Abstract
We performed transposon mutagenesis of a two-color fluorescent reporter strain to identify new regulators of the porin genes ompF and ompC in Escherichia coli. Screening of colonies by fluorescence microscopy revealed numerous mutants that exhibited interesting patterns of porin expression. One mutant harbored an insertion in the gene encoding the histidine kinase CpxA, the sensor for a two-component signaling system that responds to envelope stress. The cpxA mutant exhibited increased transcription of ompC and a very strong decrease in transcription of ompF under conditions in which acetyl phosphate levels were high. Subsequent genetic analysis revealed that this phenotype is dependent on phosphorylation of the response regulator CpxR and that activation of CpxA in wild-type cells results in similar regulation of porin expression. Using DNase I footprinting, we demonstrated that CpxR binds upstream of both the ompF and ompC promoters. It thus appears that two distinct two-component systems, CpxA-CpxR and EnvZ-OmpR, converge at the porin promoters. Within the context of envelope stress, outer membrane beta-barrel proteins have generally been associated with the sigma E pathway. However, at least for the classical porins OmpF and OmpC, our results show that the Cpx envelope stress response system plays a role in regulating their expression.
The classical porins OmpF and OmpC are major constituents of the Escherichia coli outer membrane and account for approximately 2% of the total protein content of the cell (52). These proteins allow for the passive diffusion of solutes across the outer membrane. Many environmental factors have been identified that alter OmpF and OmpC expression, including osmolarity, temperature, pH, nutrient availability, and various toxins (23, 40, 54). The importance of this complex regulation for the cell is not understood, although it has been suggested that the differential regulation of the two porins may provide a means of balancing the competing needs of access to nutrients and protection from toxins (52).
The complex environmental regulation of the porins is implemented by a similarly complex regulatory network, whose components include the EnvZ-OmpR two-component system (reviewed in references 19, 34, and 63), the small RNAs MicF and MicC (9, 14), the sigma factors σS and σE (45, 55), the global regulator Lrp (21), and the histone-like protein IHF (30, 59). The EnvZ-OmpR system is a central element of this network, since phosphorylated OmpR (OmpR-P) is absolutely required for OmpF and OmpC expression. OmpR-P levels are modulated by the histidine kinase EnvZ in response to unknown stimuli. Low levels of OmpR-P activate transcription of ompF, while high levels repress ompF and activate ompC (22, 36, 61). In vitro studies revealed that OmpR binds to three sites (designated C1, C2, and C3) upstream of the ompC promoter (41, 66) and four sites (one distal site, F4, and three proximal sites, F1, F2, and F3) upstream of the ompF promoter (28, 29, 48, 66). However, despite this detailed characterization, the mechanism underlying the differential regulation of ompF and ompC by OmpR is not understood (25, 34).
Here we describe the results of a genetic screen and a subsequent analysis which indicated that a second two-component system, the CpxA-CpxR system, also regulates porin expression. The Cpx system responds to conditions associated with envelope stress, such as alkaline pH and overproduction of secreted proteins (10, 37, 64), and also to attachment of cells to surfaces (53). The previously characterized members of the Cpx regulon include proteins involved in the folding or degradation of misfolded proteins in the periplasm (18, 57) and in the assembly of structures on the cell surface (17, 26, 31, 39, 50, 51). Regulation and monitoring of porin status, however, have been associated with a separate envelope stress-responsive system controlled by σE (reviewed in references 1, 2, and 57). In particular, overexpression of OmpF or OmpC activates σE (45) but does not activate the Cpx pathway (12). We demonstrate here that both the ompF and ompC porin genes are also members of the Cpx regulon. However, whereas activation of σE results in decreased expression of both ompF and ompC (60), activation of Cpx results in a strong decrease in ompF expression and an increase in ompC expression.
MATERIALS AND METHODS
Growth media.
