Abstract
Although many stress response genes have been characterized in Oenococcus oeni, little is known about the regulation of stress response in this malolactic bacterium. The expression of eubacterial stress genes is controlled both positively and negatively at the transcriptional level. Overall, negative regulation of heat shock genes appears to be more widespread among gram-positive bacteria. We recently identified an ortholog of the ctsR gene in O. oeni. In Bacillus subtilis, CtsR negatively regulates expression of the clp genes, which belong to the class III family of heat shock genes. The ctsR gene of O. oeni is cotranscribed with the downstream clpC gene. Sequence analysis of the O. oeni IOB 8413 (ATCC BAA-1163) genome revealed the presence of potential CtsR operator sites upstream from most of the major molecular chaperone genes, including the clp genes and the groES and dnaK operons. Using B. subtilis as a heterologous host, CtsR-dependent regulation of O. oeni molecular chaperone genes was demonstrated with transcriptional fusions. No alternative sigma factors appear to be encoded by the O. oeni IOB 8413 (ATCC BAA-1163) genome. Moreover, apart from CtsR, no known genes encoding regulators of stress response, such as HrcA, could be identified in this genome. Unlike the multiple regulatory mechanisms of stress response described in many closely related gram-positive bacteria, this is the first example where dnaK and groESL are controlled by CtsR but not by HrcA.
In order to respond to stressful situations, all organisms have developed adaptive networks, including stress responses. Organisms undergo complex programs of differential gene expression, involving a rapid increase in the concentrations of specific sets of proteins, such as heat shock proteins (HSPs). Some HSPs, such as GroEL, DnaK, small HSP, and several Clp ATPases, are molecular chaperones that facilitate the proper folding of cellular proteins. Others, such as the Clp ATP-dependent protease, degrade incorrectly folded proteins (25). In Escherichia coli, HSP synthesis is controlled at the transcriptional level by two alternative sigma factors: σ32 and σ24 (58). In contrast to E. coli, the heat shock response in the model gram-positive bacterium Bacillus subtilis involves at least three different classes of heat-inducible genes distinguished by their regulatory mechanisms (28). Genes of class I (groESL and dnaK) are regulated by the HrcA repressor, which binds to the palindromic operator sequence CIRCE (for “controlling inverted repeat of chaperone expression”). Expression of class II heat shock genes requires the alternative sigma factor σB (43, 44). Class III genes are controlled by the class three stress gene repressor, CtsR, which recognizes a tandemly repeated heptad operator sequence (16). Class IV genes comprise heat shock genes of unknown regulation, suggesting the existence of other thermoregulatory mechanisms. However, the model organism B. subtilis does not reflect all of the mechanisms of stress response regulation in gram-positive bacteria. Thus, in Streptomyces albus, a mycelium-forming gram-positive soil bacterium, two negative regulators in addition to HrcA, HspR (for “heat shock protein repressor”) and RheA (for “repressor of hsp18”), control HSP synthesis. The HspR repressor binds to an inverted-repeat sequence called HAIR (for “HspR-associated inverted repeat”) and represses the dnaK operon (27). The hsp18 gene is controlled by the negative regulator RheA, which binds to an inverted-repeat sequence (53). The groESL operon is controlled by HrcA (26). Although the induction of hsp genes is a universal response, organisms have diverse regulatory mechanisms for controlling HSP synthesis. Comparative genomics allows us to predict the regulation of heat shock genes by CtsR and/or HrcA. Thus, the dnaK and groESL operons of the lactic acid bacterium Lactococcus lactis contain CIRCE elements in their promoter regions, suggesting that these genes may be regulated by HrcA, whereas CtsR regulates clp gene expression (56). Some bacteria simultaneously use more than one strategy to ensure the well-adjusted production of heat shock proteins under harsh conditions. For example, heat shock regulation processes mediated by CIRCE and σ32 coexist in some bacteria, including Agrobacterium tumefaciens, Bordetella pertussis, Caulobacter crescentus, and Zymomonas mobilis (2, 3, 20, 39, 40, 45, 46, 50, 51). Dual heat shock regulation by HrcA and CtsR has been demonstrated for the Staphylococcus aureus dnaK and groESL operons and for the Streptococcus salivarius clpP gene (10, 11). These dual regulatory mechanisms are probably not redundant but may act together synergistically to maintain low levels of expression in the absence of stress and to ensure that synthesis of different HSPs is tightly coordinated under adverse environmental conditions.
The lactic acid bacterium Oenococcus oeni, mostly responsible for malolactic fermentation in wine, is able to survive and grow under very harsh conditions. Malolactic fermentation, occurring after the completion of alcoholic fermentation, lowers wine acidity by converting malic acid into lactic acid and improves the taste (18, 36). After alcoholic fermentation, wine is a hostile medium for bacterial growth because of nutritional starvation, low pH, and the presence of sulfites and of high ethanol concentrations. Because of its ability to grow in such a hostile medium, O. oeni is a good model for studies of stress response in lactic acid bacteria. Among O. oeni stress response genes, hsp18, trxA, clpX, and clpP have been previously characterized (4, 30, 31). Expression of these genes was followed during growth and under several stress conditions. All these genes are heat inducible, but differential expression was observed during the growth phase. clpX is preferentially expressed at the beginning of the exponential phase, clpP was expressed during all stages of growth at a high basal level and reached its maximum in the exponential phase, and hsp18 mRNA was detected only at the end of the exponential phase. The trxA gene was expressed during all stages of growth with no significant difference in the level of expression. An understanding of the regulatory mechanisms controlling stress gene expression is therefore essential in studying the ability of O. oeni to survive and grow under unfavorable environmental conditions. The determination of the complete genome sequence of O. oeni strain IOB 8413 (ATCC BAA-1163) was carried out by our laboratory in collaboration with the Laboratoire de Biotechnologie et Microbiologie Appliquée (UMR 1219, INRA-Université Victor Segalen Bordeaux 2), the Centre de Bioinformatique de Bordeaux (CbiB, Université Victor Segalen Bordeaux 2), and GENOME Express (Grenoble, France) (33). The genome assembly of O. oeni currently consists of 33 contigs. This project has revealed numerous gene systems that are likely to be important for our understanding of the physiology of this lactic acid bacterium. Here, we report the identification of a CtsR ortholog in O. oeni. Potential CtsR operator sites were found upstream from the clp genes and the groESL and dnaK operons, and we show that CtsR controls the expression of most of the O. oeni molecular chaperone genes. In contrast to the diversity of stress response mechanisms described in many gram-positive bacteria, no gene encoding an alternative sigma factor or any other known regulator of stress response, such as HrcA, could be identified in the O. oeni IOB 8413 (ATCC BAA-1163) genome. This is the first example of dnaK and groESL operons under the exclusive control of CtsR without dual regulation by both HrcA and CtsR.
MATERIALS AND METHODS
Bacterial strains, growth media, and transformation conditions.
