Abstract
DNA adenine methylase (Dam−) mutants of Salmonella enterica serovar Typhimurium contain reduced levels of FinP RNA encoded on the virulence plasmid. Dam methylation appears to regulate finP transcription, rather than FinP RNA stability or turnover. The finP promoter includes canonical −10 and −35 modules and depends on the σ70 factor. Regulation of finP transcription by Dam methylation does not require DNA sequences upstream from the −35 module, indicating that Dam acts at the promoter itself or downstream. Unexpectedly, a GATC site overlapping with the −10 module is likewise dispensable for Dam-mediated regulation. These observations indicate that Dam methylation regulates finP transcription indirectly and suggest the involvement of a host factor(s) responsive to the Dam methylation state of the cell. We provide evidence that one such factor is the nucleoid protein H-NS, which acts as a repressor of finP transcription in a Dam− background. H-NS also restrains transcription of the overlapping traJ gene, albeit in a Dam-independent fashion. Hence, the decreased FinP RNA content found in Dam− hosts of S. enterica appears to result from H-NS-mediated repression of finP transcription.
The F sex factor and its relatives have served as a paradigm of bacterial conjugation for six decades (62). Early studies carried out with F paved the way for the discovery of plasmids in other bacterial genera. For instance, a plasmid initially called “cryptic” and later found to contain virulence genes was discovered in the standard strain LT2 of Salmonella enterica serovar Typhimurium (51, 52). Virulence plasmids are also found in other strains of S. enterica serovar Typhimurium, such as the mouse virulent strains SL1344 and 14028, and in other host-adapted serotypes of S. enterica except the human-adapted serovar Typhi (46).
The virulence plasmid of S. enterica strain LT2 (pSLT) is self-transmissible (1). Unlike F, pSLT undergoes conjugation at low frequencies (1, 5), and the genetic mechanism that maintains conjugal repression can cross-regulate F transfer (48, 51, 59). Repression of mating is the norm for most F-like plasmids, and it relies on the so-called fertility inhibition system (17). Transcription of the main transfer operon (tra), which encodes functions for pilus synthesis and DNA processing during conjugal transfer, requires activation by the TraJ protein, encoded on a nearby monocistronic transcriptional unit (18, 19, 41). Synthesis of the TraJ transcriptional activator is prevented by a small, untranslated RNA encoded by an overlapping gene, finP (20, 41). Pairing between FinP RNA and the traJ mRNA leader triggers degradation of the RNA duplex by RNase III (29), a phenomenon reminiscent of the RNA interference mechanism described in eukaryotes (38). FinP RNA is a short-lived molecule unless it is stabilized by the FinO protein (30). FinO protects FinP RNA from degradation by RNase E (29) and acts as an RNA chaperone that facilitates RNA-RNA interactions (2). The F sex factor is a FinO− mutant (8), and the ability of pSLT to repress F transfer relies on providing trans-acting FinO protein (48). In certain F-like plasmids, activation of the main tra promoter, traY, is also activated by transcription of the plasmid-borne gene traM. Transcripts originating at the traM promoter appear to enter the neighboring traJ gene, thereby providing a two-cistron mRNA that can unbalance the finP/traJ transcriptional ratio and yield TraJ product for the activation of the traY promoter (11).
Besides these plasmid-encoded regulators of tra operon expression, chromosomal regulators of mating are also known (39). The list includes the transcriptional regulators ArcA (55, 57) and CRP (53), the nucleoid protein H-NS (54, 61), the leucine-responsive regulatory protein (5), and DNA adenine methylase (Dam) methylation (59). A postranscriptional control mechanism acting on the TraJ product has been also described (23, 50).
Regulation of conjugal transfer by Dam methylation occurs in F (59), pSLT (5), and R100 (7). In all cases, Dam methylation acts as a repressor of mating, and the increase in conjugation frequency detected in Dam− donors varies from plasmid to plasmid (5, 7, 59). The increased conjugal ability of Dam− mutants was initially correlated with lower levels of FinP RNA (59). Further studies have shown that repression of pSLT transfer by Dam methylation actually involves two independent, concerted actions: activation of FinP RNA synthesis and repression of traJ transcription (6).
In this study, we have investigated the cause of the lowered FinP RNA levels found in DNA adenine methylase mutants of S. enterica. We show that transcription of finP is repressed in a Dam− background by an unusual, indirect mechanism that involves the nucleoid protein H-NS (13). Our findings explain the ability of Salmonella and Escherichia coli Dam− mutants to undergo conjugation at elevated frequencies (5, 59), and they raise the possibility of a hitherto unknown relationship between Dam methylation and H-NS activity.
MATERIALS AND METHODS
Bacterial strains, plasmids, and strain construction.
Unless otherwise indicated, the strains used in this study were derived from Salmonella enterica serovar Typhimurium strain LT2 (Table 1). TR5878, a transformable strain of S. enterica, was obtained from J. R. Roth, Section of Microbiology, University of California, Davis. The hns::Kmr allele (15) was provided by N. P. Higgins, University of Alabama, Birmingham. E. coli strains P90A5 and UQ295 (CGSC 6328) were obtained from J. L. Ramos, Estación Experimental del Zaidín, CSIC, Granada, Spain. The source of the lexA(Ind−) allele used in this study was strain DA6522, obtained from D. I. Andersson, Swedish Institute for Infectious Disease Control, Solna, Sweden. Transductional crosses using phage P22 HT (49) were used for strain construction operations involving chromosomal markers and for transfer of plasmids among Salmonella strains. Plasmid pIC552 is a promoter-probe vector that permits the construction of transcriptional lac fusions (33). pMD1405, obtained from M. Drummond (John Innes Institute, Norwich, England), is a ColE1 derivative engineered for the construction of translational fusions with a lacZ gene that lacks the first eight codons. pIZ860 contains a 0.6-kb EcoRI-NaeI insert that includes pSLT DNA from −298 to +362 relative to the tra gene start site cloned on pBluescript II SK(+). This insert contains a complete finP gene but lacks the traJ promoter. pIZ879 contains a 301-bp HinfI-BamHI fragment from pSLT cloned on BamHI-EcoRV-digested pBluescript II SK(+) (59). pIZ880 is a pIC552 derivative that contains a transcriptional finP::lac fusion and lacks the traJ promoter of pSLT (59). pIZ899 carries a 326-bp EcoRV fragment of pSLT (which includes a complete finP gene) cloned onto pBluescript II SK(+) (59). pIZ900 is a pMD1405 derivative containing the same 326-bp EcoRV fragment carried by pIZ899 (59).
TABLE 1.
Strain list
| Designation | Genotype | Reference or sourcea |
|---|---|---|
| P90A5b | lacZ4 arg75 λ− F− | J. L. Ramos |
| UQ285b | lacZ4 rpoD285 argG75 λ− F− | 28 |
| SV3000 | dam-201::Tn10dTc | 58 |
| SV3081 | pSLT− | 59 |
| SV3083 | dam-201::Tn10dTc pSLT− | 59 |
| SV4107 | LT2/pIZ900 | 59 |
| SV4109 | dam-201::Tn10dTc/pIZ900 | 59 |
| SV4761 | φ(traJ′-lacZY) | 6 |
| SV4762 | φ(finP′-lacZY) | |
| SV4763 | φ(traJ′-lacZY) dam-201::Tn10dTc | 6 |
| SV4764 | φ(finP′-lacZY) dam-201::Tn10dTc | |
| SV5017 | φ(finP′-lacZY) ΔmutH501 | |
| SV5019 | φ(finP′-lacZY) ΔmutH501 lexA(Ind) | |
| SV5024 | hns::Km pSLT− | |
| SV5025 | hns::Km dam-201::Tn10dTc pSLT− | |
| SV5048 | hns::Cm | |
| SV5052 | φ(finP′-lacZY) hns::Cm | |
| SV5053 | φ(finP′-lacZY) hns::Cm dam-201::Tn10dTc | |
| SV5054 | φ(traJ′-lacZY) hns::Cm | |
| SV5055 | φ(traJ′-lacZY) hns::Cm dam-201::Tn10dTc | |
| TR5878 | galE496 r(LT2)− m(LT2)− r(S)+ilv-542 metA22 trpB2 Fels2−fliA66 strA120 xyl-404 metE551 hspL56 hspS29 | J. R. Roth |
| TT10487 | hisO1242/pGE108 | J. R. Roth |
Omitted for strains described in this study.