Minimal medium consisted of 60 mM K2HPO4, 33 mM KH2PO4, 7.6 mM (NH4)2SO4, 1.7 mM sodium citrate, and 1 mM MgSO4 (46) and was supplemented with 0.2% glucose or glycerol as indicated below. Luria-Bertani (LB) agar was obtained from Difco (Becton Dickinson, Sparks, MD). Medium A agar consisted of 7 g/liter nutrient broth (Difco), 1 g/liter yeast extract (Difco), 0.2% glycerol, 20 mM K2HPO4, 10 mM KH2PO4, and 1.5% agar (Difco) (33). Ampicillin, when needed for plasmid maintenance, was added to a concentration of 50 μg/ml.
Strains and plasmids.
The strains and plasmids used in this study are described in Table 1. P1 transductions were performed using P1kc by following standard protocols (46).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Strain construction | Reference or source |
|---|---|---|---|
| Strains | |||
| MC4100 | F−araD139 Δ(argF-lac)169 λ−flhD5301 fruA25 relA1 rpsL150(Strr) rbsR22 deoC1 | 8 | |
| MG1655 | λ−rph-1 | E. coli Genetic Stock Center, CGSC no. 7740 | |
| PIR2 | F− Δlac169 rpoS(Am) robA1 creC510 hsdR514 endA recA1 uidA(ΔMlu 1)::pir | Invitrogen (Carlsbad, CA) | |
| MDG147 | MG1655 φ(ompF+-yfp+)30 φ(ompC+-cfp+)31 | 5 | |
| EPB54-012 | MDG147 cpxA61::kan | This study | |
| PAD368 | cpxR1::spc | T. J. Silhavy | |
| EPB62 | MDG147 cpxR1::spc | MDG147 × PAD368 | This study |
| EPB28 | MDG147 Δ(ackA-pta)728::cat | This study | |
| EPB81 | EPB54-012 Δ(ackA-pta)728::cat | This study | |
| MDG131 | MC4100 φ(ompF+-yfp+)30 φ(ompC+-cfp+)31 | 4 | |
| EPB55-012 | MDG131 cpxA61::kan | MDG131 × EPB54-012 | This study |
| EPB80 | MDG131 cpxR1::spc | MDG131 × PAD368 | This study |
| EPB10 | MDG131 Δ(ackA-pta)728::cat | This study | |
| EPB83 | EPB55-012 Δ(ackA-pta)728::cat | This study | |
| FR195 | ompR101 zhf37::Tn10 | 61 | |
| EPB31 | MDG147 ompR101 zhf37::Tn10 | MDG147 × FR195 | This study |
| JMS5477 | Δ(micF-ompC)178 zei198::Tn10 | 62 | |
| EPB84 | MDG147 Δ(micF-ompC)178 zei198::Tn10 | MDG147 × JMS5477 | This study |
| EPB89 | EPB84 cpxA61::kan | EPB84 × EPB54-012 | This study |
| PAD359 | cpxRD51A zii::Tn10 | T. J. Silhavy | |
| EPB128 | MDG147 cpxRD51A zii::Tn10 | MDG147 × PAD359 | This study |
| EPB134 | MDG147 ΔcpxA62::cat | This study | |
| EPB218 | EPB128 ΔcpxA62::cat | This study | |
| EPB91 | MDG147 ΔenvZ407 | This study | |
| EPB97 | EPB91 cpxA61::kan | EPB91 × EPB54-012 | This study |
| Plasmids | |||
| pRL27 | oriγR6K, Tn5-RL27, Kmr | 38 | |
| pKD3 | oriγR6K, bla, FRT-cat-FRT | 13 | |
| pKD46 | 13 | ||
| pBR322 | bla | 7 | |
| pLD404 | pBR322 Ptrc-nlpE | 64 | |
| pTrc99a | lacIq, Ptrc-MCS, bla | 3 | |
| pEB50 | pTrc99a Ptrc-cpxA | This study | |
| pAS09b | pTrc99a Ptrc-cpxA63 | Goulian lab stock |
Transposon mutagenesis and colony screening.
For the transposon mutagenesis and colony screening procedures see the supplemental material.
Disruption of ackA-pta, cpxA, and envZ.
