, a phosphatase which can dephosphorylate RsbV can be activated. Dephosphorylated RsbV can then form a complex with RsbW, leading to the release of σB.
There is considerable variation in the biochemical makeup of the phosphatases, which can dephosphorylate RsbV in the different bacteria (4, 28). A common theme is that they all have a C-terminal PP2C phosphatase domain, which is responsible for the dephosphorylation of RsbV. In B. subtilis, there are two PP2C phosphatases that act on phosphorylated RsbV (RsbV∼P), which were termed RsbU and RsbP. RsbU has an N-terminal domain that can bind an upstream regulator (RsbT) (4). The second is RsbP, which is unique for B. subtilis and has an N-terminal PAS (Per-ARNT-Sim) domain (29). The RsbU homolog in the B. cereus group has an N-terminal CheY-like domain. The CheY domain is a widespread regulatory domain in prokaryotes. It is named after the single-domain CheY protein, which is involved in chemotaxis, but in many bacteria, the CheY domain is coupled with a C-terminal effector domain, which can have a wide variety of functions (8, 24, 30). We have earlier proposed the name RsbY for the RsbU homolog of B. cereus to reflect its structural differences with other PP2C phosphatases which perform the crucial role of dephosphorylating RsbV∼P in the σB activation pathway in other bacteria (26).
In this study, we set out to characterize the roles of RsbV, RsbW, and RsbY in regulating the σB response of B. cereus. The bacterial strains, culture conditions, and genetic methods used in this study were described previously (26, 27). The oligonucleotides used are listed in Table 1.
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide | Sequence (5′-3′)a |
|---|---|
| OERsbV-PagI-F | GGGCGGAAATCATGAATTTGGCAATAAA |
| OERsbV-XhoI-R | CTCCCTCGAGCCTTCTTTCTACTTTTTCAA |
| OERsbW-NcoI-F | GGTGCCATGGAGAGATTTGAAAAGATAG |
| OERsbW-XhoI-R | GTGGCTCGAGGTAAGATTCGTAGGTTGAGATTG |
| KORsbY-XbaI-F | GTTCTAGAGATTATGGATGCG |
| KORsbY-EcoRI-R | GGAGGAATTCCAATGCCAAATGATAAGGAAAAA |
| Erycas-SacI-F | CCCAGAGCTCGGTCCGCAAAAGAAAAAC |
| Erycas-EcoRI-R | CCACGAATTCCATACCTAATAATTTATCTAC |
| PEOrf4-R | TGTCCTTGTTCATCACTAAT |
| PrSigB-F | GAAATCGCAAATCATTTAGG |
| qPCRrsbY-F | TGCCTGAAATTGATGGACTTGA |
| qPCRrsbY-R | CGGCCAATTTATTTGCATCC |
| qPCRtufA-F | GCCCAGGTCACGCTGACTAT |
| qPCRtufA-R | TCACGTGTTTGAGGCATTGG |
| GSP1-rsbY | TGATCTTCTCTTAATGGGCTACTT |
| GSP2-rsbY | GATTTTCTTCTTGCTCTTTATGC |
| ComprsbY-HindIII-F | GGAGAAGCTTGCAGCGAAATTAAATATGACAGAG |
| ComprsbY-BamHI-R | CACCGGATCCACCCAATTTAATCCTAGTGAACAA |
Underlined nucleotides indicate introduced restriction sites.
Functional analysis of RsbV and RsbW of B. cereus.
The functions of RsbV and RsbW in B. cereus were determined by performing in vitro transcription and phosphorylation reactions. For the in vitro study of the function of RsbV and RsbW of B. cereus, the genes encoding these proteins were cloned into pET28-b (by using the OERsbV and OERsbW primer pairs, respectively), resulting in a C-terminal His6 tag. Further purification of the proteins was performed as described previously for σB (26), with the exception that RsbV and RsbW were dialyzed against 10 mM Tris-HCl (pH 8), 50 mM KCl, 10 mM MgCl2, 0.4 mM dithiothreitol, and 20% glycerol (17). In vitro transcription reactions including purified B. cereus RNA polymerase (RNAP) at 30 nM, σB at 60 nM, RsbW at 0.3 μM, and RsbV at 1.5 μM were essentially performed as described previously (27). As a template for the in vitro transcription reaction, a PCR product generated with primers BcSigBF and PEOrf4, which contains the σB-dependent promoter upstream of orf4, was used at a concentration of 30 nM. For the reactions, the template, nucleotides, σB, and the regulators RsbV and RsbW were mixed and incubated at 30°C for 5 min before B. cereus core RNAP was added.
