Abstract
A-factor (2-isocapryloyl-3R-hydroxymethyl-γ-butyrolactone) triggers streptomycin production by inducing the transcription of strR, encoding the pathway-specific transcriptional activator, through signal transduction in the A-factor regulatory cascade in Streptomyces griseus. AdpA, one of the key transcriptional activators in the cascade, bound two upstream activation sites, approximately at nucleotide positions −270 and −50 with respect to the transcriptional start point of strR, as determined by gel mobility shift assays and DNase I footprinting. Transcriptional analysis of the strR promoter with mutated AdpA-binding sites showed that both sites were required for full transcriptional activation of strR by AdpA. Potassium permanganate footprinting showed that AdpA assisted RNA polymerase in forming an open complex at an appropriate position for transcriptional initiation of strR. Nine transcriptional units within the streptomycin biosynthesis gene cluster, including the strR-aphD operon, depended on StrR, indicating that StrR is the pathway-specific transcriptional activator for the whole gene cluster. Consistent with this, expression of strR under the control of a constitutively expressed promoter in an adpA null mutant caused the host to produce streptomycin.
The ability to produce a wide variety of secondary metabolites and a mycelial growth that develops into chains of spores are two aspects characteristic of the gram-positive, filamentous, soil-dwelling bacterial genus Streptomyces. A-factor (2-isocapryloyl-3R-hydroxymethyl-γ-butyrolactone) is a representative of the γ-butyrolactone autoregulators that control secondary metabolism, morphological development, or both in actinomycetes, mainly including members of the genus Streptomyces (5, 21). It acts as a chemical signaling molecule, or a microbial hormone, for secondary metabolism and morphological differentiation at a concentration as low as 10−9 M in streptomycin-producing Streptomyces griseus (6). A-factor was originally discovered by Khokhlov et al. (14) in the 1960s as an autoregulatory factor that restored streptomycin production and spore formation in a mutant of S. griseus. In the 1980s, we confirmed the pioneer work of Khokhlov et al. and have studied A-factor regulation since then.
We have so far revealed the A-factor regulatory cascade as follows (5, 21). A-factor is gradually accumulated in a growth-dependent manner by the action of AfsA, probably condensing a glycerol derivative and a β-keto acid. When the concentration of A-factor reaches a critical level at or just before the decision point, it binds an A-factor-specific receptor, ArpA, which has bound and repressed the promoter of adpA, and dissociates ArpA from the promoter, thus leading to transcription and translation of adpA. adpA is a sole significant target of ArpA (11). The transcriptional activator AdpA then activates a variety of genes that are required for secondary metabolite formation and morphological differentiation (22). Members of the AdpA regulon so far identified are strR, the pathway-specific transcriptional activator for streptomycin biosynthesis (20, 24); an open reading frame encoding a probable pathway-specific regulator of a polyketide compound (29); adsA, encoding an extracytoplasmic function sigma factor of RNA polymerase essential for aerial mycelium formation (26); ssgA, encoding a small acidic protein essential for spore septum formation (27); amfR, encoding a regulatory protein essential for aerial mycelium formation (28); sgmA, encoding a metalloendopeptidase probably involved in apoptosis of substrate mycelium during aerial mycelium development (12); two trypsin genes (10); and three chymotrypsin genes (our unpublished data). Although the phenotype of streptomycin production was a start of the A-factor study by Khokhlov et al. and our group, no detailed study as to the molecular mechanism of strR regulation by AdpA has been accomplished.
We previously detected an A-factor-responsive protein that bound an upstream activation sequence (UAS) of strR (24) and later purified the protein and named it AdpA (20). AdpA bound the region approximately at nucleotide position −270, with respect to the transcriptional start point of strR, as determined by gel mobility shift assay (20, 24). In the present study, we determined the exact AdpA-binding site by DNase I footprinting in order to reveal how AdpA activates the transcription of strR. During these studies, we have identified an additional AdpA-binding site at nucleotide position −50 in front of the strR promoter and found that both AdpA-binding sites are necessary for full transcriptional activation of strR by AdpA. Concerning the regulation of the streptomycin biosynthesis genes by StrR, Retzlaff and Distler (23) showed strict dependence of strB1 on StrR and predicted the dependence of some transcriptional units because of StrR binding to their upstream regions (see Fig. 5 for the organization of the streptomycin biosynthesis gene cluster). We therefore determined the dependence of nine transcriptional units within the streptomycin biosynthesis gene cluster on StrR and showed that all the streptomycin biosynthesis genes are under the control of StrR.
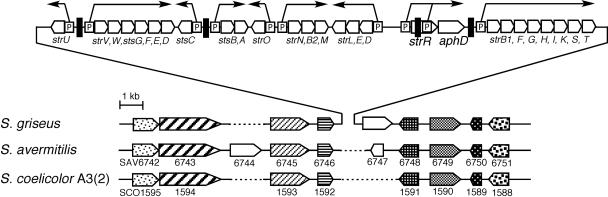
FIG. 5.
Synteny analysis of the streptomycin biosynthesis gene cluster in S. griseus. The orthologues in S. griseus, S. avermitilis, and S. coelicolor A3(2) are discriminated by different markings. The StrR-binding sequences predicted by Retzlaff and Distler (23) are indicated by black bars. Promoters (P) and transcriptional units (arrows) are also indicated.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
S. griseus IFO13350 was obtained from the Institute of Fermentation, Osaka (IFO), Japan. The S. griseus ΔadpA mutant was described previously (20). Streptomyces strains were grown in YMPD medium (20). YMPD agar contained 2% agar. Bennett agar medium (yeast extract, 0.1%; meat extract, 0.1%; NZ amine, 0.2%; agar, 2%; pH 7.2) was used for streptomycin production. R2YE medium (15) was used for the regeneration of protoplasts. Neomycin (5 μg/ml) was added when necessary. Escherichia coli JM109 and pUC19 for DNA manipulation were purchased from Takara Biochemicals. E. coli JM110 containing dam and dcm mutations was used for preparing nonmethylated Streptomyces DNA for gene disruption. Histidine-tagged AdpA (AdpA-H) was purified from E. coli BL21(DE3) harboring pET-adpA as described previously (26). Media and growth conditions for E. coli were described earlier (1). Ampicillin (50 μg/ml) and kanamycin (50 μg/ml) were used when necessary.
General recombinant DNA studies.
Restriction enzymes, T4 DNA ligase, and other DNA-modifying enzymes were purchased from Takara Biochemicals. [γ-32P]ATP (220 TBq/mmol) for end labeling at 5′ ends with T4 polynucleotide kinase was purchased from Amersham Pharmacia Biotech. Digoxigenin-labeled probes for Southern hybridization were prepared with the BcaBest DIG labeling kit (Takara). DNA was manipulated in Streptomyces (15) and in E. coli (1, 17) as described earlier. Nucleotide sequences were determined by the dideoxy chain termination method with a Thermo Sequenase fluorescence-labeled primer cycle sequencing kit (Amersham). Chromosomal DNA of S. griseus or an appropriate plasmid was used for a template in the PCR.