To delete ackA-pta, a chloramphenicol resistance cassette was amplified from pKD3 (13) using primers 5′-GGTACTTCCATGTCGAGTAAGTTAGTACTGGTTCTGGTGTAGGCTGGAGCTGCTTC-3′ and 5′-CCAGTGCGCCACGGGACAGGTCGTTAACCGGCTTGCCATATGAATATCCTCCTTAG-3′ (the underlined regions are sequences that flank the ackA-pta locus in the E. coli chromosome). The PCR product was inserted into the chromosome by Lambda Red-mediated allele replacement (13). The correct insertion was verified by PCR. The deletion (which contained an insertion of cat) was then moved into MDG131, MDG147, EPB54-012, and EPB55-012 by P1 transduction, resulting in strains EPB10, EPB28, EPB81, and EPB83, respectively. Similarly, to delete cpxA, we used primers 5′-TTTAAAACCTTGCGTGGTCGCGGCTATCTGATGGTTTCTGCTTCATGATAGTGTAGGCTGGAGCTGCTTC-3′ and 5′-TAAACGCCTTATCCTGCCTGCAAATGCGAAGTTTAACTCCGCTTATACAGCATATGAATATCCTCCTTAG-3′ (the underlined regions are sequences flanking cpxA in the E. coli genome). The cat insertion in cpxA was moved into MDG147 by P1 transduction, resulting in EPB134. To construct EPB218, a strain containing both the cpxRD51A allele and a deletion of cpxA, the ΔcpxA::cat PCR product obtained with the primers described above was electroporated into EPB128/pKD46 with subsequent curing of plasmid pKD46 by growth at 42°C, as described by Datsenko and Wanner (13). To delete envZ, we used PCR primers 5′-TGGGCTACGTCTTTGTACCGGACGGCTCTAAAGCATGAGGGTGTAGGCTGGAGCTGCTTC-3′ and 5′-ACCTTCGCCTCCCGTTTATTTACCCTTCTTTTGTCGTGCCCATATGAATATCCTCCTTAG-3′ (the underlined regions are sequences flanking envZin the E. coli genome). The PCR product was used to insert cat into envZ, as described above, and the resulting insertion was transduced into MDG147. Strain MDG147 envZ::cat was then transformed with pCP20 to remove the cat gene (13) and then cured of the plasmid by growth at 42°C. This resulted in strain EPB91.
Fluorescence assay.
Single colonies were grown to saturation in the appropriate medium at 37°C with aeration. Cultures were then diluted 1:1,000 into the same prewarmed medium and incubated further at 37°C with aeration. When these cultures reached an optical density at 600 nm (OD600) of ∼0.2, they were rapidly chilled in an ice-water slurry. Fluorescence was measured with a QuantaMaster-4/2003 spectrofluorometer (Photon Technology International, Lawrenceville, NJ). The excitation/emission wavelengths for cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP) measurements were 434 nm/475 nm and 505 nm/527 nm, respectively, with 0.5-mm slit widths for the monochromators (2-nm band pass).
Porin protein analysis.
Two-milliliter cultures were grown to saturation in glucose minimal medium. They were then subcultured by 1:1,000 dilution into 50 ml of the same prewarmed medium and grown at 37°C with aeration. When the cultures reached an OD600 of ∼0.2, cells were harvested, and the cell envelopes were isolated and analyzed by urea sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and staining with Coomassie brilliant blue as described previously (4). For quantification, gels were digitized using a flatbed scanner with a no. 12 Wratten filter (Kodak, Rochester, NY) to improve the contrast.
DNase I footprinting.
