Abstract
Xylella fastidiosa is a xylem-limited nonflagellated bacterium that causes economically important diseases of plants by developing biofilms that block xylem sap flow. How the bacterium is translocated downward in the host plant's vascular system against the direction of the transpiration stream has long been a puzzling phenomenon. Using microfabricated chambers designed to mimic some of the features of xylem vessels, we discovered that X. fastidiosa migrates via type IV-pilus-mediated twitching motility at speeds up to 5 μm min−1 against a rapidly flowing medium (20,000 μm min−1). Electron microscopy revealed that there are two length classes of pili, long type IV pili (1.0 to 5.8 μm) and short type I pili (0.4 to 1.0 μm). We further demonstrated that two knockout mutants (pilB and pilQ mutants) that are deficient in type IV pili do not twitch and are inhibited from colonizing upstream vascular regions in planta. In addition, mutants with insertions in pilB or pilQ (possessing type I pili only) express enhanced biofilm formation, whereas a mutant with an insertion in fimA (possessing only type IV pili) is biofilm deficient.
Xylella fastidiosa is a gram-negative, nonflagellated bacterium that causes economically important plant diseases, including Pierce's disease of grapevine, citrus variegated chlorosis, phony peach disease, and others (22, 25). Limited to colonizing the water-conducting xylem vessels of plants (Fig. 1), the bacterium develops biofilms that contribute to the blockage of sap flow, resulting in plant stress and disease. Various insect leafhopper species serve as vectors of X. fastidiosa. As the insects probe plant tissues in search of the vascular xylem elements and their watery sap contents, they often take up X. fastidiosa and subsequently transmit it to healthy plants (22). How the bacteria are disseminated in the xylem vessels from the feeding sites has long been a particularly puzzling and important question (15, 22). Long-distance intraplant migration of the bacteria (27) is even more perplexing since xylem sap flow is always down the pressure gradient, viz., with the transpiration stream that flows toward the leaf. Even under nocturnal conditions, when leaf stomates are mostly closed, cuticular transpiration maintains sap flow toward the leaf (acropetal), albeit at lower rates. Sap flow is seldom stagnant and rarely, if ever, moves in a reverse direction away from the leaves (basipetal). Since X. fastidiosa is a nonflagellated bacterium, one hypothesis for its ability to migrate against the normal flow of the plant's vascular system has been through the slow and incremental expansion of the bacterial colony through repeated cell division along xylem vessel walls (16). Another possibility is that occasional cavitation of the water column causes momentary reversal and short-distance flow of the sap, thereby carrying the bacteria down the xylem elements. Neither of these scenarios satisfactorily explains the colonization of upstream xylem regions. Since analysis of the X. fastidiosa genome indicates that a number of gene orthologs that may encode proteins involved in biogenesis and the function of type IV pili (25) are present, we hypothesized that the bacterium actively migrates via twitching motility. Twitching is a type of motility that some gram-negative bacteria, e.g., Pseudomonas aeruginosa, Neisseria gonorrhoeae, and Myxococcus xanthus, use to colonize new surfaces and can be important in the development of biofilms (14, 18, 29). It is facilitated by type IV pili that are generally located at one cell pole (8, 20) and is characterized by jerky, intermittent movements that facilitate surface colonization (8, 14). To date, all direct observations of twitching by bacteria have been focused mainly on mammalian pathogens or saprophytic bacteria, such as M. xanthus. There is strong evidence for twitching in the phytopathogenic bacterium Ralstonia solanacearum based on colony morphologies (12). However, twitching motility of plant-pathogenic bacteria has not yet been directly observed in situ. Moreover, evidence for the presence of pili on X. fastidiosa cells has been scant, based primarily on two brief notations of “fimbria-like fibrils” (5, 30). There are no reports of colony morphological features indicative of twitching, much less any consideration that twitching motility has a role in this pathogen.
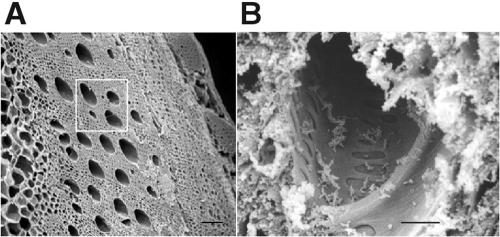
FIG. 1.
Scanning electron micrographs of tangential sections through grapevine and associated xylem vessels. (A) Water-conducting xylem vessels (several of which are noted within the white square) of a grapevine cross section serve as the habitat for X. fastidiosa cells. Bar, 200 μm. (B) View into a sectioned xylem vessel with X. fastidiosa cells attached to the vessel walls. Bar, 10 μm.
The spread of X. fastidiosa in infected grapevines has been studied mostly by destructive sampling and culturing techniques, with the presence of bacteria in any one sampled region correlated with the distance from the original site of introduction (16, 22, 27). This approach precludes our ability to follow the progression of the bacterial population temporally within the same plant and does not provide information regarding the mechanism of bacterial translocation. To circumvent these shortcomings, we fabricated microfluidic devices, constructed of silicone elastomer, that permit long-term spatial and temporal observations of individual bacteria and populations of bacteria in an environment that models xylem vessels. Using these microfluidic chambers, we examined our hypothesis that X. fastidiosa cells are able to migrate, via twitching motility, against a fluid current. In addition, we examined movement in grapevines based on twitching activities.
MATERIALS AND METHODS
Xylella fastidiosa culture.
Wild-type Xylella fastidiosa Temecula (ATCC 700964) was maintained at 28°C on modified PW agar (4) (without phenol red and with 3.5 instead of 6.0 g liter−1 bovine serum albumin [Sigma, St. Louis, MO]). Transposon mutants of X. fastidiosa were maintained on PW medium containing 50 mg liter−1 kanamycin.
Mutagenesis.
