Abstract
The role of the serine/threonine kinase PknH in the physiology and virulence of Mycobacterium tuberculosis was assessed by the construction of a pknH deletion mutant. Deletion of the pknH gene did not affect sensitivity to the antimycobacterial drug ethambutol, although it was previously thought to be involved in regulating expression of emb genes encoding arabinosyl transferases, the targets of ethambutol. Nevertheless, transcription analyses revealed that genes associated with mycobacterial cell wall component synthesis, such as emb and ini operons, are downstream substrates of the PknH signaling cascade. In vitro survival studies revealed that a mutant with a deletion of the pknH gene displayed increased resistance to acidified nitrite stress, suggesting that nitric oxide is one of the potential environmental triggers for PknH activation. The effect of pknH deletion on mycobacterial virulence was investigated in BALB/c mice. In this model, the ΔpknH mutant was found to survive and replicate to a higher bacillary load in mouse organs than its parental strain and the pknH-complemented strain. In contrast, another closely related kinase mutant, the ΔpknE mutant, obtained from the same parental strain, was not affected in its virulence phenotype. Infection of THP-1 cells or in vitro growth studies in 7H9 medium did not reveal a significant in vitro growth advantage phenotype for the ΔpknH mutant. In conclusion, we propose that the serine/threonine kinase PknH plays a role in regulating bacillary load in mouse organs to facilitate adaptation to the host environment, possibly by enabling a regulated chronic infection by M. tuberculosis.
Despite its discovery a century ago, Mycobacterium tuberculosis, the causative agent of tuberculosis, continues to kill more people than any other bacterial pathogen. It is estimated that one-third of the global population is infected with M. tuberculosis and that approximately 8 million new cases of tuberculosis arise annually, with 2 million people dying from the disease (57). The pathogen enters the host, usually by inhalation of an infected aerosol, and the bacilli are phagocytosed by alveolar macrophages. Intracellular replication of the bacterium results in a primary lesion, and this is followed by lymphohematogenous dissemination and the formation of secondary lesions in the lungs and other organs (15). Uncontrolled M. tuberculosis growth in its site of infection is associated with extensive lung damage, ultimately leading to the death of the host. However, in most individuals, disease progression is arrested at this stage by the acquired immune response resulting in the formation of granulomatous lesions, and a clinically latent state ensues. Postprimary disease arises from the subsequent reactivation of dormant bacilli (15, 19, 52).
The establishment of a persistent infection demands that microbes evade and subvert various immune mechanisms that are meant to eliminate pathogens. The host mounts a strong immune response that contains but does not eliminate the infection. The ability of the organism to survive in the face of a robust host response clearly implicates a series of evasion mechanisms by the pathogen (24, 27, 30, 44). The pathogen, therefore, should be able to sense its environment and respond in a coordinated manner by modulating the expression of its adaptive genes. Identifying the various components involved in these processes is central to our understanding of the pathogenesis of tuberculosis.
Protein phosphorylation is the principal mechanism by which extracellular signals are translated into cellular responses. In bacteria, signal transduction events are mediated by two-component regulatory systems (23, 53) and protein kinases and phosphatases (5, 14, 48). M. tuberculosis possesses 11 protein serine/threonine kinases (3, 12), of which 8 members of the kinases, including PknH kinase, have been shown to possess the catalytic enzyme activity in vitro (4, 8, 13, 22, 26, 32, 33, 47). Except in the case of PknF and PknH, the identities of the intracellular target proteins of protein serine/threonine kinases of M. tuberculosis are yet to be identified (33, 34). The M. tuberculosis PknH kinase was shown to phosphorylate in vitro the mycobacterial endogenous substrate EmbR through recognition of a Forkhead-associated domain in the protein (33). EmbR is a putative transcriptional regulator of embAB genes encoding arabinosyl transferases which are involved in the biosynthesis of arabinogalactans, a key component of the mycobacterial cell wall (6, 17, 54, 58). The embAB gene products were identified as the major target of the antimycobacterial drug ethambutol (6). Resistance to ethambutol may arise by overexpression of emb products, point mutation in the embB gene, or both (51, 54). However, it is not known whether the PknH phosphorylation of EmbR could affect the sensitivity of M. tuberculosis to ethambutol by altering the expression levels of the M. tuberculosis embCAB operon. The enzyme activities of the embC and embAB gene products contribute to the arabinosylation of lipoarabinomannans (LAM) and arabinogalactans (AG), respectively (17, 58). The AG and LAM form key structural components and play important roles in the modulation of host response during infection (reviewed in reference 7). It has been shown that the expression of the pknH gene in M. tuberculosis is downregulated upon exposure to low pH and heat shock, suggesting a role for the kinase in adaptation to environmental changes (47).
In order to assess the role of PknH in M. tuberculosis physiology and virulence, we created a pknH gene knockout mutant and found that the deletion of the pknH gene conferred altered sensitivity to in vitro treatments causing nitrosative and oxidative stresses. Significantly, upon infection of mice, the M. tuberculosis ΔpknH mutant survived to a higher load in the mouse organs, especially during the chronic stage of infection, indicating that in the wild type, the PknH kinase-mediated signaling pathway contributes to the regulation of bacillary load during the infection process.
MATERIALS AND METHODS
Construction of the pknH-targeting vector.
A 5.7-kb DNA fragment containing the pknH coding region along with approximately 1.9-kb flanking regions was amplified by PCR using Pfu Turbo DNA polymerase (Stratagene) and cloned into pBluescript vector to construct pKP165. A major part of the pknH coding sequence (1.6 kb of the 1.88 kb) was deleted, and a unique cloning site for BglII was simultaneously introduced by inverse PCR and religation of the PCR product to create pKP169. A kanamycin resistance gene from pUC4K was then inserted into this site to construct pKP178. The disrupted fragment containing the flanking regions and the kan gene was excised from this plasmid and cloned at the XbaI site of the Escherichia coli vector pBS-PacI to produce pKP181. The PacI cassette of pGOAL19 containing hyg-lacZ-sacB genes (38) was then cloned at the unique PacI site carried on the vector part of the plasmid pKP181 to make the final pknH knockout construct pKP183. This plasmid carried kan gene (located between the pknH-flanking regions) for positive selection and the sacB gene to facilitate counterselection. In addition, the lacZ and hyg markers located on the PacI cassette served in the screening for the loss of vector.
Isolation of the pknH deletion mutant of M. tuberculosis.
The pknH deletion mutant was isolated by a sequential two-step selection protocol involving positive selection for kanamycin resistance and counterselection on medium containing 2% sucrose (38, 39). The pknH-targeting vector was electroporated into M. tuberculosis strain H37Rv, and transformants were selected on 7H11 plates with kanamycin (Kan). Colonies displaying the expected phenotype for single crossovers (Kanr colonies displaying expression of the lacZ and hyg genes) were grown further and counterselected on 7H11 plates with Kan and 2% sucrose. Putative recombinants with the desired phenotypes (Sucr Kanr Hygs LacZ−) were further screened by PCR (not shown) using pairs of oligonucleotides, one located in the kan gene and the other on the genomic DNA external to the cloned flanking regions. Based on the PCR analysis, two isolates were chosen for Southern hybridization analysis. Genomic DNAs isolated from the parental wild type and the two isolates of the pknH mutant strain were subjected to Southern hybridization analysis to confirm that the ΔpknH mutant arose following double crossover homologous recombination.
Complementation of the ΔpknH strain.