Escherichia coli.
Construction of plasmid pIZ955.
Plasmid pIZ879 was digested with EcoRV and XhoI. A 100-bp fragment generated by this digestion was cloned on pIC552 previously digested with NcoI, Klenow end filled, and digested with XhoI. The resulting plasmid (pIZ955) carried a finP::lac transcriptional fusion and 66 bp upstream of the finP start site.
Construction of plasmid pIZ981.
Plasmid pIZ879 was used as a template for PCR amplification with the Universal primer of pBluescript and a primer designed ad hoc (DFP, 5′ CGG GAT CCA GAT TGA CTG GCC AGT GTT 3′). The DFP oligonucleotide introduces a BamHI restriction site located at its 5′ end. The 163-bp amplified fragment was purified, digested with BamHI and XhoI, and cloned on BglII-XhoI-digested pIC552. The resulting plasmid (pIZ981) carries a finP::lac transcriptional fusion and 37 bp upstream of the finP start site.
Construction of an hns::Cmr allele.
The hns gene was disrupted by adapting to S. enterica the method of Datsenko and Wanner (10). The oligonucleotides 5′ GCC GAC GGC ATT GAC CCG AAT GAA CTG CTG AAT AGC ATG GCT GCC GCT AAA TCC GGT ACC GTG TAG GCT GGA GCT GCT TC 3′ and 5′ TTT CAC CGT TTT CGT CAA CAT AGC TAT ATT TAG CCG GAC GAG CTG CGC GTT TAG CTT TGA CAT ATG AAT ATC CTC CTT AG 3′ were used to PCR amplify the chloramphenicol resistance cassette of plasmid pKD3 (10). The amplified plasmid was used to electroporate a derivative of strain TR5878 carrying plasmid pKD46 (10), selecting Cmr transformants. This procedure yielded a Cmr cassette insertion at the KpnI restriction site of the hns gene, approximately 330 bp downstream from the initiation codon. The construct was verified by PCR amplification with two oligonucleotides external to the hns gene, 5′ TAC ATT CCT GGC TAT TGCA C 3′ and 5′ TTT TTA TGC CTG GGG TCG TC 3′.
Construction of a finP::lac transcriptional fusion in the Salmonella virulence plasmid.
A deletion in the finP gene of pSLT was generated according to the method of Datsenko and Wanner (10). Primers were designed to eliminate a 100-bp DNA stretch of traJ from position +2 to position +102 (EMBL accession number AJ011572). The sequences of these oligonucleotides were 5′ GTG ACA AAT TAA GGC TGA AAA CAA TAT AGT GTC TTT GTA GGT GTA GGC TGG AGC TGC TTC 3′ and 5′ ATT GAC TGG CGA GTG TTC TTT CTC TAC GAT CCG TCG GAC ACA TAT GAA TAT CCT CC TTA G 3′. The resulting deletion eliminates the whole finP gene except 3 bp at its 5′ end. The kanamycin resistance cassette was excised by recombination with plasmid pCP20 (10). PCR amplification using primers from both sides of the pSLT finP locus identified kanamycin-sensitive isolates that carried the desired deletion. The sequences of these primers were 5′ GTA TCT TCA GAG ATG GAA AG 3′ and 5′ TAT GAT TAC ACG GAA AACGG 3′. The resulting FRT site was then used to integrate plasmid pCE37 (14), thereby generating a transcriptional finP::lac fusion.
Culture media and growth conditions.
E medium containing 0.2% glucose (60) was used as the minimal medium. The rich medium was nutrient broth (8 g/liter; Difco) with added NaCl (5 g/liter). Solid media contained agar at a 1.5% final concentration. Antibiotics and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) were used at the concentrations described previously (59).
β-Galactosidase assays.
Levels of β-galactosidase activity were assayed as described by Miller (40), using the CHCl3-sodium dodecyl sulfate (SDS) permeabilization procedure.
RNA extraction.
RNA preparations were obtained by guanidinium isothiocyanate lysis and phenol-chloroform extraction (9). Saturated cultures were immersed in liquid N2, and 1.4 ml of lysis solution (5 M guanidinium isothiocyanate, 50 mM Tris [pH 7.5], 10 mM EDTA, 8% [vol/vol] β-mercaptoethanol) was added. Each mixture was incubated at 60°C for 10 min before 0.28 ml of chloroform was added. After gentle shaking and centrifugation at 9,000 rpm for 10 min, 0.66 ml of isopropanol was added to the supernatant. The samples were incubated at −20°C for 15 min and centrifuged again at 9,000 rpm for 15 min. The pellets were then rinsed with 70% ethanol and dried. After resuspension in 75 μl of water (with 0.1% [vol/vol] diethyl pyrocarbonate), the samples were subjected to standard treatments with DNase and proteinase K, followed by extraction with phenol-chloroform-isoamyl alcohol and chloroform-isoamyl alcohol (47). The aqueous phase was precipitated with 1/10 volume of 3 M sodium acetate, pH 4.8, and 2.5 volumes of absolute ethanol. The samples were then kept at −78°C for at least 30 min, centrifuged, and washed with 70% ethanol. Finally, the precipitates were dried and resuspended in 20 to 40 μl of diethyl pyrocarbonate-water.
RNA electrophoresis in polyacrylamide gels.
Samples contained 6 μl of the RNA preparation and 4 μl of loading buffer containing 50% formamide. After 2 min of incubation at 94°C, the samples were chilled in ice. Electrophoretic separation was carried out on gels prepared with Tris-borate-EDTA (TBE) and containing 8% acrylamide and 7.5 M urea. The gels were 12 cm long and 0.75 mm thick. Electrophoretic separation was performed at 250 V.
Primer extension.
The oligonucleotide 5′ TAA ATC GCC GAT ACA GGG AG 3′, complementary to an internal region of the finP gene of pSLT (EMBL accession number AJ011572), was end labeled with [γ32P]ATP and annealed to 25 μg of total RNA prepared from S. enterica strains bearing plasmid pIZ900 (59). For annealing, 105 cpm of oligonucleotide was used. The end-labeled primer was extended with avian myeloblastosis virus reverse transcriptase (Boehringer Mannheim, Mannheim, Germany) under conditions described previously (37). The products of reverse transcription were analyzed in urea-polyacrylamide gels. For autoradiography, the gels were exposed to Kodak Biomax MR film.
RNA hybridization against DNA probes.
RNA was quantified by the A260/A280 ratio, using a Beckman DU spectrophotometer. Aliquots were loaded on a polyacrylamide gel under the conditions described above. After electrophoretic separation of RNA, the polyacrylamide gels were treated with cold TBE (0.5×) for 15 min. Transfer to nylon filters was achieved with a Transblot SD Semidry Transfer Cell system from Bio-Rad Laboratories, Richmond, Calif. Transfer was allowed to proceed for 1 h at 400 mA at intensities below 25 V. Prehybridization and hybridization were both carried out at 38°C, and formamide was not used. After transfer, the filters were stained with a solution of 0.3% methylene blue in 0.3 M sodium acetate, pH 5.2, to confirm both the efficiency of transfer and the presence of equivalent amounts of RNA per lane. As a loading control, rRNA fragments, which were readily visible in the stained gel, were quantified using Image Gauge software in a Fujifilm FLA-3000 Image System. The presence of equivalent amounts of RNA per lane was thus confirmed. Loads that showed differences below 10% were not considered significant. The 5′ TAA ATC GCC GAT ACA GGG AG 3′ probe, complementary to an internal finP region, was end labeled with [γ32P]ATP. After hybridization, the nylon filters were washed twice at room temperature with 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% SDS for 5 min. Two or more additional washes were carried out with either 0.6× SSC-0.1% SDS or 3× SSC-0.1% SDS for 15 min at room temperature. The filters were then exposed to an X-ray film for 1 to 7 days.
