Abstract
Escherichia coli cells were constructed in which the dnaA gene was moved to a location opposite oriC on the circular chromosome. In these cells the dnaA gene was replicated with significant delay relative to the origin. Consequently, the period where the newly replicated and hemimethylated oriC was sequestered no longer coincided with the period where the dnaA gene promoter was sequestered. DnaA protein synthesis was therefore expected to continue during origin sequestration. Despite a normal length of the sequestration period in such cells, they had increased origin content and also displayed asynchrony of initiation. This indicated that reinitiation occasionally occurred at some origins within the same cell cycle. The extra initiations took place in spite of a reduction in total DnaA protein concentration to about half of the wild-type level. We propose that this more efficient utilization of DnaA protein results from an increased availability at the end of the origin sequestration period. Therefore, coordinated sequestration of oriC and dnaA is required for maintaining controlled once-per-cell-cycle initiation.
Initiation of chromosomal replication is a critical and highly regulated step in the Escherichia coli cell cycle. DNA replication is controlled in such a way that each replication origin, oriC, is initiated once and only once each cell cycle at a certain cell mass per origin, the initiation mass (15). The initiation mass is mainly controlled by accumulation of the initiator protein, DnaA (26). The DnaA protein binds to its recognition sites within oriC, and in the presence of architectural proteins the DNA duplex opens in an AT-rich region (for review see reference 24). Duplex opening is stabilized by the binding of DnaA protein associated with ATP to the single-stranded regions (43). Finally, the DnaA protein loads the DnaB helicase associated with DnaC to the single-stranded region (30), DnaC leaves, and the replisomes are assembled (24).
When a threshold level of DnaA protein is reached within the cell, all origins are initiated virtually simultaneously, i.e., in synchrony (40). Synchronous initiation is generally explained by sequestration (inactivation [12, 29, 46]) of newly replicated and hemimethylated origins by the SeqA protein. This directs successive initiations to “old origins” only (25). However, sequestration of newly replicated origins persists only for about one-third of the cell cycle and is therefore not sufficient for maintaining once-per-cell-cycle initiation at oriC. It is essential that the initiation potential is reduced by other means during sequestration. At least three mechanisms, all serving to lower the activity of the DnaA protein, are proposed to be important for lowering the initiation potential (reviewed in reference 7). These are (i) sequestration of the dnaA gene promoter, which prevents de novo synthesis of DnaA protein (12, 44); (ii) titration of DnaA protein to newly replicated chromosomal elements, which provides a sink for DnaA (21, 22, 33); and (iii) regulatory inactivation of DnaA (RIDA), which accelerates the hydrolysis of ATP-DnaA, the form active for initiation, to inactive ADP-DnaA (19).
In this work we describe the construction of cells carrying either the wild-type dnaA gene or the dnaA46 gene in the λ attachment site (attB) located opposite oriC on the E. coli chromosome. Subsequent deletion of the original dnaA gene in these strains allowed us to evaluate the contribution of dnaA gene sequestration to timely once-per-cell-cycle initiation of replication.
MATERIALS AND METHODS
Bacterial strains and plasmids.
All strains used were Escherichia coli K-12 and derived from CM735 (metE46 trp-3 his-4 thi-1 galK2 lacY1 or lacZ4 mtl-1 ara-9 tsx-3 ton-1 rps-8 or rps-9 supE44 λ− [18]). CM742 is CM735 dnaA46 (17), ALO2465 is CM735 dnaA850::Tn10 dnaA::attB (this work), and ALO2073 is dnaA850::Tn10 dnaA46::attB (this work).
Plasmids pFH871 and pFH891 (2) served as sources for the dnaA and dnaA46 genes, respectively. The attP cassette used for site-specific integration into attB came from plasmid pTAC3588 (5), whereas the λ Int protein was supplied from plasmid pTAC3422 (5). Plasmid pFH539 is a pBR322-derived plasmid containing the dnaA gene and dnaA gene promoters (47), and plasmid pdnaN is a pBR322-derived plasmid containing the dnaN gene under plac control as well as the lacI gene (16). Plasmid pTAC4511 was previously described (3).
Growth conditions.
Cells were grown in AB minimal medium (13) supplemented with 0.2% glucose, 0.2% glycerol or 0.5% acetate, and 10 μg/ml thiamine. When indicated, Casamino Acids were added to a final concentration of 0.5%. Tryptophan, histidine, and methionine were added to 50 μg/ml where indicated. Ampicillin, tetracycline, chloramphenicol, and kanamycin were added to final concentrations of 100, 10, 6, and 50 μg/ml, respectively, when required. Cell growth was monitored by measuring optical density at 450 nm (OD450).
Flow cytometry.
Exponentially growing cells (OD450 = 0.15 to 0.25) were treated with rifampin (300 μg/ml; Novartis Pharma Inc.) and cephalexin (36 μg/ml; Sigma Chemical Co.) to inhibit initiation of DNA replication and cell division, respectively (6, 40). Incubation continued for 4 hours to complete ongoing rounds of replication. When initiation of DNA replication is inhibited but ongoing rounds of replication are allowed to finish, the number of fully replicated chromosomes per cell measured by flow cytometry will represent the number of origins in each cell at the time of drug addition (40). In a culture of cells with synchronous initiation, the integral number of chromosomes is two, four, or eight (i.e., 2n), but with asynchronous initiation, additional peaks representing cells with three, five, six, or seven chromosomes (i.e., ≠ 2n) appear. Cells were fixed in 70% ethanol and stained with 90 μg/ml mithramycin (Acros Organics Inc.) and 20 μg/ml ethidium bromide (Merck Inc.) in a buffer containing 10 mM Tris-HCl, pH 7.5, and 10 mM MgCl2 (26). Finally flow cytometry was performed as described previously (26) using an Apogee A10 instrument (Apogee Inc.). Numbers of origins per cell and relative cell mass were determined as described previously (26).
Southern blot analysis.
Total cellular DNA was prepared from exponentially growing cells according to the method of reference 27. DNA was digested with HindIII, and fragments were separated on an 0.7% agarose gel, transferred by capillary transfer to a Hybond-N+ membrane (Amersham Pharmacia Biotech), and probed with a 1,051-bp terC fragment and a 1,196-bp oriC fragment, which hybridize to 4.1-kb and 2.1-kb chromosomal HindIII fragments, respectively. Probes were prepared as described in reference 3 and labeled with [α-33P]dATP (Amersham Pharmacia) using the Random Primer system (Prime-a-gene; Promega Inc.).