A 2.3-kb DNA fragment containing the entire pknH coding region along with 311-bp upstream and 167-bp downstream regions was amplified by PCR using Pfu Turbo DNA polymerase and cloned between the BglII and XbaI sites of the attP vector pKP201 (21). The insert in the complementing clone was sequenced to verify that no mutation was introduced during PCR amplification of the fragment. The ΔpknH mutant was cotransformed with the complementing plasmid pKP264 and pBSint (50), a nonreplicating plasmid which provides integrase in trans but is subsequently lost from the cells, thereby reducing the chances of integrase-mediated excision of the complementing DNA (50). The transformants were selected on 7H11 medium with hygromycin, and the expression of pknH in the complemented strain was verified by reverse transcription-PCR (RT-PCR) analysis. RNAs isolated from the wild-type, ΔpknH mutant, and complemented strains were reverse transcribed using Moloney murine leukemia virus reverse transcriptase, and a 278-bp region located within the deleted region of the pknH gene was amplified by PCR using the primer pairs 1MO (5′-GCGCCGCAGCCAAGAATCC-3′) and 1MP (5′-AGCCGCCGCCCTGGTAGTA-3′).
In vitro growth determinations.
For comparison of the in vitro growth rates, the mycobacterial strains were grown in rolling culture conditions (43) in Dubos broth supplemented with 0.05% Tween, 0.2% glycerol, and 10% Dubos medium albumin.
Intracellular growth in THP-1 cells.
Published protocols were followed for the preparation and infection of monolayers of THP-1 cells (28). THP-1 cells were seeded at 5 × 105 per well in 2-cm2 24-well tissue culture plates and were differentiated by the addition of phorbol 12-myristate 13-acetate (20 ng/ml) and incubation for 20 h. Bacterial inocula were prepared by dilution of log-phase cultures (optical density at 600 nm [OD600], 0.6) grown in 7H9 broth, and the inoculum CFU was determined. The monolayers were infected with the M. tuberculosis strains at a multiplicity of infection of 1:5 (bacterium:THP-1) for 20 h, then washed with warm phosphate-buffered saline medium, and resuspended in warm RPMI medium, and the plates were incubated at 37°C. Intracellular bacteria were recovered by lysing the monolayers in 0.025% sodium dodecyl sulfate and then serially diluted and plated on 7H10 agar with oleic acid-albumin-dextrose-catalase (OADC) to determine their CFU.
Mouse infection studies.
The parental strain, the ΔpknH mutant, the ΔpknE mutant, and the pknH-complemented strain were grown in 7H9 broth containing 0.05% Tween and 10% albumin-dextrose complex (ADC) to an OD600 of approximately <0.5. The cultures were vortexed with 2-mm-diameter glass beads to break any clumps, and the bacteria were allowed to settle. Bacterial suspensions were diluted in phosphate-buffered saline, and 6- to 8-week-old female BALB/c mice were injected intravenously with approximately 5 × 105 bacteria. The survival and multiplication of the M. tuberculosis strains were determined by enumerating bacterial CFU in the lungs and spleens of four infected mice for each group (three mice at the last time point for the ΔpknH mutant-infected group).
Sensitivity of the pknH strains to acidified nitrite and oxidative stress-causing treatments.
Bacterial strains were initially grown to early log phase (OD600 ∼ 0.3) in Middlebrook 7H9-Tween-albumin-dextrose-sodium chloride (ADS) broth (25). For testing susceptibility to reactive nitrogen intermediates (RNI), bacteria were harvested, washed, and resuspended in the acidified medium (7H9-Tween-ADS broth adjusted to pH 5.4). These bacterial suspensions were diluted as necessary and divided into two 10-ml aliquots. Sodium nitrite (3 mM final concentration) was added to one of the aliquots, whereas the other aliquot served as untreated control. Following incubation in the presence of acidified nitrite stress for 48 h, viability was determined by serial dilution and plating on 7H10 agar with OADC and compared with the CFU obtained for the untreated control.
To test the effect of pknH deletion on sensitivity to peroxide- and superoxide-generating treatments, early-log-phase cultures (OD600, 0.3) were exposed to the following treatments: 10 mM H2O2 (Fisher Sciences), 50 mM paraquat (Sigma-Aldrich), or no treatment. The stresses were applied for 48 h, and the cultures were then serially diluted and plated to determine viability.
RNA extraction and real-time quantitative PCR.
Early-log-phase cultures (OD600 ∼ 0.3) of the pknH strains were divided into two aliquots; ethambutol (final concentration, 0.2 μg/ml) was added to one aliquot, whereas the other aliquot served as untreated control. After 24 h of incubation in a roller incubator, bacteria were harvested and RNAs were extracted by using FastRNA Pro Blue kit (Qbiogene). Cells were lysed in the presence of glass beads in a FastPrep instrument (Qbiogene) according to the instructions provided with the FastRNA Pro kit. After extraction with chloroform and precipitation with ethanol, RNA was dissolved in diethyl pyrocarbonate-treated water. Contaminating DNA was removed by digestion with RNase-free Turbo DNase (Ambion) according to the supplier's instructions, and the RNeasy minikit (QIAGEN) was used for the subsequent cleanup procedures. Removal of DNA was confirmed by performing PCR using an aliquot of the DNase-treated RNA as a template. Reverse transcription reactions were carried out in 20-μl volume containing 0.5 μg RNA, random primers, and the buffer and enzyme components of the RevertAid H-Minus first-strand cDNA synthesis kit (MBI Fermentas) according to the supplied protocol. Reactions in which the reverse transcriptase was omitted served as controls for DNA contamination. Following cDNA synthesis, the RT enzyme was inactivated by incubating at 75°C for 10 min, and the volume of the RT reaction was made up to 50 μl. Control PCR amplifications for the expressions of sigA- and sigC-specific mRNAs were performed on the cDNA templates from the parental strain, the ΔpknH mutant, and the complement to confirm that the cDNAs from the three strains served as templates for PCR. Real-time PCR analysis was carried out on the DNA Engine Opticon instrument (MJ Research) using the PCR master mix containing SYBR green dye (Finnzymes). The 20-μl PCRs consisted of PCR master mix (Finnzymes), 300 nM concentrations of each primer, and 4 μl of cDNA template. The sequences of the primers used in the real-time PCR are given in Table 1. In each case, the test gene and the normalizing gene (sigA) were assayed along with a set of standard samples (genomic DNA), and the amounts of gene-specific mRNA were normalized to the amount of sigA mRNA.
TABLE 1.
Nucleotide sequences of the primers used for real-time quantitative PCR
| Gene | Rv no. | Forward primer (5′-3′) | Reverse primer (5′-3′) | Product size (bp) |
|---|---|---|---|---|
| sigA | Rv2703 | CTCGGTTCGCGCCTACCTCA | GCGCTCGCTAAGCTCGGTCA | 130 |
| embC | Rv3793 | ATCACCGAGCTGCTGATG | TGCGAGTCACCGTTCCTA | 145 |
| embB | Rv3795 | GACGAGTCCTGGCATCAA | CATGGCGTATTCGAGCAC | 250 |
| iniB | Rv0341 | GTCGAGCATGGCTTGGTCCT | GTACGACGGCAGTTCGATGG | 285 |
| iniA | Rv0342 | TGGCGGTGTCTCTAGGTTCC | TCCACGTCAGCAGTCAGGTC | 172 |
Statistical analysis.
The significance of the differences between the experimental groups was determined by two-tailed, unpaired Student's t test. Differences with a P value of <0.05 were considered significant.
RESULTS
Isolation of the M. tuberculosis ΔpknH mutant.