The EZ::TN transposome system (7) was used to generate X. fastidiosa mutants. Electrocompetent cells were prepared according to published procedures (7). Xylella fastidiosa was cultured in modified PW liquid medium at 28°C on a rotary shaker (180 rpm) for 7 days. The cell culture was spread onto modified PW plates and incubated for 7 days at 28°C, at which time cells were gently washed off each plate with 2 to 3 ml of modified PW medium. The cells were centrifuged (5,000 × g) at 4°C for 5 min; washed in 10 ml of cold, sterile 10% glycerol; concentrated by centrifugation; and resuspended in 1 ml cold 10% glycerol. The suspension was centrifuged again and resuspended in 10% glycerol to a final concentration of approximately 109 cells ml−1 and held on ice until electroporated with the EZ::TN transposome.
One microliter of EZ::TN <KAN-2> Tn5 transposome (20 ng of transposon DNA) was mixed with 40 μl electrocompetent cells. The mixture was placed in a 0.2-cm-gap electroporation chamber (Bio-Rad, Hercules, CA), and electroporation was conducted at 2,500 V, 200 Ω, and 25 μF for 5 ms. Electroporated cells were transferred to 1 ml modified PW medium and incubated with agitation at 28°C for 24 h. Then 50 μl of the cell suspension was plated onto PW agar containing 50 mg liter−1 kanamycin. For controls, electroporated cells were also plated onto PW medium without kanamycin, and the wild-type strain was plated onto PW agar with kanamycin. Fifteen individual colonies from the PW plates containing kanamycin were checked for Tn5 insertion by PCR. Each mutant of interest was examined by Southern analysis (23) to confirm a single Tn5 insertion; i.e., genomic DNA was cut with EcoRI, separated on a 1% agarose gel, and probed with a 0.6-kb fragment derived from the Tn5 sequence by using the DIG High Prime DNA labeling and detection starter kit I (Roche, Mannheim, Germany).
Screening for twitching mutants.
Thirteen hundred individual X. fastidiosa colonies from the transposon mutagenesis were picked with sterile toothpicks and spotted in quadruplicate onto fresh PW agar plates that prior to spotting were air dried, uncovered, in a laminar-flow hood for at least 15 min. Following incubation at 28°C for 3 days, the edge morphology of the colonies was examined using a dissecting microscope. Colonies with a peripheral fringe were designated as having a wild-type twitching phenotype. Colonies lacking a peripheral fringe were designated as having a twitching defect. These mutants were screened a second time to verify the morphological trait. From 1,300 mutants, we identified 10 that lacked a peripheral fringe and several with a reduced fringe.
Screening for biofilm-modified mutants.
Mutants were screened for biofilm formation (19) in PD2 medium (4) on polystyrene microtiter plates. Wild-type X. fastidiosa and liquid PD2 medium were used as positive and negative controls, respectively. Plates were incubated for 7 days at 28°C. The assay was repeated at least twice to confirm the biofilm phenotype for specific mutants. Quantification of biofilm was performed according to published protocols (19).
Growth rate analysis.
Growth rates of wild-type and mutant X. fastidiosa were determined by transferring 5- to 7-day-old cells grown on PW agar into liquid PW medium. The initial optical density for each culture was adjusted to an absorbance of 0.05 at 600 nm. Three milliliters of each culture was added to a 13- by 100-mm glass tube and incubated at 28°C with continuous agitation at 175 rpm. Cell density was determined at regular intervals by measuring optical density at an A600.
Sequencing of mutants.
EcoRI or HindIII was used to digest the genomic DNA of mutants. Fragments were ligated into pUC18 for sequencing. DNA fragments flanking the Tn5 insertion were sequenced, edited with DNASTAR (University of Wisconsin), and analyzed with BLAST against the genome sequence of X. fastidiosa Temecula (http://aeg.lbi.ic.unicamp.br/world/xfpd/).
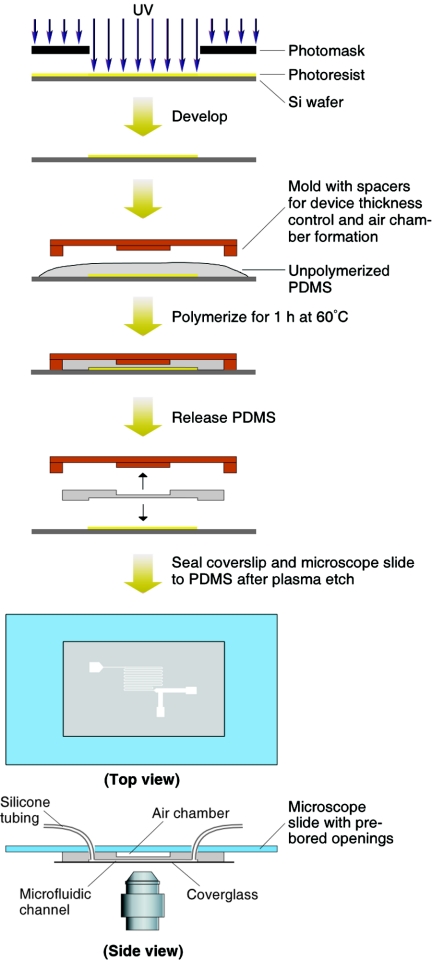
Fabrication of microfluidic chambers.
Polydimethylsiloxane (PDMS) (Sylgard 184; Dow Corning, Midland, MI) microfluidic chambers were created from silicon wafer masters on which negative photoresist (SU-8 50; MicroChem, Newton, MA) was patterned using standard lithographic methods (31). For preparation of the chambers, unpolymerized PDMS was poured over the SU-8 and cured at 60°C for 1 h. Spacers, as depicted in Fig. 2, were used to maintain the thickness control of the overall chamber dimensions and to create an air chamber above the microfluidic channels. Following curing, the PDMS device was released from the photoresist wafer master. Openings were bored through the PDMS at the patterned channel ends, which were later used to fit supply tubing. Prior to assembly of the chamber, the PDMS replica and a glass coverslip (no. 11/2, 22 by 40 mm; Corning) were exposed to an air plasma at 30 W for 2 min (3). To provide additional stability to the otherwise flexible PDMS chamber, the underside of the chamber was similarly plasma sealed to a 51- by 76-mm microscope slide previously bored with circular openings to be aligned over the inlet/outlet positions. Silicone rubber tubing (5.1-mm outside diameter, 2.1-mm inside diameter, 0.8-mm wall; Cole-Parmer, Vernon Hills, IL) was inserted into each opening of the PDMS replica and sealed with unpolymerized PDMS, cured, and reinforced with Torr Seal (Varian, Inc., Lexington, MA). The tubing was then connected to the barbed end of plastic luer connectors (Cole-Parmer) that were in turn connected to a waste reservoir or to a 5-ml gastight syringe (Hamilton Company, Reno, NV) controlled with a syringe pump (Pump 11 Pico Plus; Harvard Apparatus, Holliston, MA).