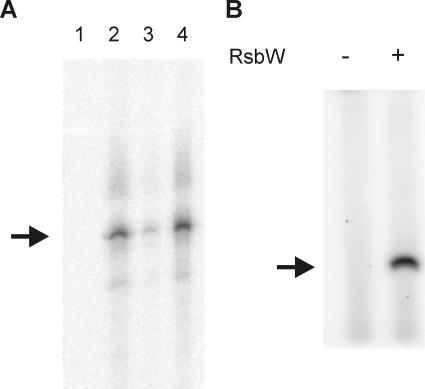
The in vitro transcription experiments confirmed the predicted functions of RsbW as an anti-sigma factor and RsbV as an anti-sigma factor antagonist (Fig. 1A). In B. subtilis, RsbW acts as a kinase on RsbV (7, 31). This property of RsbV and RsbW from B. cereus was also tested in an in vitro phosphorylation assay, which was performed according to previously described methodology (16, 31). In short, 1 μM RsbV, 1 μM RsbW, 40 μCi [γ-32P]ATP (3,000 Ci/mmol), and 20 μM non-radioactively labeled ATP were mixed in kinase buffer (50 mM Tris-HCl, pH 7.6, 50 mM KCl, 10 mM MgCl2, 1 mM dithiothreitol, 0.1 mM EDTA) and incubated at 30°C for 30 min, and the reaction was terminated by the addition of 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer and heating at 85°C for 5 min. Control reactions in which RsbW was omitted were also performed. Samples were separated on an 18% polyacrylamide gel. This revealed that also in B. cereus, RsbW can phosphorylate RsbV (Fig. 1B). In conclusion, the basic functions of RsbV and RsbW in B. cereus are essentially identical to the B. subtilis homologs, even though their primary sequence homologies at the amino acid level are relatively limited (43% for RsbV and 56% for RsbW [26]).
FIG. 1.
Functional analysis of RsbV and RsbW of B. cereus. (A) In vitro transcription assays for the determination of the function of RsbV and RsbW. A PCR template containing the σB-dependent promoter site 5′ of orf4 was used in the in vitro transcription reactions including B. cereus core RNAP (lane 1); core RNAP and σB (lane 2); core RNAP, σB, and RsbW (lane 3); and core RNAP, σB, RsbW, and RsbV (lane 4). After electrophoresis, runoff transcription products were visualized by autoradiography. The size of the σB-dependent transcription product is indicated with the arrow. (B) Phosphorylation of RsbV by RsbW. The phosphorylation reaction mixture contained 40 μCi [γ-32P]ATP (3,000 Ci/mmol), 1 μM RsbV, and where indicated, 1 μM RsbW. Non-radioactively labeled ATP was added at a concentration of 20 μM. Proteins were separated on an 18% SDS-PAGE gel, and phosphorylated proteins were visualized by autoradiography. The position of RsbV (determined by running a sample of purified RsbV in parallel to the phosphorylation reaction mixtures) is indicated by the arrow.
RsbY has a crucial role in regulating σB activity of B. cereus.