Cloning of a part of the streptomycin biosynthesis gene cluster.
A 0.4-kb fragment (nucleotide positions −225 to +218 with respect to the transcriptional start point of strR) was amplified by PCR with primers pb-F and gs-R (Table 1) and labeled with digoxigenin. By standard DNA manipulation including colony hybridization using this DNA fragment as a probe, a 5.4-kb KpnI-PvuII fragment containing five complete genes (strD, strR, aphD, strB1, and strF) and two truncated genes (strE and strG) was cloned between the KpnI and HincII sites of pUC19.
TABLE 1.
Primers used in this study
| Gene and primer | Positionsa | Sequence (5′ to 3′) |
|---|---|---|
| strR | ||
| gs-F | −482 to −463 | GCGGCACGTATGGCCTCCAG |
| gs-R | +218 to +199 | CGACATCCTCGCCGGCACTG |
| fAa-F | −184 to −162 | AATGAAATCGCTGACAGGCGGTG |
| fAa-R | +19 to −4 | AGAGCAATGCTTTCGCACTTCGC |
| fAs-F | −127 to −107 | GTTCCGGTCCTCTCCGCCCTG |
| fAs-R | +77 to +53 | TGTTCCCTGAAATATGCTCCATTAC |
| fBa-F | −394 to −373 | TGAGGCGGGTTCCTGTGCCGCC |
| fBa-R | −200 to −223 | AACCGCAGTTTGATTGCCGAATAC |
| fBs-F | −344 to −324 | TCCTCGCGTGGTCTTGGGCCG |
| fBs-R | −138 to −158 | AGCCAGCGCCTGCGCCCGATC |
| pb-F | −225 to −202 | ATGTATTCGGCAATCAAACTGCGG |
| pf-R | +68 to +44 | AAATATGCTCCATTACACACCCTTC |
| hrdB | ||
| hrdB-F | −121 to −98 | TCGGCCCATTTCGTCACGTATGAG |
| hrdB-R | +193 to +170 | TCGATGAGCGCCATCACAGACTCG |
| adpA | ||
| adpA-F | −223 to −200 | AGCCCCCGCATCCCTCCGCGGCGA |
| adpA-R | +54 to +31 | ACTCGCGAAGCGCACAGGGAAGTG |
| strB1 | ||
| sB1-F | −198 to −176 | AGCCCTGAACTCCTGAAGCACTG |
| sB1-R | +177 to +156 | AAGGCTCGACTCGGCAGTGCTC |
| strD | ||
| sD-F | −428 to −404 | TGTTCCCTGAAATATGCTCCATTAC |
| sD-R | −8 to −28 | TCCTCGCGTGGTCTTGGGCCG |
| strON | ||
| sON-F | −252 to −231 | ACGGTACGACCAACTGTCCTCG |
| sON-R | +158 to +138 | AACAGGGCCTTCTCCACGGCG |
| stsB | ||
| sB-F | −244 to −220 | ATCCATGATTTCCCTCGATTTCGAG |
| sB-R | +148 to +138 | TCGGCTCCCTGTTCGGCCAGC |
| stsC | ||
| sC-F | −250 to −230 | ACTGCCGGGCATCGCTGACTC |
| sC-R | +158 to +137 | TACGAATCCCGGCCCTGATAGG |
| strV | ||
| sV-F | −250 to −230 | AAGCGCGAGGCTAGGTCGGTC |
| sV-R | +155 to +135 | ACCACCGGGATGAGCGCCTGC |
| strU | ||
| sU-F | −450 to −431 | AAGCGGCCAACCCGGCCGTC |
| sU-R | −34 to −54 | AATGCGACCCGTCGCAGCGGG |
The nucleotide positions for strR, hrdB, adpA, and strB1 are given taking their transcriptional start points as +1. Those for strD, stsB, stsC, strV, and strU are given taking the first letters of their start codons as +1. The nucleotide positions for strO and strN are given taking the first letter of the start codon of strO as +1.
Gel mobility shift assay.
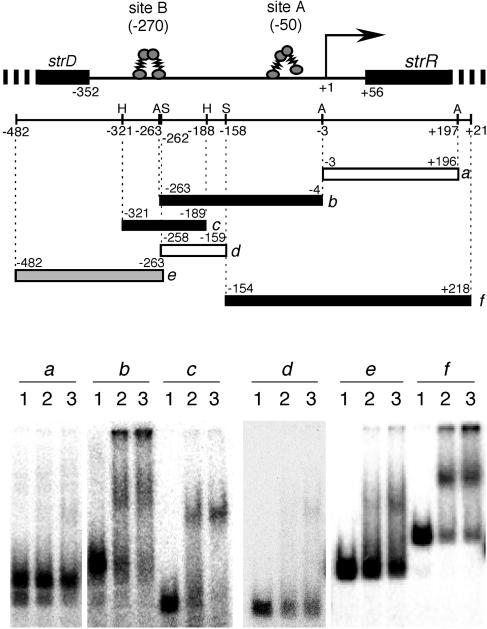
Purification of AdpA-H from E. coli BL21(DE3) and gel mobility shift assays were described previously (26). For gel mobility shift assays with AdpA-H and various regions in front of strR, a 700-bp DNA fragment containing the intervening region between strR and strD was amplified by PCR with primers gs-F and gs-R (Table 1). Six DNA fragments, a to f (see Fig. 1), were prepared by digestion of the amplified fragment with appropriate restriction enzymes and 32P labeled with T4 polynucleotide kinase.
FIG. 1.
Gel mobility shift assays for determination of AdpA-binding sites upstream of strR. Six different probes, a to f, were used. The restriction enzymes are abbreviated as follows: A, AccII; H, HaeIII; S, Sau3AI. The numbers below the restriction map indicate the positions, taking the transcriptional start point of strR as +1. The amounts of AdpA-H used in the gel mobility shift assays were 0 μg (lane 1), 0.1 μg (lane 2), and 0.4 μg (lane 3). Approximate AdpA-binding sites A and B are shown above the restriction map.
DNase I footprinting.
The method of DNase I footprinting was described previously (26). For analysis of AdpA-binding sites, a 32P-labeled fragment was prepared by PCR with a pair of 32P-labeled (indicated with an asterisk) and unlabeled primers listed in Table 1: fAa-F and fAa-R* for the antisense strand of site A, fAs-F* and fAs-R for the sense strand of site A, fBa-F and fBa-R* for the antisense strand of site B, and fBs-F* and fBs-R for the sense strand of site B.
Alterations of the AdpA-binding sequences by PCR.