DNA sequencing.
Sequencing reactions were carried out with the Sequenase 2.0 sequencing kit (U.S. Biochemical Corp., Cleveland, Ohio). The manufacturer's instructions were followed. Additionally, 1 μl of unlabeled dATP (10 μM) was added to the reaction mixtures. Sequencing gels were prepared in TBE and contained 6% acrylamide and 500 g/liter urea. The gels were run in a Poker Face SE1500 sequencer (Hoeffer Scientific Instruments, San Francisco, Calif.); dried in a Slab Gel Dryer, model SE1160 (Hoeffer Scientific Instruments); and developed by exposure to X-ray film.
Determination of plasmid copy number by quantitative PCR.
A method of DNA extraction that does not modify the chromosome/plasmid DNA ratio was adapted from Brandi et al. (4). Five-milliliter overnight cultures were washed once with 1 ml of 1 M NaCl and three times with 1 ml of TE (10 mM Tris-HCl, pH 8, 1 mM EDTA). The cells were finally resuspended in 200 μl TE and lysed by three rounds of freeze-thawing followed by 5 min if boiling. After appropriate dilutions, aliquots of the crude cell extracts were subjected to quantitative PCR (QPCR) to establish the relative chromosomal and plasmid DNA contents. Approximately 10 ng total DNA, determined by agarose gel electrophoresis and ethidium bromide staining, was used for each amplification. A Lightcycler-FastStartDNA Master SYBR Green I kit (Roche Diagnostics, Sant Cugat del Vallès, Spain) was used for real-time PCR according to the manufacturer's instructions. The oligonucleotides used were 5′ TAC ACC TGC CAT CTG GCT GC 3′ and 5′ ATT ATA CTG GGC TTC TCC GG 3′ for plasmid quantification and 5′ CTT ACA TTG CCA AAC AAC GTC AA AT 3′ and 5′ GAA AGC TCA AAC GCA TCT TCC AGT 3′ for the chromosome. The concentration of oligonucleotides in every reaction was 1 μM. Amplifications were performed in an iCycler (Bio-Rad Laboratories, Alcobendas, Spain) and analyzed with iCycler IQ Software (version 3.0). The QPCR conditions were 600 s at 95°C for enzyme activation and 40 cycles of 30 s at 95°C, 30 s at 50°C, and 30 s at 72°C. An additional step starting from 60 to 95°C (0.5°C/s) was performed to establish the denaturation curve specific for each amplification. The observation of a unique fluorescence fall at the fusion temperature of each PCR product certified amplification specificity. Fluorescence was measured at the end of each polymerization step (excitation, 450 to 495 nm; emission, 505 to 537 nm). Fluorescence versus the reaction cycle was plotted, and the PCR baseline and threshold cycles were automatically determined. Calibration curves were established for a concentration range from 0.6 ng to 12 ng of pSLT DNA. For this purpose, serial dilutions of plasmid DNA obtained with a QIAprep Spin Miniprep extraction kit (QIAGEN-Izasa, Barcelona, Spain) were used as templates to determine QPCR calibration curves. Each point was replicated twice.
Site-directed mutagenesis of the finP promoter.
A modified version of the procedure described previously (26) was used for site-directed mutagenesis of the finP promoter. A 326-bp fragment from pIZ899 was PCR amplified using the Universal and Reverse primers of pBluescript II SK(+) as external primers. For the GATC→GATG substitution, the internal primers were 5′ TTC TCA CGA TGC GTC GGA CAC AT 3′ and 5′ ATG TGT CCG ACG CAT CGT AGA GAA 3′. For the GATC→GCTC substitution, the internal primers were 5′ TTC TCT ACG CTC CGT CGG ACA CAT 3′ and 5′ ATG TGT CCG ACG GAG CGT AGA GAA 3′. For the GATC→GGTC substitution, a 655-bp fragment from pIZ860 was amplified using the Universal and Reverse primers of pBluescript II SK(+) as external primers and the oligonucleotides 5′ TTC TCT ACG GTC CGT CGG ACA CAT 3′ and 5′ ATG TGT CCG ACG GAC CGT AGA GAA 3′ as internal primers. The initial amplification was carried out at a hybridization temperature of 54°C. The amplification products were purified, and 1 μl of each product, previously diluted 1/10, was used as a template for a second amplification (cycles 1 to 10 at 63°C; cycles 11 to 25 at 55°C). In all cases, site-directed mutagenesis of the finP GATC destroyed a Sau3A restriction site; the absence of this site was used to confirm that the second amplification had generated the product expected. The product was then cloned on pBluescript II SK(+) and sequenced.
Gel retardation assays with H-NS protein.
Gel retardation assays were performed as described previously (34). The DNA fragment used was a PCR amplification product obtained with primers 5′ AAG CAG CTC CAT GTC CAA G 3′ and 5′ GGT ATA GGA TCT CTC AGC C 3′. The template DNA was plasmid pIZ860 (59).
RESULTS
Lack of Dam does not alter pSLT copy number.
A previous study showed that the pSLT plasmid undergoes higher frequencies of conjugal transfer from a Dam− donor (5). Because a well- known role of Dam methylation is the regulation of DNA replication, both in the bacterial chromosome and in certain plasmids (36), we considered the possibility that a dam mutation might alter the copy number of plasmid pSLT. If such were the case, the altered copy number might provide a simple explanation for the distinct donor abilities of Dam+ and Dam− hosts. Fertility inhibition relies on tight regulation of the level of the TraJ protein, and any imbalance in the ratio of the finP and traJ transcripts can potentially increase TraJ synthesis and derepress conjugal transfer (18). On these grounds, we compared the copy numbers of plasmid pSLT in Dam− and Dam+ isogenic strains (SV3000 and LT2, respectively) using a quantitative-PCR protocol. Reverse and forward oligonucleotide primers were designed to amplify a 500-bp fragment adjacent to the chromosomal replication origin (oriC) and a 681-bp fragment containing the pSLT-borne finO gene. The DNA sizes of these fragments are in the range for SYBR Green detection according to the manufacturer's instructions. The PCR efficiency (E) for each primer was calculated from amplification slopes. They were found to be similar (−0.44 < log E < 0.39), allowing the comparison of results. Threshold cycles were normalized, indicating the relative copy number of pSLT per chromosome. Amplifications were performed in quadruplicate. The Dam− strain SV3000 showed an average of 1.003 pSLT plasmids per oriC equivalent, with a standard deviation of 0.13 (13%). The parental Dam+ strain showed an average of 1.02 plasmids per oriC equivalent, with a standard deviation of 0.204 (20%). Thus, according to real-time PCR results, the Dam genetic background does not influence the pSLT copy number.
Half-lives of pSLT-encoded FinP RNA in Dam+ and Dam− hosts.