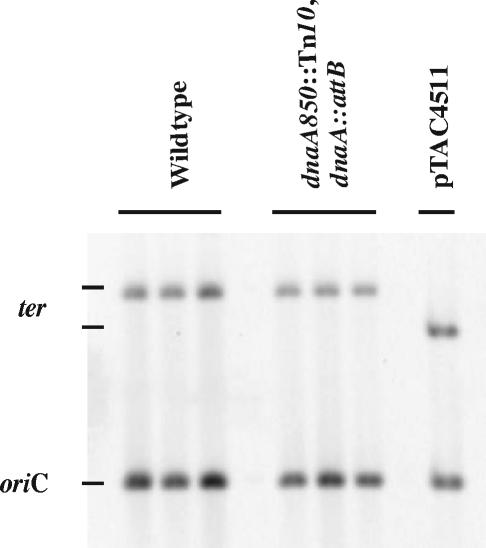
The oriC/terC ratio was determined by measuring the intensities of hybridization to the 2.14- and the 4.1-kb fragments using ImageQuant version 5.2 software (Molecular Dynamics Inc.). Hybridization signals were normalized to the signals of control plasmid pTAC4511 (3) included on the gel, where oriC and terC bands are present in a 1:1 ratio.
Microscopy.
Cultures were grown in glucose-Casamino Acids medium to an OD450 of 0.25, and cells were fixed in 70% ethanol. Cells were applied to glass slides coated with a 0.1% solution of poly-l-lysine (Sigma). To visualize the nucleoid content, cells were stained with a 4′,6-diamidino-2-phenylindole (DAPI)-containing buffer (Pierce Biotechnology Inc.) at a final concentration of 1 μg/ml.
Microscopy was performed using a Leica DM5000B phase-contrast/fluorescence microscope (Leica Microsystems A/S) equipped with a 100× PL Fluotar numerical aperture 1.3 objective. Pictures were taken using a DC 480 camera (Leica Microsystems A/S) that was connected to a computerized image analysis system (IM 50; Leica Microsystems A/S).
Immunoblotting.
Exponentially growing cells were harvested for Western blot analysis at an OD450 of 0.3 to 0.4. Cells were resuspended in 10 mM Tris-HCl, pH 8, 10 mM MgCl2, and the total protein content was determined using a colorimetric assay (28) in order to adjust all samples to the same protein level. Samples of 5 to 10 μg of protein were applied to the gel. Proteins were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Criterion-Precast Gel; 10 to 20% Tris-HCl; Bio-Rad Inc.) and transferred to a polyvinylidene difluoride membrane, 0.2 μm (Millipore), with a semidry blotting apparatus (Bio-Rad Inc.). All the following incubations were at room temperature. The membrane was blocked overnight in Tris-buffered saline (TBSa) plus 2% Tween as main blocking agent (2% Tween 20, 150 mM NaCl, 50 mM Tris-HCl, pH 10), rinsed with TBSa plus 0.05% Tween (0.05% Tween 20, 150 mM NaCl, 50 mM Tris-HCl, pH 10) for 5 min, incubated for 2 h with polyclonal rabbit anti-DnaA antiserum (obtained from K. Skarstad), and washed with TBSa plus 0.05% Tween. The membrane was further incubated for 1.5 h in the presence of porcine anti-rabbit immunoglobulin G antibody conjugated to alkaline phosphatase (DAKO A/S) and washed with TBSa plus 0.05% Tween. The membrane was scanned on a Storm 840 imaging system (Molecular Dynamics Inc.), and quantification was carried out using ImageQuant version 5.2 software (Molecular Dynamics Inc.).
Measurement of the replication and division periods.
The replication time, C, for cells grown in rich medium was determined from the origin/terminus ratio (O/T) obtained from Southern blotting. For a culture having a doubling time τ, and a chromosomal replication time C, the stoichiometry between origins O and termini T is given by the formula O/T = 2C/τ (10). After the C period and the number of origins per cell is determined, the time following termination of replication until cell division, D, can be calculated using the formula ori/cell = 2(C + D)/τ (10). Note that these determinations of C and D periods are valid even for cells that initiate replication asynchronously (10).
For slow-growing cells the O/T-based method proved not to be sufficiently sensitive for an accurate determination of C and D periods. These periods were therefore determined by computer simulation of DNA histograms obtained by flow cytometry of exponentially growing cells (31).
Calculation of gene dosage.
Gene dosage of the dnaA gene was calculated for wild-type cells and dnaA850::Tn10 cells carrying the dnaA gene in attB using the following formula (10): XC = 2{[C*(1 − m) + D]/τ}, where m determines the location of the specific gene (X) relative to the origin of replication. As the distance between the E. coli origin and terminus is 2.32 Mb, mwt = 0.043/2.32 = 0.02, for the wild-type dnaA gene located 43 kb from oriC. The attB region is located 1.51 Mb from oriC, and mattB is 1.51/2.32 = 0.65. τ is the doubling time. C is the time of replication (from initiation to termination of the chromosome). C was determined as described above. D is the time following termination of replication until cell division. D was determined as described above. Note that this calculation of gene dosage is valid even for cells that initiate replication asynchronously (10).
RESULTS
Cells with the dnaA gene located at attB.
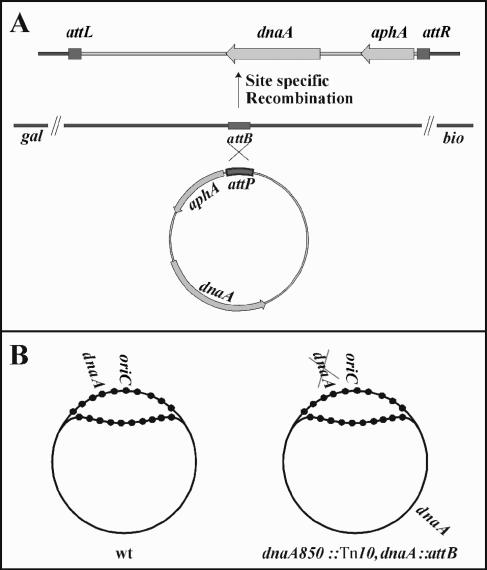
In E. coli the dnaA gene is located about 43 kb away from the replication origin. Consequently the dnaA gene is replicated shortly (less than 1 minute) after initiation at oriC has taken place. dnaA and oriC are therefore hemimethylated and sequestered largely simultaneously (12) (Fig. 1B), and no or very little de novo DnaA protein synthesis takes place during origin sequestration (44). Furthermore both regions remain sequestered for a large fraction of the cell cycle (12). We constructed cells where only one functional copy of the dnaA gene was inserted into the lambda attachment site attB located about 1.5 Mb away from the origin. Briefly, DNA fragments containing the dnaA or dnaA46 gene were ligated to a fragment containing the λ attP site in such a way that the resultant circular molecules did not contain replication origins (Fig. 1A). Site-specific recombination with the bacterial attB in the presence of lambda integrase (5) resulted in cells containing the original dnaA gene at its normal position as well as a second copy of either dnaA or dnaA46 in the attB site. The original dnaA gene was subsequently replaced by the dnaA850::Tn10 allele by P1 transduction (Fig. 1B). In the resultant cells, oriC and dnaA are separated by 1.5 Mb. Because the distance between oriC and the dnaA gene now corresponds to approximately two-thirds of the distance from the origin to the terminus of replication, replication of the dnaA gene will be delayed with approximately two-thirds of a replication time relative to the origin at any given growth rate. Consequently, the origin region and the dnaA gene promoter are not expected to be sequestered simultaneously in these cells (Fig. 1B). Cells carrying the dnaA gene in the attB site grew slower than their wild-type counterparts (Table 1). The growth defect could be complemented by a plasmid carrying dnaA but not by a dnaN-carrying plasmid (Table 1) and therefore resulted from altered expression of the dnaA gene in its new location rather than polar effects of the dnaA850::Tn10 on the dnaN, recF, and gyrB genes. Because the Tn10 is inserted about 60 bp following the start codon of dnaA (23), the downstream genes were transcribed from promoters within or downstream of dnaA (38).