Mutations were introduced by PCR into the AdpA-binding sites in front of strR. The TTTCGGTC sequence in the consensus AdpA-binding sequence at site A (see Fig. 3A) was changed to CCTCTAGA, containing an XbaI recognition sequence, as follows. A DNA fragment upstream of the mutation point was amplified by PCR with primers 5′-CGggatccATCCCGGCGGCACGTATGGCC-3′ (primer bs-F; positions −488 to −468; the lowercase letters indicate a BamHI site) and 5′-GgaattcTCTAGAGGAGCGACCTCGAAATAGGAGGGC-3′ (positions −64 to −85; the lowercase and italic letters indicate EcoRI and XbaI sites, respectively, and the underlining indicates the nucleotides to be changed). Similarly, a DNA fragment downstream of the mutation point was amplified with 5′-CGggatccTCTAGAATGCGGACAGCTTTACTTGGCCG-3′ (positions −55 to −33; the lowercase and italic letters indicate BamHI and XbaI sites, respectively) and 5′-GgaattcTGCATGCCGTCGATGACGCGC-3′ (primer bs-R; positions +306 to +286; the lowercase letters indicate an EcoRI site). After the absence of PCR errors had been checked by nucleotide sequencing, the two amplified fragments were connected via the common XbaI site and placed between the BamHI and EcoRI sites of pUC19, resulting in pUC19-RmA. The 32P-labeled probe containing the XbaI mutation at site A was prepared by PCR with primers fAa-F and fAs-R (Table 1) with pUC19-RmA as the template.
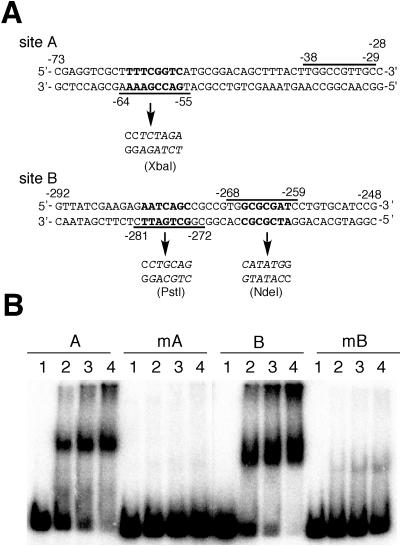
FIG. 3.
Mutational analysis of the two AdpA-binding sites. (A) Mutations introduced in the AdpA-binding sites. An XbaI site generated at site A and a PstI and an NdeI site generated at site B are shown by italic letters. The nucleotides changed are indicated by boldface letters. The AdpA-binding sequences, positions −64 to −55 and positions −38 to −29 at site A and positions −281 to −272 and positions −268 to −259 at site B, are shown. (B) Gel mobility shift assays for determination of AdpA binding to the mutated sites. Probe A (positions −184 to +77) contained the intact AdpA-binding site, and probe mA contained mutated site A. Probe B (positions −394 to −138) contained the intact AdpA-binding sites, and probe mB contained mutated site B. The amounts of AdpA-H used in lanes 1 to 4 were 0, 0.06, 0.12, and 0.24 μg.
For introduction of a mutation into the two AdpA-binding sequences at site B, we first created an NdeI mutation (see Fig. 3A) in the same way as for the XbaI mutation at site A. The GCGCGAT sequence in the consensus AdpA-binding sequence at site B (see Fig. 3A) was changed to CATATGG, containing an NdeI recognition sequence, as follows. The upstream region was amplified by PCR with primers bs-F and 5′-GgaattcCATATGCACGGCGGCTGATTCTCTTCGA-3′ (positions −267 to −288; the lowercase and italic letters indicate EcoRI and NdeI sites, respectively; the underlining indicates the nucleotides to be changed). The downstream region was amplified with primers 5′-CGggatccCATATGGCCTGTGCATCCGTGTAAGGGGC-3′ (positions −259 to −238; the lowercase and italic letters indicate BamHI and NdeI sites, respectively) and bs-R. The two amplified fragments were connected via the common NdeI site and placed between the BamHI and EcoRI sites of pUC19, resulting in pUC19-RmB1. Next, we further introduced a PstI mutation (see Fig. 3A) into the other AdpA-binding sequence at site B. The AATCAGC sequence in the other AdpA-binding sequence at site B (see Fig. 3A) was changed to CCTGCAG, containing a PstI recognition sequence, as follows. The upstream region was amplified with bs-F and 5′-GgaattcCTGCAGGCTCTTCGATAACTGTTCGGAAGAAC-3′ (positions −281 to −305; the lowercase and italic letters indicate EcoRI and PstI sites, respectively; the underlining indicates the nucleotides to be changed). The downstream region was amplified with 5′-CGggatccCTGCAGCGCCGTGCATATGGCCTGTGC-3′ (positions −273 to −254; the lowercase and italic letters indicate BamHI and PstI sites, respectively; the mutated nucleotides are indicated by underlining) and bs-R using pUC19-RmB1 as the template. The two amplified fragments were connected via the common PstI site placed between the BamHI and EcoRI sites of pUC19, resulting in pUC19-RmB. Similarly, using pUC19-RmA as the template, pUC19-RmAB containing XbaI, NdeI, and PstI mutations was constructed. The 32P-labeled probe containing the NdeI and PstI mutations at site B was prepared by PCR with primers fBa-F and fBs-R (Table 1) with pUC19-RmB as the template.
Construction of expression plasmids for strR and aphD with a mutated AdpA-binding site(s).
The strR coding sequence, together with aphD, was amplified by PCR with primers 5′-GCtctagaCATATGGAGCATATTTCAGGGAACAGC-3′ (positions +56 to +79; the lowercase and italic letters indicate an XbaI and NdeI sites, respectively; the ATG in the NdeI site is the start codon of strR) and 5′-GgaattcATGCGTTCGACTGCGTGTGATCG-3′ (positions +2381 to +2359; the lowercase letters indicate an EcoRI site) and placed between the XbaI and EcoRI sites of pUC19. After the absence of PCR errors had been checked by nucleotide sequencing, the SphI-EcoRI fragment (positions +300 to +2381; an SphI site is present at position +305) was excised. This SphI-EcoRI fragment and the BamHI-SphI fragment from pUC19-RmA were placed between the BamHI and EcoRI sites of pUC19 by three-fragment ligation. The HindIII-EcoRI fragment containing the XbaI mutation at site A and the strR-aphD region was then excised from the pUC19 plasmid and placed between the HindIII and EcoRI sites of pKUM20 with its copy number of one to two per chromosome (28), resulting in pRmA (see Fig. 4C). pRmB containing the PstI and NdeI mutations at site B and pRmAB containing the mutations at both site A and site B were similarly constructed using the BamHI-SphI fragment from pUC19-RmB and pUC19-RmAB, respectively.