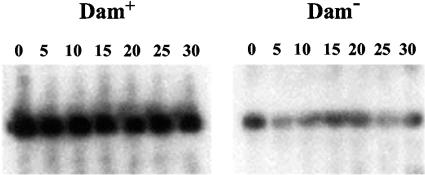
Preliminary evidence that Dam methylation regulates finP transcription rather than FinP RNA stability or turnover had been provided by an earlier study (59). A transcriptional finP::lac fusion constructed on a multicopy promoter-probe plasmid vector showed different activities in Dam+ and Dam− hosts. Since the fusion contained only 28 bp of finP transcript, the possibility of postranscriptional regulation seemed unlikely (59). Despite this evidence, we carried out a direct test to compare the half-lives of pSLT-encoded FinP RNA in Dam+ and Dam− strains of S. enterica. Late-exponential cultures of strains SV4107 and SV4109 were prepared in minimal medium and treated with rifampin (250 μg/liter) to block transcription initiation. Samples for RNA analysis were periodically collected, immersed in liquid N2, and centrifuged at 4°C. Because Dam− mutants contain smaller amounts of FinP RNA (59), samples from the Dam+ strain contained 20 μg of total RNA, while those from the Dam− strain contained 60 μg of total RNA. Electrophoretic separation in polyacrylamide gels was followed by Northern blot analysis. Larger amounts of FinP RNA were consistently found in the Dam+ strain, and densitometric analysis indicated that the difference was around two- to threefold (data not shown). Since the RNA concentration was threefold higher in the samples from the Dam− strain, the actual difference in FinP RNA content must be around six- to ninefold. No difference in FinP RNA decay was detected between the wild type and the Dam− mutant, suggesting that Dam methylation does not regulate FinP RNA turnover (Fig. 1). A side observation was that pSLT-encoded FinP RNA appears to be long-lived, even in a Dam− host. Because SV4107 and SV4109 both carry pSLT, the stability of FinP RNA may reflect the activity of the pSLT-encoded FinO product.
FIG. 1.
Chemical half-lives of pSLT-encoded FinP RNA in Dam+ and Dam− hosts. Samples for RNA analysis were extracted at the times indicated (in minutes) on the top of each panel and hybridized against a finP probe. Time zero was the time of rifampin addition.
Activity of a pSLT-borne finP::lacZ fusion in Dam+ and Dam− hosts.
To rule out the possibility of an artifact related to the use of a high-copy-number vector, a finP::lac fusion was constructed on pSLT itself, using a procedure described previously (14). When the resulting fusion was tested in Dam+ and Dam− backgrounds (strains SV4762 and SV4764), its β-galactosidase activity proved to be significantly higher in a Dam+ host: 178 ± 39 Miller units compared to 44 ± 9 Miller units in a Dam− mutant (average of five independent experiments). Aside from confirming previous observations (5, 59), these experiments supported the view that higher finP expression in a Dam+ background reflected elevated activity of the finP promoter: in the finP::lac fusion constructed, most of the finP transcript (all but 3 nucleotides downstream of the transcription start site) had been eliminated. Thus, it seemed likely that the difference in the FinP RNA contents found in Dam+ and Dam− hosts might reflect a difference in finP promoter activity.
Identification of the finP gene start site.
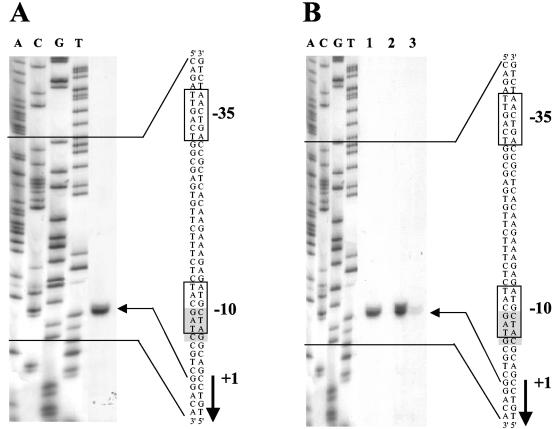
Primer extension was used to map the 5′ terminus of the finP transcript (Fig. 2A). For this purpose, a strain of S. enterica carrying plasmid pIZ900 (SV4107) was grown in minimal medium until late exponential phase; total RNA was then prepared. For the extension reaction, a labeled oligonucleotide complementary to the finP coding sequence was used. The extension products were separated in a polyacrylamide gel containing 6% urea. A sequencing reaction was run in parallel and used as a size marker; DNA sequencing was primed by the same oligonucleotide employed for primer extension. The finP transcript was found to start at the location predicted by sequence data (59).
FIG. 2.
(A) Extension of a FinP RNA primer with avian myeloblastosis virus reverse transcriptase. The extension product is indicated by an arrow. The DNA sequence of the finP promoter region of plasmid pSLT, the transcription start site, and the GATC site that overlaps the −10 module are also shown. (B) Reverse transcriptase extension of a FinP RNA primer, from a wild-type finP promoter (lane 1) and from a GATC-less finP promoter (GATC→GATG, carried on pIZ922). Primer extension of the latter was examined in a Dam+ background (lane 2) and a Dam− background (lane 3). The extension products are indicated by an arrow. The DNA sequence, the transcription start site, and the GATG site are also shown.
Transcription of finP is σ70 dependent.
DNA sequence analysis in silico suggests that the finP gene may be transcribed from a canonical σ70-dependent promoter that contains a GATC site overlapping with its −10 module (Fig. 2A). In search of experimental evidence that might support σ70 dependence, we tested the effect of a thermosensitive rpoD mutation on the expression of a finP::lac fusion. For this purpose, plasmid pIZ880 was electroporated into the isogenic E. coli strains P90AE and UQ285 (28). Overnight cultures grown at 30°C in LB-ampicillin were used to start fresh cultures, which were grown at 30°C (permissive temperature) and 42°C (restrictive temperature) in LB-ampicillin. After 14 h of incubation, aliquots were extracted and cooled down in ice; β-galactosidase activities were then measured. In an RpoD+ background, the activities of the finP::lac fusion were similar at 42°C and 30°C. In the RpoD(Ts) strain, however, the activity of the finP::lac fusion was 15-fold higher at 30°C than at 42°C (Table 2). These observations provide evidence that transcription of the finP gene requires the rpoD-encoded sigma factor, σ70.
TABLE 2.
Effect of an rpoD mutation on finP transcription
| Strain | Temp of assay (°C) | β-Galactosidase activity (Miller units) |
|---|---|---|
| P90A5 | 30 | 2,934 ± 240 |
| 42 | 3,373 ± 185 | |
| UQ285 | 30 | 3,565 ± 388 |
| 42 | 256 ± 47 |
Site-directed mutagenesis of the finP promoter.
The presence of a GATC site in the finP promoter of pSLT suggested the possibility that the methylation state of the −10 module might directly regulate the finP promoter (59). We also speculated that such a regulatory mechanism might be shared by other F-like plasmids, because the GATC site that overlaps the −10 module is also found in F and R100 (59), and these plasmids, like pSLT, undergo derepression in a Dam− background (7, 59). To examine this hypothesis, the GATC site present in the finP promoter was replaced with GATG (plasmid pIZ922), GCTC (pIZ1515), and GGTC (pIZ1529) using a method described previously (26). The effects of these changes on finP transcription were then analyzed in Dam+ (SV3081) and Dam− (SV3083) hosts. A primer extension experiment using the GATG-containing finP promoter is shown in Fig. 2B. The GATC→GATG mutation did not alter the transcription start site, but the amounts of finP transcript detected in three independent experiments were consistently reduced in a Dam− host. The GCTC mutation had similar effects (data not shown). Hence, these experiments provided evidence that a GATC-less finP promoter was still regulated by Dam methylation. The GGTC mutation produced an inactive finP promoter (data not shown). The latter observation is in agreement with data reported for other F-like plasmids (16, 30).
The failure of GATC→GATG and GATC→GCTC mutations to make finP transcription Dam independent was further supported by their effects on the expression of a traJ::lac translational fusion (Table 3). Neither mutation altered the expression pattern typical of the traJ::lac fusion (always higher in a Dam− host, indicative of reduced FinP RNA content). We thus reached the unanticipated conclusion that the GATC located on the finP promoter was not involved in Dam-mediated regulation. A corollary was that Dam methylation regulated finP transcription in an indirect fashion via a trans-acting regulator, which was in turn regulated by Dam methylation.