FIG. 1.
Construction of strains with dnaA or dnaA46 in the chromosomal attB site. (A) The dnaA/dnaA46 gene was ligated to a nonreplicating attP-carrying cassette (bottom). Lambda Int protein-dependent site-specific recombination with the chromosomal attB site resulted in cells carrying an additional dnaA gene inserted in attB. Subsequently the dnaA gene was removed from its normal location. (B) In wild-type cells dnaA is replicated shortly after oriC. Consequently, both oriC and dnaA are hemimethylated and sequestered by the SeqA protein (•) in the same part of the cell cycle (left). In cells carrying a dnaA gene in attB only, this is replicated with significant delay relative to oriC. Consequently, the dnaA gene is not sequestered for the same part of the cell cycle as the origin (right).
TABLE 1.
Cell cycle parameters for fast-growing cells
| Strain | Growth medium | Doubling time (min) | No. of origins/cell | Relative cell mass | Relative cell mass/origin |
|---|---|---|---|---|---|
| CM735 (wt) | Glucose + Casamino Acids | 38 | 4.9 | 1.0 | 1.0 |
| ALO2465 (dnaA850::Tn10 dnaA::attB) | Glucose + Casamino Acids | 60 | 6.9 | 1.6 | 1.1 |
| ALO2465/pFH539a | Glucose + Casamino Acids | 41 | NDe | ND | ND |
| ALO2465/pdnaNb | Glucose + Casamino Acids | 59 (−IPTG), 77 (+IPTG)d | ND | ND | ND |
| CM735 | Acetate | 220 | 2.0 | 1.0 | 1.0 |
| ALO2465 | Acetate | 219 | 2.2 | 1.0 | 0.9 |
| CM742 (dnaA46)c | Glucose + Casamino Acids | 58 | 4.2 | 1.0 | 1.2 |
| ALO2073 (dnaA850::Tn10 dnaA46::attB)c | Glucose + Casamino Acids | 66 | 3.9 | 1.2 | 1.5 |
| CM742c | Glycerol | 107 | 2.0 | 1.0 | 1.0 |
| ALO2073c | Glycerol | 124 | 2.5 | 1.5 | 0.8 |
pFH539 is a pBR322-derived plasmid containing the dnaA gene and dnaA gene promoters (47).
pdnaN is a pBR322-derived plasmid containing the dnaN gene under plac control as well as the lacI gene (16).
Growth temperature was 30°C.
IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 1 mM.
ND, not determined.
DnaA synthesis following initiation does not shorten the sequestration period.
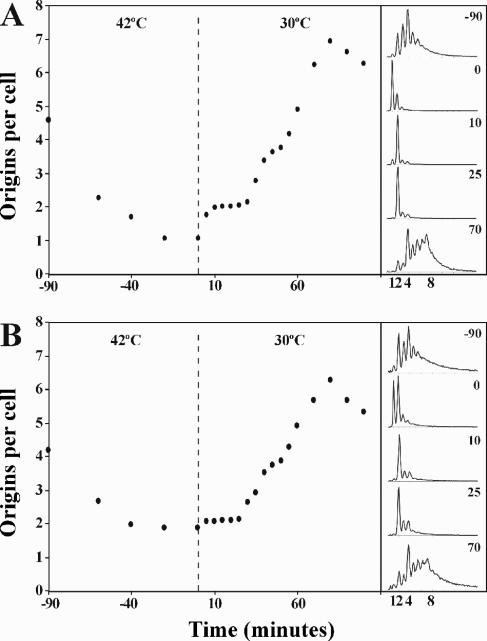
The minimal time between successive initiations was measured for strains CM742 (dnaA46) and ALO2073 (dnaA850::Tn10 dnaA46::attB) as previously described (45). Cells were grown exponentially at a permissive temperature (30°C) for five to six generations, shifted to 42°C, and incubated at this nonpermissive temperature for 90 min before being shifted back to a permissive temperature (30°C). The average number of origins in both strains decreased during incubation at the nonpermissive temperature (Fig. 2), indicating that initiations ceased while cells continued to divide. After 90 min at 42°C, CM742 cells contained mainly one fully replicated chromosome, whereas a significant number of ALO2073 cells contained two chromosomes. Upon the shift back to 30°C, resulting in reactivation of the DnaA protein, all cells of CM742 initiated replication within the first 5 min and again after approximately 30 min. This period of 25 min thus represents the minimal interreplication time for these cells (Fig. 2A). For strain ALO2073 the situation was similar. All cells containing one origin of replication initiated immediately after the shift back to permissive temperature, whereas cells containing two chromosomes for unknown reasons did not (Fig. 2B). A second round of initiation was observed after 30 min. The minimal time between successive initiations in these cells was therefore 25 min as well.
FIG. 2.
Reinitiation in cells carrying dnaA in attB. Cells of strain CM742 (A) and CM742 dnaA850::Tn10 dnaA46::attB (ALO2073) (B) were grown at 30°C in minimal medium supplemented with glucose, Casamino Acids, and tryptophan. At time T = −90 the culture was shifted to 42°C, kept at this nonpermissive temperature for 90 min, and shifted back to 30°C at time T = 0. At the times indicated cell samples were collected for treatment with rifampin and cephalexin for 4 h prior to flow cytometry analysis. The median (the value above and below which 50% of the distribution can be found) was used as a robust measure of the central tendency of individual cells (39) and is plotted as origins per cell. The panels on the right-hand side of the figure show selected DNA histograms for rifampin-treated cultures.