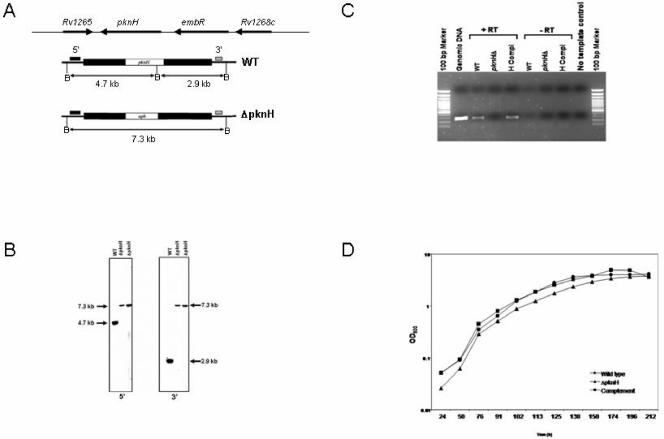
To assess the role of PknH in M. tuberculosis physiology and virulence, we created an M. tuberculosis mutant with a deletion of the gene encoding the PknH kinase by allelic exchange. The isolation of the mutant involved an approach based on electroporation with a nonreplicating targeting vector and a two-step selection procedure consisting of a positive selection step for the isolation of a single crossover strain which was then subjected to counterselection on sucrose medium. Genomic DNAs isolated from the parental wild-type strain and two isolates of the pknH mutant strain were subjected to Southern hybridization analysis to confirm that the ΔpknH mutant arose following double crossover homologous recombination as outlined in Fig. 1A. The genotype of the ΔpknH strain was confirmed by Southern hybridization (Fig. 1B). To produce a complemented strain, the ΔpknH mutant was transformed with an integrating vector carrying the wild-type pknH gene expressed from its native promoter. RT-PCR analysis confirmed that the pknH RNA was produced in the parental and complemented strains but not in the ΔpknH mutant (Fig. 1C).
FIG. 1.
Genotype and in vitro phenotype of the M. tuberculosis ΔpknH mutant strain. (A) Structure of the M. tuberculosis pknH locus. Black bars correspond to the pknH-flanking regions that were cloned into a nonreplicating vector to make the knockout construct. Allelic exchange resulted in the replacement of pknH with a kanamycin resistance gene in the ΔpknH mutant strain. Locations of the hybridizing probes (small solid bars) and the expected sizes of the hybridizing fragments of BamHI (B) digests for each of the 5′ and 3′ Southern blots are indicated. (B) Southern blot analysis. Blots of BamHI-digested genomic DNAs from the wild type and two ΔpknH mutant isolates were hybridized to 5′ and 3′ DNA probes. The sizes of the hybridizing fragments determined from the migration distances of the molecular size markers confirmed the genomic context expected for the ΔpknH strain. (C) RT-PCR analysis. A 278-bp PCR product corresponding to an internal fragment of the pknH gene was amplified by PCR from the cDNAs of the parental wild-type strain (WT) and the complemented strain (H comp) but not from the ΔpknH mutant and the negative control reactions. Expression of the housekeeping gene sigA was found in all three strains, indicating that the lack of pknH expression in the ΔpknH strain was not due to degradation of the isolated RNA sample (data not shown). (D) pknH deletion did not affect in vitro growth of M. tuberculosis. Growth of the wild type (•), the ΔpknH mutant (▴), and the complemented strain (▪) was monitored in Dubos broth cultures. Similar growth patterns were observed during incubation in 7H9 with OADC and Proskauer and Beck medium supplemented with 0.05% Tween 80 (not shown).
In order to find out whether the deletion of the pknH gene affected growth of the bacterium in vitro, we compared the growth of the mutant with its parent and the pknH-complemented strain. No differences in in vitro growth rates were observed between the ΔpknH mutant and its parental strain in axenic cultures (Fig. 1D), whether grown in Dubos broth, 7H9 broth with OADC, or Proskauer and Beck medium supplemented with 0.05% Tween 80 and whether grown in rolling or static cultures (data not shown). Thus, the deletion of the pknH gene does not affect the ability of the bacterium to take up and metabolize nutrients required for in vitro growth.
Role of PknH kinase in regulating the expression of the ini and emb operons.
The M. tuberculosis PknH kinase was previously shown to phosphorylate EmbR (33), which in turn was suggested to regulate the expression of the embAB operon encoding the mycobacterial arabinosyl transferases which are involved in the biosynthesis of arabinogalactan and lipoarabinomannans (6, 58). Genes of the emb operon as well as the ini operon which encode proteins associated with the mycobacterial cell wall were previously shown to be induced by treatment with the antituberculosis drugs ethambutol and isoniazid (1, 2). The availability of the M. tuberculosis ΔpknH mutant enabled us to assess the role of PknH in the regulation of specific gene transcriptions related to synthesis of these cell wall-associated components. Therefore, we carried out real-time PCR analysis to investigate whether pknH deletion affected the expression of emb and ini genes. Expressions of these genes were not significantly affected in uninduced bacteria. However, treatment with a sublethal concentration of ethambutol (0.2 μg/ml) for 24 h induced the expression of genes belonging to both the emb and ini operons in the parental strain but downregulated expression of these genes in the ΔpknH mutant strain (Table 2), indicating that the emb and ini operons belong to the PknH signaling cascade. This indicates that PknH kinase mediates a regulated expression of these genes which encode cell wall-associated products. Since the deletion of pknH affected ethambutol-induced expressions of embCAB genes, we wanted to determine whether the ΔpknH mutant displayed altered susceptibility to the antituberculosis drug ethambutol. Survival assays based on growth in liquid medium containing serial dilutions of the antibiotic as well as on 7H10 plates containing various concentrations of the drug did not reveal any differences in susceptibility between the strains. The ethambutol sensitivity of the ΔpknH mutant was found to be similar to that of its parental strain (MIC, 0.4 μg/ml) as determined by the 1% proportion survival method (6).
TABLE 2.
Real-time PCR analysis of gene expression following ethambutol treatmenta
| Gene | Ratio of gene/sigA
|
||
|---|---|---|---|
| Wild type | ΔpknH mutant | Complement | |
| embC | 1.96 ± 0.21 | 0.60 ± 0.15 | 1.91 ± 0.37 |
| embB | 1.33 ± 0.30 | 0.56 ± 0.02 | 2.03 ± 0.09 |
| iniB | 1.91 ± 0.60 | 0.16 ± 0.05 | 2.40 ± 0.24 |
| iniA | 1.40 ± 0.42 | 0.52 ± 0.16 | 2.09 ± 0.41 |
Real-time PCR analysis was used to quantify the amounts of gene-specific RNAs obtained from untreated and ethambutol-treated cultures (0.2 μg/ml ethambutol for 24 h), and the values obtained were normalized to the levels of sigA RNA. Data presented are mean ± standard deviation (n = 3) induction (n-fold) of gene expression following ethambutol treatment. The values obtained for the ΔpknH mutant strain are significantly different (P < 0.01) from those of the wild type and the complement.
Increased tolerance of the ΔpknH mutant to acidified nitrite stress.
Even though the alterations of the levels of emb gene expression were detected following exposure in vitro to sublethal concentrations of ethambutol, the in vivo signals that trigger phosphorylation by the PknH kinase are likely dependent on the host's intracellular environment encountered by the pathogen. Upon infection of macrophages, intracellular bacteria encounter many signals, including reactive oxygen and nitrogen intermediates. Nitric oxide, produced in response to activation of the host cells, plays a crucial role in controlling M. tuberculosis intracellular growth and infection (35, 46). Therefore, we wanted to investigate whether any alterations mediated by the deletion of the pknH gene conferred altered sensitivity to RNI- and reactive oxygen intermediate-producing treatments.
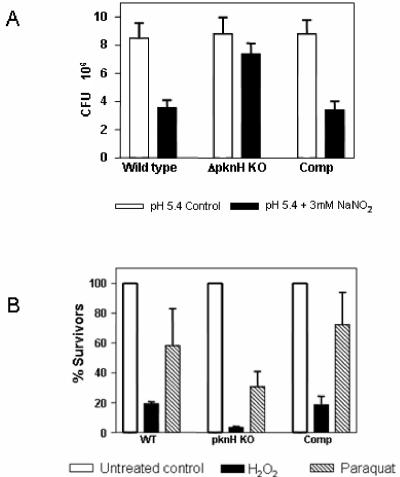
The survival of the parental strain, the ΔpknH mutant, and the complement was assayed following in vitro exposure of bacteria to acidified sodium nitrite (3 mM). Under these conditions, the ΔpknH strain survived significantly better than its parental wild-type strain (Fig. 2A). CFU determinations following exposures for 48 h revealed that approximately 84% of mutant bacteria survived the treatment compared to 42% of the parental strain. Though it seems relatively modest, the twofold-increased resistance of the ΔpknH mutant strain to acidified nitrite observed in this study was statistically significant and consistently obtained over independent experiments. The survival of the complement was comparable (39%) to that of the parental strain (Fig. 2A). In contrast to the in vitro resistance observed for nitrite, the ΔpknH mutant strain was more sensitive than its parental strain to in vitro peroxide- and superoxide-producing treatments included in this study (Fig. 2B). The survival of the complemented strain was similar to that of the parental strain, confirming that it was the deletion of the pknH gene which led to the consequent alterations in the sensitivity phenotypes reported in this work. These assays indicate that M. tuberculosis probably uses PknH kinase to sense free radicals as environmental cues and trigger responses that contribute to its survival.