FIG. 2.
Fabrication process and mounting of the microfluidic chamber (100 μm deep, 100 μm wide, and 14 cm long). Si, silicon.
Growth of X. fastidiosa in microfluidic chambers.
Modified PW medium (without phenol red and without bovine serum albumin) was flushed through the flow chamber via the syringe pump, followed by the perfusion of X. fastidiosa cells. The flow was arrested until cells bound to ∼1% of the area of chamber walls. Unbound cells were flushed from the chamber, after which the flow was decreased to the desired rate prior to image capture.
Light microscopy.
Temporal and spatial studies of bacterial attachment and migration via twitching were conducted in the microfluidic chambers mounted on an inverted microscope using 40× phase-contrast optics. Time-lapse images were recorded with a SPOT-RT digital camera (Diagnostic Instruments, Inc., Sterling Heights, MI) controlled by MetaMorph image software (Universal Imaging, Downington, PA). The migration displacement of X. fastidiosa cells exhibiting twitching motility was measured by mapping cell positions in Cartesian coordinates with respect to time, with the origin being the starting position of the cell. The frame-by-frame velocities of the twitching cells were measured by calculating the centroid position of the cell at each time interval. All experiments were conducted at room temperature (23 ± 2°C).
Electron microscopy.
Specimens were prepared for scanning electron microscopy by chemical fixation in 1% aqueous osmium tetroxide at 4°C for 18 h, after which they were rinsed, dehydrated, critical-point dried, and sputter coated with gold. For transmission electron microscopy, cells were obtained from either the peripheral fringe of 2-day-old colonies or by scraping cells from the agar surface spread with cells 24 h earlier. Cells deposited on Formvar-coated grids were negatively stained with phosphotungstic acid. Pilus lengths were determined from digitized micrograph negatives by using the MetaMorph image analysis software.
Basipetal movement in plants.
Grape plants (Vitis vinifera cv. Chardonnay) were needle inoculated (9) at the seventh internode from the shoot apex with cell suspensions of wild-type and mutant (1A2, 5A7, and 6E11) X. fastidiosa cells in a succinate-citrate-phosphate buffer, pH 7.0 (10). Plants were maintained on a cycle of 12 hours of light and 12 hours of darkness at 28°C in a greenhouse. After 11 weeks, vines were cut from the main trunk and surface sterilized, and 1-cm sections were aseptically excised at measured distances basipetally from the original point of inoculation. The 1-cm sections were crushed in sterile polycarbonate bags containing 100 μl of sterile distilled water. The triturate was spread onto PW agar (for wild-type X. fastidiosa) or PW agar containing kanamycin (for mutant X. fastidiosa). Following 6 days at 28°C, the plates were examined for the presence of X. fastidiosa. PCR was used to confirm the identities of the isolates. In addition, the twitching phenotype was confirmed microscopically. To determine an approximate maximum distance for basipetal transport due to cavitation of the grape sap column at the onset of the incubation period, 0.2-μm-diameter fluorescent beads (Molecular Probes, Inc., Eugene, OR) were similarly introduced into grape shoots, which were subsequently cross-sectioned and examined for the presence of the beads.
RESULTS
X. fastidiosa colony morphology indicative of twitching motility.
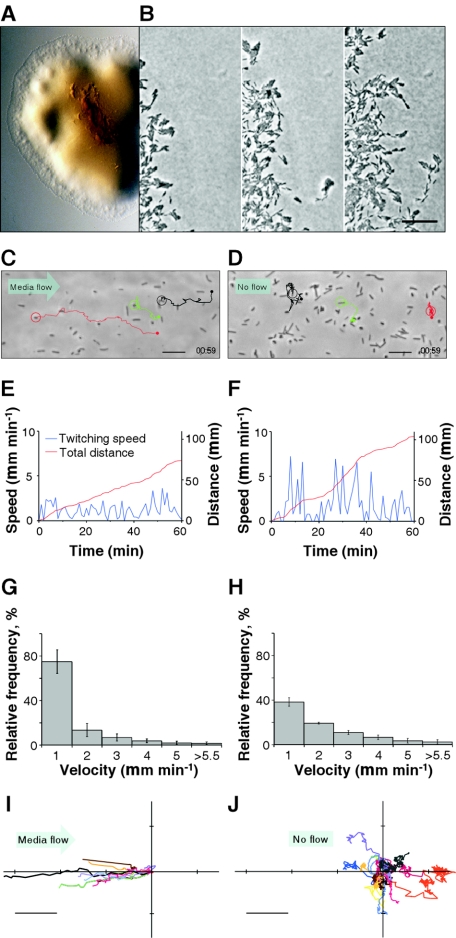
Xylella fastidiosa exhibited a colony morphology consistent with twitching motility, viz., a fringed margin (Fig. 3A) that is observed in other bacterial species (2, 6, 11, 12, 24). Microscopic examination of the fringed margin by using time-lapse imaging methods revealed that both individual bacteria and aggregates of bacteria migrated between 0.01 and 0.32 μm min−1, generally in a direction away from the colony periphery (Fig. 3B; see movie 1A in the supplemental material; also see expanded versions and additional movies at http://www.nysaes.cornell.edu/pp/faculty/hoch/movies/).
FIG. 3.