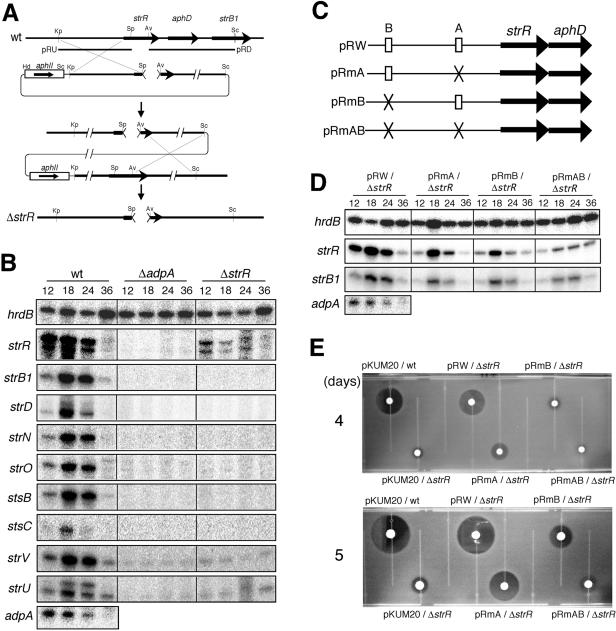
FIG. 4.
Requirement of both site A and site B for full activation of strR by AdpA. (A) Schematic representation of the procedure used for construction of the ΔstrR mutant. The restriction enzymes are abbreviated as follows: Av, AviII; Kp, KpnI; Pv, PvuII; Sc, SacI; Sp, SphI. (B) Low-resolution S1 nuclease mapping of strR and other promoters within the streptomycin biosynthetic gene cluster (strB1, strD, strN, strO, stsB, stsC, strV, and strU) in S. griseus ΔadpA and ΔstrR mutants. RNA was prepared from cells grown on Bennett agar medium at 28°C for the indicated number of hours. (C) Schematic representation of the plasmids used for promoter assays. (D) Low-resolution S1 mapping of strR and strB1 in the ΔstrR mutant harboring pRW, pRmA, pRmB, or pRmAB. (E) Streptomycin production by the ΔstrR mutant harboring pRW, pRmA, pRmB, or pRmAB. As controls, the wild-type (wt) strain harboring the vector pKUM20 and the ΔstrR mutant harboring pKUM20 were also assayed. These strains were grown on Bennett agar medium at 28°C for 4 or 5 days, and soft agar containing spores of B. subtilis was overlaid. The plates were incubated at 37°C overnight, and streptomycin production was detected by inhibition of the growth of the indicator.
Disruption of strR.
A 2.0-kb KpnI-SphI (position +305) fragment containing a 5′ portion of strR was cloned between the KpnI and SphI sites of pUC19 (plasmid pRU). A 2.6-kb AviII (position +942)-SacI fragment containing a 3′ portion of strR was cloned between the HincII and SacI sites of pUC19 (plasmid pRD). The SacI-SphI fragment excised from pRU by use of the restriction sites in the multicloning site, the SphI-EcoRI fragment from pRD, and a HindIII-SacI fragment carrying the kanamycin (neomycin) resistance gene from Tn5 were placed between the EcoRI and HindIII sites by four-fragment ligation. This plasmid was introduced into S. griseus IFO13350, and neomycin (5 μg/ml)-resistant transformants containing the whole plasmid sequence as a result of a single crossover were selected. Neomycin-sensitive colonies as candidates of the ΔstrR mutant were then isolated after a neomycin-resistant transformant had been cultured in the absence of neomycin. Correct strR-disrupted mutants were selected by Southern hybridization with the kanamycin (neomycin) resistance gene and a 0.4-kb fragment amplified by PCR with pb-F and gs-R (Table 1) as digoxigenin-labeled probes (data not shown).
S1 nuclease mapping.
Total RNA was isolated with ISOGEN (Nippon Gene) from cells grown on cellophane on the surface of Bennett agar medium. The method of S1 nuclease mapping was described previously (13). Hybridization probes were prepared by PCR with a pair of 32P-labeled and nonlabeled primers. Table 1 lists the forward (F) and reverse (R) primer sequences for preparing these probes: hrdB-F and hrdB-R* for hrdB; adpA-F and adpA-R* for adpA; fAa-F and gs-R* for strR; sB1-F and sB1-R* for strB1; sD-F and sD-R* for strD; sON-F and sON-R* for strO; sON-F* and sON-R for strN; sB-F and sB-R* for stsB; sC-F and sC-R* for stsC; sV-F and sV-R* for strV; and sU-F and sU-R* for strU. The primers with an asterisk were 32P labeled at the 5′ end with T4 polynucleotide kinase before PCR.
For detection of the transcription of strR on plasmids pRW, pRmA, pRmB, and pRmAB, the probe with 32P at the 5′ end which was deleted from the chromosome of the ΔstrR mutant was prepared as follows. pRA containing the DNA fragment (positions +56 to +2381) between the XbaI and EcoRI sites of pUC19 was constructed, and the SphI-EcoRI fragment (positions +300 to +2381) was inserted into pUC19, generating pRA-SE. By using pRA-SE as the template, a DNA fragment containing a portion of the pUC19 sequence (230 bp) and a portion of the strR coding sequence (positions +300 to +470) was amplified by PCR with 5′-TACGCAAACCGCCTCTCCCCG-3′ (positions 225 to 205 upstream of the HindIII site of pUC19) and *5′-TCTCCAGGACACGGGTCGCCG-3′ (positions +470 to +450). It was used as the probe to detect the strR transcript from +300 to +470.
Potassium permanganate footprinting.
The methods of potassium permanganate footprinting and RNA polymerase preparation were described previously (28). The 32P-labeled DNA fragment containing the strR promoter and the two AdpA-binding sites was prepared by PCR with primers gs-F and pf-R* (Table 1).
RESULTS AND DISCUSSION
Identification of two AdpA-binding sites in front of the strR promoter.
Because AdpA binds multiple sites of some target genes (29), we performed gel mobility shift assays by using various probes covering a long region at and around the transcriptional start point of strR (Fig. 1). In addition, it was hard to predict the actual AdpA-binding sites in silico because the consensus AdpA-binding sequence is rather generous (29). For convenience, we used histidine-tagged AdpA with the structure AdpA-Leu-Glu-His6. AdpA gave a strong shift signal for probe b (nucleotide positions −263 to −4) and probe f (positions −154 to +218), in addition to probe c (positions −321 to −189). Probe c contained the originally identified AdpA-binding site at position −270. No significant shift signal for probe d (positions −258 to −159) and probe a (positions −3 to +196) was observed, which indicated that AdpA bound an additional site located between positions −158 and −4. Probe e (positions −482 to −263) gave a faint signal, probably because the right end of this probe lacked a portion of the AdpA-binding site at the originally identified site at −270 in probe c. The originally detected site was named site B, and the newly detected site was named site A.
DNase I footprinting analysis of the two AdpA-binding sites.