TABLE 3.
Effects of mutations that eliminate the GATC site in the finPpromoter on the activity of a translational traJ::lac fusion
| Sequence at finP promoter | Background | β-Galactosidase activity (Miller units) |
|---|---|---|
| GATC (wt)a | Dam+ | 1,293 ± 331 |
| GATC (wt) | Dam− | 10,043 ± 1521 |
| GATG | Dam+ | 952 ± 282 |
| GATG | Dam− | 12,031 ± 2809 |
| GCTC | Dam+ | 2,294 ± 237 |
| GCTC | Dam− | 18,600 ± 3000 |
wt, wild type.
DNA regions upstream from the finP promoter are not required for Dam-mediated regulation.
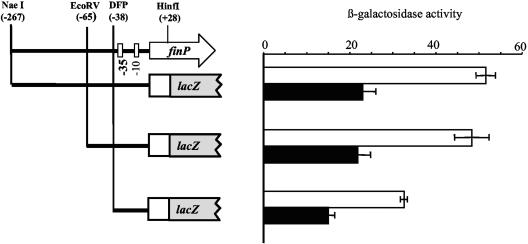
The possibility that a trans-acting factor might bind at or near the finP promoter led us to define potential upstream regulatory elements that might serve as targets for such regulation. For this purpose, deletions were generated on plasmid pIZ880. The resulting plasmids were introduced into Dam+ and Dam− isogenic strains (SV3081 and SV3083, respectively). None of the deletions rendered finP transcription Dam independent (Fig. 3). Because the deletion generated with the DFP primer carried only 3 bp upstream from the −35 module, we concluded that no DNA sequence upstream from this module was necessary for Dam-mediated regulation of finP transcription, Hence, we hypothesized that a Dam-sensitive regulator of finP transcription might act at the promoter itself or downstream. Such sites of action are not uncommon among regulators of σ70-dependent genes (27).
FIG. 3.
Effects of deletions upstream of the finP promoter on Dam-mediated regulation of finP transcription. The traJ::lac fusion starting at the NaeI restriction site was carried on plasmid pIZ880. Its deletion derivative starting at the EcoRV site was carried on pIZ955, and the DFP-generated fusion was carried on pIZ981. These plasmids were introduced into isogenic Dam+ and Dam− strains that lacked pSLT (SV3081 and SV3083, respectively). The activities (in tens of Miller units) of the fusions were measured in both backgrounds (Dam+, white bars; Dam−, black bars). The data shown are the means and standard deviations of six independent experiments.
Expression of the finP gene is not affected by SOS induction.
Regulation of the finP gene by Dam methylation follows a pattern opposite to that of genes of the SOS regulon: expression of finP is lowered in a Dam− host, while LexA-regulated genes show increased expression in a Dam− background (36, 44, 58). However, the evidence that finP transcription was regulated by Dam in an indirect fashion left open the possibility that an SOS-related regulator might control finP transcription. This possibility was examined in two kinds of experiments.
(i) The expression of a finP::lac transcriptional fusion (carried by strain SV4762) was examined in the presence of nalidixic acid, a well known SOS-inducing agent. LB plates containing X-Gal were seeded with >107 cells of SV4762 (which formed a lawn after growth). A small amount of nalidixic acid powder was placed in the center of each plate, and the plates were incubated overnight at 37°C. As a control, the same test was carried out with TT10897, a strain that contained an SOS-inducible cea::lac fusion (44). Blue (Lac+) halos were clearly observed in the latter, but not with SV4762. Hence, these experiments provided evidence that expression of the finP gene remained unaltered upon SOS induction.
(ii) The expression levels of the same finP::lac transcriptional fusion were compared in LexA+ and LexA(Ind−) backgrounds (strains SV5017 and SV5019, respectively). The LexA(Ind−) background prevents SOS induction, because the LexA mutant protein is unable to undergo autodigestion upon RecA activation (56). Dam-mediated regulation of finP was still observed in a LexA(Ind−) background (data not shown). Hence, we concluded that the Dam-sensitive regulator of finP transcription was not a product encoded by the SOS regulon.
Effect of H-NS on finP and traJ transcription.
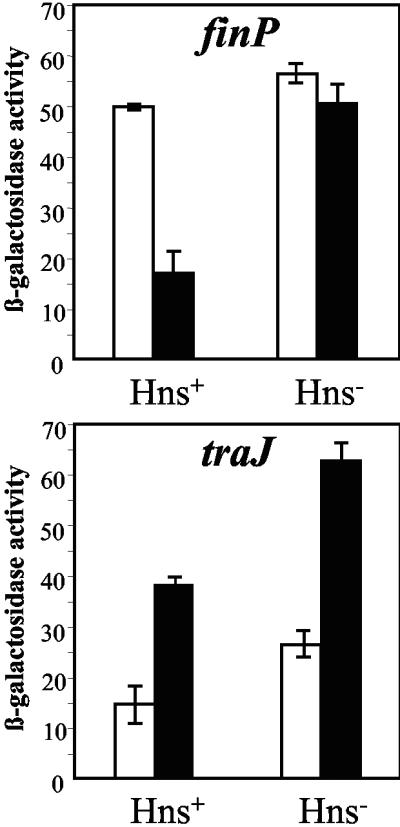
A search for the Dam-sensitive regulator of finP transcription was carried out using previously described genetic screens (5, 58) based on the detection of mutations that altered the expression of finP::lac transcriptional fusions. One such screen revealed that a finP::lac fusion, which is Lac+ in a Dam+ host and Lac− in a Dam− host, became Lac+ in a Dam− Hns− mutant, suggesting that the nucleoid protein H-NS repressed finP transcription in a Dam− background. Because certain S. enterica hns mutations appeared to cause impaired growth if combined with a dam mutation (data not shown), two well-characterized hns alleles were used: an hns::Kmr insertion allele (15) and an hns::Cmr allele constructed ad hoc by the method of Datsenko and Wanner (10). Both alleles proved to be compatible with a dam mutation. The effect of hns gene disruption on finP transcription was assayed using two different finP::lac constructs: (i) a finP::lac fusion carried on pIZ880, a multicopy plasmid introduced into strains SV3081, SV3083, SV5024, and SV5025; (ii) a finP::lac fusion constructed in pSLT itself (strains SV4762, SV4764, SV5052, and SV5053). Both assays confirmed that the decrease in finP transcription typical of Dam− mutants was relieved in the presence of an hns mutation. In contrast, the effect of H-NS observed in a wild-type (Dam+) background was very small, if any (Fig. 4).
FIG. 4.
β-Galactosidase activities (in tens of Miller units) of single-copy finP::lac and traJ::lac transcriptional fusions (carried on pSLT) in Dam+ (white bars) and Dam− (black bars) hosts. The host strains used were SV4762 (Dam+ Hns+), SV4764 (Dam− Hns+), SV5052 (Dam+ Hns−), and SV5053 (Dam− Hns−). The data shown are the means and standard deviations of four independent experiments.