This experiment clearly demonstrates that the minimal time between initiations at the same origin is not shortened in ALO2073 cells which have continued DnaA protein synthesis during origin sequestration. The DnaA protein synthesized during incubation at nonpermissive temperature in these cells will reduce but not turn off the dnaA transcription when renatured (2, 9). It therefore seems that the duration of the sequestration period is insensitive to smaller changes in the DnaA protein concentration.
Occasional reinitiation of replication in cells carrying dnaA in attB.
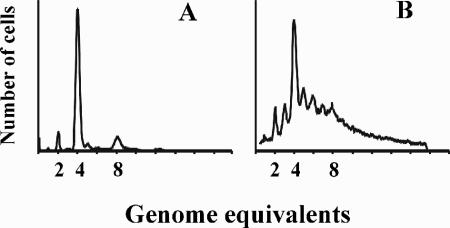
We analyzed the cell cycle of CM735 (wt) and CM742 (dnaA46) cells as well as their isogenic counterparts carrying either the wild-type dnaA gene inserted in attB (dnaA850::Tn10 dnaA::attB; ALO2465) or the dnaA46 gene inserted in attB (dnaA850::Tn10 dnaA46::attB; ALO2073). When analyzed by flow cytometry, wild-type cells contained mainly four origins of replication while some contained two and some contained eight origins (Fig. 3A). This indicates that origins were initiated in synchrony (40). Cells of ALO2465 carrying the dnaA gene in attB also contained mainly four fully replicated origins, but in addition a number of cells had three, five, six, and seven origins (Fig. 3B), indicating that initiations occurred asynchronously in these cells. The number of origins per cell was increased from an average of 4.9 for the wild type to 6.9 in cells carrying dnaA in attB (Table 1). Because the cell size was also somewhat increased for the latter, the cell mass per origin was about the same for the two cell types (Table 1). The asynchrony observed for strain ALO2465 did not result from polar effects of the dnaA850::Tn10 on dnaN, recF, or gyrB gene expression because cells in which the normal dnaA gene was inactivated by a nonpolar dnaA allele were similar to ALO2465 cells with respect to synchrony (not shown). Similarly, strain ALO2465 continued to initiate asynchronously when additional DnaN protein was supplied from a plasmid (not shown).
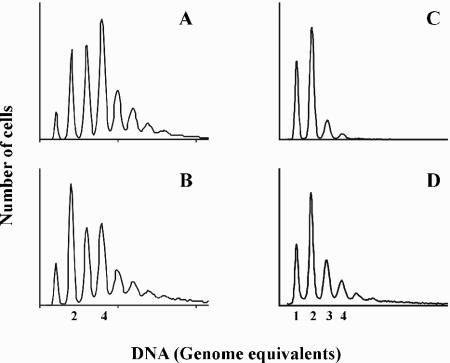
FIG. 3.
Initiation of replication is affected by the dnaA gene location. DNA histograms of CM735 (wt) (A) and ALO2465 (dnaA850::Tn10 dnaA::attB) (B). Cells were grown at 37°C in minimal medium supplemented with glucose, Casamino Acids, and tryptophan and treated with rifampin and cephalexin prior to flow cytometry analysis. Distinct peaks represent the accumulation of cells with an integral number of chromosomes which correspond to the number of origins at the time of drug addition (40). The scale on the number of cells is arbitrary and represents a minimum of 60,000 cells in all panels.
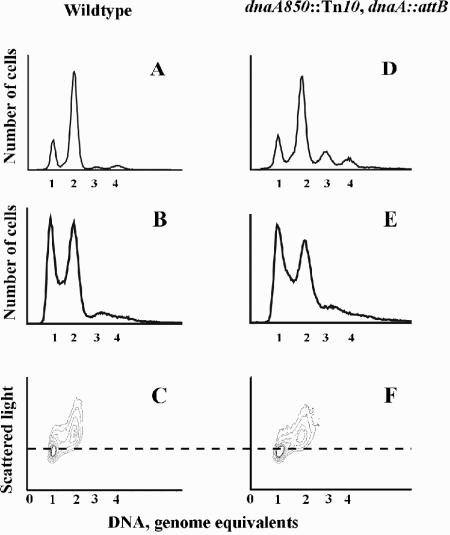
When grown with acetate as sole carbon source, ALO2465 cells grew with the same doubling time as wild-type cells (∼220 min) but contained more origins per cell, although the difference was not as pronounced as that observed at fast growth (Fig. 4; Table 1). The average cell mass was similar to the mass of wild-type cells, and it therefore contained an increased number of origins per mass. Because initiations took place on one origin only, the cell mass at initiation can be read off the three-parameter contour plots (Fig. 4C and F) (8). Wild-type cells and ALO2465 cells carrying dnaA in attB had a similar initiation mass.
FIG. 4.
The initiation mass of slow-growing cells carrying dnaA in attB. Flow cytometry histograms of wild-type (CM735) and dnaA850::Tn10 cells carrying dnaA in attB (ALO2465) grown under steady-state conditions at 37°C in minimal medium supplemented with acetate, tryptophan, histidine, and methionine. The parameters measured were number of origins in rifampin- and cephalexin-treated cells (A and D) and DNA content of exponentially growing cells (B and E). Three-parameter contour plots of exponentially growing cells are shown in panels C and F. The isocontour lines are drawn through points with the same number of cells. Thus, the collection of almost concentric rings at the one- or two-chromosome position represents a “mountain” of cells. The dashed line represents the cell mass at which initiation takes place (8). The scale on number of cells and scattered light is arbitrary and represents a minimum of 100,000 cells in all panels.
We suggest that continued synthesis of wild-type DnaA protein during origin sequestration led to reinitiation at the same origin in some cells. These extra initiations caused an increase in origin content per cell. It is conceivable that the extra initiations occurred at the end of the sequestration period, as this was not affected by continued DnaA synthesis.
The increased number of origins in cells carrying dnaA in attB was not a consequence of initiations becoming asynchronous per se because asynchronous dnaA46 mutant cells grown in rich medium (Fig. 5) had their origin content reduced relative to wild-type cells (Table 1). Cells carrying the dnaA46 gene in attB were also asynchronous (Fig. 5) but contained slightly fewer origins than cells carrying the dnaA46 gene in its normal location (compare Fig. 5A and B; Table 1). Cell size was increased which resulted in an increased cell mass/origin ratio, indicating an increased initiation mass in these cells. When strain CM742 (dnaA46) was grown in glycerol minimal medium with a doubling time of 107 min, cells contained mainly one or two replication origins (Fig. 5C; Table 1), indicating that initiations took place only once on a single copy of oriC. Cells having the dnaA46 gene in attB (ALO2073) grew slower, and a considerable fraction of cells contained three or four replication origins. Therefore, some origins were reinitiated in the same cell cycle under slow growth. Because the cell size was also increased, the cell mass per origin was about the same as for wild-type cells.