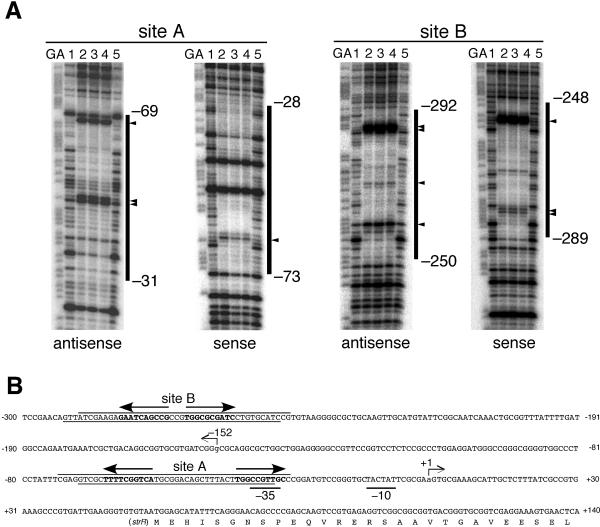
The two AdpA-binding sites upstream of strR were determined by DNase I footprinting with 32P-labeled probes (Fig. 2). AdpA-binding site A was protected from DNase I digestion at nucleotide positions −31 to −69 of the antisense strand and positions −73 to −28 of the sense strand (Fig. 2A). Several positions on the antisense strand at which enhanced DNase I digestion was observed, −46, −47, and −68, were probably exposed on the surface of the AdpA-DNA complex. On the sense strand, position −67 was also sensitive to DNase I. AdpA-binding site B was determined to be at positions −250 to −292 of the antisense strand and positions −289 to −248 of the sense strand. Positions −257, −268, −285, and −286 on the antisense strand and −255, −283, and −284 on the sense strand were hypersensitive to DNase I.
FIG. 2.
DNase I footprinting for determination of the two AdpA-binding sites upstream of strR. (A) DNase I footprinting assays were performed on the antisense and sense strands of sites A and B. The amounts of AdpA-H used in lanes 1 to 5 were 0, 0.6, 1.2, 2.4, and 0 μg, respectively. The DNase I digests were run with the same probes that were chemically cleaved (GA lanes). The nucleotide sequences protected from DNase I digestion, together with the nucleotide positions of their boundaries, are indicated. The nucleotides that are hypersensitive to DNase I are indicated by triangles. (B) Nucleotide sequence of the strR promoter and AdpA-binding sites. The arrow at position +1 indicates the transcriptional start point of strR. The arrow at position −152 indicates the transcriptional start point of strD. The DNA sequences protected from DNase I digestion are also shown, together with the AdpA-binding sequences (in boldface letters and with arrows) at sites A and B. The arrows indicate the AdpA-binding sequences (5′-TGGCSNGWWY-3′) that go from the 5′ to the 3′ side.
The consensus AdpA-binding sequence is 5′-TGGCSNGWWY-3′ (S is G or C, W is A or T, Y is T or C, and N is any nucleotide) (29). Site A contains two consensus AdpA-binding sequences, 5′-TGACCGAAAA-3′ and 5′-TGGCCGTTGC-3′, as a divergent repeat with a space of 16 bp (Fig. 2B). The AdpA-binding sites of this type are called type I (29). The AdpA-binding site for adsA (26), site 1 and site 3 of ssgA (27), and site 1 of amfR (28) are examples. According to our speculative model (29), a dimer of AdpA supposedly binds type I sites by anchoring one AdpA-binding sequence with the two helix-turn-helix DNA-binding motifs in one subunit and the other AdpA-binding sequence with the other DNA-binding motifs in the other subunit. Because of a very flexible linker between the N-terminal dimerization domain and the C-terminal DNA-binding domain, AdpA binds type I sites with various spaces, with its optimum spaces of 13 to 14 bp and 2 bp (29). The manner of AdpA binding to site A will be discussed below because the AdpA-binding sequence near the transcriptional start point completely overlaps the −35 element of the strR promoter.
Site B contains two AdpA-binding sequences, 5′-CGGCTGATTC-3′ and 5′-TGGCGCGATC-3′, as a divergent repeat with a space of 3 bp. AdpA binds sites of type I with optimal spaces of 13 to 14 bp and 2 bp. Since the AdpA-binding site of type I with a space of 2 bp has been named type I′, as is found for site 1 for ssgA (29), we can also group site B in type I′ and predict the manner of AdpA binding to this site, as shown in Fig. 1.
Alterations of the two AdpA-binding sequences.
We generated nucleotide changes at sites A and B to determine the importance of the AdpA-binding sequences at these sites in AdpA binding. An XbaI site was created in one of the AdpA-binding sequences at site A so that the number of nucleotides was unchanged (Fig. 3A). Gel mobility shift assays with the 32P-labeled probe containing the XbaI mutation at site A showed that AdpA-H no longer bound this mutated site A (Fig. 3B).
For introduction of a mutation into the two AdpA-binding sequences at site B, we first created an NdeI mutation (Fig. 3A) in one of the AdpA-binding sequences at site B. Because site B containing only the NdeI mutation still showed significant affinity for AdpA (data not shown), a PstI mutation (Fig. 3A) was further introduced into the other AdpA-binding sequence at site B. The 32P-labeled probe containing both the NdeI and PstI mutations showed very weak affinity for AdpA-H (Fig. 3B).
Disruption of one of the two AdpA-binding sequences at site A abolished the affinity for AdpA, whereas disruption of both AdpA-binding sequences at site B was required for abolishment of the affinity. We assume that two AdpA-binding sequences are required, in principle, for AdpA to bind type I and type I′ sites. However, it is apparent that one of the two AdpA-binding sequences in some type I and type I′ sites has enough affinity for AdpA to bind, like a single AdpA-binding sequence in type II sites (29).
Importance of the two AdpA-binding sites for transcriptional activation of strR.
We constructed an strR null mutant (the ΔstrR mutant) by double crossover to use this mutant as a host for determining the importance of the two AdpA-binding sites in the transcriptional activation of strR (Fig. 4A). The ΔstrR mutant constructed in this way contained an in-frame deletion from His-84 to Val-295 of StrR (nucleotide positions +305 to +940). As expected, the ΔstrR mutant grew normally and formed spores in the same time course as the wild-type strain. We constructed strR expression plasmids containing the mutations at site A and/or site B to determine whether one or both of the AdpA-binding sites are required for transcriptional activation of the strR promoter (Fig. 4C). aphD, encoding a streptomycin resistance determinant (streptomycin-6-phosphotransferase), was included in this construction because aphD is transcribed mainly by readthrough from the strR promoter (25). The DNA fragment containing the strR-aphD region together with the upstream region was cloned into pKUM20 with its copy number of one to two per chromosome (28), resulting in pR series plasmids. pRmA had an XbaI mutation at site A. pRmB had NdeI and PstI mutations at site B. pRmAB had all mutations at both sites. pRW was a control plasmid with no mutations. These four plasmids were introduced by transformation into the ΔstrR mutant.
(i) Effects of the mutations at sites A and B on in vivo transcription.