Twitching motility characteristics of wild-type X. fastidiosa. (A) Light micrograph of a colony with a peripheral fringe. (B) Sequential images (0 min, 7 h 18 min, and 14 h 20 min, left to right) depicting progressive migration of twitching cells (see movie 1A in the supplemental material; also see expanded versions and additional movies at http://www.nysaes.cornell.edu/pp/faculty/hoch/movies/). Bar, 20 μm. (C) Light micrograph depicting paths of three bacteria (circled) twitching against a flow of 20,000 μm min−1 (see movie 2 in the supplemental material and the expanded versions and additional movies at the above-cited website). Bar, 10 μm. (D) Similar depiction as in panel C, but with no flow (see movie 3 in the supplemental material and the expanded versions and additional movies at the above-cited website). Bar, 10 μm. (E and F) Twitching speed and cumulative distance traveled by the red-circled cells in panels C and D, respectively. (G and H) Histograms depicting twitching speeds for cells under flow (20,000 μm min−1) (G) and no-flow (H) conditions. Error bars represent standard errors of the mean (n = 17). (I and J) Twitching paths (starting at coordinates 0,0) measured in a 1-h period for cells under conditions of flow (20,000 μm min−1) (I) and no flow (J). Bar, 20 μm.
Upstream twitching of X. fastidiosa in a microfluidic chamber.
In the microfluidic chamber, the cells were either prostrate on the substratum or erect and attached at one pole once they bound to the surface (Fig. 3C and D and 4A; see movies 2 and 3 in the supplemental material and the expanded versions and additional movies at the above-noted website). Twitching movements propelled individual cells in various directions depending on the rate and direction of medium flow (Fig. 3C to J). Under stagnant no-flow conditions, the path of bacterial movement was confined to relatively small regions with no directional preference for migration (Fig. 3D and J). At a flow velocity of 20,000 μm min−1, a rate comparable to grapevine xylem sap flow under high transpiration conditions (21), the bacteria migrated predominantly against the direction of flow (Fig. 3C and I). Bacteria that moved passively with the flow were either free-floating or intermittently attached to the substratum. The maximum twitching velocity for X. fastidiosa cells under no-flow conditions was 25.3 ± 3.5 μm min−1 (average ± standard error, n = 17) and randomly oriented, whereas under rapid-flow conditions it was 4.9 ± 1.1 μm min−1 (average ± standard error, n = 17) and predominantly upstream. While the cumulative distance traveled by twitching X. fastidiosa cells was greater under no-flow conditions (∼100 μm h−1) than under rapid-flow conditions (∼70 μm h−1) (Fig. 3E and F), the total displacement (from point of origin to final cell position) by actively migrating cells was always greater under flow conditions (Fig. 3I and J). Cells migrating under rapid-flow conditions spent significantly more time (75%) traveling at speeds of ≤1.5 μm min−1 (Fig. 3G), whereas cells under no-flow conditions spent only 40% of the time traveling at this rate (Fig. 3H). This can be attributed to the increase in resistance to movement at higher fluid flows.
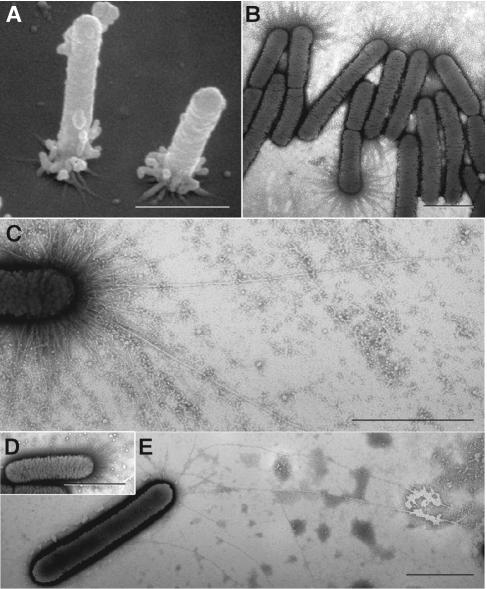
FIG. 4.
Pili of wild-type and mutant X. fastidiosa. (A) Scanning electron micrograph of wild-type cells attached to the substratum at the pilus-bearing polar ends. Many pili degenerated into globular masses during specimen preparation. (B and C) Transmission electron micrographs of negatively stained (phosphotungstic acid) wild-type cells depicting an abundance of short pili (B), and fewer long type IV pili (C). (D and E) Negatively stained preparations of mutants 1A2 and 6E11 depicting only short pili and only longer (type IV) pili, respectively. Pilus lengths are noted in Fig. 8. Bars, 1 μm.
pilB and pilQ mutants.
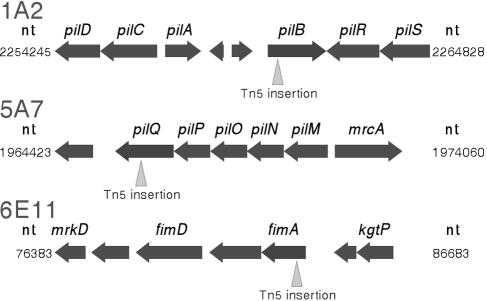
Twitch-minus mutants were generated to facilitate the identification of genes associated with the underlying twitching mechanism. Twitching mutants were selected by observing bacterial colonies for the presence or absence of a twitching-associated peripheral fringe (6, 11, 12, 24). A number of twitch-minus mutants were identified, two of which (1A2 and 5A7) are reported in this paper. Colony peripheries of 1A2 and 5A7 were well demarcated without a peripheral fringe (and without bacteria being distinctly separated from the main colony mass) (Fig. 5). Colony expansion for these two mutants occurred through repeated cell division and gradual spread as the cell mass increased (see movie 1B in the supplemental material; also see expanded versions and additional movies at http://www.nysaes.cornell.edu/pp/faculty/hoch/movies/). When we examined mutants 1A2 (see movie 4 in the supplemental material and the expanded versions and additional movies at the above-cited website) and 5A7 (not shown) in the microfluidic chambers, no cell migration occurred with or without medium flow. Sequence analysis of mutants 1A2 and 5A7 indicated that transposon insertion occurred in open reading frames PD1927 and PD1691 of the X. fastidiosa genome, corresponding to orthologs of pilB and pilQ of Escherichia coli, respectively (Fig. 6), genes that are involved in the formation of type IV pili (1, 28, 29).