FIG. 2.
Deletion of M. tuberculosis pknH increases resistance to acidified nitrite stress. (A) Survival (CFU) of the pknH strains following exposure to 3 mM NaNO2 in acidified medium (pH 5.4) for 48 h was determined and compared with the CFU obtained from pH 5.4 medium lacking NaNO2. Error bars represent standard deviations obtained for triplicate cultures of each strain. The values obtained for the ΔpknH mutant strain are significantly different (P < 0.0001) from those for the wild type and the complement. (B) Effect of oxidizing stresses on viability of the pknH strains. Bacteria were grown in 7H9-Tween-ADS medium, exposed to different stressing reagents (10 mM H2O2 or 50 mM paraquat) for 48 h, and plated to determine viability. The bars represent mean percent survival (% CFU treated/untreated) for each treatment, and the error bars indicate standard deviations obtained from triplicate cultures.
Increased bacterial load of the ΔpknH mutant in mouse organs.
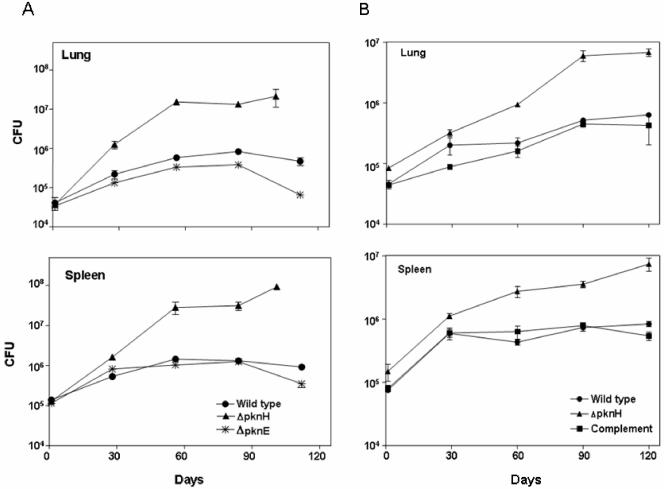
To investigate whether the deletion of the pknH gene affects M. tuberculosis virulence, the ΔpknH mutant strain was compared with its parental strain in BALB/c mice. As seen in Fig. 3A, even though initial depositions of the inocula in the organs were approximately equal, much higher numbers of bacteria were recovered from mice infected with the ΔpknH strain at all time points. By day 56, approximately 26-fold more bacilli were recovered from the lungs and 20-fold more bacilli from the spleens of mice infected with the ΔpknH mutant than from mice infected with the parental strain (P < 0.005). Since one of the ΔpknH mutant-infected mice had died before day 101 and the other three mice were considered too ill to prolong the experiment any further, the remaining three mice were sacrificed for CFU counts 11 days prior to those infected with the parental and ΔpknE mutant strains. By day 101, nearly 45-fold more bacteria were recovered from the lungs and 100-fold more bacteria were recovered from the spleens of mice infected with the ΔpknH mutant than were recovered on day 112 from mice infected with the parental strain (Fig. 3A). In contrast to the ΔpknH mutant, the survival and growth of another kinase mutant, the ΔpknE mutant, which was isolated from the same parental strain as the ΔpknH mutant, displayed an in vivo survival phenotype similar to that of the parental strain except at the last time point, when the recovered CFU of the ΔpknE mutant was less than that of the parental strain (Fig. 3A). The measurement of the organ weights at the last time point indicated that the ΔpknH mutant-infected mice had a 2-fold increase in the weight of the spleen and 1.8-fold increase in the weight of the lung compared to the mice infected with the parental strain (P < 0.05). Thus, infection with the ΔpknH mutant leads to an increased organ mass associated with severe pathogenic state.
FIG. 3.
Deletion of the pknH gene leads to increased bacillary load in BALB/c mice. Following intravenous route of infection, the bacillary load in the lungs and spleen was determined in two experiments (A and B), and the mean CFU counts for the wild-type (•), ΔpknH mutant (▴), ΔpknE mutant (✻), and pknH-complemented (▪) strains were plotted. The error bars indicate standard deviation. The CFU means obtained for the ΔpknH mutant strain after day 30 are significantly greater than those of the wild type as measured by the two-tailed Student t test analysis for groups of unequal variance (P < 0.01).
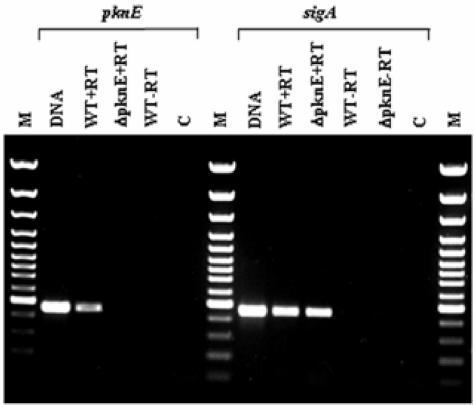
In order to confirm that the increased in vivo growth and survival of the ΔpknH mutant strain were indeed due to the targeted deletion of the pknH gene, the pknH-complemented strain was included in a second mouse infection experiment and the progress of the infection was determined. As illustrated in Fig. 3B, the survival and growth of the complemented strain in the mouse organs were similar to those of the parental strain at all time points, indicating that the increased bacterial load of the ΔpknH mutant strain was indeed due to the deletion of the pknH gene. After 4 weeks of infection in this experiment (Fig. 3B), the ΔpknH mutant continued to grow at a higher rate than the parental strain, and the differences in CFU were statistically significant (P < 0.005). At least 10-fold more mutant bacilli were recovered from the lungs on days 90 and 120, mirroring the increased bacillary load observed in the first experiment. In this experiment also, one of the ΔpknH mutant-infected mice had died earlier, indicating the pathogenic status of ΔpknH mutant-infected mice. Even though the parental strain CFU loads in the lungs did not increase by 100-fold within 28 days, as normally seen upon infection by aerosol route or following initial lower CFU deposition in lungs after intravenous injection, there was a statistically significant increase in bacterial CFU loads specifically in the organs of ΔpknH mutant-infected mice. The less-than-expected increase in the CFU between day 1 and day 28 in the organs of mice infected with the parental strain could possibly be due to the higher load of bacterial deposition in the organs upon injection or due to an altered virulence property of the parental strain. M. tuberculosis can display diminished virulence if subcultured continuously in medium. In this study, however, due to the two-step procedure used in the isolation of the mutants, the ΔpknH mutant strain had been streaked and grown for more generations on 7H11 medium than the parental strain, which was kept frozen. Nevertheless, the ΔpknH mutant strain displayed increased survival in mouse organs compared to the parental strain and the ΔpknE mutant isolated from the same parental strain. Reverse transcription analysis was used to confirm that the ΔpknE mutant strain used in the mouse infection experiment did not express pknE RNA (Fig. 4). Significantly, reintroduction of the functional pknH gene into the ΔpknH mutant strain restored the virulence phenotype to the parental levels, confirming that the increased bacterial load in ΔpknH mutant-infected mice was indeed caused by the absence of the pknH gene.
FIG. 4.