O. oeni strain IOB 8413 (ATCC BAA-1163) was grown at 30°C in FT80 medium (pH 5.3) (9). For stress experiments, cells grown to the mid-exponential phase were shocked for half an hour by transfer to 42°C and addition of ethanol (11% [vol/vol]) or HCl (1 M) to pH 3.6. B. subtilis 168 (trpC2) and QB4991 [trpC2 amyE::(′lacZ aphA3)ΔctsR] (16) were used as heterologous hosts to measure the activities of transcriptional fusions. E. coli ER2738 [F′ lacIq Δ(lacZ)M15 zzf::Tn10(Tetr)/fhuA2 supE thi Δ(lac-proAB) Δ(hsdMS-mcrB)] (New England Biolabs) was used for cloning experiments. E. coli was grown at 37°C in Luria-Bertani (LB) medium. Electroporation was used for E. coli transformation, with selection on LB plates supplemented with ampicillin (100 μg/ml). B. subtilis was grown at 37°C in LB medium and transformed as described previously (42). Transformants were selected on LB plates supplemented with chloramphenicol (5 μg/ml).
RNA extraction and analysis.
RNA extraction was performed using Tri Reagent (Sigma) according to the manufacturer's instructions and 0.4 g of glass beads (70 to 100 μm) to disrupt cells with a FastPrep cell disintegrator (Bio 101, Inc.). Samples were then treated as recommended by the manufacturer and used for Northern blotting, primer extension analysis, reverse transcription-PCR (RT-PCR), or quantitative RT-PCR (QRT-PCR) experiments. Northern blotting was carried out as described by Sambrook et al. (48). A DNA fragment corresponding to the O. oeni ctsR gene was amplified by PCR using oligonucleotides olcg1 and ctsR1 (Table 1). This fragment was radiolabeled with [α-32P] dATP (Perkin-Elmer) using a random-primer DNA-labeling kit (Invitrogen) and used as a probe in Northern hybridization experiments. Primer extensions were performed by incubating 5 μg of RNA, 20 pmol of oligonucleotide, 92 GBq of [α-32P]dATP (111 TBq/mmol; Perkin-Elmer), and 100 U of SuperScript II reverse transcriptase (Invitrogen). For each primer extension experiment, two oligonucleotides were chosen to hybridize approximately 100 bp downstream from the translation initiation codon (Table 1). The corresponding DNA-sequencing reactions were carried out by using the same oligonucleotides and PCR-amplified DNA fragments carrying the respective promoter regions. RT-PCR and QRT-PCR were performed with O. oeni RNA (2 μg) treated with 2 units of DNase (Invitrogen). cDNAs were synthesized with random hexamers (50 ng) using the Superscript II reverse transcriptase. PCRs were performed using cDNAs with appropriate primers (Table 1). hsp mRNA levels were quantified by QRT-PCR assays using qPCR Mastermix and a SYBR Green I kit (Eurogentec). The ldhD gene (AJ831540) was chosen as an internal control gene (4, 17). Amplifications were performed on a Bio-Rad I-cycler with the SYBR Green system. Thermal-cycling conditions included the following steps: initial denaturation at 95°C for 10 min, followed by 45 cycles at 95°C for 15 s and 60°C for 30 s. Fluorescence measurements were recorded during each annealing step. Four dilutions of cDNAs were performed. The specificity of each primer pair was determined with a melting curve. The efficiency of real-time amplification was determined by running a standard curve with serial dilutions of cDNA. A PCR that amplifies the target sequence with 100% efficiency (E) can double the amount of PCR products in each cycle. The efficiency, E, is calculated by the formula E = [10−(1/s) − 1] × 100, where s is the slope of the standard curve. The results were analyzed using the comparative critical-threshold method, in which the amount of target RNA is adjusted to a reference relative to an internal calibrated target RNA.
TABLE 1.
Oligonucleotides used in this study
| Name | Sequence (5′ → 3′)a | Created restriction site | Plasmid construction and function |
|---|---|---|---|
| Transcriptional fusion constructions | |||
| olcg11 | GGGGAATTCAATTGAGTGACGGACAAAAGAT | EcoRI | pDLctsR |
| olcg12 | GGGGGATCCCGCTTCGTTTAATCTCGGCG | BamHI | pDLctsR and primer extension on ctsR |
| olcg16 | ATCGGCGGATCCTATCAAATACCTCCTATTAACTAA | BamHI | pDLhsp18 |
| olcg20 | GCCCGAATTCTAAATTAATCGAAGCCTTTTGAC | EcoRI | pDLhsp18 |
| olcg3 | GGGGAATTCCCATTGGGGAGAGGCCGGC | EcoRI | pDLgrpE |
| olcg4 | GGGAAGCTTGCTTGGATCCTTTCACTGCT | HindIII | pDLgrpE and primer extension on grpE |
| olcg14 | CCCGAATTCTTTCGTTGATAATTTTGAAAACATT | EcoRI | pDLgroES and pDLgroESmut |
| olcg15 | CCCGGATCCTATGTAAAACCTCCTTTTAATTTG | BamHI | pDLgroES and pDLgroESmut |
| groES4 | CTACGGCTGCAGAAAAGGTCAAACTTTTTGACAAAG | PstI | pDLgroESmut |
| groES5 | CGGACTCTGCAGAATTGCGTTATAATTTTACGCGG | PstI | pDLgroESmut |
| clpE1 | CCCGAATTCGACAATTTGAGAATCTCTGACCA | EcoRI | pDLclpL2 |
| clpE2 | CCCGGATCCCTGAACCGTTATTTGCTTGTTG | BamHI | pDLclpL2, QRT-PCR and primer extension on clpL2 |
| olcg7 | GGGGAATTCCCCGGAGGCCAGTTTAAGC | EcoRI | pDLclpP |
| olcg8 | GGGGGATCCCCTTGAACCAAGATAATTCGG | BamHI | pDLclpP |
| olcg9 | GGGGAATTCGGATGCTTTTGAAGATGCCG | EcoRI | pDLclpX |
| olcg40 | CCCGGATCCTGATTCTACGTCCATGCTTTTT | BamHI | pDLclpX |
| olcg17 | ATCGGCGAATTCATAAGATAATTTTATCTCTTTTATAAG | EcoRI | pDLmleA |
| olcg18 | ATCGGCGGATCCATTTTCTCCTCAAGAACCACAT | BamHI | pDLmleA |
| RNA analysis | |||
| olcg1 | ATGCATGCCATGGCAGAAGCTAATATTTCAG | RT-PCR, QRT-PCR on ctsR and DNA probe | |
| ctsR1 | AAACGGGTGTTGATTACATAATT | RT-PCR, QRT-PCR, primer extension on ctsR and DNA probe | |
| clpCtsR2 | TTTCATTTCTAATATCATCGTCG | RT-PCR on ctsR-clpC | |
| clpE3 | ATTATAATGACGATCCCTTCGT | QRT-PCR on clpL2 | |
| clpE4 | TGTCCAAACGGATAACCTCC | Primer extension on clpL2 | |
| groES2 | TTCCGCCAACCTTTTCTTCTT | Primer extension on groES | |
| groES3 | TAACAGACTTGGGAGCTTTTG | Primer extension on groES | |
| groES6 | GCCACAACAGAACCCATCACTGGTT | QRT-PCR on groES | |
| groES7 | GGCGATCGAATTGTTCTTAGTAT | QRT-PCR on groES | |
| grpE7 | CGCAGGCAGAAAAGAACAATC | QRT-PCR on grpE | |
| grpE8 | GCTGAAGACGAAGCAGTTGC | QRT-PCR and primer extension on grpE | |
| ldhD1 | GCCGCAGTAAAGAACTTGATG | QRT-PCR on ldhD | |
| ldhD2 | TGCCGACAACACCAACTGTTT | QRT-PCR on ldhD |
Specific restriction sites are underlined.
DNA manipulations and analysis.