FIG. 5.
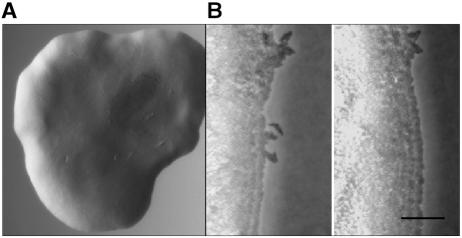
Xylella fastidiosa mutant characteristics. (A) Light micrograph of X. fastidiosa mutant 1A2 colony on agar medium with a well-demarcated fringeless periphery. (B) Sequential images (0 min and 20 h 19 min, respectively) depicting progressive expansion of the 1A2 colony (see movie 1B in the supplemental material; also see expanded versions and additional movies at http://www.nysaes.cornell.edu/pp/faculty/hoch/movies/). Bar, 20 μm.
FIG. 6.
Genomic organization of genes flanking Tn5 insertions in X. fastidiosa mutants 1A2, 5A7, and 6E11. nt, nucleotide.
fimA mutant.
From the initial screen of mutants, one (6E11) which retained the peripheral colony fringe and exhibited active twitching mobility was subsequently found to be biofilm deficient (see below). Sequence analysis indicated that transposon insertion occurred in open reading frame PD0062, corresponding to the fimA gene of E. coli (Fig. 6).
Biofilm formation.
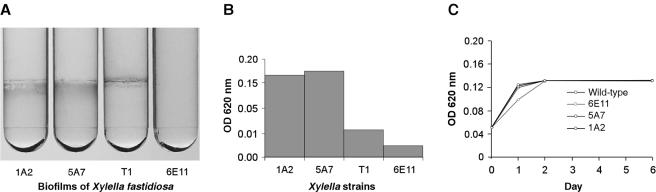
Since type IV pili have been implicated in biofilm development (18) and the ability of X. fastidiosa to cause disease, we sought to characterize biofilm formation in the wild type and mutants. Both mutants 1A2 and 5A7 had greater biofilm production than the wild-type strain (Fig. 7A and B). Mutant 6E11, which retained the peripheral-colony fringe and twitching motility phenotypes (see movie 5 at http://www.nysaes.cornell.edu/pp/faculty/hoch/movies/), was biofilm deficient (Fig. 7A and B). All three X. fastidiosa mutants (1A2, 5A7, and 6E11) grew at a rate similar to that of the wild type (Fig. 7C).
FIG. 7.
Biofilm production and growth curves of the X. fastidiosa wild type and mutants. (A) Biofilm formation by the X. fastidiosa wild type and mutants 1A2, 5A7, and 6E11 following 7-day growth at 28°C. Biofilms were stained with 0.1% crystal violet. (B) The crystal violet stain was subsequently removed from the tubes with dimethyl sulfoxide and quantified for optical density at 620 nm. OD, optical density. (C) Growth curves for the X. fastidiosa wild type and mutants 1A2, 5A7, and 6E11 in PW medium at 28°C.
Two classes of pili in X. fastidiosa.
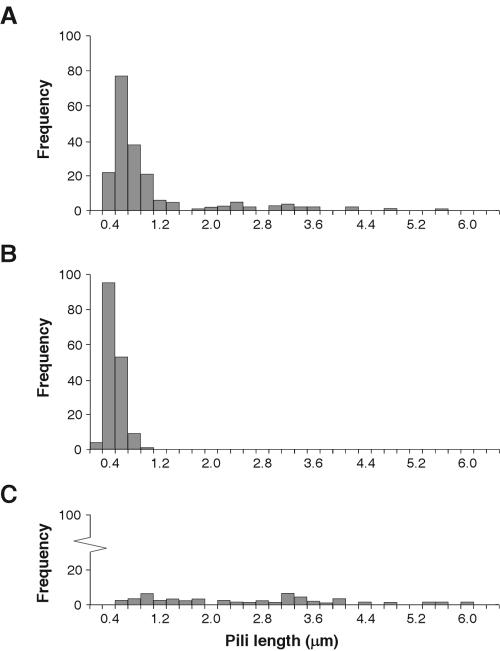
We examined wild-type and mutant X. fastidiosa cells for the presence of pili. Scanning and transmission electron microscopy revealed major differences between wild-type and mutant X. fastidiosa cells with regard to the type of pili present (Fig. 4), which partly explains the observed differences in motility and biofilm development. Negative staining of transmission electron microscopy preparations of the wild-type strain revealed an abundance of pili, the majority of which were 0.4 to 1.0 μm in length with many additional filaments of 1.0 to 5.8 μm in length (Fig. 4B and C and 8A). Mutants 1A2 and 5A7 had only the shorter class of pili (Fig. 4D and 8A), whereas mutant 6E11 had predominantly long pili (Fig. 4E and 8C).
FIG. 8.
Pilus measurement in X. fastidiosa. Histograms of pilus length distributions for the wild type (A) and mutants 1A2 (B) and 6E11 (C). Number of cells represented were 11 (A), 12 (B), and 14 (C).
Inhibition of basipetal movement in planta.
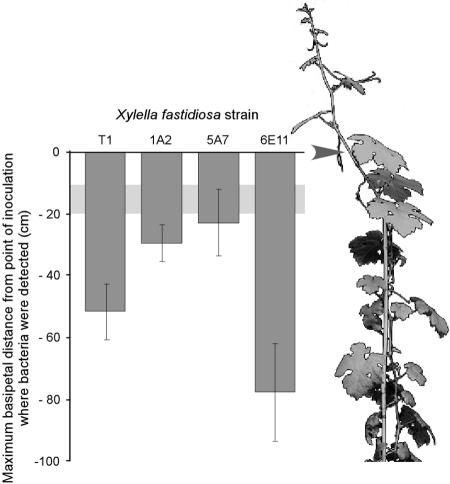
To further test the hypothesis that twitching motility may be involved in the basipetal translocation of X. fastidiosa in planta, we inoculated grapevines with X. fastidiosa. Wild-type bacteria and the 6E11 mutant were recovered from grapevine sections that were considerably more upstream from the sites of inoculation and higher than in the nontwitching mutants 1A2 and 5A7 (Fig. 9). All strains were recovered from vine regions within 2 to 3 cm of the apex (data not shown). Fluorescent latex beads similarly introduced into grapevines were observed in xylem vessels 10 to 20 cm upstream of the introduction sites after 2 h, indicative of passive transport following cavitation of the xylem water column.