Previously, we have identified the rsbY gene, which is directly downstream of the sigB operon (26). Its C-terminal PP2C-domain, its close proximity to the sigB gene, and the absence of other genes with obvious homology to the important regulatory protein RsbU in B. subtilis, L. monocytogenes, and S. aureus already suggested that RsbY has a role in regulating σB activity of B. cereus. To check this hypothesis, a deletion mutant of rsbY was constructed by allelic replacement of the rsbY gene with an erythromycin resistance cassette. A 3.2-kb product was amplified by PCR with the primers KORsbY-XbaI-F and KORsbY-EcoRI-R, digested with the appropriate restriction enzymes, and cloned into pATΔS28 (18), resulting in pATΔrsbY. Note that the XbaI site in this PCR product is a natural restriction site which lies in the sigB gene. Restriction of this site did not generate mutations in the sigB gene, as was confirmed by subsequent sequencing of the sigB gene in the rsbY deletion mutant. Subsequently, the plasmid pATΔrsbY was digested with SacI and MunI. These enzymes cut in the rsbY gene at positions 110 and 980, respectively (the complete rsbY gene is 1,143 bases long). Subsequently, the erythromycin resistance cassette of pUC18ERY (25) was amplified with the primers Erycas-SacI-F and Erycas-EcoRI-R and, upon restriction with SacI and EcoRI, cloned into the digested pATΔrsbY vector (note that digestion with MunI and EcoRI results in compatible sites), resulting in pATΔrsbYery. This plasmid was then transformed to E. coli HB101/pRK24, and the resulting strain was used in conjugation experiments with B. cereus to generate the rsbY deletion mutant B. cereus FM1401, according to previously described methodology (26). Subsequently, the activation of σB under various stress conditions was studied by immunoblotting with σB antiserum. Cultures of B. cereus ATCC 14579 and FM1401 in the mid-exponential growth phase (OD600, 0.4 to 0.5) were stressed by a heat shock from 30°C to 42°C and by the addition of ethanol or NaCl to a final concentration of 4% (vol/vol) or 2.5% (wt/vol), respectively. Proteins were extracted before and 2.5, 5, 10, and 30 min after the stress exposure and immunoblotted anti-σB antiserum as described previously (26), with the modification that the resulting immunocomplexes were visualized by using the SuperSignal West Pico chemiluminescent substrate (Perbio, Etten-Leur, The Netherlands).
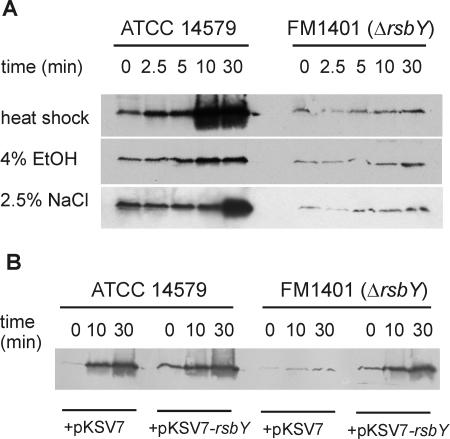
Under all tested conditions, an almost completely abolished σB response was observed in the rsbY null mutant (Fig. 2A). This clearly shows that RsbY is the key regulator of σB activity in B. cereus. The slight σB-activating effect upon stress exposure that remains in B. cereus FM1401 may be due to an as yet unidentified regulatory mechanism of minor importance. During stress exposure, proteins were isolated at regular intervals, and this allowed the study of the time course of σB activation. It is clear that in B. cereus, this response can be extremely rapid: already after 2.5 min, upon a heat shock, an increase in σB levels can be noted, and after 10 min, σB reached maximal levels. Also, during ethanol stress, there is a rapid increase of σB levels. The response of σB to osmotic stress is slower: only after 30 min can a strong activation of σB be observed.
FIG. 2.
The effect of deletion of rsbY on the stress-induced activation of σB in B. cereus. (A) Cellular σB levels in B. cereus ATCC 14579 and its rsbY deletion mutant FM1401 upon stress exposure. Bacterial proteins were extracted from cultures in the mid-exponential growth phase (time, 0 min) and upon exposure to the indicated stress for 2.5, 5, 10, and 30 min. Forty micrograms of protein of each sample was loaded on a 15% SDS-PAGE gel. σB was detected by immunoblotting with anti-σB antiserum as described in the text. EtOH, ethanol. (B) In trans complementation of the rsbY deletion mutant restores the activation of σB under stress. B. cereus ATCC 14579 and its rsbY deletion mutant FM1401 carrying the vector pKSV7 or pKSV7-rsbY (which contains full-length rsbY under the control of its natural promoter) were grown until mid-exponential growth phase and heat shocked from 30°C to 42°C. Proteins were extracted at the indicated times. Electrophoresis and the detection of σB were performed as described above.
The important role of RsbY in the σB activation pathway of B. cereus was confirmed by complementation of the rsbY deletion mutant in trans with a wild-type copy of rsbY expressed under the control of its own σA-dependent promoter (see below for the description of the promoter of rsbY). A PCR fragment, generated with the primers ComprsbY-HindIII-F and ComprsbY-BamHI-R, was cloned into the gram-positive shuttle vector pKSV7 (23), resulting in pKSV7-rsbY. B. cereus ATCC 14579 and FM1401 were transformed with the plasmids pKSV7 and pKSV7-rsbY by electroporation (1). Immunoblotting with anti-σB antiserum showed that the activation of σB upon a heat shock was restored in the rsbY deletion mutant upon complementation with the wild-type copy of rsbY (Fig. 2B).