RT-PCR analysis for the expression of the M. tuberculosis pknE gene. A 447-bp PCR product corresponding to the internal DNA fragment located between positions 1188 and 1634 bp of the pknE coding sequence was amplified by PCR from the cDNAs of the wild type (WT) but not from the ΔpknE mutant and the negative control reactions. Expression of a 451-bp sigA-specific PCR product, corresponding to the 542- to 992-bp region of the sigA gene, was amplified from the RNA of both the ΔpknE mutant and its parental strain.
Increased growth of the ΔpknH mutant strain is not due to better infectivity.
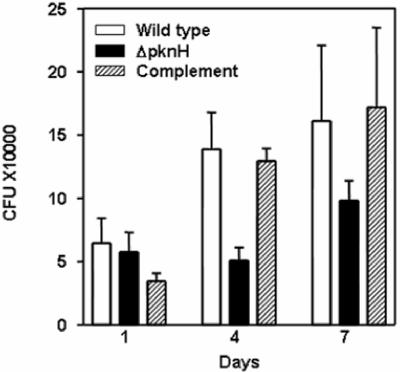
In order to assess whether the increased growth rate of the ΔpknH mutant strain is due to an enhanced phagocytosis by macrophages or a result of better intracellular survival and replication, the mutant was investigated in phorbol 12-myristate 13-acetate-differentiated THP-1 cells. Monolayers of THP-1 cells were infected with the parental strain, the ΔpknH mutant, and the pknH-complemented strain for 20 h, and the survival of the intracellular bacteria was determined at various time points. As seen in Fig. 5, the ΔpknH mutant was found to grow at a slower rate than its parental wild-type strain until day 7 (P < 0.05). However, it was obvious that the uptake and survival of the ΔpknH mutant was similar to that of the parent strain on day 1, and the mutant did not display a higher intracellular replication capacity soon after phagocytosis.
FIG. 5.
Intracellular growth of the pknH strains in THP-1 cells. Monolayers of differentiated THP-1 cells were infected with the pknH strains for 20 h, and CFU of intracellular bacteria were determined on the indicated days postinfection. The bars represent CFU means and standard deviations obtained from triplicate infections. The mean CFU differences between the ΔpknH mutant and its parental wild-type strain for days 4 and 7 are statistically significant (P < 0.05).
Since the ΔpknH mutant strain displayed in vitro growth rates in 7H9 growth medium similar to those of its parental strain, the higher bacillary load of the mutant bacteria in the mouse organs reflects the increased ability of the ΔpknH mutant bacteria to survive the host response. Alternatively, in the wild type, PknH kinase-mediated alterations contribute, by as-yet-unidentified mechanisms, to a steady-state chronic infection leading to persistence of the pathogen in its host.
DISCUSSION
The intracellular pathogen M. tuberculosis is able to establish long-term persistent infection in its host despite the induction of host-activated inflammatory and antimicrobial responses. This demands that M. tuberculosis cells sense and respond to various host-induced stress signals. In this study, we examined the role of the PknH serine/threonine kinase in mycobacterial virulence, since the kinase was predicted to be involved in the regulation of genes that contribute to the synthesis of cell wall-associated components, such as arabinogalactan and lipoarabinomannan (3, 7, 58). An M. tuberculosis mutant with a deletion of the pknH gene was isolated by allelic exchange, which is consistent with the finding that it is not an essential gene for in vitro growth (45). The pknH gene is the second serine/threonine kinase gene to be knocked out in M. tuberculosis. We reported the first targeted deletion of pknG last year (13). In contrast to the ΔpknG mutant, which displayed altered in vitro growth phenotypes during the stationary phase of growth and in a nutrient minimal medium, deletion of the pknH gene did not affect the in vitro growth properties of the bacterium. Mouse infection studies revealed that, compared to its parental strain, the ΔpknH mutant survived to a higher load in the organs of BALB/c mice, especially during the late stages of infection. In contrast, the virulence phenotype of the ΔpknE mutant, which was isolated from the same parental strain, was similar to that of the parental strain, indicating that the observed increased bacillary load in the ΔpknH mutant-infected mouse organs was a gene-specific phenotype resulting from the deletion of the pknH gene. This was further confirmed by complementing the mutant phenotype by introducing a functional wild-type pknH gene expressed from an integrated vector into the mutant and assaying its virulence property. The ΔpknH mutant did not display increased survival upon infection of differentiated THP-1 cells, indicating that the deletion of the pknH gene does not lead to higher infectivity of the macrophages.
Based on these results, we suggest that M. tuberculosis uses, either directly or indirectly, a PknH kinase-mediated mechanism to confer a regulated growing profile as a survival strategy within its host. It is likely that slow-growing pathogens have evolved diverse mechanisms to regulate their growth inside hosts. Uncontrolled replication inside the host may not be ideal if it contributes to rapid death of the host. Alternatively, formation of granulomas and containment of bacterial replication lead to persistence of the pathogen and survival of the host. Targeted gene knockout studies carried out by other groups also have revealed that diverse gene products of M. tuberculosis can contribute to the control of bacterial growth and virulence in mice. For example, gene disruptions in four specific two-component regulators (37) or in the mce1 operon (49) lead to hypervirulence. The M. tuberculosis ΔdevR mutant, with a deletion of the two-component system DevR, displayed increased bacterial load in the organs of DBA mice, with levels similar to those obtained for the ΔpknH mutant strain in this study. It was reported that the production of nitric oxide and tumor necrosis factor alpha was induced in ΔdevR mutant-infected murine macrophages and that the ΔdevR mutant displayed increased growth in gamma interferon-activated macrophages 24 h after induction, but this growth advantage was lost by 72 h. However, the mutant did not display increased resistance to oxidative and acidic stress treatments in vitro (37). In the case of mce1, virulence studies indicated that the mce1 operon deletion mutant of the M. tuberculosis Erdman strain was unable to enter into the stable persistent infection stage in mouse lungs due to the increased bacterial replication leading to the premature death of the mice infected with the mutant. The authors correlated the increased bacterial load of the Δmce1 mutant with the absence of organized granuloma formation seen in histological examination of the infected lung tissues. Another strain of M. tuberculosis, the clinical variant HN878, displays a “hyperlethal” phenotype in mice due to the production of a specific phenolic glycolipid encoded by the pks1-15 gene cluster which is inactivated in the wild-type H37Rv strain (40). The hypervirulent phenotype of the HN878 strain was attributed to the suppression of specific host immune response mechanisms. In the case of the mce1 operon mutant, the hypervirulence was attributed to the decreased production of tumor necrosis factor alpha, interleukin-6, monocyte chemoattractant protein-1, and nitric oxide in murine macrophages infected with the Δmce1 mutant (49).
In the case of the ΔpknH mutant reported in this work, even though histopathology examinations could not be performed, the mice infected with the ΔpknH mutant had increased lung weight compared to the mice infected with the parental strain, indicating an advanced disease state. Moreover, the proliferation of the ΔpknH mutant bacteria had resulted in the death of one of the four mice prior to the last scheduled time point in both experiments. These observations lead us to suggest that the deletion of the pknH gene leads to hypervirulence in BALB/c mice; however, we could not perform time-to-death mouse infection experiments and histological examination of infected tissues to confirm the pathogenesis phenotype.