Molecular biology techniques were carried out using standard methods (48). Double-stranded plasmid DNA was purified with the QIAprep spin miniprep kit (QIAGEN). Nucleotide sequences were determined by the dideoxy chain termination method using the DNA sequencing cycle Reader kit (MBI Fermentas). PCR products and DNA restriction fragments were purified using the QIAquick PCR purification kit (QIAGEN). Restriction endonucleases (Invitrogen), DNA ligase (Invitrogen), and Taq polymerase (Qbiogen) were used according to the manufacturers' specifications.
Plasmids and plasmid constructions.
Plasmid pDL (57) was used to construct transcriptional fusions with the Bacillus stearothermophilus bgaB gene, encoding a thermostable β-galactosidase (29), with subsequent integration at the amyE locus. bgaB transcriptional fusions were constructed using EcoRI-BamHI (or EcoRI-HindIII) fragments generated by PCR using the oligonucleotide pairs olcg17/olcg18, olcg11/olcg12, olcg16/olcg20, olcg9/olcg40, olcg7/olcg8, clpE1/clpE2, olcg3/olcg4, and olcg14/olcg15 (Table 1), corresponding to the O. oeni mleA (X82326), ctsR (AJ890338), hsp18 (AJ250422), clpX (Y15953), clpP (AJ606044), clpL2 (AJ890337), grpE (AJ890339), and groES (AJ890340) promoter regions, respectively. These fragments were digested with EcoRI and BamHI (or HindIII) and inserted into EcoRI- and BamHI (or HindIII)-digested pDL to generate pDLmleA, pDLctsR, pDLhsp18, pDLclpX, pDLclpP, pDLclpL2, pDLgrpE, and pDLgroES, respectively. To examine the effect of site-directed mutagenesis of the CtsR operator sequence on the expression of a groES′-bgaB fusion, a PstI site was introduced between nucleotides −22 and −27 of the groES regulatory region in pDLgroES to generate pDLgroESmut through PCR using the oligonucleotide pairs olcg15/groES4 and groES5/olcg14 (Table 1).
β-Galactosidase assays.
Overnight cultures of B. subtilis grown on LB medium supplemented with chloramphenicol (5 μg/ml) were diluted to an optical density at 600 nm of 0.05 in fresh medium and grown at 37°C until an optical density at 600 nm of 0.5 was reached, after which one-half of the culture was shifted to 48°C. Samples were taken each hour to determine β-galactosidase activity as described previously (41, 42), and the activity was expressed as Miller units per mg cellular protein. Protein concentrations were determined using the Bradford method (Bio-Rad [Richmond, Calif.] reagent) with bovine serum albumin as the standard (8).
Database comparisons and sequence analysis.
Sequence comparisons with the GenBank database were accomplished using the National Center for Biotechnology Information BLAST (1) network service. Multiple alignments were performed with the CLUSTAL W program (13). A systematic search using regular expressions was performed to find CtsR-binding sites (GTCAANNNNGGTC) in the O. oeni genome. O. oeni sequences were from the O. oeni IOB 8413 (ATCC BAA-1163) sequencing project (CONSRTM Laboratoire de Microbiologie-Université de Bourgogne, GENOME Express, Institut National de la Recherche Agronomique [INRA], Laboratoire de Microbiologie et Biotechnologique Appliquée-Faculté d'Oenologie de Bordeaux, and Centre de Bioinformatique de Bordeaux-Université Bordeaux 2). The genome sequence data will be available though a dedicated website.
Nucleotide sequence accession numbers.
The DNA sequence data described in this work have been deposited in GenBank with the following accession numbers: AJ890337 (clpL2), AJ890340 (groES), AJ890339 (grpE), and AJ890338 (ctsR-clpC).
RESULTS
Characterization of a ctsR-like gene in O. oeni.
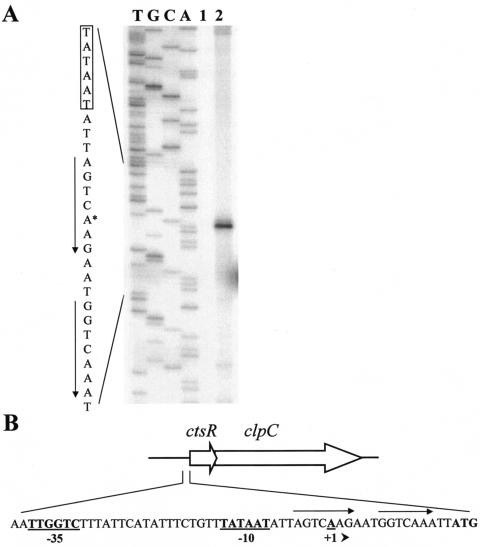
Analysis of the O. oeni IOB 8413 (ATCC BAA-1163) genome revealed a 480-bp open reading frame (ORF) encoding an 18-kDa protein sharing 50% amino acid sequence identity with CtsR of B. subtilis, which negatively regulates the transcription of several stress-regulated genes (16, 28). Thus, this O. oeni ORF was named ctsR. The O. oeni CtsR protein contains the predicted helix-turn-helix DNA binding motif characterized by Derré et al. (16). Analyzing the DNA region downstream of ctsR, we identified a 2.47-kb ORF encoding an 823-amino-acid polypeptide with a predicted molecular mass of 91.5 kDa. The amino acid sequence of this predicted protein has strong amino acid sequence identity with members of the HSP100/Clp ATPase family. The similarity is particularly striking in the ATP binding domains. Comparison with the microbial genome database (http://www.ncbi.nlm.nih.gov/BLAST/) revealed that the O. oeni Clp ATPase shares 58%, 51%, and 50% identical amino acid residues with the ClpC proteins of Enterococcus faecalis (NP 816879), B. subtilis (P37571), and Listeria innocua (NP 469609), respectively, and it was therefore designated ClpC. The stop codon (TAA) of the O. oeni ctsR gene overlaps the probable start site of clpC by 8 nucleotides, as previously described in Lactococcus lactis (56). Downstream of ctsR, we did not find an inverted-repeat structure typical of Rho-independent transcriptional terminators, suggesting that ctsR and clpC are organized as an operon. The 5′-end mRNA of ctsR was determined by primer extension analysis using RNA extracted from an exponentially growing bacterial culture either before or after heat shock treatment (Fig. 1). The transcriptional start site was identified at nucleotide position −15, with reference to the presumed translational start codon. A −35 (TTGGTC) and a −10 (TATAAT) hexamer separated by 18 nucleotides, highly similar to the consensus of O. oeni housekeeping promoters, were identified at an appropriate distance from the transcriptional start site. No extension signal was detected using RNA extracted from cells harvested before stress treatment. These results show that ctsR expression is induced by heat shock. Three nucleotides upstream of the transcriptional start site, we found a 17-bp sequence (AGTCAAGAATGGTCAAA) closely resembling the consensus sequence A/GGTCAAANANA/GGTCAAA of the CtsR-binding site in gram-positive bacteria (16). Expression of O. oeni ctsR was investigated by Northern blotting using a 150-bp ctsR-specific PCR fragment as a probe. A 3.6-kb transcript, likely corresponding to a ctsR and clpC cotranscript, was detected in RNA extracted from the shocked culture (data not shown). Additionally, RT-PCR analysis using appropriate primers confirmed the presence of a ctsR-clpC cotranscript (data not shown).
FIG. 1.