Expression analysis of rsbY.
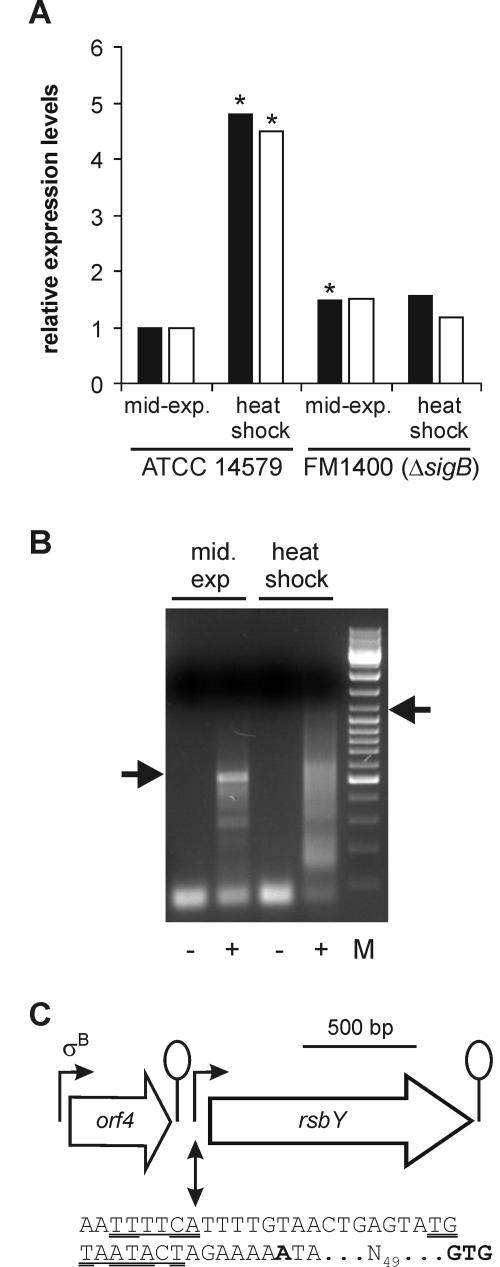
The levels of expression of rsbY in B. cereus ATCC 14579 and its sigB deletion mutant (B. cereus FM1400) during the mid-exponential growth phase and upon a heat shock from 30°C to 42°C were determined. Northern analysis was first used to asses the expression of rsbY, but no transcripts could be visualized (data not shown), and consequently, real-time PCR was employed in follow-up experiments. RNA was extracted from two independent cultures of mid-exponential phase and heat-shocked B. cereus cells by using RNAwiz (Ambion, Huntingdon, United Kingdom). Residual DNA from the RNA preparations was enzymatically removed by using TURBO DNA-free (Ambion), and cDNA was synthesized from RNA by using Superscript III reverse transcriptase (Invitrogen, Breda, The Netherlands), 2 pmol of a rsbY gene-specific primer (qPCRrsbY-R), each deoxynucleoside triphosphate at a concentration of 0.5 mM, and 1 μg of total RNA. Reverse transcription and quantitative PCRs on the synthesized cDNAs by using an ABI Prism 7700 with SYBR green technology (PE Applied Biosystems, Nieuwekerk a/d IJssel, The Netherlands) were performed as described previously (2). Relative transcript levels were calculated by using the relative expression software tool (REST) (19). The expression of the tufA gene was used as a reference for the determination of induction levels. The expression of rsbY was found to be upregulated approximately 4.6-fold upon a heat shock in B. cereus ATCC 14579 but not in the sigB deletion mutant, which indicates that the expression of rsbY is upregulated under a heat shock in a σB-dependent fashion (Fig. 3A). Note that in one experiment, a small but borderline significant difference in the expression of rsbY in the mid-exponential growth between B. cereus ATCC 14579 and B. cereus FM1400 was observed. This effect could not be reproduced in the second experiment, leading to the conclusion that in the mid-exponential growth phase, the expression of rsbY is essentially identical in both strains.
FIG. 3.