Nevertheless, the increased bacterial load of the ΔpknH strain recovered from the mouse organs suggests an enhanced in vivo survival and/or replication of the mutant. This was particularly observed during the stage at which replication of the wild-type bacteria is maintained at a steady state which leads to chronic infection. It is known that the control of M. tuberculosis infections is mediated by M. tuberculosis-specific CD4+ and CD8+ T cells via secretion of gamma interferon and other Th1 cytokines that activate antimycobacterial mechanisms of infected macrophages (18). The production of nitric oxide in response to cytokines or pathogen-derived molecules is an important host defense mechanism that controls intracellular infections (10, 36). Upon the onset of acquired immune response, activated macrophages induce nitric oxide synthase, which catalyzes generation of nitric oxide and RNI from l-arginine. Evidence for the significance of nitric oxide in controlling mycobacterial infection comes from a murine model of tuberculosis showing progressive infection in animals unable to produce the inducible isoform of nitric oxide synthase and in animals treated with a nitric oxide synthase inhibitor (9, 29). However, mycobacteria have evolved effective mechanisms to counteract the host response. Recently, it was shown that Mycobacterium bovis BCG prevents colocalization of inducible nitric oxide synthase with mycobacterial phagosomes and thereby ensures the survival of the bacteria inside macrophages (30). However, whether this inhibition of colocalization can persist during the entire course of infection is not known. Therefore, it is advantageous for the pathogen to possess additional protective mechanisms. It is known that nitric oxide treatment can induce specific genes of M. tuberculosis that play a role in enabling adaptive response. A low concentration of nitric oxide signal elicited by treatment with diethylenetriamine/nitric oxide adduct was shown to modulate expression of a set of 48 genes, many of which were implicated in the bacterial adaptive response leading to dormancy (55). Furthermore, 29 M. tuberculosis proteins comprising enzymes involved in intermediary metabolism, lipid metabolism, and antioxidant defense were identified as the targets of S nitrosylation reaction initiated by treating bacteria with an inhibitory concentration of RNI (41). These reports imply that responding to the nitric oxide stress could be critical to the progress and outcome of the infection. Some clinical isolates of M. tuberculosis display altered sensitivity to RNI-mediated cytotoxicity (20, 42), and it was thought that this might affect the virulence of these strains. We investigated whether the deletion of the pknH gene resulted in altered sensitivity to acidified nitrite and found that the ΔpknH mutant displayed increased survival compared to the parental and complemented strains. The observed increase in the tolerance of the ΔpknH mutant, as reflected by a twofold-increased survival of mutant bacteria, although seeming relatively modest, was consistently observed in independent experiments and was similar to that reported for the New York outbreak clinical strain CB3.3 (20). While the ΔpknH mutant strain is more tolerant to acidified nitrite stress in vitro, whether the deletion of the pknH gene causes increased tolerance to RNI in vivo remains to be confirmed, although it has been reported that at pH 5.5, even 0.5 mM nitrite in 0.5 ml generates as much nitric oxide as 3 × 105 activated macrophages over 24 h (16). In contrast to the resistance to acidified nitrite, the ΔpknH mutant displayed increased sensitivity to peroxide- and superoxide-producing treatments. These results suggest that nitric oxide and peroxide radicals are potential triggers of opposing effects on PknH kinase. Alternatively, modulations of gene expressions resulting from the absence of the PknH kinase activity contribute to the lower sensitivity of the ΔpknH mutant to reactive nitrogen intermediates and higher susceptibility to superoxide stresses investigated in this study.
In order to identify the downstream components of the PknH signaling cascade, we carried out transcriptional analysis of genes in the embCAB operon that were predicted to be regulated by the PknH substrate EmbR, since it was suggested that the phosphorylation of EmbR is likely to affect the expressions of the embCAB genes encoding the mycobacterial arabinosyl transferases (3, 6, 7, 33). We also investigated the expression of iniBAC genes, which encode proteins associated with the cell wall and were shown to be induced by lethal concentrations of the antituberculosis drugs ethambutol and isoniazid (2). Real-time PCR analyses indicated that in vitro treatment with a sublethal concentration of ethambutol induced the expression of the genes belonging to both the embCAB and iniBAC operons in the parental strain. However, the antibiotic treatment downregulated expressions of these genes in the ΔpknH mutant, indicating that the regulation of these genes was dependent on a signaling cascade mediated by the PknH kinase. The emb genes encode arabinosyl transferases which are involved in the synthesis of AG and LAM, whereas the iniA gene is thought to be associated with a multidrug resistance-like efflux pump activity (11). In spite of the downregulation of the emb operon in the ΔpknH mutant, both the mutant and the parental strain displayed similar MICs for ethambutol. Recently, it has been shown that alterations in the expression levels of iniA either by overexpression or by gene deletion did not alter the MIC for isoniazid (MIC, 0.2 μg/ml) (11). However, when exposed to a specific sublethal concentration of isoniazid (0.05 μg/ml), the iniA deletion mutant displays a defect in tolerance phenotype as assessed by a daily growth index analysis. The authors suggested that even though IniA is associated with an efflux pump activity, the sublethal concentration of isoniazid induces a tolerance mechanism unrelated to antibiotic transport in M. tuberculosis (11). In the present study, the deletion of the pknH gene resulted in the downregulation of ini operon expression following treatment with a sublethal concentration of ethambutol. However, the signals mediating similar responses during in vivo growth are yet to be identified. It is possible that in the wild-type bacteria, PknH kinase senses a host signal and induces the expression of iniA and other genes whose gene products facilitate the transport of molecules that play a role in regulating intracellular growth. Therefore, in the absence of the PknH kinase activation, this control cannot be enabled, resulting in the increased replication of the ΔpknH mutant bacteria.
In contrast to the phenotype of the ΔpknH mutant, deletion of the pknG gene in M. tuberculosis results in attenuation of virulence in mice (13). PknG kinase is required for growth under stationary conditions, is a sensor of nutritional stress, and plays a crucial role in regulating cellular glutamine/glutamate levels. PknG also contributes to the survival of mycobacteria within macrophages by preventing fusion of phagosomes with lysosomes (56). Thus, the two serine/threonine kinases of M. tuberculosis, PknG and PknH, play distinct roles to facilitate mycobacterial virulence: PknG plays a crucial role in the establishment of infection, whereas the PknH kinase regulates intracellular bacterial growth at later stages of the infection process, particularly during the chronic phase. Phenotypes of the ΔpknH, ΔdevR, and Δmce1 mutants suggest that to help regulate its in vivo growth, M. tuberculosis uses diverse signaling pathways to induce appropriate responses, resulting in a “balanced” phase of chronic infection. Modulation of growth requiring activities of a signaling kinase has been proposed recently to be one of the strategies employed by bacteria to ensure survival. For example, upon exposure to β-lactam antibiotics, Escherichia coli uses the dpiBA-mediated two-component signal transduction system to temporarily halt cell division, thereby limiting the bactericidal effect of the antibiotics (31).
In conclusion, we propose that the PknH kinase of M. tuberculosis mediates a host signal and triggers a response that contributes to the in vivo survival and/or growth of the bacterium, perhaps to facilitate a “balanced growth” which leads to chronic infection. The nature of the in vivo inducing signal and the identity of the various components of the PknH-mediated signaling pathway involved in this adaptive gene response still need to be resolved.
Acknowledgments
We thank BCCDC for providing access to a Containment Level 3 facility, Evangelos Stavropoulos for murine infection experiments at NIMR, and Rick Stokes for valuable advice.
Funding for this research was provided by the National Centres of Excellence, the Canadian Bacterial Disease Network (Y.A.-G.), the Canadian Institute of Health Research (CIHR) grant MOP-68857 (Y.A.-G.), and the Medical Research Council UK (M.J.C. and E.O.D.). Y.A.-G. is a Canadian Institute of Health Research and British Columbia Lung Association Scholar.
Footnotes
This paper is dedicated to the memory of Jo Colston, a friend, colleague, and mentor we miss greatly.