We first examined the transcription of strR in the ΔadpA and ΔstrR mutants by S1 nuclease mapping (Fig. 4B). RNA was isolated with Isogen (Nippon Gene) from cells grown on cellophane on the surface of Bennett agar medium. hrdB encoding a principal σ factor of RNA polymerase was used to check the purity and amount of the RNA used, as described previously (26). In wild-type strain IFO13350, strR was transcribed in the same time course as adpA. Of the two transcripts for strR, the large transcript corresponds to that reported previously (25), and therefore the small one is a degraded transcript. On the other hand, no strR transcript was detected in the ΔadpA mutant, as observed previously (20). An unexpected finding was that the strR transcription was greatly reduced in the ΔstrR mutant. This suggests that StrR acts as an activator of its own transcription, although there is no plausible StrR-binding sequence, 5′-GTTCGActG(N)11CagTcGAAc-3′ (highly conserved nucleotides are in capitals, and conserved nucleotides are in lowercase letters; N is any nucleotide) (23), in front of its transcriptional start point.
Under the culture conditions used, strain IFO13350 and the ΔstrR mutant grew as substrate mycelium until 24 h and as aerial mycelium thereafter. An additional unexpected observation was that transcription of strR occurred rather early in the cells from which the RNA was isolated. As described below, streptomycin production was observed at 3 days of growth. This may be ascribed to the difference in the inoculation of the cells; the RNA was prepared from the cells grown on agar medium by spreading a lump of mycelium, which supposedly contains A-factor in a larger amount than in the cells grown by inoculating a small amount of mycelium with a toothpick for streptomycin assays.
We next determined the transcription of strR on plasmids pRW, pRmA, pRmB, and pRmAB by using the probe with 32P at the 5′ end which was deleted from the chromosome of the ΔstrR mutant. This means that the strR transcripts in Fig. 4D originated from the strR promoters on these plasmids. pRW containing native sites A and B and the strR-aphD region (Fig. 4C) allowed the transcription of plasmid-borne strR in the ΔstrR mutant (Fig. 4D). However, the transcription of strR in the ΔstrR mutant harboring pRmA or pRmB was much less active than that from the intact strR promoter on pRW. The strR transcription in the ΔstrR mutant harboring pRmAB was severely impaired. The detectable transcription of strR in the ΔstrR mutant harboring pRmA or pRmB may be ascribed to the ability of AdpA to bind to the mutated sites with very low affinity. These findings suggest that both site A and site B are required for full activation of strR by AdpA.
(ii) Effects of the mutations at sites A and B on streptomycin production.
The wild-type strain S. griseus IFO13350 harboring the vector pKUM20 produced 9.0 ± 2.9 μg and 200 ± 46 μg of streptomycin per colony on day 4 and day 5, respectively (Fig. 4E), as determined by a bioassay using Bacillus subtilis as an indicator (7). The amount of the streptomycin produced was calculated by using a calibration curve, obtained with authentic streptomycin. Consistent with the idea that StrR is the pathway-specific activator for streptomycin biosynthesis, almost no streptomycin was produced by the ΔstrR mutant harboring pKUM20. A tiny halo around the colony was probably due to some other anti-B. subtilis substance produced by S. griseus. This is in agreement with the observation that in the ΔstrR mutant no transcription of streptomycin biosynthesis genes was detected (see below). The ΔstrR mutant harboring pRW produced 4.7 ± 2.1 μg and 100 ± 61 μg of streptomycin on day 4 and day 5, respectively, showing that strR transcription occurred to almost the same extent as in the wild-type strain. On the other hand, the ΔstrR mutant harboring pRmA produced 1.6 ± 0.4 μg and 26.5 ± 11.2 μg of streptomycin per colony on days 4 and 5, respectively. Likewise, the ΔstrR mutant harboring pRmB produced 0.6 ± 0.2 μg and 2.8 ± 0.5 μg of streptomycin per colony on days 4 and 5, respectively. In addition, the ΔstrR mutant harboring pRmAB produced almost no streptomycin, just like the ΔstrR mutant harboring the vector pKUM20. The amounts of streptomycin produced by these mutants reflected the strR promoter activities on the plasmids (Fig. 4D). Furthermore, the transcriptional assays and streptomycin assays showed that site B contributed more to the transcriptional activation by AdpA than site A.
(iii) Effects of the mutations at sites A and B on the transcription of strB1, one of the streptomycin biosynthesis genes.
Retzlaff and Distler (23) reported that strB1, encoding aminocyclitol amidinotransferase, one of the streptomycin biosynthesis enzymes, possesses an StrR-binding sequence, 5′-GTTCGActG(N)11CagTcGAAc-3′, and is activated by StrR. We confirmed their observation; in the wild-type strain, strB1 was transcribed actively at 18 and 24 h whereas no strB1 transcription occurred in the ΔstrR mutant or in the ΔadpA mutant (Fig. 4B). We next determined the strB1 transcription in the ΔstrR mutant harboring pRmA, pRmB, and pRmAB (Fig. 4D). strB1 transcription was restored by strR on pRW containing the intact strR gene to a significant level. The degrees of restoration of strB1 transcription by pRmA, pRmB, and pRmAB reflected, in principle, those of strR transcription (Fig. 4D). The large difference in the yields of streptomycin does not reflect the rather small difference in strB1 transcription, which is an example to show that the level of transcription does not always correspond to the level of translation.
Transcription of all the streptomycin biosynthesis genes is dependent on StrR.
Retzlaff and Distler (23) showed that StrR binds the intervening region between stsC and stsB and predicted the presence of an StrR-binding sequence between strU and strV (see Fig. 5), although no transcriptional analysis other than strB1 and strR has yet been performed. We determined the dependence of the transcription of these genes on StrR by low-resolution S1 mapping (Fig. 4B). In the wild-type strain, these four genes were transcribed in the same time course as strB1. The approximate transcriptional start points of strU, stsC, and stsB were 215, 25, and 70 nucleotides upstream of the respective start codons. The transcriptional start point of strV was near the first nucleotide of the ATG start codon. However, no transcription of these genes occurred in the ΔadpA or ΔstrR mutant, indicating that these four transcriptional units depend on StrR. Since introduction of pRW into the ΔstrR mutant restored streptomycin production, the transcription of all these units was supposedly restored by strR.
There is no apparent StrR-binding sequence upstream of strO, strN, or strD (23). However, the transcription of these genes also depended on StrR; no transcription occurred in the ΔstrR or ΔadpA mutant, whereas these genes were transcribed in the wild-type strain in the same time course as strB1 (Fig. 4B). The transcriptional start point of strO was near the first nucleotide of the ATG start codon. The transcriptional start points of strN and strD were approximately 20 and 200 nucleotides upstream from the respective start codons. These findings showed that these three transcriptional units were also dependent on StrR, either directly or indirectly. We assume that StrR binds to the upstream regions of the respective promoters, despite the absence of an apparent StrR-binding sequence, because no DNA-binding proteins other than StrR are encoded within the streptomycin biosynthetic gene cluster, as described below. Further analysis of the StrR-controlled transcription of these genes is required.
strR as a unique regulator in the streptomycin biosynthesis gene cluster.