Expression analysis of rsbY. (A) Real-time PCR quantification of rsbY transcript levels. Cultures of B. cereus ATCC 14579 and the sigB deletion mutant FM1400 were grown until the mid-exponential growth phase and exposed to a mild heat shock from 30°C to 42°C for 10 min. The results of RNA extraction from two independently treated cultures (black and white bars) are shown. The expression level of rsbY was set at 1 during the mid-exponential growth phase in B. cereus ATCC 14579, and the other conditions were compared to that condition by using REST (19). Expression levels of rsbY significantly different (P < 0.05) from the levels of expression of rsbY during mid-exponential growth phase in B. cereus ATCC 14579 are indicated with an asterisk. (B) 5′ RACE mapping of rsbY promoter sites. The RNA used in the 5′ RACE reactions was isolated from B. cereus ATCC 14579 under the same growth conditions as those described above. Agarose gel electrophoresis of PCR-amplified untailed (negative controls, indicated by the minus symbols) and poly(dC)-tailed (plus symbols) cDNA is shown. The marker (M) shown is the GeneRuler DNA Mix marker from MBI Fermentas GmbH, St. Leon-Rot, Germany. Fragments that were cloned and sequenced are indicated with the arrows. (C) Overview of the transcription of rsbY in B. cereus ATCC 14579. The σA-dependent promoter site directly upstream of rsbY is shown with the identified −35 and −10 regions underlined.Double-underlined residues indicate matches with the B. subtilis σA promoter consensus sequence (11). Bold type indicates the mapped transcriptional start site. The spacing to the GTG start codon of rsbY is also indicated. The orf4 reading frame with its σB-dependent promoter upstream of rsbY is indicated. Lollipops indicate stem-loop structures.
The 5′ ends of the rsbY transcripts were mapped by using 5′ rapid amplification of cDNA ends (RACE) performed on RNA samples isolated from B. cereus ATCC 14579 with the 5′ RACE system (Invitrogen) using the GSP RsbY primers (Table 1) according to the manufacturer's instructions. During exponential growth, rsbY is transcribed from a promoter that is situated directly upstream from rsbY (Fig. 3B). This promoter is probably σA dependent, even though its sequence is somewhat different from the σA promoter consensus sequence from B. subtilis. These apparent mismatches may result in the low-level transcription of the rsbY gene under exponential growth conditions. With RNA that was isolated upon a heat shock, a smear was observed in the 5′ RACE reaction, which may indicate degradation of the mRNA. Upon the cloning and sequencing of an approximately 1.0-kb product, which appeared to be the largest fragment visible upon electrophoresis, a second promoter site was mapped. The 5′ RACE reaction product mapped to a position in orf4, the open reading frame 5′ of rsbY. However, the fact that no clear σB-dependent promoter site exists at this position leads to the conclusion that this is probably a degradation product of a transcript that originates from the σB-dependent promoter site upstream of orf4. The fact that transcripts originating from orf4 contribute to rsbY expression is remarkable because directly downstream of orf4, a stem-loop structure with a calculated free energy of formation of −9.4 kcal/mol exists. Previous Northern analysis of the transcription of orf4 did not show transcription proceeding beyond this structure (26). However, by use of a more sensitive PCR-based method, it was shown that transcription through this stem-loop structure occurs and that it contributes significantly to the upregulation of rsbY transcription under stress conditions.
A model for the regulation of σB activity in B. cereus.
On the basis of the results obtained in this study, a model for the regulation of σB activity in B. cereus can be drawn up (Fig. 4). The main conclusions of this study are that RsbV and RsbW of B. cereus have functions identical to those of their homologs in other gram-positive bacteria and that the unique regulator RsbY is crucial for regulating σB activity in B. cereus. The N-terminal CheY response regulator domain of RsbY suggests that RsbY is activated through a mechanism which involves phosphorylation of a conserved aspartate residue in the CheY domain by an as yet unidentified kinase. The coupling of a CheY domain to a PP2C phosphatase domain in itself is not unique, but it is a rare occurrence, as most CheY domains are coupled to a C-terminal binding DNA output domain that activates or represses transcription of specific target genes (8). If the activation of σB in B. cereus is directly coupled with a sensor kinase, as in a classical two-component signal transduction cascade, this would mean a major difference with the σB activation pathway of B. subtilis in which more partner-switching units and large protein complexes form important parts of the sensing and signaling cascade (3, 14, 20). So it appears that even though B. cereus and B. subtilis are relatively closely related bacteria, two different sensing and signaling pathways leading to the activation of the σB have evolved. In this respect, it is also noteworthy that the rsbY gene is partially under transcriptional control of σB. In other gram-positive bacteria, the PP2C phosphatase that is responsible for the dephosphorylation of RsbV is constitutively transcribed. It remains to be determined if this positive feedback effect of σB levels on the expression of rsbY contributes to the process of σB activation in B. cereus.