(A) Determination of the transcription initiation site of the ctsR gene by primer extension analysis. Total RNA was extracted from O. oeni cells harvested in the exponential phase before (lane 1) or after (lane 2) a 30-min shift to 42°C. Primer extension products corresponding to the ctsR gene are shown alongside DNA-sequencing reaction products (lanes T, G, C, and A). The corresponding nucleotide sequence is shown on the left. The transcriptional start site is indicated by an asterisk, the −10 sequence is boxed, and arrows indicate the likely CtsR operator sites. (B) Organization of the ctsR-clpC operon and nucleotide sequence of the ctsR promoter region. The putative −10 and −35 sequences are underlined and boldface, arrows indicate the likely CtsR operator sites, and the initiation codon (ATG) is in boldface.
Stress induction of the O. oeni ctsR-clpC operon.
To examine whether expression of ctsR is induced by stress in O. oeni, we exposed exponential-phase cultures to various stress conditions: upshift from 30°C to 42°C, addition of ethanol (11% [vol/vol]), or shift to pH 3.6. Total RNAs were isolated before or 30 min after stress treatment. A quantitative RT-PCR experiment was set up using the constitutive ldhD gene as an internal control (4, 17). Intragenic fragments of ldhD and ctsR were amplified using the ldhD1/ldhD2 and olcg1/ctsR1 oligonucleotide pairs, respectively (Table 1). As expected, higher levels of ctsR mRNA were detected in stressed cells than in unstressed cells (Table 2, row 1). The amount of ctsR transcript increased fivefold after heat shock or the addition of ethanol and fourfold after downshift to pH 3.6.
TABLE 2.
Relative mRNA expression of O. oeni stress genes as determined by QRT-PCR
| Gene | Increase (n-fold) in ratio of stress gene expressiona
|
||
|---|---|---|---|
| 42°C | Ethanol (11% [vol/vol]) | pH 3.6 | |
| ctsR | 5 | 5 | 4 |
| clpL2 | 12 | 75 | 5 |
| grpE | 16 | 5 | 3 |
| groES | 6 | 5 | 1 |
Thirty minutes after stress treatment. Induction ratios were calculated relative to the level of transcripts detected in unstressed cells.
Prediction of CtsR regulon members by scanning the IOB 8413 (ATCC BAA-1163) genome.
Invariance of the DNA recognition helix motif of the CtsR sequence is in agreement with the high conservation of the target nucleotide sequence (16). A similar sequence was identified in the promoter regions of the hsp18, clpX, and clpP genes of O. oeni (4, 30, 32). To identify members of the O. oeni CtsR regulon, a detailed DNA motif analysis of the O. oeni IOB 8413 (ATCC BAA-1163) genome was carried out using the CtsR operator consensus sequence. Additional CtsR operator sites were found upstream from the clpL2 gene, as well as the dnaK and groESL operons encoding the major cell chaperones. No CtsR operator site was found upstream from the temperature-induced ftsH gene, encoding an AAA-type metalloprotease (7). Alignment of the seven potential CtsR-binding sites identified in the O. oeni genome produced the following consensus sequence: (A/G)GTCAA(A/G)AANGGTCAA(A/G) (Fig. 2), which is very similar to the consensus sequence defined by Derré et al., (A/G)GTCAAAAN(A/G)GTCAAA (16). As previously pointed out by Jobin et al. (32), although a potential binding site is also found upstream from the clpX gene (32), the number of nucleotides between the repeats is not conserved, suggesting that this sequence may be vestigial. We note that three direct heptanucleotide sequences are found upstream from the grpE gene, the first gene of the dnaK operon, where the second repeat is complementary to the −35 sequence of the promoter. Three motifs have previously been reported upstream from the clpP gene and four motifs upstream from the hsp18 gene (4, 16, 30).
FIG. 2.
Alignment of CtsR-binding sites identified upstream from ctsR, hsp18, clpX, grpE, clpP, groES, and clpL2 genes of O. oeni IOB 8413 (ATCC BAA-1163). Identical nucleotides are shaded. The numbers indicate positions relative to the transcriptional start site. GenBank accession numbers for database sequences are as follows: clpL2 (AJ890337), groES (AJ890340), grpE (AJ890339), ctsR (AJ890338), hsp18 (AJ250422), clpX (Y15953), and clpP (AJ606044).
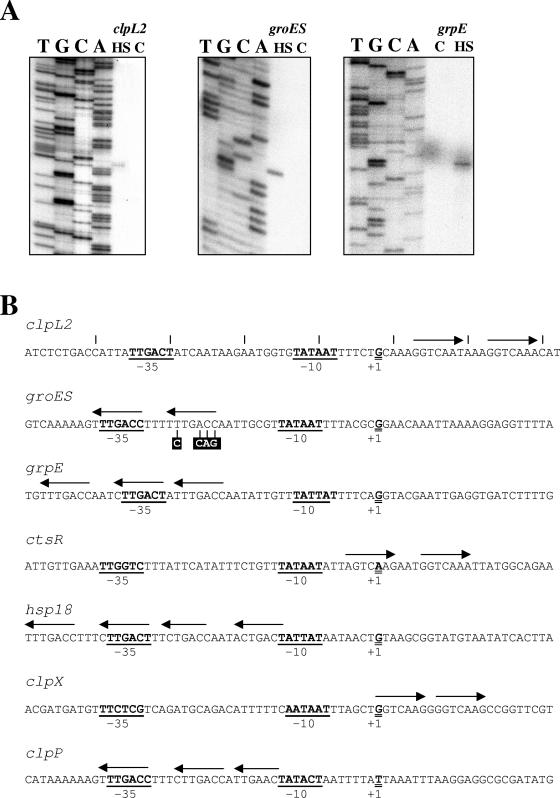
Expression of the clpL2 gene and the groESL and dnaK operons is induced by stress.
The clpL2 gene of O. oeni encodes a second ClpL protein showing 48% identical amino acid residues with the product of the previously characterized clpL gene (4). The O. oeni dnaK operon is comprised of three genes, grpE, dnaK, and cbpA, whereas the groES and groEL genes constitute the groESL operon. The organization of the major cell chaperone genes in O. oeni did not differ significantly from that described in many gram-positive bacteria (52). However, instead of dnaJ, the O. oeni dnaK operon presents a cbpA gene. The CbpA protein is an analog of the DnaJ molecular chaperone, which lacks the 69-amino-acid cysteine-rich zinc finger domain of DnaJ (55). Mapping of the transcriptional start sites was investigated for the clpL2 gene and for the groES and dnaK operons (Fig. 3A). Transcription start points are preceded by appropriately spaced −10 sequences and −35 sequences, which share strong similarity with sequences previously described for O. oeni promoters (Fig. 3B). Moreover, putative CtsR operator sequences overlap or are near the −35 and −10 sequences of the identified transcriptional start sites, which strongly suggests CtsR-dependent expression of these genes. The induction of hsp18, clpX, and clpP expression under stress conditions has previously been reported (4, 30, 32). Characterization of the clpL2 gene and the groESL and dnaK operons was investigated to determine if these genes are also stress response genes. In order to determine the level of induction of the clpL2 gene and the dnaK and groESL operons after stress treatment, a quantitative RT-PCR experiment was set up. For the dnaK and groESL operons, expression of the first gene of each operon, grpE and groES, respectively, was followed. Induction ratios were calculated relative to the level of transcripts detected in unstressed cells (Table 2). Intragenic fragments of the clpL2 (AJ831552), groES, and grpE genes were amplified using the clpE2/clpE3, groES6/groES7, and grpE7/grpE8 oligonucleotide pairs, respectively. As shown in Table 2, the amounts of clpL2, grpE, and groES transcripts were increased 12-fold, 16-fold, and 6-fold, respectively, 30 min after a temperature upshift from 30 to 42°C. After addition of ethanol (11% [vol/vol]), the induction factors were 75 for clpL2 and 5 for grpE and groES. After a shift to pH 3.6, the amounts of clpL2 and grpE transcripts increased fivefold and threefold, respectively. No increase in the amount of groES transcripts was observed under these conditions. These results show that the expression of genes preceded by a likely CtsR operator site is stress inducible and probably depends on the CtsR repressor.