FIG. 5.
Initiation of replication is affected by the dnaA46 gene location at slow growth. DNA histograms of CM742 (dnaA46) (A and C) and ALO2073 (dnaA850::Tn10 dnaA46::attB) (B and D) cells. Cells were grown at 30°C in minimal medium supplemented with glucose, Casamino Acids, and tryptophan (A and B) or with glycerol, tryptophan, histidine, and methionine (C and D) and treated with rifampin and cephalexin prior to flow cytometry analysis. Distinct peaks represent the accumulation of cells with an integral number of chromosomes which correspond to the number of origins at the time of drug addition (40).The scale on number of cells is arbitrary and represents a minimum of 60,000 cells in all panels.
Cell cycle periods of cells carrying dnaA in attB.
The cellular number of origins is a function of the replication and division periods (C and D) as well as cellular growth rate. The increase in number of origins in cells carrying the wild-type dnaA gene in attB could therefore result from an increase in the C or D period of these cells or both (10).
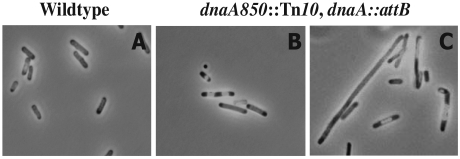
We determined the origin-to-terminus ratio for fast-growing wild-type cells and dnaA850::Tn10 cells carrying the dnaA gene in attB by Southern blot hybridization (Fig. 6). We included a sample of plasmid pTAC4511 containing both oriC and terC on the blot in order to correct for differential labeling efficiencies of the oriC and terC probes (3). In wild-type cells the ori/ter ratio was found to be 2.5. Because this ratio depends only on the cellular growth rate and length of the C period, the latter could be determined to be approximately 45 min (Table 2). Knowing the number of origins per cell (Fig. 3; Table 1), we could determine the D period to be approximately 42 min. These durations of C and D periods are in good agreement with what was previously determined for this strain (1). Strain ALO2465 carrying the dnaA gene in attB grew slower than its wild-type counterpart but had a slightly higher ori/ter ratio of 3.0. The C and D periods of this strain were 90 and 77 min, respectively (Table 2), i.e., both were extended considerably relative to wild-type cells. A microscopic inspection revealed that cells carrying the dnaA gene in attB were larger than wild type, heterogeneous, and frequently filamentous with irregular positioning of the nucleoids (Fig. 7).
FIG. 6.
The ori/ter ratio is increased in cells carrying dnaA in attB. Three individual cultures of wild-type (CM735) and dnaA850::Tn10 dnaA::attB (ALO2465) cells were grown exponentially at 37°C in minimal medium supplemented with glucose, Casamino Acids, and tryptophan. DNA was extracted and digested with HindIII, and Southern hybridization was performed with an oriC probe and a ter probe (Materials and Methods). A sample of plasmid pTAC4511 digested with HindIII was also included on the blot. This sample contains oriC and terC in a 1:1 proportion.
TABLE 2.
Replication and division periods for cells carrying dnaA in attB
| Strain | Growth medium | Doubling time (min) | ori/ter ratio | C period (min) | D period (min) |
|---|---|---|---|---|---|
| CM735 (wt) | Glucose + Casamino Acids | 38 | 2.5 | 45 ± 8 | 42 |
| ALO2465 (dnaA850::Tn10 dnaA::attB) | Glucose + Casamino Acids | 60 | 3.0 | 90 ± 13 | 77 |
| CM735 (wt) | Acetate | 220 | NDa | 79 | 103 |
| ALO2465 (dnaA850::Tn10 dnaA::attB) | Acetate | 219 | ND | 92 | 90 |
ND, not determined.
FIG. 7.
Cells carrying dnaA in attB are heterogeneous with respect to cell size and nucleoid positioning. Strains CM735 (wt) (A) and CM735 dnaA850::Tn10 dnaA::attB (ALO2465) (B and C). Panel B shows the morphology of the majority of ALO2465 cells, whereas the less frequent filamentous cells are shown in panel C. Cells were grown at 37°C in minimal medium supplemented with glucose, Casamino Acids, and tryptophan. Cells were fixed and stained prior to microscopic analysis as described in Materials and Methods.
For slow-growing cells, the cell cycle periods were determined from simulations of the DNA histograms of exponentially growing cells (31). Under these growth conditions wild-type cells had C and D periods of 79 and 103 min, respectively, whereas dnaA850::Tn10 cells carrying the dnaA gene in attB had C and D periods of 92 and 90 min, respectively (Table 2). Microscopic inspection of slow-growing cells revealed no gross differences between wild-type cells and dnaA850::Tn10 cells carrying the dnaA gene in attB (not shown).
We may conclude that, in fast-growing dnaA850::Tn10 cells carrying the dnaA gene in attB, the increased number of origins per cell is accompanied by an increase in duration of both C and D periods. Because cells are quite heterogeneous, these average numbers are likely to cover large cell-to-cell differences, and individual cells in such a culture presumably do not undergo identical cell cycles. The increased C period could result from overinitiation at oriC such as previously observed (4, 26, 34, 42) or from a slight shortage of DnaN protein due to the Tn10 insertion in dnaA. The dnaN gene encodes the ring-shaped β-subunit dimer of the polymerase III holoenzyme—the so-called sliding clamp—which stabilizes the binding of polymerase III to DNA strands to enable processive replication over lengths of several kilobases (36, 48). At slow growth differences between wild-type cells and dnaA850::Tn10 cells carrying dnaA in attB were smaller, with less than 15% difference in the duration of their C and D periods.
The level of DnaA protein is decreased in cells carrying the dnaA gene in attB.
The increase in origin content for cells carrying the wild-type dnaA gene in attB might result from an increased level of DnaA protein relative to wild-type cells. Such differences in expression of the same gene in different chromosomal contexts have previously been observed (37).