REFERENCES
- 1.Alland, D., I. Kramnik, T. R. Weisbrod, L. Otsubo, R. Cerny, L. P. Miller, W. R. Jacobs, Jr., and B. R. Bloom. 1998. Identification of differentially expressed mRNA in prokaryotic organisms by customized amplification libraries (DECAL): the effect of isoniazid on gene expression in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 95:13227-13232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alland, D., A. J. Steyn, T. Weisbrod, K. Aldrich, and W. R. Jacobs, Jr. 2000. Characterization of the Mycobacterium tuberculosis iniBAC promoter, a promoter that responds to cell wall biosynthesis inhibition. J. Bacteriol. 182:1802-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Av-Gay, Y., and M. Everett. 2000. The eukaryotic-like Ser/Thr protein kinases of Mycobacterium tuberculosis. Trends Microbiol. 8:238-244. [DOI] [PubMed] [Google Scholar]
- 4.Av-Gay, Y., S. Jamil, and S. J. Drews. 1999. Expression and characterization of the Mycobacterium tuberculosis serine/threonine protein kinase PknB. Infect. Immun. 67:5676-5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bakal, C. J., and J. E. Davies. 2000. No longer an exclusive club: eukaryotic signalling domains in bacteria. Trends Cell Biol. 10:32-38. [DOI] [PubMed] [Google Scholar]
- 6.Belanger, A. E., G. S. Besra, M. E. Ford, K. Mikusova, J. T. Belisle, P. J. Brennan, and J. M. Inamine. 1996. The embAB genes of Mycobacterium avium encode an arabinosyl transferase involved in cell wall arabinan biosynthesis that is the target for the antimycobacterial drug ethambutol. Proc. Natl. Acad. Sci. USA 93:11919-11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Briken, V., S. A. Porcelli, G. S. Besra, and L. Kremer. 2004. Mycobacterial lipoarabinomannan and related lipoglycans: from biogenesis to modulation of the immune response. Mol. Microbiol. 53:391-403. [DOI] [PubMed] [Google Scholar]
- 8.Chaba, R., M. Raje, and P. K. Chakraborti. 2002. Evidence that a eukaryotic-type serine/threonine protein kinase from Mycobacterium tuberculosis regulates morphological changes associated with cell division. Eur. J. Biochem. 269:1078-1085. [DOI] [PubMed] [Google Scholar]
- 9.Chan, J., K. Tanaka, D. Carroll, J. Flynn, and B. R. Bloom. 1995. Effects of nitric oxide synthase inhibitors on murine infection with Mycobacterium tuberculosis. Infect. Immun. 63:736-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan, J., Y. Xing, R. S. Magliozzo, and B. R. Bloom. 1992. Killing of virulent Mycobacterium tuberculosis by reactive nitrogen intermediates produced by activated murine macrophages. J. Exp. Med. 175:1111-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colangeli, R., D. Helb, S. Sridharan, J. Sun, M. Varma-Basil, M. H. Hazbón, R. Harbacheuski, N. J. Megjugorac, W. R. Jacobs, Jr., A. Holzenburg, J. C. Sacchettini, and D. Alland. 2005. The Mycobacterium tuberculosis iniA gene is essential for activity of an efflux pump that confers drug tolerance to both isoniazid and ethambutol. Mol. Microbiol. 55:1829-1840. [DOI] [PubMed] [Google Scholar]
- 12.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 13.Cowley, S., M. Ko, N. Pick, R. Chow, K. J. Downing, B. G. Gordhan, J. C. Betts, V. Mizrahi, D. A. Smith, R. W. Stokes, and Y. Av-Gay. 2004. The Mycobacterium tuberculosis protein serine/threonine kinase PknG is linked to cellular glutamate/glutamine levels and is important for growth in vivo. Mol. Microbiol. 52:1691-1702. [DOI] [PubMed] [Google Scholar]
- 14.Cozzone, A. J. 1998. Post-translational modification of proteins by reversible phosphorylation in prokaryotes. Biochimie 80:43-48. [DOI] [PubMed] [Google Scholar]
- 15.Dannenberg, A. M., Jr., and G. A. W. Rook. 1994. Pathogenesis of pulmonary tuberculosis: an interplay of tissue damaging and macrophage-activating immune responses—dual mechanisms that control bacillary multiplication, p. 459-483. In B. R. Bloom (ed.), Tuberculosis, pathogenesis, protection and control. American Society for Microbiology, Washington, D.C.
- 16.Ehrt, S., D. Schnappinger, S. Bekiranov, J. Drenkow, S. Shi, T. R. Gingeras, T. Gaasterland, G. Schoolnik, and C. Nathan. 2001. Reprogramming of the macrophage transcriptome in response to interferon-gamma and Mycobacterium tuberculosis: signaling roles of nitric oxide synthase-2 and phagocyte oxidase. J. Exp. Med. 194:1123-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Escuyer, V. E., M. A. Lety, J. B. Torrelles, K. H. Khoo, J. B. Tang, C. D. Rithner, C. Frehel, M. R. McNeil, P. J. Brennan, and D. Chatterjee. 2001. The role of the embA and embB gene products in the biosynthesis of the terminal hexaarabinofuranosyl motif of Mycobacterium smegmatis arabinogalactan. J. Biol. Chem. 276:48854-48862. [DOI] [PubMed] [Google Scholar]
- 18.Flynn, J. L., and J. Chan. 2001. Immunology of tuberculosis. Annu. Rev. Immunol. 19:93-129. [DOI] [PubMed] [Google Scholar]
- 19.Flynn, J. L., and J. Chan. 2001. Tuberculosis: latency and reactivation. Infect. Immun. 69:4195-4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedman, C. R., G. C. Quinn, B. N. Kreiswirth, D. C. Perlman, N. Salomon, N. Schluger, M. Lutfey, J. Berger, N. Poltoratskaia, and L. W. Riley. 1997. Widespread dissemination of a drug-susceptible strain of Mycobacterium tuberculosis. J. Infect. Dis. 176:478-484. [DOI] [PubMed] [Google Scholar]
- 21.Frota, C. C., K. G. Papavinasasundaram, E. O. Davis, and M. J. Colston. 2004. The AraC family transcriptional regulator Rv1931c plays a role in the virulence of Mycobacterium tuberculosis. Infect. Immun. 72:5483-5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gopalaswamy, R., P. R. Narayanan, and S. Narayanan. 2004. Cloning, overexpression, and characterization of a serine/threonine protein kinase pknI from Mycobacterium tuberculosis H37Rv. Protein Expr. Purif. 36:82-89. [DOI] [PubMed] [Google Scholar]
- 23.Gross, R., B. Arico, and R. Rappuoli. 1989. Families of bacterial signal-transducing proteins. Mol. Microbiol. 3:1661-1667. [DOI] [PubMed] [Google Scholar]
- 24.Honer zu Bentrup, K., and D. G. Russell. 2001. Mycobacterial persistence: adaptation to a changing environment. Trends Microbiol. 9:597-605. [DOI] [PubMed] [Google Scholar]
- 25.Jacobs, W. R., Jr., G. V. Kalpana, J. D. Cirillo, L. Pascopella, S. B. Snapper, R. A. Udani, W. Jones, R. G. Barletta, and B. R. Bloom. 1991. Genetic systems for mycobacteria. Methods Enzymol. 204:537-555. [DOI] [PubMed] [Google Scholar]
- 26.Koul, A., A. Choidas, A. K. Tyagi, K. Drlica, Y. Singh, and A. Ullrich. 2001. Serine/threonine protein kinases PknF and PknG of Mycobacterium tuberculosis: characterization and localization. Microbiology 147:2307-2314. [DOI] [PubMed] [Google Scholar]
- 27.Koul, A., T. Herget, B. Klebl, and A. Ullrich. 2004. Interplay between mycobacteria and host signalling pathways. Nat. Rev. Microbiol. 2:189-202. [DOI] [PubMed] [Google Scholar]
- 28.Lukey, P., and E. Hooker. 2001. Macrophage virulence assays, p. 271-280. In T. Parish and N. G. Stoker (ed.), Methods in molecular medicine: Mycobacterium tuberculosis protocols, vol. 54. Humana Press Inc., Totowa, N.J. [DOI] [PubMed] [Google Scholar]