FIG. 3.
(A) Primer extension analysis of clpL2, groES, and grpE mRNAs. Total RNA was extracted from O. oeni cells harvested in the exponential phase (lane C) or after a 30-min shift to 42°C (lane HS). Primer extension products are shown alongside DNA-sequencing reaction products (lanes T, G, C, and A). (B) Nucleotide sequences of the clpL2, groESL, grpE, ctsR, hsp18, clpX, and clpP promoter regions. Potential −35 and −10 sequences are underlined and boldface, transcriptional start points are indicated by +1, and CtsR heptad direct-repeat operator sequences are indicated by arrows. Mutation of the groES promoter performed by insertion of a PstI site in the repeated sequence of the CtsR operator site is indicated.
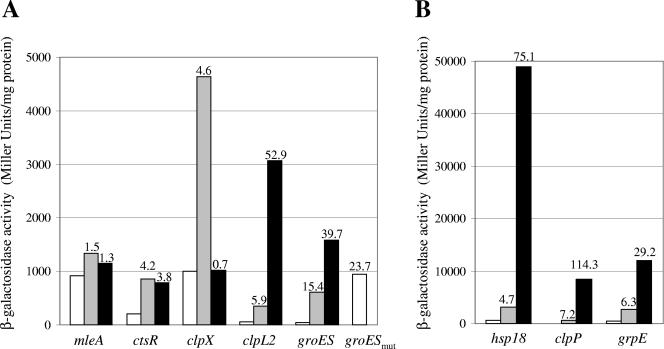
Apart from clpX, all O. oeni genes preceded by a likely CtsR operator site are derepressed in a B. subtilis ΔctsR mutant.
Because no genetic tool adapted to carry out gene inactivation in O. oeni is yet available, CtsR-dependent regulation of O. oeni genes was examined using B. subtilis as a heterologous host. Transcriptional fusions between the promoter regions of the O. oeni ctsR, hsp18, clpX, clpP, clpL2, grpE, and groES genes (Table 1) and the bgaB gene of B. stearothermophilus, which encodes a thermostable β-galactosidase, were integrated as single copies at the amyE locus of B. subtilis. The mleA gene, whose promoter region contains no CtsR operator site, was used as a negative control. The β-galactosidase activities of strains carrying bgaB transcriptional fusions were monitored during growth in Luria broth medium. As expression of fusions followed similar patterns (data not shown), expression levels were compared in the mid-exponential phase. As shown in Fig. 4, expression of transcriptional fusions was low in the wild-type strain at 37°C and expression was increased 4- to 15-fold when cells were shifted to 48°C. As expected, no increase in the expression of the mleA′-bgaB fusion was observed. These results indicate that the tested expression of O. oeni stress response genes is inducible under heat shock conditions in the heterologous host, B. subtilis. To test whether CtsR does indeed play a role in controlling expression of O. oeni genes, the expression of these fusions was tested in B. subtilis strain QB4991, in which the entire B. subtilis ctsR gene is deleted (16) (Fig. 4). In a ΔctsR background, expression of ctsR′-bgaB, clpL2′-bgaB, groES′-bgaB, hsp18′-bgaB, clpP′-bgaB, and grpE′-bgaB fusions was derepressed, whereas expression of mleA′-bgaB and clpX′-bgaB fusions was unchanged. Apart from expression of the clpX′-bgaB fusion, expression levels of fusions were not significantly different at 37°C and 48°C in the ΔctsR mutant (data not shown). These results strongly suggest that the O. oeni hsp18 and clpL2 genes and the ctsR-clpC, dnaK, and groESL operons are negatively controlled by CtsR in O. oeni, whereas clpX expression does not seem to depend on this regulator. We note that, apart from clpX and ctsR genes, the derepressed expression levels are higher in the ΔctsR mutant than in the wild-type strain after a temperature upshift from 37°C to 48°C. Similar results were reported by Derré et al. in experiments to follow heat shock induction of the B. subtilis clpP gene (16). The residual inductions observed in the ΔctsR mutant could be the consequence of a complex pleiotropic role played by the CtsR regulator. Moreover, in a heterologous host, the expression of fusions may not be completely derepressed because of differences in the affinities of the B. subtilis CtsR protein for the O. oeni direct-repeat CtsR operator sites.
FIG. 4.
CtsR negatively regulates hsp gene expression. Expression of the mleA′-bgaB, ctsR′-bgaB, clpX′-bgaB, clpL2′-bgaB, groES′-bgaB, and groESmut′-bgaB (A) and the hsp18′-bgaB, clpP′-bgaB, and grpE′-bgaB (B) transcriptional fusions was measured in the wild-type strain (white and gray bars) or in the ΔctsR mutant (QB4991) (black bars). Cells were grown in LB medium at 37°C. Expression of bgaB fusions was compared in the mid-exponential phase before (white and black bars) or after (gray bars) transfer to 48°C. The numbers indicate induction factors, which were calculated relative to the expression level of each fusion measured in the wild-type strain grown at 37°C.
Site-directed mutagenesis of the groESL promoter.
To confirm that the CtsR operator site of the hsp genes is the key element recognized by CtsR in B. subtilis, site-directed mutagenesis of the groES promoter was performed. The mutated promoter sequence was obtained by insertion of a PstI site in the repeated sequence of the CtsR operator site. In this way the TTGACC sequence (located at positions −22 to −27) was mutated to CTGCAG (Fig. 3B). This mutated promoter sequence was cloned upstream from the bgaB gene, and the groESmut′-bgaB transcriptional fusion was then integrated as a single copy at the amyE locus of B. subtilis. The activity of the groESmut′-bgaB fusion was measured in the wild-type ctsR background. Compared to that of groES′-bgaB, expression of the groESmut′-bgaB fusion was 24-fold higher. These data demonstrated that B. subtilis CtsR negatively regulates O. oeni hsp expression by specific recognition of the predicted direct-repeat CtsR operator site located upstream from these genes.