The finding that H-NS acted as a repressor of finP transcription in a Dam− background raised the question of whether H-NS might likewise regulate traJ transcription. Furthermore, a recent study had shown that the sudden decrease in F transfer observed upon entry into stationary phase (21) is caused by H-NS-mediated repression of traJ transcription (61). To compare traJ transcription in Dam+ and Dam− hosts, the hns::Kmr and hns::Cmr alleles were used, and the assays were carried out in multicopy (pIZ898) and single-copy (pSLT) traJ::lac fusions. The host strains for the multicopy construct were SV3081, SV3083, SV5024, and SV5025, all lacking pSLT; the single-copy fusion was in turn introduced into SV4761, SV4763, SV5054, and SV5055. The results obtained in both series of experiments were similar and can be summarized as follows. (i) In a wild-type background, expression of a traJ::lac transcriptional fusion increased in the presence of an hns mutation, indicating that H-NS is a repressor of traJ (Fig. 4). H-NS-mediated repression was, however, partial and did not prevent traJ expression under any of the conditions tested (see below). (ii) Because Dam methylation represses traJ transcription (6), the effect of an hns mutation was also tested in a Dam− background. Derepression of traJ expression by an hns mutation was detected in both Dam+ and Dam− hosts (Fig. 4). Hence, unlike finP, the traJ gene is repressed by H-NS regardless of the Dam methylation state of the cell. (iii) Increase of traJ expression in the presence of an hns mutation was also observed in a Lrp− background. An lrp mutation caused a strong decrease in traJ transcription, as previously described (5). However, the activity of a traJ::lac transcriptional fusion was higher in an Lrp− Hns− mutant than in an Lrp− mutant (see Fig. S1 in the supplemental material). Hence, Lrp-mediated activation and H-NS-mediated repression of traJ transcription appear to be independent phenomena.
Additional experiments indicated that H-NS-mediated repression of traJ transcription occurred in both exponential- and stationary-phase cultures (see Fig. S1 in the supplemental material). This observation suggests that H-NS-mediated regulation of the pSLT traJ gene is different from that shown in F: while in the latter H-NS acts as a growth phase-dependent traJ repressor (61), in pSLT it merely restrains traJ transcription. Such restraint is growth phase independent, a difference consistent with the ability of pSLT to undergo transfer from stationary-phase cultures (5) (see Table S1 in the supplemental material).
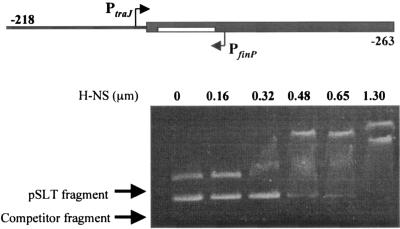
Analysis of H-NS binding to the traJ-finP region of the Salmonella virulence plasmid.
In the F sex factor, HN-S has been shown to bind a DNA stretch that encompasses the traJ-finP region (61). To analyze whether H-NS was likewise able to bind the homologous traJ-finP region in plasmid pSLT, a 589-bp fragment of pIZ860 was PCR amplified. This fragment contained 218 bp upstream of the traJ start site and 236 bp upstream of the finP start site (shown in the diagram in Fig. 5, top). H-NS binding was assayed in the presence of competitor DNA: a 427-bp fragment from the E. coli hemolysin operon previously shown to lack specificity for H-NS binding (34). Preferential retardation of the DNA fragment that contained the pSLT traJ-finP region was clearly observed as the H-NS concentration increased. Therefore, the ability of H-NS to restrain traJ transcription may be tentatively correlated with its ability to bind the traJ-finP region of plasmid pSLT. These observations, however, also raise an intriguing question: if the region bound by H-NS includes the finP promoter, why is the latter repressed by H-NS in a Dam− background only? As discussed below, this effect cannot be explained by a local effect of Dam methylation on H-NS binding, because the only GATC site found in the region is dispensable for Dam-mediated regulation (Table 3).
FIG. 5.
Binding of H-NS to a 589-bp fragment of the traJ-finP region of pSLT, assayed by gel retardation analysis. A diagram of the DNA fragment under study is shown at the top. The coordinates at each fragment end indicate the distance from the traJ start site (left) and from the finP start site (right). The concentrations of H-NS used are also shown.
DISCUSSION
Genes whose expression is affected by Dam methylation have been described in both E. coli and S. enterica (32, 36). Such Dam-regulated genes were often discovered in genetic or microarray screens that compared gene expression in Dam+ and Dam− isogenic strains (31, 42, 58). It must be noted, however, that a difference in gene expression between Dam+ and Dam− hosts does not imply a direct involvement of Dam methylation in gene regulation. For instance, in both E. coli and Salmonella, genes of the SOS regulon show increased expression in a Dam− background (36, 44, 58). The effect is, however, indirect: in the absence of Dam-directed strain discrimination, the mismatch repair MutHLS complex produces double-strand DNA breaks, and SOS induction occurs (22). The example of the SOS gene network is unlikely to be unique, given the enormous differences found in the proteomes of Dam+ and Dam− strains (42) and the smaller number of genes that appear to be directly regulated by Dam methylation (31, 42). Aside from this caveat, examples of genes whose transcription is regulated by the methylation state of critical GATC sites have been known for two decades and can be classified into two classes. (i) The first class contains genes whose expression is coupled to the cell cycle, such as the IS10 transposase gene (45) and the E. coli trpR gene (35). Promoters of this class contain GATC sites whose methylation states regulate RNA polymerase binding. (ii) The second class comprises genes that contain GATC sites in upstream regulatory regions. In such cases, the methylation state of one or more GATC sites regulates binding of specific transcription factors. Examples of this class are the papBA promoter of uropathogenic E. coli (25), the mom promoter of phage Mu (3), the Ag43 promoter of E. coli (24), and the traJ gene of the Salmonella virulence plasmid (6).
Here, we describe a case of transcriptional regulation by Dam methylation that does not fit in these two classes. The finP gene of the Salmonella virulence plasmid is transcribed at higher levels in a Dam+ background. However, Dam-mediated regulation of finP transcription does not require the presence of GATC sites at or near the promoter: in fact, a mutant finP promoter devoid of the GATC site that overlaps its −10 module is still regulated by Dam. Hence, Dam methylation regulates finP transcription in an indirect fashion, a trait that is reminiscent of the LexA-regulated SOS gene network. Unlike SOS genes, however, finP transcription is not regulated by DNA damage and remains unaltered in a LexA (Ind−) background. On these grounds, we postulated that finP was regulated by an unknown, Dam-sensitive, trans-acting factor (either a positive effector of transcription active in a Dam+ background or a negative effector active in a Dam− background).
Trials for the Dam-sensitive finP regulator identified H-NS as a repressor of finP transcription. H-NS is a nucleoid-associated protein that participates in the organization of bacterial chromatin and usually acts as a repressor of gene expression (13). At the finP promoter, however, repression by H-NS is observed only in a Dam− background. This difference cannot be explained by a direct effect of Dam methylation upon H-NS binding, because Dam-mediated repression is still observed in a GATC-less finP promoter. The involvement of upstream DNA sequences is likewise ruled out by deletion analysis. Hence, H-NS-mediated repression of finP must reflect a condition or state that occurs in Dam− mutants but not in the wild type. For instance, a tentative explanation may be that a higher H-NS concentration exists in Salmonella Dam− mutants, as reported in E. coli (42). Although no evidence exists that the H-NS level in the cell is directly regulated by Dam methylation, one speculation may be that higher H-NS content is required to compensate for nucleoid distortions caused by lack of adenosine methylation. N6 methylation at individual GATC sites is known to influence local DNA structure (12, 43), and the cumulative effect of nonmethylation at the 20,000 GATC sites present in the genome might have a major impact on nucleoid topology.
In the F sex factor, H-NS has been shown to silence traJ and traM expression during entry into stationary phase (61). Binding of H-NS to a region that encompasses the traM and traJ promoters suggests that silencing is a regional effect (61). In pSLT, H-NS is also a repressor of traJ transcription, but repression is only partial and does not increase in stationary-phase cultures. This difference is consistent with the observation that transfer of the Salmonella virulence plasmid is largely insensitive to the growth stage. It must be noted that F-like plasmids share a general regulatory design, but individual regulatory traits are also found. An example concerns H-NS, identified here as a repressor of mating as previously shown for F (61) but described instead as an activator of mating functions in pRK100 (54). Another example involves the leucine-responsive regulatory protein, which is an activator of traJ transcription in the Salmonella virulence plasmid (5) but not in pRK100 (54). Plasmid-specific regulatory features may reflect distinct responses to physiological and environmental signals, and perhaps distinct strategies for plasmid spread in bacterial populations.