FIG. 9.
Basipetal translocation of X. fastidiosa in planta. Maximum distances for wild-type and mutant X. fastidiosa cells were recovered from grapevine regions upstream of the inoculation sites (represented by 0 on the y axis; arrow of illustrated vine) after 11 weeks. The light-gray horizontal band is the zone representing the maximum distance that 0.2-μm fluorescent latex beads traveled passively. Error bars represent standard errors of the means.
DISCUSSION
Using microfluidic chambers, we observed unmistakable long-distance migration of individual as well as small aggregates of X. fastidiosa cells. The fact that migration is directionally controlled, viz., against rapidly flowing currents of growth medium, is particularly interesting and unique. Such directional migration of nonflagellated bacteria helps explain not only how X. fastidiosa cells might colonize grape xylem vessels upstream from the initial site of infection, thus enhancing intraplant spread of the bacteria, but also how other bacterial pathogens may migrate in plant vascular systems. For example, Ralstonia solanacearum, the causal agent of numerous vascular wilt diseases of plants, may possess a similar twitching-mediated migration mechanism since it was previously shown to exhibit colony morphologies characteristic of twitching behavior (12).
What induces X. fastidiosa to migrate upstream in a flowing medium? We hypothesize that twitching motility against a stream is related to the physical forces of flow. Accordingly, X. fastidiosa cells attach to the substratum via the short class of pili located predominately at one pole (Fig. 4A and B), and under flow conditions, the tethered cells tilt downstream, thus angling the attached pole toward the upstream direction. Since the majority of the type IV pili are extruded from a symmetrical region at this pole through PilQ-mediated and -gated pores, most will “reach” forward and anchor to the substratum in the upstream regions. Few of these type IV pili will extend to downstream regions. As the pili are retracted, the cells will move in an incremental step upstream. The direction of flow will keep the cells more or less slanted downstream; thus, cell movement will be oriented directly upstream. With repeated extension and retraction of additional pili, such twitching motility will culminate in long-distance upstream travel. Under no-flow conditions, we predict that the type IV pili will be extruded and attach in random directions from the attached position of the cell, resulting in nonoriented twitching motility. If the directional migration facilitated by type IV pili observed in X. fastidiosa cells is due to the physical forces of flow, then such directional movements would also occur with other type IV piliated, rod-shaped bacteria, such as P. aeruginosa and Vibrio cholerae (14, 20). In fact, recent observations in our laboratory with a nonflagellated strain of P. aeruginosa (26) indicate that they, too, migrate preferentially against the flow of growth medium in the microfluidic channels (unpublished data; see additional movies at http://www.nysaes.cornell.edu/pp/faculty/hoch/movies/).
In the microfluidic channel with mean flow velocities of 20,000 μm min−1, X. fastidiosa cells migrated against the direction of flow at ca. 5 μm min−1. While 20,000 μm min−1 is the mean flow velocity for the microfluidic channel, the actual flow velocity 1 to 2 micrometers away from the channel wall in the zone affecting the attached bacteria, based on a Reynolds number of 0.03, is between 1,584 and 3,136 μm min−1, which is equivalent to a shear force of about 82 fN. Pilus-induced tension in N. gonorrhoeae was previously shown to be about 110 pN (13), which greatly exceeds the shear force calculated above. If type IV pili are of comparable strengths in all bacteria, it would not be surprising to find that X. fastidiosa is able to migrate upstream against the flow rates used in this study. In the present system, 20,000 μm min−1 represents a continuous mean flow velocity, and although this rate is within the range of flow for xylem sap in grapevines under high transpiration conditions (21), it would not be sustained through diurnal cycles where transpiration would fluctuate. Furthermore, the rate of long-distance migration of X. fastidiosa is unimpeded in our artificial microfluidic system, whereas in planta, xylem pit membranes and other xylem vessel obstacles would conceivably lower the rate of spread.
Under no-flow conditions in the microfluidic channels, X. fastidiosa cells exhibited twitching motilities of ca. 25 μm min−1, but unlike under flow conditions, total twitching distance and displacement were limited, and the direction of travel was randomly oriented (Fig. 3D, F, and J). This twitching speed is comparable to that of other twitching bacteria, e.g., P. aeruginosa, under no-flow conditions, whose twitching speed has been observed to be ca. 30 μm min−1 (26).
In this study, we demonstrated that X. fastidiosa cells possess two morphologically distinct classes of pili, shorter 0.4- to 1.0-μm pili and longer 1.0- to 5.8-μm pili (Fig. 4B and C and 8). We believe that the longer pili are equivalent to type IV pili based on their length, their polar position, and the fact that disruption of the pilB and pilQ genes in X. fastidiosa resulted in cells that bore only the short pili and did not twitch (Fig. 4E). The gene products PilB and PilQ have been well studied in a number of other bacterial species. PilB is known to function as a nucleotide binding protein supplying energy for pilin subunit translocation and assembly, whereas PilQ is a multimeric outer membrane protein that forms gated pores, through which the pilus is extruded (1, 28, 29). Mutants deficient in these proteins have smooth-colony edge phenotypes, do not twitch, and are generally devoid of type IV pili (1, 11, 12, 28). Genomic data indicate that the fimA gene ortholog in X. fastidiosa (5) and the fimA gene in E. coli (17) code for an essential type I pilus protein that functions in surface attachment and biofilm formation. The presence of both long and short pili on the wild-type X. fastidiosa strain, the occurrence of only long pili on the twitching, biofilm-deficient mutant (6E11), and the absence of long pili on the twitch-minus, biofilm-enhanced mutants (1A2 and 5A7) clearly indicate distinct functional roles for the two length classes of pili. Moreover, the correlation between enhanced biofilm formation in mutants with only short pili (1A2 and 5A7) and decreased biofilm formation in the mutant with no short pili (6E11) strongly suggests that short pili are important in surface attachment and biofilm development. Furthermore, disruption of the fimA gene in X. fastidiosa mutant 6E11 resulted in cells bearing only long pili (Fig. 4E). Mutant 6E11 is motile at a rate not significantly different than that for the wild-type strain (data not shown).