DISCUSSION
Little is known about the regulation of stress response in O. oeni, although many stress response genes have been characterized in the bacterium. Analysis of the complete O. oeni IOB 8413 (ATCC BAA-1163) genome sequence indicates the existence of an ortholog of the CtsR regulator, as well as several potential target genes. In this work, we have characterized the ctsR gene of O. oeni IOB 8413 (ATCC BAA-1163). As in most low-G+C% gram-positive bacteria studied so far, ctsR is the first gene of the clpC operon in O. oeni. Expression of the ctsR-clpC locus is induced by stress, and the transcription start site, located upstream from ctsR, appears to be dependent on CtsR. In S. aureus and the gram-positive rod-shaped bacteria (Bacillus, Clostridium, and Listeria), the ctsR gene is the first gene of a four-cistron operon in which the last gene encodes the ClpC protein and the mcsA and mcsB genes encode modulators of CtsR (10, 34, 35, 47, 49). In L. lactis, three genes, ctsR, clpC, and orf55, constitute the operon (56), whereas in the Streptococcus group there are only two genes, ctsR and clpC, expressed via the same transcript driven from the promoter upstream of ctsR (49). Furthermore, genome sequence analysis allowed us to predict that the ctsR and clpC genes are organized as an operon in Lactobacillus plantarum (NC_004567). The absence of mcsA-like and mcsB-like genes appears to be specific to the Streptococcus group and lactic acid bacteria and suggests a mechanism to modulate CtsR different from those described for the model organism B. subtilis. Initial attempts to complement the B. subtilis ΔctsR mutant by expressing the heterologous O. oeni ctsR gene in B. subtilis were unsuccessful. The O. oeni ctsR gene, integrated at the thrC locus under the control of the PxylA xylose-inducible promoter, was transcribed in the heterologous host in the presence of xylose. Nevertheless, repression of transcriptional fusions was not observed. A similar result was obtained with constructions including the ribosome binding site motif of the B. subtilis ctsR gene. This result could be explained by a positive effect of a Clp ATPase, such as the O. oeni ClpL protein, on CtsR. Indeed, previous work suggests a positive role of the B. subtilis ClpC protein on CtsR by activating CtsR and promoting DNA binding (15). The O. oeni CtsR protein may be rapidly degraded in the B. subtilis ΔctsR mutant, because of the absence of a specific cochaperon.
In addition to the likely CtsR operator sites previously identified upstream from the clpX and hsp18 genes, analysis of the O. oeni IOB 8413 (ATCC BAA-1163) genome database allowed us to predict CtsR-binding sites upstream from the clpL2 gene, as well as the ctsR-clpC, groESL, and dnaK operons. The CtsR-binding sites overlap or are near the σ70-dependent promoters, suggesting that CtsR probably acts by competing or interfering with RNA polymerase E-σ70 binding. Repression may also occur throughout a roadblock mechanism preventing RNA polymerase progression along the DNA. Using B. subtilis as a heterologous host, we showed that the O. oeni hsp18, clpP, and clpL2 genes and ctsR-clpC, dnaK, and groESL operons are repressed by CtsR from B. subtilis. Moreover, site-directed mutagenesis of the CtsR operator site located in the groESL promoter region led to derepression of the expression of the groESL operon. Taken together, these results indicate that CtsR acts as the master regulator of molecular chaperone gene expression in O. oeni.
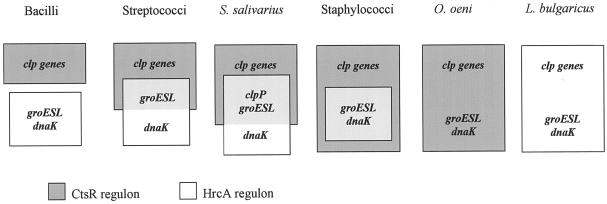
Heat shock gene regulation in gram-positive bacteria is mediated by the alternative sigma factor σB and/or by transcriptional repressors, such as CtsR, HrcA, or HspR (10-12, 16, 21, 23, 56). However, the numbers of sigma factors found in bacterial genomes differ greatly, ranging from 18 in B. subtilis (37) to only 3 in the L. lactis genome (6), emphasizing the complexity and diversity of genetic regulatory mechanisms in bacteria. The O. oeni IOB 8413 (ATCC BAA-1163) genome contains only one housekeeping sigma factor gene (rpoD), and to the best of our knowledge no genes for alternative sigma factors, such as σB. Moreover, the promoters of stress genes characterized in this study are preceded by typical σ70 −35 and −10 boxes, suggesting that they all depend on the housekeeping form of RNA polymerase for expression. Regulation of clp genes of gram-positive bacteria is often described as CtsR dependent, whereas groESL and dnaK operons are often controlled by HrcA. In B. subtilis and closely related species (Bacillus anthracis, B. stearothermophilus, Bacillus halodurans, Clostridium acetobutylicum, Clostridium difficile, Clostridium perfringens, Listeria monocytogenes, and L. innocua), the two regulons are clearly distinct (11), whereas in the streptococcal group (Streptococcus pneumoniae, Streptococcus pyogenes, Streptococcus mutans, Streptococcus agalactiae, L. lactis, and S. salivarius), the HrcA and CtsR regulons partially overlap (Fig. 5). In the latter group, the groESL operon presents both the CtsR target site and the CIRCE sequence organized in tandem (12). In S. aureus, HrcA and CtsR act together to control the expression of the dnaK and groESL operons (10). In this work, we demonstrate that the CtsR repressor is the major regulator of molecular chaperone gene expression in O. oeni. Interestingly, we have been unable to identify either an hrcA gene or CIRCE operator sequences in the O. oeni IOB 8413 (ATCC BAA-1163) genome sequence so far. In contrast, genome sequence analysis indicates the existence of a predominant HrcA control of stress gene expression in Lactobacillus bulgaricus (S. Penaud and E. Maguin, personal communication), suggesting the absence of combined regulation by CtsR and HrcA in both of these lactic acid bacteria. Chastanet et al. (12) have shown that CtsR and HrcA act together synergistically to maintain low-level expression of the dnaK and groESL operons in the absence of stress, suggesting that this dual regulation is probably not redundant. In fact, the predominant CtsR control of molecular chaperone gene expression in O. oeni, like the probable major role of HrcA in L. bulgaricus, may play a role in coordinating synthesis of HSPs during stress response. Under optimal growth conditions, CtsR would prevent the synthesis of unnecessary stress proteins until environmental changes (ethanol, acid, nutritional stresses, etc.) strongly induce transcription of stress genes, including the clp genes and major cell chaperone operons (groESL and dnaK), thus enhancing the adaptability of these lactic acid bacteria under adverse environmental conditions by coordinating the synthesis of HSPs.
FIG. 5.
Dual regulation by CtsR and HrcA in different gram-positive bacteria. In many gram-positive bacteria, the CtsR and the HrcA regulons coexist. In B. subtilis and closely related bacilli (B. anthracis, B. stearothermophilus, B. halodurans, C. acetobutylicum, C. difficile, C. perfringens, L. monocytogenes, and L. innocua), the two regulons are distinct, whereas in the streptococcal group (S. pneumoniae, L. lactis, and S. salivarius), they partially overlap, and the HrcA regulon is entirely embedded within the CtsR regulon in S. aureus. O. oeni and L. bulgaricus have original heat shock gene regulation with a predominant control of molecular chaperone genes by either CtsR or HrcA, respectively (adapted from Chastanet et al. [11]).
Apart from the clpX gene, all of the tested O. oeni molecular chaperone genes are dependent on CtsR. The expression of clpX has been shown to be heat inducible in different organisms (22, 24, 32, 54). The clpX gene of B. subtilis belongs to class IV heat shock genes, controlled by an unknown mechanism (23). In this work, we have noted the presence of a vestigial CtsR-binding site upstream from the clpX gene promoter of O. oeni, since the spacing between the direct repeats is not conserved and CtsR is not able to repress clpX. This site may be an evolutionary remnant, suggesting that this gene was once under CtsR regulation. Furthermore, quantitative RT-PCR analysis of genes under the control of CtsR revealed that induction factors are different depending on the gene and type of stress treatment. There may be other mechanisms, perhaps at the posttranscriptional level, such as mRNA stability, to ensure regulation of molecular chaperone synthesis under adverse environmental conditions. Previous studies have identified a long untranslated sequence at the 5′ end of clpX mRNA (5′ untranslated region) and suggested that it could be involved in stability and/or control of translation (32), as described previously in different organisms (14, 19, 38). Future work will involve studying the role of this 5′ untranslated region in the stability of mRNA under stress conditions, which could constitute an additional level of heat shock gene regulation in O. oeni.