Synteny analysis of the streptomycin biosynthesis gene cluster, in a total of 32.6 kb (Fig. 5), shows that this gene cluster has been inserted into the genome of S. griseus by horizontal transfer, as suggested previously by Egan et al. (4). Comparison of the genome structures of Streptomyces coelicolor A3(2) (3) and S. griseus (our unpublished data) reveals that the streptomycin biosynthesis gene cluster, together with a gene for a hypothetical membrane protein, was inserted into the S. griseus genome between the genes corresponding to SCO1592 and SCO1591. At this site in Streptomyces avermitilis (8), a functionally unknown gene, SAV6747, is inserted. An important implication of the synteny analysis is that the streptomycin biosynthesis gene cluster contains strR as a unique transcriptional regulator; no transcriptional regulator-like proteins other than StrR are encoded within the horizontally transferred DNA fragment.
Open-complex formation facilitated by AdpA.
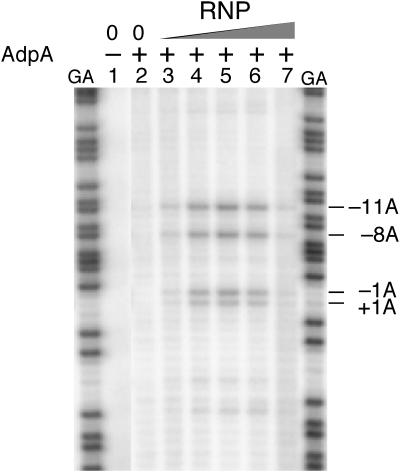
The AraC/XylS family proteins increase the affinity of RNA polymerase for their target genes and facilitate the formation of an open complex (18, 19). As demonstrated for AraC (9), MelR (2), SoxS (16), and Rns (19), we previously showed that AdpA recruits RNA polymerase to the promoter region of amfR, adsA, and ssgA and facilitates the isomerization of the RNA polymerase-DNA complex into an open complex competent for transcriptional initiation (28, 29). We examined whether AdpA facilitates open-complex formation at the strR promoter by potassium permanganate footprinting (Fig. 6). This method is based on the characteristic of potassium permanganate to react preferentially with thymidines on unpaired nucleotides. Briefly, KMnO4-reacted nucleotides on a single strand, formed as a result of open-complex formation, were chemically cleaved with piperidine and detected on a sequencing gel. RNA polymerase was prepared from exponentially growing cells of S. griseus IFO13350 as described previously (28). Incubation of the DNA fragment (positions −482 to +68) containing the strR promoter in the presence of RNA polymerase alone gave no open complex (data not shown), as was found for amfR (28), ssgA (29), and adsA (29). However, addition of both RNA polymerase and AdpA-H revealed several thymidine residues, from position −11 to +1 on the antisense strand, that were modified and detected by the footprinting. When the concentration of AdpA-H was fixed at 20 nM, open-complex formation was detected depending on the concentration of RNA polymerase. These results show that AdpA facilitates open-complex formation at an appropriate position for transcriptional initiation of strR.
FIG. 6.
Dependence of open-complex formation at the strR promoter on AdpA, as determined by potassium permanganate footprinting. The 32P-labeled antisense strand of the strR promoter region (positions −482 to +68) was used. The concentration of AdpA-H in lanes 2 to 7 was 20 nM. Ten nanomolar AdpA equals 8.8 ng of AdpA in a total volume of 20 μl. The concentrations of RNA polymerase (RNP) in lanes 3 to 7 were 5, 15, 50, 150, and 300 nM, respectively. The positions of the thymidines reacted with KMnO4, corresponding to adenines of the sense strand, were determined using the G+A sequencing ladder (lane GA) as a reference. The nucleotide numbers are shown taking the transcriptional start point as +1.
Signal transduction from A-factor to the streptomycin biosynthesis gene cluster.
Some target genes of AdpA, such as amfR (28), sgmA (12), and sprA (our unpublished data), contain two UASs to which AdpA binds. The two UASs are both necessary for transcriptional activation by AdpA, the molecular mechanism of which still remains to be elucidated. It should be noted that one of the AdpA-binding sequences at site A overlaps with the −35 element of the strR promoter. If the DNA-binding domain in one subunit of the AdpA dimer sat at this position, it would prevent RNA polymerase from binding to the strR promoter properly. Since the AdpA dimer bound near the promoter supposedly interacts with RNA polymerase, we speculate that, on binding RNA polymerase, AdpA binds site A by anchoring the AdpA-binding sequence of the 5′ side with only one of the DNA-binding domains so as to make room for RNA polymerase to bind the promoter. This is consistent with the observation that only the XbaI mutation at site A was sufficient to avoid the affinity of AdpA (Fig. 3B).
We show here that the pathway-specific transcriptional activator gene for streptomycin biosynthesis, strR, is activated by two molecules of AdpA that separately bind two UASs located at nucleotide positions −270 and −50. In addition, strR appears to activate its own transcription because the strR transcription in the ΔstrR mutant was greatly repressed. Due to this autoactivation system, the A-factor signal starting from A-factor to strR, via ArpA and AdpA, to trigger streptomycin production must lead to a rapid increase in the amount of StrR, which in turn leads to rapid and simultaneous transcription of other streptomycin biosynthetic genes within the cluster. Concerning the streptomycin biosynthesis genes, strB1 was under the control of StrR, as reported by Retzlaff and Distler (23). They also showed that stsB and stsC, having an strR-binding sequence upstream of their promoters, are bound by StrR. Their prediction that StrR activates the transcription of these genes is true since the present study shows that all four of these genes are activated by StrR in the same manner as strB1. We have also shown that the remaining three promoters for strO, strN, and strD in the cluster are similarly activated by StrR although these promoters do not contain an apparent StrR-binding sequence (23). All these results suggest that StrR, as a sole regulator within the gene cluster, serves as a transcriptional activator for all the streptomycin biosynthesis genes. Consistent with this idea, strR (together with aphD) under the control of the hrdB promoter on pKUM20 caused the ΔadpA mutant to produce streptomycin, although its growth was significantly impaired, probably due to some inhibitory effect of strR overexpression (data not shown).
Biosynthesis of secondary metabolites is controlled by a variety of external signals, such as nutrient conditions, including carbon, nitrogen, and phosphate, and physiological conditions. Since strR is a unique regulator in the streptomycin biosynthesis gene cluster, these signals must be gathered to the promoter of strR. We previously detected several different proteins that bind the upstream region of the strR promoter by gel mobility shift assay (24). These proteins may represent a certain signal and regulate strR in either a positive or a negative way, thus controlling the whole streptomycin biosynthesis genes. The structure of the streptomycin biosynthesis gene cluster is simple and makes a simple target to study secondary metabolite formation in response to external signals.