We inoculated grapevines with wild-type and mutant X. fastidiosa cells to confirm in vitro observations with pilus-mediated basipetal movement as it might occur in planta. Translocation of X. fastidiosa from inoculation sites has been documented mostly quantitatively for acropetal movement through destructive sampling and culturing techniques, as well as through histological examination (16, 22, 27). Quantitative data on basipetal, i.e., upstream, movement of this pathogen are not readily available. Initial translocation of X. fastidiosa artificially inoculated into grapevines by using the “needle technique” is complicated by the fact that embolism of the xylem water column usually accompanies the procedure and transports bacteria both basipetally and acropetally from the inoculation site. In the present study, fluorescent latex beads were used to obtain an estimate of the relative distance these beads were carried by the cavitated water column. This distance was assumed to be representative of the initial distance bacteria would have been transported following embolism and was used as the starting point in evaluating subsequent migration of bacteria. Only the bacteria which possess longer type IV pili (i.e., the wild type and the 6E11 mutant) were able to migrate substantial distances basipetally (Fig. 9). Calculations made from these distances after 11 weeks reveal that they migrated at estimated speeds of 3 to 5 μm min−1, a rate within the 0.1- to 25-μm min−1 range observed in vitro. Furthermore, mutant 6E11 exhibited migration distances greater than that of the wild type, which may be due to the absence of the shorter type I pili, thus suggesting their role in adhesion (5).
Our results present the first direct observation of twitching motility by a plant-pathogenic bacterium. This phenomenon may very well explain how X. fastidiosa and other nonflagellated gram-negative xylem-limited bacteria are able to spread within plants against the prevailing direction of xylem sap flow to previously noninvaded regions. These findings on twitching-mediated motility present a newly observed phenomenon for cell migration in fluid systems and should be applicable to future studies of the X. fastidiosa-grape system as well as in other bacterial-plant and bacterial-mammalian systems.
Supplementary Material
Acknowledgments
We extend our thanks to A. Stroock for his helpful suggestions concerning microfluidics and to S. Lindow and K. Newman for their advice and suggestions regarding X. fastidiosa biology during the course of this investigation. We also thank H. Berg for supplying the nonflagellated P. aeruginosa strain (PAKfliC::Gmr).
The research was supported, in part, by grants from the Nanobiotechnology Center (NBTC), an STC program of the National Science Foundation under agreement no. ECS-9876771, and from the USDA/CSREES administered through the University of California Pierce's Disease Research Grants Program. This work was also performed, in part, at the Cornell Nanofabrication Facility (a member of the National Nanofabrication Users Network) which is supported by the National Science Foundation under grant ECS-9731293, its users, Cornell University, and Industrial Affiliates.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Alm, R. A., and J. S. Mattick. 1997. Genes involved in the biogenesis and function of type-4 fimbriae in Pseudomonas aeruginosa. Gene 192:89-98. [DOI] [PubMed] [Google Scholar]
- 2.Bradley, D. E. 1980. A function of Pseudomonas aeruginosa PAO polar pili: twitching motility. Can. J. Microbiol. 26:146-154. [DOI] [PubMed] [Google Scholar]
- 3.Chaudhury, M. K., and G. M. Whitesides. 1991. Direct measurement of interfacial interactions between semispherical lenses and flat sheets of poly(dimethylsiloxane) and their chemical derivatives. Langmuir 7:1013-1025. [Google Scholar]
- 4.Davis, M. J., W. J. French, and N. W. Schaad. 1981. Axenic culture of the bacteria associated with phony disease of peach and plum leaf scald. Curr. Microbiol. 6:309-314. [Google Scholar]
- 5.Feil, H., W. S. Feil, J. C. Detter, A. H. Purcell, and S. E. Lindow. 2003. Site-directed disruption of the fimA and fimF genes of Xylella fastidiosa. Phytopathology 93:675-682. [DOI] [PubMed] [Google Scholar]
- 6.Glessner, A., R. S. Smith, B. H. Iglewski, and J. B. Robinson. 1999. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of twitching motility. J. Bacteriol. 181:1623-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guilhabert, M. R., L. M. Hoffman, D. A. Mills, and B. C. Kirkpatrick. 2001. Transposon mutagenesis of Xylella fastidiosa by electroporation of Tn5 synaptic complexes. Mol. Plant-Microbe Interact. 14:701-706. [DOI] [PubMed] [Google Scholar]
- 8.Henrichsen, J. 1983. Twitching motility. Annu. Rev. Microbiol. 37:81-93. [DOI] [PubMed] [Google Scholar]
- 9.Hill, B. L., and A. H. Purcell. 1995. Multiplication and movement of Xylella-fastidiosa within grapevine and 4 other plants. Phytopathology 85:1368-1372. [Google Scholar]
- 10.Hopkins, D. L. 1984. Variability of virulence in grapevine among isolates of Pierce's disease bacterium. Phytopathology 74:1395-1398. [Google Scholar]