The identification of members of the CtsR regulon is an essential step toward a more comprehensive understanding of the role of this regulon in stress adaptation. Analysis of the effects of mutations in individual CtsR-dependent genes on stress adaptation is required to address whether and to what extent CtsR-dependent genes contribute to survival in wine. These studies will provide an essential contribution to the understanding of the cell physiology of O. oeni. In order to carry out gene inactivation in O. oeni, future studies will focus on the construction of an integrative vector for gene disruption derived from the conjugative plasmid pGID052 (5).
Acknowledgments
We are grateful to P.S.D.M. for helpful discussion and proofreading the manuscript and D. Garmyn for constant interest in this work. We thank T. Msadek for providing B. subtilis strains and the pDL vector.
This work was supported by the Ministère de l'Education Nationale de la Recherche et de la Technologie, the Université de Bourgogne, and the Institut National de la Recherche Agronomique.
REFERENCES
- 1.Altschul, S., T. Madden, A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avedissian, M., and S. Lopes Gomes. 1996. Expression of the groESL operon is cell-cycle controlled in Caulobacter crescentus. Mol. Microbiol. 19:79-89. [DOI] [PubMed] [Google Scholar]
- 3.Barbosa, M. F., L. P. Yomano, and L. O. Ingram. 1994. Cloning, sequencing and expression of stress genes from the ethanol-producing bacterium Zymomonas mobilis: the groESL operon. Gene 148:51-57. [DOI] [PubMed] [Google Scholar]
- 4.Beltramo, C., C. Grandvalet, F. Pierre, and J. Guzzo. 2004. Evidence for multiple levels of regulation of Oenococcus oeni clpP-clpL locus expression in response to stress. J. Bacteriol. 186:2200-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beltramo, C., M. Oraby, G. Bourel, D. Garmyn, and J. Guzzo. 2004. A new vector, pGID052, for genetic transfer in Oenococcus oeni. FEMS Microbiol. Lett. 236:53-60. [DOI] [PubMed] [Google Scholar]
- 6.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bourdineaud, J. P., B. Nehme, S. Tesse, and A. Lonvaud-Funel. 2003. The ftsH gene of the wine bacterium Oenococcus oeni is involved in protection against environmental stress. Appl. Environ. Microbiol. 69:2512-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bradford, M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 9.Cavin, J., H. Prevost, J. Lin, P. Schmitt, and C. Diviès. 1989. Medium for screening Leuconostoc oenos strains defective in malolactic fermentation. Appl. Environ. Microbiol. 55:751-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chastanet, A., J. Fert, and T. Msadek. 2003. Comparative genomics reveal novel heat shock regulatory mechanisms in Staphylococcus aureus and other Gram-positive bacteria. Mol. Microbiol. 47:1061-1073. [DOI] [PubMed] [Google Scholar]
- 11.Chastanet, A., and T. Msadek. 2003. ClpP of Streptococcus salivarius is a novel member of the dually regulated class of stress response genes in Gram-positive bacteria. J. Bacteriol. 185:683-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chastanet, A., M. Prudhomme, J. P. Claverys, and T. Msadek. 2001. Regulation of Streptococcus pneumoniae clp genes and their role in competence development and stress survival. J. Bacteriol. 183:7295-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chenna, R., H. Sugawara, T. Koike, R. Lopez, T. J. Gibson, D. G. Higgins, and J. D. Thompson. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31:3497-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chowdhury, S., C. Ragaz, E. Kreuger, and F. Narberhaus. 2003. Temperature-controlled structural alterations of an RNA thermometer. J. Biol. Chem. 278:47915-47921. [DOI] [PubMed] [Google Scholar]
- 15.Derré, I., G. Rapoport, and T. Msadek. 2000. The CtsR regulator of stress response is active as a dimer and specifically degraded in vivo at 37°C. Mol. Microbiol. 38:335-347. [DOI] [PubMed] [Google Scholar]
- 16.Derré, I., G. Rapoport, and T. Msadek. 1999. CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in Gram-positive bacteria. Mol. Microbiol. 31:117-131. [DOI] [PubMed] [Google Scholar]
- 17.Desroche, N., C. Beltramo, and J. Guzzo. 2005. Determination of an internal control to apply reverse transcription quantitative PCR to study stress response in the lactic acid bacterium Oenococcus oeni. J. Microbiol. Methods 60:325-333. [DOI] [PubMed] [Google Scholar]
- 18.Dicks, L. M., F. Dellaglio, and M. D. Collins. 1995. Proposal to reclassify Leuconostoc oenos as Oenococcus oeni [corrig.] gen. nov., comb. nov. Int. J. Syst. Bacteriol. 45:395-397. [DOI] [PubMed] [Google Scholar]
- 19.Emory, S. A., and J. G. Belasco. 1990. The ompA 5′ untranslated RNA segment functions in Escherichia coli as a growth-rate-regulated mRNA stabilizer whose activity is unrelated to translational efficiency. J. Bacteriol. 172:4472-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernandez, R. C., and A. A. Weiss. 1995. Cloning and sequencing of the Bordetella pertussis cpn10/cpn60 (groESL) homolog. Gene 158:151-152. [DOI] [PubMed] [Google Scholar]
- 21.Gerth, U., E. Kruger, I. Derré, T. Msadek, and M. Hecker. 1998. Stress induction of the Bacillus subtilis clpP gene encoding a homologue of the proteolytic component of the Clp protease and the involvement of ClpP and ClpX in stress tolerance. Mol. Microbiol. 28:787-802. [DOI] [PubMed] [Google Scholar]
- 22.Gerth, U., A. Wipat, C. R. Harwood, N. Carter, P. T. Emmerson, and M. Hecker. 1996. Sequence and transcriptional analysis of clpX, a class-III heat-shock gene of Bacillus subtilis. Gene 181:77-83. [DOI] [PubMed] [Google Scholar]
- 23.Gertz, S., S. Engelmann, R. Schmid, A. K. Ziebandt, K. Tischer, C. Scharf, J. Hacker, and M. Hecker. 2000. Characterization of the sigma (B) regulon in Staphylococcus aureus. J. Bacteriol. 182:6983-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gottesman, S., W. P. Clark, V. de Crecy-Lagard, and M. R. Maurizi. 1993. ClpX, an alternative subunit for the ATP-dependent Clp protease of Escherichia coli. Sequence and in vivo activities. J. Biol. Chem. 268:22618-22626. [PubMed] [Google Scholar]
- 25.Gottesman, S., S. Wickner, and M. R. Maurizi. 1997. Protein quality control: triage by chaperones and proteases. Genes Dev. 11:815-823. [DOI] [PubMed] [Google Scholar]
- 26.Grandvalet, C., G. Rapoport, and P. Mazodier. 1998. hrcA, encoding the repressor of the groEL genes in Streptomyces albus G, is associated with a second dnaJ gene. J. Bacteriol. 180:5129-5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grandvalet, C., P. Servant, and P. Mazodier. 1997. Disruption of hspR, the repressor gene of the dnaK operon in Streptomyces albus G. Mol. Microbiol. 23:77-84. [DOI] [PubMed] [Google Scholar]