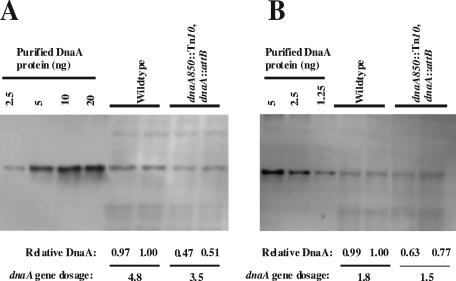
We therefore quantified the cellular content of DnaA protein in wild-type (CM735) cells and cells carrying the dnaA gene in attB only (ALO2465). The immunoblot revealed that the concentration of DnaA protein in cells carrying the dnaA gene in attB was reduced to about half of the wild-type level during growth in rich medium (Fig. 8A), whereas less change relative to wild type was observed when growth was slow (Fig. 8B). Because attB is located fairly close to the chromosomal terminus of replication, the gene dosage of dnaA, when inserted here, must be lower than the gene dosage of the dnaA gene at its normal origin-proximal location. We calculated the gene dosage of dnaA (10) in the two different strains, using the previously determined C and D periods. In rich medium the dnaA gene dosage for wild-type cells was found to be 4.8, which is in good agreement with the average number of origins per cell (Table 1), whereas it was reduced to 3.5 in cells carrying dnaA in attB. During slow growth the dnaA gene dosage was reduced from 1.8 in wild-type cells to 1.5 in cells carrying dnaA in attB (Fig. 8).
FIG. 8.
Cellular DnaA protein content. Strains CM735 (wt) and CM735 dnaA850::Tn10 dnaA::attB (ALO2465) were grown at 37°C in minimal medium supplemented with glucose, Casamino Acids, and tryptophan (A) or in minimal medium supplemented with acetate, tryptophan, histidine, and methionine (B). Samples were taken, and the total protein content was adjusted to the same amount using a Lowry analysis prior to loading on a sodium dodecyl sulfate-polyacrylamide gel. For fast-growing cells 8 μg of total protein was applied to the gel (A) whereas only 5 μg was used for slow-growing cells (B). Following blotting, the filter was probed with a polyclonal DnaA antibody. The control lanes contain the indicated amounts of purified DnaA protein. The relative amount of DnaA protein was quantified for each sample and normalized to wild-type cells. The gene dosage of dnaA was calculated as described in Materials and Methods.
We can conclude that the additional initiations observed for cells carrying the dnaA gene in the attB site did not result from an increased level of DnaA protein. In contrast, the DnaA protein level of these cells was reduced relative to the wild type primarily at fast growth (Fig. 8). Because the dnaA gene dosage in ALO2465 cells was also reduced relative to wild type, the dnaA gene was expressed in a gene dosage-dependent manner. This may indicate that autoregulation plays little role for dnaA gene expression in normally growing cells such as previously described (4) or that dnaA gene expression, despite being autoregulated, shows a dampened dependence on gene dosage (14).
DISCUSSION
We have constructed cells where the dnaA gene was moved from its normal location adjacent to oriC to the attB locus, 1.5 Mb away from the origin. In these cells the dnaA gene is replicated, hemimethylated, and sequestered with significant delay relative to the origin. Consequently, synthesis of DnaA protein is assumed to continue during origin sequestration in these cells. This led to occasional reinitiation at some origins within the same cell cycle, manifested as an increased number of replication origins per cell and asynchronous initiation.
Timely sequestration of the dnaA gene is required for controlled initiation of replication.
In E. coli, the dnaA gene promoter is sequestered by a mechanism similar to that of oriC and for the same fraction of the cell cycle (12). Sequestration of the dnaA gene promoter prevents de novo DnaA protein synthesis (44), and this mechanism was proposed to reduce the amount of the initiator DnaA protein to a level that does not sustain any further initiations within the same cell cycle (reviewed in reference 7).
In wild-type cells the initiation mass is normally set by accumulation of DnaA protein (26). It was therefore surprising to find that fast-growing cells carrying their only copy of the dnaA gene at attB had cell mass per origin similar to that of wild-type cells, despite a 50% reduction in DnaA protein concentration. In order for cells to accumulate sufficient DnaA protein for initiation to take place, they must grow to a larger size before they initiate replication at the same amount of DnaA per origin as wild-type cells, which explains the observed increase in cell size and in generation time. However, once initiation(s) occurs at a given origin it is not limited to once per cell cycle, which explains why cells that carry the dnaA gene at attB had increased origin content and initiated asynchronously. Such occasional reinitiation at an origin within the same cell cycle results in a culture where individual cells undergo different cell cycles (32). In agreement with this, cells carrying dnaA in attB were quite heterogeneous with respect to both cell size and nucleoid positioning.
There are at least two scenarios for how origins may be reinitiated in cells carrying dnaA in attB. First, the activity of the DnaA protein itself could be increased, i.e., by having a larger fraction in the ATP-bound form. This is not likely since cells are wild type with respect to the RIDA system (19, 20). Second, the timing of DnaA protein synthesis within the cell cycle could be altered in such a way that the protein becomes available for initiation in a part of the cell cycle where initiations do not normally occur. In the period following initiation at oriC, the origin is hemimethylated and sequestered whereas the dnaA gene, now located in attB and therefore replicated with a delay of two-thirds of the C period, remains fully methylated and continues to synthesize DnaA protein. Some of this newly synthesized DnaA protein is expected to be in the ATP-bound form and active for initiation. At the end of the oriC sequestration period, where origins again become available for initiation, the level of DnaA protein is elevated, and we suggest that this leads to reinitiation at some origins within the same cell cycle. The net result of such occasional reinitiation is a culture of cells that are asynchronous and contain an elevated number of origins per cell (Fig. 3B; Table 1). The increased replication time in cells carrying dnaA in attB may well be a consequence of occasional reinitiation at oriC; seqA mutant cells reinitiate replication at an origin within the same cell cycle due to lack of sequestration (29), and DnaA protein-overproducing cells reinitiate at the end of the sequestration period (34). In both cases untimely reinitiation at an origin leads to a considerable increase in the replication period.
In dnaA46 mutant cells initiations occur throughout most of or the entire cell cycle, and some origins are reinitiated while others are not initiated at all (41). Therefore, in fast-growing cells containing multiple origins of replication, initiation at one origin only leads to replication and sequestration of the dnaA46 gene located near the initiated origin, whereas the remainder of the dnaA46 genes carried within the same cell are not sequestered. Consequently, DnaA46 protein synthesis occurs continuously throughout the cell cycle even when the gene is in its normal position. Therefore, there is little effect of moving the dnaA46 gene to the attB position with respect to initiation synchrony. The lower dnaA46 gene dosage is, however, reflected in a slightly reduced number of origins per cell and increase in initiation mass (Fig. 5A and B; Table 1). In slow-growing dnaA46 cells initiations take place at a single origin, and this leads to sequestration of oriC as well as the dnaA46 gene, i.e., the gene is expressed in a cell-cycle-specific manner. This explains why we observe that cells carrying dnaA46 in attB overinitiate replication at slow growth (Fig. 5; Table 1); DnaA46 protein synthesis continues through origin sequestration so that origins are frequently reinitiated in the cell cycle.