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas from Monkasho and the Bio Design Program of the Ministry of Agriculture, Forestry, and Fisheries of Japan.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingstone, D. O. Moore, J. S. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Belyaeva, T. A., J. T. Wade, C. L. Webster, V. J. Howard, M. S. Thomas, E. I. Hyde, and S. J. W. Busby. 2000. Transcription activation at the Escherichia coli melAB promoter: the role of MelR and the cyclic AMP receptor protein. Mol. Microbiol. 36:211-222. [DOI] [PubMed] [Google Scholar]
- 3.Bentley, S. D., K. F. Chater, A.-M. Cerdeño-Tárraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C.-H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M.-A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 4.Egan, S., P. Wiener, D. Kallifidas, and E. M. H. Wellington. 1998. Transfer of streptomycin biosynthesis gene clusters within streptomycetes isolated from soil. Appl. Environ. Microbiol. 64:5061-5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horinouchi, S. 2002. A microbial hormone, A-factor, as a master switch for morphological differentiation and secondary metabolism in Streptomyces griseus. Front. Biosci. 7:d2045-d2057. [DOI] [PubMed] [Google Scholar]
- 6.Horinouchi, S., and T. Beppu. 1994. A-factor as a microbial hormone that controls cellular differentiation and secondary metabolism in Streptomyces griseus. Mol. Microbiol. 12:859-864. [DOI] [PubMed] [Google Scholar]
- 7.Horinouchi, S., Y. Kumada, and T. Beppu. 1984. Unstable genetic determinant of A-factor biosynthesis in streptomycin-producing organisms: cloning and characterization. J. Bacteriol. 158:481-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ikeda, H., J. Ishikawa, A. Hanamoto, M. Shinose, H. Kikuchi, T. Shiba, Y. Sakaki, M. Hattori, and S. Omura. 2003. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21:526-531. [DOI] [PubMed] [Google Scholar]
- 9.Johnson, C. M., and R. F. Schleif. 2000. Cooperative action of the catabolite activator protein and AraC in vitro at the araFGH promoter. J. Bacteriol. 182:1995-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kato, J., W.-J. Chi, Y. Ohnishi, S.-K. Hong, and S. Horinouchi. 2005. Transcriptional control by A-factor of two trypsin genes in Streptomyces griseus. J. Bacteriol. 187:286-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato, J., I. Miyahisa, M. Mashiko, Y. Ohnishi, and S. Horinouchi. 2004. A single target is sufficient to account for the biological effects of the A-factor receptor protein of Streptomyces griseus. J. Bacteriol. 186:2206-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kato, J., A. Suzuki, H. Yamazaki, Y. Ohnishi, and S. Horinouchi. 2002. Control by A-factor of a metalloendopeptidase gene involved in aerial mycelium formation in Streptomyces griseus. J. Bacteriol. 184:6016-6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelemen, G. H., P. Brian, K. Flärdh, L. Chamberlin, K. F. Chater, and M. J. Buttner. 1998. Developmental regulation of transcription of whiE, a locus specifying the polyketide spore pigment in Streptomyces coelicolor A3(2). J. Bacteriol. 180:2515-2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khokhlov, A. S., I. I. Tovarova, L. N. Borisova, S. A. Pliner, L. A. Schevchenko, E. Y. Kornitskaya, N. S. Ivkina, and I. A. Rapoport. 1967. A-factor responsible for the biosynthesis of streptomycin by a mutant strain of Actinomyces streptomycini. Dokl. Akad. Nauk SSSR 177:232-235. [PubMed] [Google Scholar]
- 15.Kieser, H., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 16.Li, Z., and B. Demple. 1996. Sequence specificity for DNA binding by Escherichia coli SoxS and Rob proteins. Mol. Microbiol. 20:937-945. [DOI] [PubMed] [Google Scholar]
- 17.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 18.Martin, R. G., and J. L. Rosner. 2001. The AraC transcriptional activators. Curr. Opin. Microbiol. 4:132-137. [DOI] [PubMed] [Google Scholar]
- 19.Munson, G. P., and J. R. Scott. 2000. Rns, a virulence regulator within the AraC family, requires binding sites upstream and downstream of its own promoter to function as an activator. Mol. Microbiol. 36:1391-1402. [DOI] [PubMed] [Google Scholar]
- 20.Ohnishi, Y., S. Kameyama, H. Onaka, and S. Horinouchi. 1999. The A-factor regulatory cascade leading to streptomycin biosynthesis in Streptomyces griseus: identification of a target gene of the A-factor receptor. Mol. Microbiol. 34:102-111. [DOI] [PubMed] [Google Scholar]
- 21.Ohnishi, Y., J.-W. Seo, and S. Horinouchi. 2002. Deprogrammed sporulation in Streptomyces. FEMS Microbiol. Lett. 216:1-7. [DOI] [PubMed] [Google Scholar]
- 22.Ohnishi, Y., H. Yamazaki, J. Kato, A. Tomono, and S. Horinouchi. 2005. AdpA, a central transcriptional regulator in the A-factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces griseus. Biosci. Biotechnol. Biochem. 69:431-439. [DOI] [PubMed] [Google Scholar]
- 23.Retzlaff, L., and J. Distler. 1995. The regulator of streptomycin gene expression, StrR, of Streptomyces griseus is a DNA binding activator protein with multiple recognition sites. Mol. Microbiol. 18:151-162. [DOI] [PubMed] [Google Scholar]
- 24.Vujaklija, D., S. Horinouchi, and T. Beppu. 1993. Detection of an A-factor-responsive protein that binds to the upstream activation sequence of strR, a regulatory gene for streptomycin biosynthesis in Streptomyces griseus. J. Bacteriol. 175:2652-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vujaklija, D., K. Ueda, S.-K. Hong, T. Beppu, and S. Horinouchi. 1991. Identification of an A-factor-dependent promoter in the streptomycin biosynthetic gene cluster of Streptomyces griseus. Mol. Gen. Genet. 229:119-128. [DOI] [PubMed] [Google Scholar]
- 26.Yamazaki, H., Y. Ohnishi, and S. Horinouchi. 2000. An A-factor-dependent extracytoplasmic function sigma factor (σAdsA) that is essential for morphological development in Streptomyces griseus. J. Bacteriol. 182:4596-4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamazaki, H., Y. Ohnishi, and S. Horinouchi. 2003. Transcriptional switch on of ssgA by A-factor, which is essential for spore septum formation in Streptomyces griseus. J. Bacteriol. 185:1273-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamazaki, H., Y. Takano, Y. Ohnishi, and S. Horinouchi. 2003. amfR, an essential gene for aerial mycelium formation, is a member of the AdpA regulon in the A-factor regulatory cascade in Streptomyces griseus. Mol. Microbiol. 50:1173-1187. [DOI] [PubMed] [Google Scholar]
- 29.Yamazaki, H., A. Tomono, Y. Ohnishi, and S. Horinouchi. 2004. DNA-binding specificity of AdpA, a transcriptional activator in the A-factor regulatory cascade in Streptomyces griseus. Mol. Microbiol. 53:555-572. [DOI] [PubMed] [Google Scholar]