FIG. 4.
Model for the regulation of σB activity in B. cereus. Under nonstress conditions, σB is kept in an inactive state by its anti-sigma factor, RsbW. This protein also functions as a kinase, which can phosphorylate (P) the anti-sigma factor antagonist, RsbV. When RsbV is dephosphorylated by the action of the RsbY phosphatase, RsbV can bind to RsbW. This leads to the release of σB from its complex with RsbW and, upon association of σB to core RNAP, to the transcription of σB-dependent genes. RsbY has an N-terminal CheY response regulator domain. This strongly suggests that an as-yet-unidentified kinase (shown in gray) can phosphorylate this domain, leading to the activation of the phosphatase.
The identification of mechanisms that regulate the activity of RsbY is currently under way in our laboratory and may provide important mechanistic clues on how stress conditions are sensed and signaled leading to the activation of σB in B. cereus and in closely related bacteria like B. anthracis and B. thuringiensis. This may provide indications on how the activation pathway of σB can be perturbed. A disruption of the stress response may lead to the sensitization of bacteria under stress conditions. This approach may then be used to counter the growth and survival of bacteria from the B. cereus group during food processing or pathogenesis.
REFERENCES
- 1.Bone, E. J., and D. J. Ellar. 1989. Transformation of Bacillus thuringiensis by electroporation. FEMS Microbiol. Lett. 49:171-177. [DOI] [PubMed] [Google Scholar]
- 2.Bron, P. A., M. Marco, S. M. Hoffer, E. Van Mullekom, W. M. de Vos, and M. Kleerebezem. 2004. Genetic characterization of the bile salt response in Lactobacillus plantarum and analysis of responsive promoters in vitro and in situ in the gastrointestinal tract. J. Bacteriol. 186:7829-7835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, C. C., R. J. Lewis, R. Harris, M. D. Yudkin, and O. Delumeau. 2003. A supramolecular complex in the environmental stress signalling pathway of Bacillus subtilis. Mol. Microbiol. 49:1657-1669. [DOI] [PubMed] [Google Scholar]
- 4.Delumeau, O., S. Dutta, M. Brigulla, G. Kuhnke, S. W. Hardwick, U. Volker, M. D. Yudkin, and R. J. Lewis. 2004. Functional and structural characterization of RsbU, a stress signaling protein phosphatase 2C. J. Biol. Chem. 279:40927-40937. [DOI] [PubMed] [Google Scholar]
- 5.de Vries, Y. P., L. M. Hornstra, W. M. de Vos, and T. Abee. 2004. Growth and sporulation of Bacillus cereus ATCC 14579 under defined conditions: temporal expression of genes for key sigma factors. Appl. Environ. Microbiol. 70:2514-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drobniewski, F. A. 1993. Bacillus cereus and related species. Clin. Microbiol. Rev. 6:324-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dufour, A., and W. G. Haldenwang. 1994. Interactions between a Bacillus subtilis anti-sigma factor (RsbW) and its antagonist (RsbV). J. Bacteriol. 176:1813-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galperin, M. Y. 2004. Bacterial signal transduction network in a genomic perspective. Environ. Microbiol. 6:552-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaur, A. H., C. C. Patrick, J. A. McCullers, P. M. Flynn, T. A. Pearson, B. I. Razzouk, S. J. Thompson, and J. L. Shenep. 2001. Bacillus cereus bacteremia and meningitis in immunocompromised children. Clin. Infect. Dis. 32:1456-1462. [DOI] [PubMed] [Google Scholar]
- 10.Hecker, M., and U. Volker. 2001. General stress response of Bacillus subtilis and other bacteria. Adv. Microb. Physiol. 44:35-91. [DOI] [PubMed] [Google Scholar]