- 11.Huang, B., C. B. Whitchurch, and J. S. Mattick. 2003. FimX, a multidomain protein connecting environmental signals to twitching motility in Pseudomonas aeruginosa. J. Bacteriol. 185:7068-7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu, H. L., Y. W. Kang, S. Genin, M. A. Schell, and T. P. Denny. 2001. Twitching motility of Ralstonia solanacearum requires a type IV pilus system. Microbiology 147:3215-3229. [DOI] [PubMed] [Google Scholar]
- 13.Maier, B., M. Koomey, and M. P. Sheetz. 2004. A force-dependent switch reverses type IV pilus retraction. Proc. Natl. Acad. Sci. USA 101:10961-10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mattick, J. S. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56:289-314. [DOI] [PubMed] [Google Scholar]
- 15.Newman, K. L., R. P. P. Almeida, A. H. Purcell, and S. E. Lindow. 2004. Cell-cell signaling controls Xylella fastidiosa interactions with both insects and plants. Proc. Natl. Acad. Sci. USA 101:1737-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newman, K. L., R. P. P. Almeida, A. H. Purcell, and S. E. Lindow. 2003. Use of a green fluorescent strain for analysis of Xylella fastidiosa colonization of Vitis vinifera. Appl. Environ. Microbiol. 69:7319-7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orndorff, P. E., A. Devapali, S. Palestrant, A. Wyse, M. L. Everett, R. R. Bollinger, and W. Parker. 2004. Immunoglobulin-mediated agglutination of and biofilm formation by Escherichia coli K-12 require the type 1 pilus fiber. Infect. Immun. 72:1929-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 19.O'Toole, G. A., L. A. Pratt, P. I. Watnick, D. K. Newman, V. B. Weaver, and R. Kolter. 1999. Genetic approaches to study biofilms. Methods Enzymol. 310:91-109. [DOI] [PubMed] [Google Scholar]
- 20.Ottow, J. C. G. 1975. Ecology, physiology, and genetics of fimbriae and pili. Annu. Rev. Microbiol. 29:79-108. [DOI] [PubMed] [Google Scholar]
- 21.Peuke, A. D. 2000. The chemical composition of xylem sap in Vitis vinifera L. cv. Riesling during vegetative growth on three different franconian vineyard soils and as influenced by nitrogen fertilizer. Am. J. Enol. Vitic. 51:329-339. [Google Scholar]
- 22.Purcell, A. H., and D. L. Hopkins. 1996. Fastidious xylem-limited bacterial plant pathogens. Annu. Rev. Phytopathol. 34:131-151. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook, J., and D. W. Russel. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Semmler, A. B. T., C. B. Whitchurch, and J. S. Mattick. 1999. A re-examination of twitching motility in Pseudomonas aeruginosa. Microbiology 145:2863-2873. [DOI] [PubMed] [Google Scholar]
- 25.Simpson, A. J. G., F. C. Reinach, P. Arruda, F. A. Abreu, M. Acencio, R. Alvarenga, L. M. C. Alves, J. E. Araya, G. S. Baia, C. S. Baptista, M. H. Barros, E. D. Bonaccorsi, S. Bordin, J. M. Bove, M. R. S. Briones, M. R. P. Bueno, A. A. Camargo, L. E. A. Camargo, D. M. Carraro, H. Carrer, N. B. Colauto, C. Colombo, F. F. Costa, M. C. R. Costa, C. M. Costa-Neto, L. L. Coutinho, M. Cristofani, E. Dias-Neto, C. Docena, H. El-Dorry, A. P. Facincani, A. J. S. Ferreira, V. C. A. Ferreira, J. A. Ferro, J. S. Fraga, S. C. Franca, M. C. Franco, M. Frohme, L. R. Furlan, M. Garnier, G. H. Goldman, M. H. S. Goldman, S. L. Gomes, A. Gruber, P. L. Ho, J. D. Hoheisel, M. L. Junqueira, E. L. Kemper, J. P. Kitajima, J. E. Krieger, E. E. Kuramae, F. Laigret, M. R. Lambais, L. C. C. Leite, E. G. M. Lemos, M. V. F. Lemos, S. A. Lopes, C. R. Lopes, J. A. Machado, M. A. Machado, A. Madeira, H. M. F. Madeira, C. L. Marino, M. V. Marques, E. A. L. Martins, E. M. F. Martins, A. Y. Matsukuma, C. F. M. Menck, E. C. Miracca, C. Y. Miyaki, C. B. Monteiro-Vitorello, D. H. Moon, M. A. Nagai, A. Nascimento, L. E. S. Netto, A. Nhani, F. G. Nobrega, L. R. Nunes, M. A. Oliveira, M. C. de Oliveira, R. C. de Oliveira, D. A. Palmieri, A. Paris, B. R. Peixoto, G. A. G. Pereira, H. A. Pereira, J. B. Pesquero, R. B. Quaggio, P. G. Roberto, V. Rodrigues, A. J. D. Rosa, V. E. de Rosa, R. G. de Sa, R. V. Santelli, H. E. Sawasaki, A. C. R. da Silva, A. M. da Silva, F. R. da Silva, W. A. Silva, J. F. da Silveira, et al. 2000. The genome sequence of the plant pathogen Xylella fastidiosa. Nature 406:151-157. [DOI] [PubMed] [Google Scholar]
- 26.Skerker, J. M., and H. C. Berg. 2001. Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. USA 98:6901-6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stevenson, J. F., M. A. Matthews, L. C. Greve, J. M. Labavitch, and T. L. Rost. 2004. Grapevine susceptibility to Pierce's disease II: the progression of anatomical symptoms. Am. J. Enol. Vitic. 55:238-245. [Google Scholar]
- 28.Strom, M. S., and S. Lory. 1993. Structure-function and biogenesis of the type IV pili. Annu. Rev. Microbiol. 47:565-596. [DOI] [PubMed] [Google Scholar]
- 29.Wall, D., and D. Kaiser. 1999. Type IV pili and cell motility. Mol. Microbiol. 32:1-10. [DOI] [PubMed] [Google Scholar]
- 30.Wells, J. M., B. C. Raju, H.-Y. Hung, W. G. Weisburg, L. Mandelco-Paul, and D. J. Brenner. 1987. Xylella fastidiosa gen. nov., sp. nov.: gram-negative, xylem-limited, fastidious plant bacteria related to Xanthomonas spp. Int. J. Syst. Bacteriol. 37:136-143. [Google Scholar]
- 31.Xia, Y. N., and G. M. Whitesides. 1998. Soft lithography. Annu. Rev. Mater. Sci. 28:153-184. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.