It is likely that cells carrying dnaA in attB sequester their dnaA gene following passage of the replication fork, i.e., continue to express dnaA in a cell-cycle-specific manner that is altered relative to wild type. This poses the main difference between this and a previous report showing that cell-cycle-specific expression of dnaA is not important for once-per-cell-cycle initiation (26). In the earlier report, the dnaA gene was expressed from a lac promoter on a plasmid, whereas the chromosome carried a dnaA46 mutation. At elevated temperature active DnaA protein was presumably expressed from the plasmid only in a non-cell-cycle-specific manner, yet initiations were synchronous. It is not clear to us why our data differ from this report. The data presented here are on the other hand in agreement with several recent reports indicating that constitutive and elevated DnaA protein expression, from a plasmid, leads to asynchronous initiations (4, 34, 41).
Two mechanisms were previously proposed to lower the amount of the initiator DnaA protein during origin sequestration. RIDA converts DnaA associated with ATP to the ADP-associated form inactive for initiation (19, 20), and replication generates new DnaA protein binding sites that serve to bind (titrate) free DnaA protein away from the origin (21, 22). The relative contribution of these two mechanisms to maintaining once-per-cell-cycle initiation has been assessed by preventing RIDA from taking place (11) and by deletion of the datA region (22). In both cases, reinitiation at origins within the same cell cycle was observed, resulting in initiation asynchrony and an elevated number of origins per cell (11, 22). However, it has recently been reported that deletion of datA mainly results in early initiation at a reduced mass per origin (35), suggesting that this locus mainly controls the accumulation of DnaA protein necessary for an initiation event. Our data add the timely sequestration of the dnaA gene promoter to RIDA and possibly datA as necessary elements for maintaining once-per-cell-cycle initiation of replication.
Acknowledgments
We thank C. P. Nielsen for excellent technical assistance; T. Atlung, F. G. Hansen, and E. C. Guzman for providing strains and plasmids; F. G. Hansen for help with the computer simulations; R. Bugge Jensen for help with microscopy; and K. Skarstad for providing the anti-DnaA antiserum and the purified DnaA protein.
This work was supported by grants from the Danish Natural Sciences Research Council, the Danish Medical Research Council, and The Novo Nordisk Foundation.
REFERENCES
- 1.Allman, R., T. Schjerven, and E. Boye. 1991. Cell cycle parameters of Escherichia coli K-12. J. Bacteriol. 173:7970-7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atlung, T., E. Clausen, and F. G. Hansen. 1985. Autoregulation of the dnaA gene of Escherichia coli. Mol. Gen. Genet. 200:442-450. [DOI] [PubMed] [Google Scholar]
- 3.Atlung, T., and F. G. Hansen. 1999. Low-temperature-induced DnaA protein synthesis does not change initiation mass in Escherichia coli K-12. J. Bacteriol. 181:5557-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atlung, T., and F. G. Hansen. 1993. Three distinct chromosome replication states are induced by increasing concentrations of DnaA protein in Escherichia coli. J. Bacteriol. 175:6537-6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atlung, T., L. J. Rasmussen, L. J. Nellemann, and F. Holm. 1991. A versatile method for integration of genes and gene fusions into the L(lambda) attachment site of Escherichia coli. Gene 107:11-17. [DOI] [PubMed] [Google Scholar]
- 6.Boye, E., and A. Løbner-Olesen. 1991. Bacterial growth control studied by flow cytometry. Res. Microbiol. 142:131-135. [DOI] [PubMed] [Google Scholar]
- 7.Boye, E., A. Løbner-Olesen, and K. Skarstad. 2000. Limiting DNA replication to once and only once. EMBO Rep. 1:479-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boye, E., T. Stokke, N. Kleckner, and K. Skarstad. 1996. Coordinating DNA replication initiation with cell growth: differential roles for DnaA and SeqA proteins. Proc. Natl. Acad. Sci. USA 93:12206-12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braun, R. E., K. O'Day, and A. Wright. 1985. Autoregulation of the DNA replication gene dnaA in E. coli. Cell 40:159-169. [DOI] [PubMed] [Google Scholar]
- 10.Bremer, H., and G. Churchward. 1977. An examination of the Cooper-Helmstetter theory of DNA replication in bacteria and its underlying assumptions. J. Theor. Biol. 69:645-654. [DOI] [PubMed] [Google Scholar]
- 11.Camara, J. E., K. Skarstad, and E. Crooke. 2003. Controlled initiation of chromosomal replication in Escherichia coli requires functional Hda protein. J. Bacteriol. 185:3244-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campbell, J. L., and N. Kleckner. 1990. E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell 62:967-979. [DOI] [PubMed] [Google Scholar]
- 13.Clark, D. J., and O. Maaløe. 1967. DNA replication and the division cycle in Escherichia coli. J. Mol. Biol. 23:99-112. [Google Scholar]
- 14.Das, N., M. Valjavec-Gratian, A. N. Basuray, R. A. Fekete, P. P. Papp, J. Paulsson, and D. K. Chattoraj. 2005. Multiple homeostatic mechanisms in the control of P1 plasmid replication. Proc. Natl. Acad. Sci. USA 102:2856-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donachie, W. D. 1968. Relationship between cell size and time of initiation of DNA replication. Nature 219:1077-1079. [DOI] [PubMed] [Google Scholar]
- 16.Grigorian, A. V., R. B. Lustig, E. C. Guzman, J. M. Mahaffy, and J. W. Zyskind. 2003. Escherichia coli cells with increased levels of DnaA and deficient in recombinational repair have decreased viability. J. Bacteriol. 185:630-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen, E. B., T. Atlung, F. G. Hansen, O. Skovgaard, and K. von Meyenburg. 1984. Fine structure genetic map and complementation analysis of mutations in the dnaA gene of Escherichia coli. Mol. Gen. Genet. 196:387-396. [DOI] [PubMed] [Google Scholar]
- 18.Hansen, F. G., and K. von Meyenburg. 1979. Characterization of the dnaA, gyrB and other genes in the dnaA region of the Escherichia coli chromosome on specialized transducing phages lambda-tna. Mol. Gen. Genet. 175:135-144. [DOI] [PubMed] [Google Scholar]
- 19.Katayama, T., T. Kubota, K. Kurokawa, E. Crooke, and K. Sekimizu. 1998. The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E. coli chromosomal replicase. Cell 94:61-71. [DOI] [PubMed] [Google Scholar]
- 20.Kato, J., and T. Katayama. 2001. Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J. 20:4253-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kitagawa, R., H. Mitsuki, T. Okazaki, and T. Ogawa. 1996. A novel DnaA protein-binding site at 94.7 min on the Escherichia coli chromosome. Mol. Microbiol. 19:1137-1147. [DOI] [PubMed] [Google Scholar]
- 22.Kitagawa, R., T. Ozaki, S. Moriya, and T. Ogawa. 1998. Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein. Genes Dev. 12:3032-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kogoma, T., and K. von Meyenburg. 1983. The origin of replication, oriC, and the dnaA protein are dispensible in stable DNA replication (sdrA) mutants of Escherichia coli K-12. EMBO J. 2:463-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kornberg, A., and T. A. Baker. 1992. DNA replication. W. H. Freeman and Company, New York, N.Y.