An enigmatic aspect of mating regulation in the Salmonella virulence plasmid is that H-NS-mediated restraint of traJ transcription is insensitive to Dam methylation, while the overlapping finP gene escapes H-NS-mediated repression in a wild-type (Dam+) background. From a mechanistic point of view, this difference is intriguing, since a large DNA stretch that includes both traJ and finP appears to be bound by H-NS, as previously described for F (61). However, the finP and traJ promoters are transcribed in opposite directions, thus raising the possibility that H-NS repression might be strand specific. On the other hand, H-NS is known to bind curved DNA (13), and curvature in the traJ-finP region might have distinct effects on promoters lying on opposite strands. Aside from these speculations, the avoidance of H-NS-mediated silencing at the finP gene certainly fits in the logic of fertility inhibition: repression of traJ, but not of finP, can be viewed as a coordinated action for the repression of conjugal plasmid transfer. Mating restraint mechanisms, no matter how odd or subtle, can be expected to have a high adaptive value to avoid the burden that overexpression of mating functions would otherwise impose on the host cell (63).
Supplementary Material
Acknowledgments
This work was supported by grants BIO2001-232-CO2-02 and BIO 2004-3455-CO2-02 from the Spanish Ministry of Education and Science.
Several strains used in this work were kindly provided by N. Patrick Higgins, Juan L. Ramos, and John Roth. We are grateful to Manuel Espinosa, Fernando Govantes, and Matxalen Llosa for helpful discussions.
REFERENCES
- 1.Ahmer, B. M. M., M. Tran, and F. Heffron. 1999. The virulence plasmid of Salmonella typhimurium is self-transmissible. J. Bacteriol. 181:1364-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur, D. C., A. F. Ghetu, M. J. Gubbins, R. A. Edwards, L. S. Frost, and J. N. M. Glover. 2003. FinO is an RNA chaperone that facilitates sense-antisense RNA interactions. EMBO J. 22:6346-6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolker, M., and R. Kahmann. 1989. The Escherichia coli protein OxyR discriminates between methylated and unmethylated states of the phage Mu promoter. EMBO J. 8:2403-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brandi, L., M. Falconi, and S. Ripa. 2000. Plasmid curing effect of trovalfloxacin. FEMS Microbiol. Lett. 184:297-302. [DOI] [PubMed] [Google Scholar]
- 5.Camacho, E. M., and J. Casadesús. 2002. Conjugal transfer of the virulence plasmid of Salmonella enterica is regulated by the leucine-responsive regulatory protein and DNA adenine methylation. Mol. Microbiol. 44:1589-1598. [DOI] [PubMed] [Google Scholar]
- 6.Camacho, E. M., and J. Casadesús. Regulation of traJ transcription in the Salmonella virulence plasmid by strand-specific DNA adenine hemimethylation. Mol. Microbiol., in press. [DOI] [PubMed]
- 7.Camacho, E. M., A. Serna, and J. Casadesús. Unpublished data.
- 8.Cheah, K. C., and R. Skurray. 1986. The F plasmid carries an IS3 insertion within finO. J. Gen. Microbiol. 132:3269-3275. [DOI] [PubMed] [Google Scholar]
- 9.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidine thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 10.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 90:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dempsey, W. B. 1994. Regulation of R100 conjugation requires traM in cis to traJ. Mol. Microbiol. 13:313-316. [DOI] [PubMed] [Google Scholar]
- 12.Diekmann, S. 1987. DNA methylation can enhance or induce DNA curvature. EMBO J. 6:4213-4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorman, C. J. 2004. H-NS: a universal regulator for a dynamic genome. Nat. Rev. Microbiol. 2:391-400. [DOI] [PubMed] [Google Scholar]
- 14.Ellermeier, C. D., A. Janakiraman, and J. M. Slauch. 2002. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene 290:153-161. [DOI] [PubMed] [Google Scholar]
- 15.Falconi, M., V. McGovern, C. Gualerzi, D. Hillyard, and N. P. Higgins. 1991. Mutations altering chromosomal protein H-NS induce mini-Mu transposition. New Biol. 3:615-625. [PubMed] [Google Scholar]
- 16.Finlay, B. B., L. S. Frost, W. Paranchych, and N. S. Willetts. 1986. Nucleotide sequences of five IncF plasmid finP alleles. J. Bacteriol. 167:754-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finnegan, D. J., and N. S. Willetts. 1973. The site of action of the F transfer inhibitor. Mol. Gen. Genet. 127:307-316. [DOI] [PubMed] [Google Scholar]
- 18.Firth, N., K. Ippen-Ihler, and R. A. Skurray. 1996. Structure and function of the F factor and mechanism of conjugation, p. 2377-2401. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, D.C.
- 19.Frost, L. S., K. Ippen-Ihler, and R. A. Skurray. 1994. Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol. Rev. 58:162-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frost, L. S., S. Lee, N. Yanchar, and W. Paranchych. 1989. finP and fisO mutations in FinP anti-sense RNA suggest a model for FinOP action in the repression of bacterial conjugation by the Flac plasmid JCFL0. Mol. Gen. Genet. 218:152-160. [DOI] [PubMed] [Google Scholar]
- 21.Frost, L. S., and J. Manchak. 1998. F-phenocopies: characterization of expression of the F transfer region in stationary phase. Microbiology 144:2579-2587. [DOI] [PubMed] [Google Scholar]
- 22.Glickman, B. W., and M. Radman. 1980. Escherichia coli mutator mutants deficient in methylation-instructed DNA mismatch correction. Proc. Natl. Acad. Sci. USA 77:1063-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gubbins, M. J., I. Lau, W. R. Will, J. M. Manchak, T. L. Raivio, and L. S. Frost. 2002. The positive regulator, TraJ, of the Escherichia coli F plasmid is unstable in a cpxA* background. J. Bacteriol. 184:5781-5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henderson, I. R., and P. Owen. 1999. The major phase-variable outer membrane protein of Escherichia coli structurally resembles the immunoglobulin A1 protease class of exported protein and is regulated by a novel mechanism involving Dam and OxyR. J. Bacteriol. 181:2132-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hernday, A., M. Krabbe, B. Braaten, and D. A. Low. 2003. The mechanism by which DNA adenine methylase and PapI activate the pap epigenetic switch. Mol. Cell 12:954-957. [DOI] [PubMed] [Google Scholar]
- 26.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 27.Huerta, A. M., and J. Collado-Vides. 2003. Sigma70 promoters in Escherichia coli: specific transcription in dense regions of overlapping promoter-like signals. J. Mol. Biol. 333:261-278. [DOI] [PubMed] [Google Scholar]
- 28.Isaksson, L. A., S. E. Skold, J. Skjoldebrand, and R. Takata. 1977. A procedure for isolation of spontaneous mutants with temperature sensitive of RNA and/or protein. Mol. Gen. Genet. 156:233-237. [DOI] [PubMed] [Google Scholar]
- 29.Jerome, L. J., T. van Biesen, and L. S. Frost. 1999. Degradation of FinP antisense RNA from F-like plasmids: the RNA-binding protein, FinO, protects FinP from ribonuclease E. J. Mol. Biol. 285:1457-1473. [DOI] [PubMed] [Google Scholar]
- 30.Lee, S. H., L. S. Frost, and W. Paranchych. 1992. FinOP repression of the F plasmid involves extension of the half-life of FinP RNA by FinO. Mol. Gen. Genet. 235:131-139. [DOI] [PubMed] [Google Scholar]
- 31.Løbner-Olesen, A., M. G. Marinus, and F. G. Hanssen. 2003. Role of SeqA and Dam in Escherichia coli gene expression: a global/microarray analysis. Proc. Natl. Acad. Sci. USA 100:4672-4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Low, D. A., N. J. Weyand, and M. J. Mahan. 2001. Roles of DNA adenine methylation in regulating bacterial gene expression and virulence. Infect. Immun. 69:7197-7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macián, F., I. Pérez-Roger, and M. E. Armengod. 1994. An improved vector system for constructing transcriptional lacZ fusions: analysis of regulation of the dnaA, dnaN, recF and gyrB genes of Escherichia coli. Gene 145:17-24. [DOI] [PubMed] [Google Scholar]
- 34.Madrid, C., J. M. Nieto, S. Paytubi, M. Falconi, C. O. Gualerzi, and A. Juárez. 2002. Temperature- and H-NS-dependent regulation of a plasmid-encoded virulence operon expressing Escherichia coli hemolysin. J. Bacteriol. 184:5058-5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marinus, M. G. 1985. DNA methylation influences trpR promoter activity in Escherichia coli. Mol. Gen. Genet. 200:185-186. [DOI] [PubMed] [Google Scholar]
- 36.Marinus, M. G. 1996. Methylation of DNA, p. 782-791. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, D.C.