- 29.MacMicking, J. D., R. J. North, R. LaCourse, J. S. Mudgett, S. K. Shah, and C. F. Nathan. 1997. Identification of nitric oxide synthase as a protective locus against tuberculosis. Proc. Natl. Acad. Sci. USA 94:5243-5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller, B. H., R. A. Fratti, J. F. Poschet, G. S. Timmins, S. S. Master, M. Burgos, M. A. Marletta, and V. Deretic. 2004. Mycobacteria inhibit nitric oxide synthase recruitment to phagosomes during macrophage infection. Infect. Immun. 72:2872-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller, C., L. E. Thomsen, C. Gaggero, R. Mosseri, H. Ingmer, and S. N. Cohen. 2004. SOS response induction by beta-lactams and bacterial defense against antibiotic lethality. Science 305:1629-1631. [DOI] [PubMed] [Google Scholar]
- 32.Molle, V., C. Girard-Blanc, L. Kremer, P. Doublet, A. J. Cozzone, and J. F. Prost. 2003. Protein PknE, a novel transmembrane eukaryotic-like serine/threonine kinase from Mycobacterium tuberculosis. Biochem. Biophys. Res. Commun. 308:820-825. [DOI] [PubMed] [Google Scholar]
- 33.Molle, V., L. Kremer, C. Girard-Blanc, G. S. Besra, A. J. Cozzone, and J. F. Prost. 2003. An FHA phosphoprotein recognition domain mediates protein EmbR phosphorylation by PknH, a Ser/Thr protein kinase from Mycobacterium tuberculosis. Biochemistry 42:15300-15309. [DOI] [PubMed] [Google Scholar]
- 34.Molle, V., D. Soulat, J. M. Jault, C. Grangeasse, A. J. Cozzone, and J. F. Prost. 2004. Two FHA domains on an ABC transporter, Rv1747, mediate its phosphorylation by PknF, a Ser/Thr protein kinase from Mycobacterium tuberculosis. FEMS Microbiol. Lett. 234:215-223. [DOI] [PubMed] [Google Scholar]
- 35.Nathan, C., and M. U. Shiloh. 2000. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc. Natl. Acad. Sci. USA 97:8841-8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nathan, C., and Q. W. Xie. 1994. Nitric oxide synthases: roles, tolls, and controls. Cell 78:915-918. [DOI] [PubMed] [Google Scholar]
- 37.Parish, T., D. A. Smith, S. Kendall, N. Casali, G. J. Bancroft, and N. G. Stoker. 2003. Deletion of two-component regulatory systems increases the virulence of Mycobacterium tuberculosis. Infect. Immun. 71:1134-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parish, T., and N. G. Stoker. 2000. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146:1969-1975. [DOI] [PubMed] [Google Scholar]
- 39.Pelicic, V., J. M. Reyrat, and B. Gicquel. 1996. Positive selection of allelic exchange mutants in Mycobacterium bovis BCG. FEMS Microbiol. Lett. 144:161-166. [DOI] [PubMed] [Google Scholar]
- 40.Reed, M. B., P. Domenech, C. Manca, H. Su, A. K. Barczak, B. N. Kreiswirth, G. Kaplan, and C. E. Barry III. 2004. A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature 431:84-87. [DOI] [PubMed] [Google Scholar]
- 41.Rhee, K. Y., H. Erdjument-Bromage, P. Tempst, and C. F. Nathan. 2005. S-nitroso proteome of Mycobacterium tuberculosis: enzymes of intermediary metabolism and antioxidant defense. Proc. Natl. Acad. Sci. USA 102:467-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rhoades, E. R., and I. M. Orme. 1997. Susceptibility of a panel of virulent strains of Mycobacterium tuberculosis to reactive nitrogen intermediates. Infect. Immun. 65:1189-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rickman, L., J. W. Saldanha, D. M. Hunt, D. N. Hoar, M. J. Colston, J. B. Millar, and R. S. Buxton. 2004. A two-component signal transduction system with a PAS domain-containing sensor is required for virulence of Mycobacterium tuberculosis in mice. Biochem. Biophys. Res. Commun. 314:259-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Russell, D. G. 2001. Mycobacterium tuberculosis: here today, and here tomorrow. Nat. Rev. Mol. Cell Biol. 2:569-577. [DOI] [PubMed] [Google Scholar]
- 45.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48:77-84. [DOI] [PubMed] [Google Scholar]
- 46.Scanga, C. A., V. P. Mohan, K. Tanaka, D. Alland, J. L. Flynn, and J. Chan. 2001. The inducible nitric oxide synthase locus confers protection against aerogenic challenge of both clinical and laboratory strains of Mycobacterium tuberculosis in mice. Infect. Immun. 69:7711-7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharma, K., H. Chandra, P. K. Gupta, M. Pathak, A. Narayan, L. S. Meena, R. C. D'Souza, P. Chopra, S. Ramachandran, and Y. Singh. 2004. PknH, a transmembrane Hank's type serine/threonine kinase from Mycobacterium tuberculosis is differentially expressed under stress conditions. FEMS Microbiol. Lett. 233:107-113. [DOI] [PubMed] [Google Scholar]
- 48.Shi, L., M. Potts, and P. J. Kennelly. 1998. The serine, threonine, and/or tyrosine-specific protein kinases and protein phosphatases of prokaryotic organisms: a family portrait. FEMS Microbiol. Rev. 22:229-253. [DOI] [PubMed] [Google Scholar]
- 49.Shimono, N., L. Morici, N. Casali, S. Cantrell, B. Sidders, S. Ehrt, and L. W. Riley. 2003. Hypervirulent mutant of Mycobacterium tuberculosis resulting from disruption of the mce1 operon. Proc. Natl. Acad. Sci. USA 100:15918-15923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Springer, B., P. Sander, L. Sedlacek, K. Ellrott, and E. C. Bottger. 2001. Instability and site-specific excision of integration-proficient mycobacteriophage L5 plasmids: development of stably maintained integrative vectors. Int. J. Med. Microbiol. 290:669-675. [DOI] [PubMed] [Google Scholar]
- 51.Sreevatsan, S., K. E. Stockbauer, X. Pan, B. N. Kreiswirth, S. L. Moghazeh, W. R. Jacobs, Jr., A. Telenti, and J. M. Musser. 1997. Ethambutol resistance in Mycobacterium tuberculosis: critical role of embB mutations. Antimicrob. Agents Chemother. 41:1677-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stewart, G. R., B. D. Robertson, and D. B. Young. 2003. Tuberculosis: a problem with persistence. Nat. Rev. Microbiol. 1:97-105. [DOI] [PubMed] [Google Scholar]
- 53.Stock, J. B., A. J. Ninfa, and A. M. Stock. 1989. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol. Rev. 53:450-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Telenti, A., W. J. Philipp, S. Sreevatsan, C. Bernasconi, K. E. Stockbauer, B. Wieles, J. M. Musser, and W. R. Jacobs, Jr. 1997. The emb operon, a gene cluster of Mycobacterium tuberculosis involved in resistance to ethambutol. Nat. Med. 3:567-570. [DOI] [PubMed] [Google Scholar]
- 55.Voskuil, M. I., D. Schnappinger, K. C. Visconti, M. I. Harrell, G. M. Dolganov, D. R. Sherman, and G. K. Schoolnik. 2003. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198:705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walburger, A., A. Koul, G. Ferrari, L. Nguyen, C. Prescianotto-Baschong, K. Huygen, B. Klebl, C. Thompson, G. Bacher, and J. Pieters. 2004. Protein kinase G from pathogenic mycobacteria promotes survival within macrophages. Science 304:1800-1804. [DOI] [PubMed] [Google Scholar]
- 57.World Health Organization. March. 2004, posting date. Tuberculosis: infection and transmission in 2002. Fact sheet no. 104. World Health Organization, Geneva, Switzerland. [Online.] http://www.who.int/mediacentre/factsheets/fs104/en/.
- 58.Zhang, N., J. B. Torrelles, M. R. McNeil, V. E. Escuyer, K. H. Khoo, P. J. Brennan, and D. Chatterjee. 2003. The Emb proteins of mycobacteria direct arabinosylation of lipoarabinomannan and arabinogalactan via an N-terminal recognition region and a C-terminal synthetic region. Mol. Microbiol. 50:69-76. [DOI] [PubMed] [Google Scholar]