- 25.Løbner-Olesen, A., F. G. Hansen, K. V. Rasmussen, B. Martin, and P. L. Kuempel. 1994. The initiation cascade for chromosome replication in wild-type and Dam methyltransferase deficient Escherichia coli cells. EMBO J. 13:1856-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Løbner-Olesen, A., K. Skarstad, F. G. Hansen, K. von Meyenburg, and E. Boye. 1989. The DnaA protein determines the initiation mass of Escherichia coli K-12. Cell 57:881-889. [DOI] [PubMed] [Google Scholar]
- 27.Løbner-Olesen, A., and U. von Freiesleben. 1996. Chromosomal replication incompatibility in Dam methyltransferase deficient Escherichia coli cells. EMBO J. 15:5999-6008. [PMC free article] [PubMed] [Google Scholar]
- 28.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 29.Lu, M., J. L. Campbell, E. Boye, and N. Kleckner. 1994, SeqA: a negative modulator of replication initiation in E. coli. Cell 77:413-426. [DOI] [PubMed] [Google Scholar]
- 30.Marszalek, J., and J. M. Kaguni. 1994. DnaA protein directs the binding of DnaB protein in initiation of DNA replication in Escherichia coli. J. Biol. Chem. 269:4883-4890. [PubMed] [Google Scholar]
- 31.Michelsen, O., M. J. Teixeira de Mattos, P. R. Jensen, and F. G. Hansen. 2003. Precise determinations of C and D periods by flow cytometry in Escherichia coli K-12 and B/r. Microbiology 149:1001-1010. [DOI] [PubMed] [Google Scholar]
- 32.Molina, F., and K. Skarstad. 2004. Replication fork and SeqA focus distributions in Escherichia coli suggest a replication hyperstructure dependent on nucleotide metabolism. Mol. Microbiol. 52:1597-1612. [DOI] [PubMed] [Google Scholar]
- 33.Morigen, E. Boye, K. Skarstad, and A. Lobner-Olesen. 2001. Regulation of chromosomal replication by DnaA protein availability in Escherichia coli: effects of the datA region. Biochim. Biophys. Acta 1521:73-80. [DOI] [PubMed] [Google Scholar]
- 34.Morigen, A. Løbner-Olesen, and K. Skarstad. 2003. Titration of the Escherichia coli DnaA protein to excess datA sites causes destabilization of replication forks, delayed replication initiation and delayed cell division. Mol. Microbiol. 50:349-362. [DOI] [PubMed] [Google Scholar]
- 35.Morigen, F. Molina, and K. Skarstad. 2005. Deletion of the datA site does not affect once-per-cell-cycle timing but induces rifampin-resistant replication. J. Bacteriol. 187:3913-3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Donnell, M. E. 1987. Accessory proteins bind a primed template and mediate rapid cycling of DNA polymerase III holoenzyme from Escherichia coli. J. Biol. Chem. 262:16558-16565. [PubMed] [Google Scholar]
- 37.Owen-Hughes, T. A., G. D. Pavitt, D. S. Santos, J. M. Sidebotham, C. S. Hulton, J. C. Hinton, and C. F. Higgins. 1992. The chromatin-associated protein H-NS interacts with curved DNA to influence DNA topology and gene expression. Cell 71:255-265. [DOI] [PubMed] [Google Scholar]
- 38.Quinones, A., and W. Messer. 1988. Discoordinate expression in the dnaA-dnaN operon of Escherichia coli. Mol. Gen. Genet. 213:118-124. [DOI] [PubMed] [Google Scholar]
- 39.Shapiro, H. M. 1995. Practical flow cytometry. Wiley-Liss, New York, N.Y.
- 40.Skarstad, K., E. Boye, and H. B. Steen. 1986. Timing of initiation of chromosome replication in individual E. coli cells. EMBO J. 5:1711-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skarstad, K., and A. Løbner-Olesen. 2003. Stable co-existence of separate replicons in Escherichia coli is dependent on once-per-cell-cycle initiation. EMBO J. 22:140-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skarstad, K., A. Løbner-Olesen, T. Atlung, K. von Meyenburg, and E. Boye. 1989. Initiation of DNA replication in Escherichia coli after overproduction of the DnaA protein. Mol. Gen. Genet. 218:50-56. [DOI] [PubMed] [Google Scholar]
- 43.Speck, C., and W. Messer. 2001. Mechanism of origin unwinding: sequential binding of DnaA to double- and single-stranded DNA. EMBO J. 20:1469-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Theisen, P. W., J. E. Grimwade, A. C. Leonard, J. A. Bogan, and C. E. Helmstetter. 1993. Correlation of gene transcription with the time of initiation of chromosome replication in Escherichia coli. Mol. Microbiol. 10:575-584. [DOI] [PubMed] [Google Scholar]
- 45.von Freiesleben, U., M. A. Krekling, F. G. Hansen, and A. Løbner-Olesen. 2000. The eclipse period of Escherichia coli. EMBO J. 19:6240-6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.von Freiesleben, U., K. V. Rasmussen, and M. Schaechter. 1994. SeqA limits DnaA activity in replication from oriC in Escherichia coli. Mol. Microbiol. 14:763-772. [DOI] [PubMed] [Google Scholar]
- 47.von Meyenburg, K., F. G. Hansen, T. Atlung, L. Boe, I. G. Clausen, B. van Deurs, E. B. Hansen, B. B. Jorgensen, F. Jorgensen, L. Koppes, O. Michelsen, J. Nielsen, P. E. Pedersen, K. V. Rasmussen, E. Riise, and O. Skovgaard. 1985. Facets on the chromosomal origin of replication oriC of E. coli, p. 260-281. In M. Schaechter, F. C. Neidhardt, J. Ingraham, and N. O. Kjeldgaard (ed.), The molecular biology of bacterial growth. Jones & Bartlett, Boston, Mass.
- 48.Wickner, S. 1976. Mechanism of DNA elongation catalyzed by Escherichia coli DNA polymerase III, dnaZ protein, and DNA elongation factors I and III. Proc. Natl. Acad. Sci. USA 73:3511-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]