- 11.Helmann, J. D., and C. P. Moran, Jr. 2002. RNA polymerase and sigma factors, p. 289-312. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 12.Hilliard, N. J., R. L. Schelonka, and K. B. Waites. 2003. Bacillus cereus bacteremia in a preterm neonate. J. Clin. Microbiol. 41:3441-3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jensen, G. B., B. M. Hansen, J. Eilenberg, and J. Mahillon. 2003. The hidden lifestyles of Bacillus cereus and relatives. Environ. Microbiol. 5:631-640. [DOI] [PubMed] [Google Scholar]
- 14.Kim, T. J., T. A. Gaidenko, and C. W. Price. 2004. A multicomponent protein complex mediates environmental stress signaling in Bacillus subtilis. J. Mol. Biol. 341:135-150. [DOI] [PubMed] [Google Scholar]
- 15.Kotiranta, A., K. Lounatmaa, and M. Haapasalo. 2000. Epidemiology and pathogenesis of Bacillus cereus infections. Microbes Infect. 2:189-198. [DOI] [PubMed] [Google Scholar]
- 16.Min, K. T., C. M. Hilditch, B. Diederich, J. Errington, and M. D. Yudkin. 1993. σF, the first compartment-specific transcription factor of B. subtilis, is regulated by an anti-sigma factor that is also a protein kinase. Cell 74:735-742. [DOI] [PubMed] [Google Scholar]
- 17.Miyazaki, E., J. M. Chen, C. Ko, and W. R. Bishai. 1999. The Staphylococcus aureus rsbW (orf159) gene encodes an anti-sigma factor of SigB. J. Bacteriol. 181:2846-2851.10217777 [Google Scholar]
- 18.Namy, O., M. Mock, and A. Fouet. 1999. Co-existence of clpB and clpC in the Bacillaceae. FEMS Microbiol. Lett. 173:297-302. [DOI] [PubMed] [Google Scholar]
- 19.Pfaffl, M. W., G. W. Horgan, and L. Dempfle. 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Price, C. W. 2002. General stress response, p. 369-384. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 21.Rasko, D. A., M. R. Altherr, C. S. Han, and J. Ravel. 2005. Genomics of the Bacillus cereus group of organisms. FEMS Microbiol. Rev. 29:303-329. [DOI] [PubMed] [Google Scholar]
- 22.Schoeni, J. L., and A. C. Wong. 2005. Bacillus cereus food poisoning and its toxins. J. Food Prot. 68:636-648. [DOI] [PubMed] [Google Scholar]
- 23.Smith, K., and P. Youngman. 1992. Use of a new integrational vector to investigate compartment-specific expression of the Bacillus subtilis spoIIM gene. Biochimie 74:705-711. [DOI] [PubMed] [Google Scholar]
- 24.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 25.van Kranenburg, R., J. D. Marugg, I. I. van Swamm, N. J. Willem, and W. M. de Vos. 1997. Molecular characterization of the plasmid-encoded eps gene cluster essential for exopolysaccharide biosynthesis in Lactococcus lactis. Mol. Microbiol. 24:387-397. [DOI] [PubMed] [Google Scholar]
- 26.Van Schaik, W., M. H. Tempelaars, J. A. Wouters, W. M. De Vos, and T. Abee. 2004. The alternative sigma factor σB of Bacillus cereus: response to stress and role in heat adaptation. J. Bacteriol. 186:316-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Schaik, W., M. H. Zwietering, W. M. de Vos, and T. Abee. 2004. Identification of σB-dependent genes in Bacillus cereus by proteome and in vitro transcription analysis. J. Bacteriol. 186:4100-4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Schaik, W., and T. Abee. 2005. The role of σB in the stress response of gram-positive bacteria—targets for food preservation and safety. Curr. Opin. Biotechnol. 16:218-224. [DOI] [PubMed] [Google Scholar]
- 29.Vijay, K., M. S. Brody, E. Fredlund, and C. W. Price. 2000. A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the sigmaB transcription factor of Bacillus subtilis. Mol. Microbiol. 35:180-188. [DOI] [PubMed] [Google Scholar]
- 30.Volz, K. 1993. Structural conservation in the CheY superfamily. Biochemistry 32:11741-11753. [DOI] [PubMed] [Google Scholar]
- 31.Yang, X., C. M. Kang, M. S. Brody, and C. W. Price. 1996. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev. 10:2265-2275. [DOI] [PubMed] [Google Scholar]