- 28.Hecker, M., W. Schumann, and U. Volker. 1996. Heat-shock and general stress response in Bacillus subtilis. Mol. Microbiol. 19:417-428. [DOI] [PubMed] [Google Scholar]
- 29.Hirata, H., T. Fukazawa, S. Negoro, and H. Okada. 1986. Structure of a beta-galactosidase gene of Bacillus stearothermophilus. J. Bacteriol. 166:722-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jobin, M. P., F. Delmas, D. Garmyn, C. Divies, and J. Guzzo. 1997. Molecular characterization of the gene encoding an 18-kilodalton small heat shock protein associated with the membrane of Leuconostoc oenos. Appl. Environ. Microbiol. 63:609-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jobin, M. P., D. Garmyn, C. Divies, and J. Guzzo. 1999. Expression of the Oenococcus oeni trxA gene is induced by hydrogen peroxide and heat shock. Microbiology 145:1245-1251. [DOI] [PubMed] [Google Scholar]
- 32.Jobin, M. P., D. Garmyn, C. Divies, and J. Guzzo. 1999. The Oenococcus oeni clpX homologue is a heat shock gene preferentially expressed in exponential growth phase. J. Bacteriol. 181:6634-6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klaenhammer, T., E. Altermann, F. Arigoni, A. Bolotin, F. Breidt, J. Broadbent, R. Cano, S. Chaillou, J. Deutscher, M. Gasson, M. van de Guchte, J. Guzzo, A. Hartke, T. Hawkins, P. Hols, R. Hutkins, M. Kleerebezem, J. Kok, O. Kuipers, M. Lubbers, E. Maguin, L. McKay, D. Mills, A. Nauta, R. Overbeek, H. Pel, D. Pridmore, M. Saier, D. van Sinderen, A. Sorokin, J. Steele, D. O'Sullivan, W. de Vos, B. Weimer, M. Zagorec, and R. Siezen. 2002. Discovering lactic acid bacteria by genomics. Antonie Leeuwenhoek 82:29-58. [DOI] [PubMed] [Google Scholar]
- 34.Kruger, E., T. Msadek, S. Ohlmeier, and M. Hecker. 1997. The Bacillus subtilis clpC operon encodes DNA repair and competence proteins. Microbiology 143:1309-1316. [DOI] [PubMed] [Google Scholar]
- 35.Kruger, E., D. Zuhlke, E. Witt, H. Ludwig, and M. Hecker. 2001. Clp-mediated proteolysis in Gram-positive bacteria is autoregulated by the stability of a repressor. EMBO J. 20:852-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kunkee, R. E. 1991. Some roles of malic acid in the malolactic fermentation in wine making. FEMS Microbiol. Lett. 88:55-71. [Google Scholar]
- 37.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 38.Lundberg, U., A. von Gabain, and O. Melefors. 1990. Cleavages in the 5′ region of the ompA and bla mRNA control stability: studies with an E. coli mutant altering mRNA stability and a novel endoribonuclease. EMBO J. 9:2731-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mantis, N. J., and S. C. Winans. 1992. Characterization of the Agrobacterium tumefaciens heat shock response: evidence for a sigma 32-like sigma factor. J. Bacteriol. 174:991-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Michel, G. P. 1993. Cloning and expression in Escherichia coli of the dnaK gene of Zymomonas mobilis. J. Bacteriol. 175:3228-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Msadek, T., V. Dartois, F. Kunst, M. Herbaud, F. Denizot, and G. Rapoport. 1998. ClpP of Bacillus subtilis is required for competence development, motility, degradative enzyme synthesis, growth at high temperature and sporulation. Mol. Microbiol. 27:899-914. [DOI] [PubMed] [Google Scholar]
- 43.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Volker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183:5617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Price, C. W., P. Fawcett, H. Ceremonie, N. Su, C. K. Murphy, and P. Youngman. 2001. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41:757-774. [DOI] [PubMed] [Google Scholar]
- 45.Reisenauer, A., C. D. Mohr, and L. Shapiro. 1996. Regulation of a heat shock sigma 32 homolog in Caulobacter crescentus. J. Bacteriol. 178:1919-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roberts, R. C., C. Toochinda, M. Avedissian, R. L. Baldini, S. L. Gomes, and L. Shapiro. 1996. Identification of a Caulobacter crescentus operon encoding hrcA, involved in negatively regulating heat-inducible transcription, and the chaperone gene grpE. J. Bacteriol. 178:1829-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rouquette, C., M. T. Ripio, E. Pellegrini, J. M. Bolla, R. I. Tascon, J. A. Vazquez-Boland, and P. Berche. 1996. Identification of a ClpC ATPase required for stress tolerance and in vivo survival of Listeria monocytogenes. Mol. Microbiol. 21:977-987. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook, J., E. F. Frisch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 49.Schumann, W., M. Hecker, and T. Msadek. 2002. Regulation and function of heat-inducible genes in Bacillus subtilis, p. 359-368. In A. L. Sonenshein, J. A. Hoch, and R. M. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, D.C.
- 50.Segal, G., and E. Z. Ron. 1995. The dnaKJ operon of Agrobacterium tumefaciens: transcriptional analysis and evidence for a new heat shock promoter. J. Bacteriol. 177:5952-5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Segal, G., and E. Z. Ron. 1993. Heat shock transcription of the groESL operon of Agrobacterium tumefaciens may involve a hairpin-loop structure. J. Bacteriol. 175:3083-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Segal, R., and E. Z. Ron. 1996. Regulation and organization of the groE and dnaK operons in Eubacteria. FEMS Microbiol. Lett. 138:1-10. [DOI] [PubMed] [Google Scholar]
- 53.Servant, P., C. Grandvalet, and P. Mazodier. 2000. The RheA repressor is the thermosensor of the HSP18 heat shock response in Streptomyces albus. Proc. Natl. Acad. Sci. USA 97:3538-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Skinner, M. M., and J. E. Trempy. 2001. Expression of clpX, an ATPase subunit of the Clp protease, is heat and cold shock inducible in Lactococcus lactis. J. Dairy Sci. 84:1783-1785. [DOI] [PubMed] [Google Scholar]
- 55.Ueguchi, C., M. Kakeda, H. Yamada, and T. Mizuno. 1994. An analogue of the DnaJ molecular chaperone in Escherichia coli. Proc. Natl. Acad. Sci. USA 91:1054-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Varmanen, P., H. Ingmer, and F. K. Vogensen. 2000. ctsR of Lactococcus lactis encodes a negative regulator of clp gene expression. Microbiology 146:1447-1455. [DOI] [PubMed] [Google Scholar]
- 57.Yuan, G., and S. Wong. 1995. Regulation of groE expression in Bacillus subtilis: the involvement of the sigma A-like promoter and the roles of the inverted repeat sequence (CIRCE). J. Bacteriol. 177:5427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yura, T., K. Nakahigashi, and M. Kanemori. 1996. Transcriptional regulation of stress-inducible genes in procaryotes. EXS 77:165-181. [DOI] [PubMed] [Google Scholar]
- 59.Zuber, U., and W. Schumann. 1994. CIRCE, a novel heat shock element involved in regulation of heat shock operon dnaK of Bacillus subtilis. J. Bacteriol. 176:1359-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]