- 37.Marqués, S., J. L. Ramos, and K. Timmis. 1993. Analysis of the mRNA structure of the Pseudomonas putida TOL meta fission pathway operon around the transcription initiation point, the xylTE and the xylFJ regions. Biochim. Biophys. Acta 1216:227-236. [DOI] [PubMed] [Google Scholar]
- 38.Matzke, M. A., and J. A. Birchler. 2005. RNAi-mediated pathways in the nucleus. Nat. Rev. Genet. 6:24-35. [DOI] [PubMed] [Google Scholar]
- 39.McEwen, J., and P. Silverman. 1980. Chromosomal mutations of Escherichia coli that alter expression of conjugative plasmid functions. Proc. Natl. Acad. Sci. USA 77:513-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 41.Mullineaux, P., and N. Willetts. 1985. Promoters in the transfer region of plasmid F, p. 605-614. In D. R. Helinski, S. N. Cohen, D. B. Clewell, D. A. Jackson, and A. Hollaender (ed.), Plasmids in bacteria. Plenum Publishing Corp., New York, N.Y. [DOI] [PubMed]
- 42.Oshima, T., C. Wada, Y. Kawagoe, T. Ara, M. Maeda, Y. Masuda, S. Hiraga, and H. Mori. 2002. Genome-wide analysis of deoxyadenosine methyltransferase-mediated control of gene expression in Escherichia coli. Mol. Microbiol. 45:673-695. [DOI] [PubMed] [Google Scholar]
- 43.Polaczek, P., K. Kwan, and J. L. Campbell. 1998. GATC motifs may alter the conformation of DNA depending on sequence context and N6-adenine methylation status: possible implications for DNA-protein recognition. Mol. Gen. Genet. 258:488-493. [DOI] [PubMed] [Google Scholar]
- 44.Prieto, A. I., F. Ramos-Morales, and J. Casadesús. 2004. Bile-induced DNA damage in Salmonella enterica. Genetics 168:1787-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roberts, D., B. C. Hoopes, W. R. McClure, and N. Kleckner. 1985. IS10 transposition is regulated by DNA adenine methylation. Cell 43:117-130. [DOI] [PubMed] [Google Scholar]
- 46.Rotger, R., and J. Casadesús. 1999. The virulence plasmids of Salmonella enterica. Int. Microbiol. 2:177-184. [PubMed] [Google Scholar]
- 47.Sambrook, J., and D. W. Russell. 2002. Molecular cloning, 3rd ed., vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 48.Sanderson, K. E., S. K. Kadam, and P. R. MacLachlan. 1983. Derepression of F factor function in Salmonella typhimurium. Can. J. Microbiol. 29:1205-1212. [DOI] [PubMed] [Google Scholar]
- 49.Schmieger, H. 1972. Phage P22 mutants with increased or decreased transducing abilities. Mol. Gen. Genet. 119:75-88. [DOI] [PubMed] [Google Scholar]
- 50.Silverman, P. M., L. Tran, R. Harris, and H. M. Gaudin. 1993. Accumulation of the F plasmid TraJ protein in cpx mutants of Escherichia coli. J. Bacteriol. 175:921-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith, H. R., G. O. Humphreys, N. D. F. Grindley, J. D. Grindley, and E. S. Anderson. 1973. Molecular studies of a fi+ plasmid from strains of Salmonella typhimurium LT2. Mol. Gen. Genet. 126:1443-1451. [DOI] [PubMed] [Google Scholar]
- 52.Spratt, B. G., R. J. Rowbury, and G. G. Meynell. 1973. The plasmid of Salmonella typhimurium LT2. Mol. Gen. Genet. 121:347-353. [DOI] [PubMed] [Google Scholar]
- 53.Starcic, M., D. Zgur-Bertok, B. J. Jordi, M. M. Wosten, W. Gaastra, and J. P. van Putten. 2003. The cyclic AMP-cyclic AMP receptor protein complex regulates activity of the traJ promoter of the Escherichia coli conjugative plasmid pRK100. J. Bacteriol. 185:1616-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Starcic-Erjavec, M., J. P. M. van Putten, W. Gaastra, B. J. A. M. Jordi, M. Grabnar, and D. Zgur-Bertok. 2003. H-NS and Lrp serve as positive modulators of traJ expression from the Escherichia coli plasmid pRJ100. Mol. Gen. Genom. 270:94-102. [DOI] [PubMed] [Google Scholar]
- 55.Strohmaier, H., R. Noiges, S. Kotschan, G. Sawers, G. Högenauer, E. Zechner, and G. Koraimann. 1998. Signal transduction and bacterial conjugation: characterization of the role of ArcA in regulating conjugative transfer of the resistance plasmid R1. J. Mol. Biol. 277:309-316. [DOI] [PubMed] [Google Scholar]
- 56.Sutton, M. D., B. T. Smith, V. G. Godoy, and G. C. Walker. 2000. The SOS response: recent insights into umuDC-dependent mutagenesis and DNA damage tolerance. Annu. Rev. Genet. 34:479-497. [DOI] [PubMed] [Google Scholar]
- 57.Taki, K., T. Abo, and E. Othsubo. 1998. Regulatory mechanisms in expression of the traY-I operon of sex factor plasmid R100: involvement of traJ and traY gene products. Genes Cells 3:331-345. [DOI] [PubMed] [Google Scholar]
- 58.Torreblanca, J., and J. Casadesús. 1996. DNA adenine methylase mutants of Salmonella typhimurium and a novel Dam-regulated locus. Genetics 144:15-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Torreblanca, J., S. Marqués, and J. Casadesús. 1999. Synthesis of FinP RNA by plasmids F and pSLT is regulated by DNA adenine methylation. Genetics 151:31-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vogel, H., and D. Bonner. 1956. Acetylornithase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218:97-106. [PubMed] [Google Scholar]
- 61.Will, R. W., J. Lu, and L. S. Frost. 2004. The role of H-NS in silencing F transfer gene expression during entry into stationary phase. Mol. Microbiol. 54:769-782. [DOI] [PubMed] [Google Scholar]
- 62.Willetts, N. S. 1993. Bacterial conjugation. A historical perspective, p. 1-22. In D. B. Clewell (ed.), Bacterial conjugation. Plenum Press, New York, N.Y.
- 63.Zatyka, M., and C. M. Thomas. 1998. Control of genes for conjugative transfer of plasmids and other mobile elements. FEMS Microbiol. Rev. 21:291-319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.