The ompF and ompC promoter regions were amplified by PCR using Taq polymerase (Invitrogen, Carlsbad, CA) with primer pairs 5′-CCATCAGAAACAAAATTTCCGT-3′/5′-TAAGTTCTGTCAATAAAAATTTACGG-3′ and 5′-GCCTTTATTTGCTTTTTTATGCCAC-3′/5′-CTTAAGAATAAGTTATTGATTTTAAC-3′, respectively. The F4 and ompC upstream regions were amplified by PCR with primer pairs 5′-CTAATTTAGCGTCTTCAAGAG-3′/5′-CTTTCAGACATCCAGAATGC-3′ and 5′-CTGGAAATTATGCGGATG-3′/5′-AGAATAACTCCCGCTATCATC-3′, respectively. The products were cloned into TOPO cloning vector pCR4 by following the manufacturer's directions (Invitrogen). The resulting plasmids were used as templates to amplify DNA for the footprinting reactions. For generation of radiolabeled DNA, one primer of each pair listed above was labeled with 32P using T4 polynucleotide kinase (New England Biolabs, Beverly, MA) and [γ-32P]ATP (3,000 Ci/mmol; 5 mCi/ml; Perkin-Elmer, Shelton, CT). The PCR products were passaged through a QIAquick spin column per the manufacturer's instructions (QIAGEN) and subsequently purified on a 4% Tris-acetate-EDTA polyacrylamide gel. The desired fragments were removed from the gel, and the DNA was eluted into diffusion buffer (0.5 M ammonium acetate, 10 mM magnesium acetate, 1 mM EDTA, pH 8.0, 0.1% SDS) for 1 h at 50°C. The eluted DNA was suspended in 4 volumes of QIAGEN buffer PB and passaged through a QIAquick spin column. The resulting DNA was denatured and reannealed with a gradient of 1°C per 45 s in the presence of 50 mM NaCl. CpxR was phosphorylated with phosphoramidate as described previously (42), except that the reaction was carried out at 37°C. The binding reactions were performed in binding buffer (12% glycerol, 4 mM Tris-Cl, pH 7.6, 20 mM KCl, 2 mM EDTA, 1 mM dithiothreitol) with 1 μg of poly(dI-dC) (Roche, Indianapolis, IN) and 50,000 cpm of DNA for 20 min at 37°C. Then 125 ng of DNase I (Roche) in 5 μl was added to each reaction mixture, and the reactions were stopped after 30 s with 380 mM sodium acetate, pH 5.2, 20 mM EDTA, and 2 μg glycogen (Roche). The reaction products were ethanol precipitated and resuspended in sequencing stop buffer (U.S. Biochemicals, Cleveland, OH). Then the reaction products were resolved on an 8% 7.5 M urea-Tris-borate-EDTA polyacrylamide gel and exposed to a phosphorimaging screen. DNA sequencing ladders were generated with a Thermo Sequenase cycle sequencing kit (U.S. Biochemicals) using the same labeled primers. Images were acquired with a Molecular Dynamics Storm 860 Phosphorimager using the ImageQuant software package.
RESULTS
Transposon mutagenesis identified CpxA as a regulator of porin expression.
To monitor transcription of the porin genes ompF and ompC, we used the two-color fluorescent reporter strain MDG147, which contains chromosomal operon fusions of yfp to ompF and cfp to ompC (5). We observed that colonies of MDG147 growing on medium A agar plates initially showed uniform CFP and YFP fluorescence. However, as the colonies continued to grow, the fluorescence became nonuniform; the boundaries of the colonies showed an increased level of yellow fluorescence (increased ompF transcription) and a decreased level of cyan fluorescence (decreased ompC transcription) (Fig. 1). For colonies growing on LB agar, on the other hand, the CFP and YFP fluorescence was more uniform for both early and late growth of the colonies (data not shown). The characteristic pattern of fluorescence for colonies on medium A suggested that this would be a good starting point for genetic screening to search for additional regulators of porin expression.
FIG. 1.
Ring fluorescence phenotype of colonies growing on medium A agar: YFP and CFP fluorescence images of a colony of MDG147 grown on medium A agar for 30 h. Images were obtained with a 250-ms YFP exposure and a 25-ms CFP exposure. The ratio of the two images was computed as described in the supplemental material. Images of colonies of mutants can also be found in the supplemental material.
Approximately 5,000 mini-Tn5 transposon insertions in MDG147 were selected on LB agar containing kanamycin and then spotted onto medium A agar without antibiotic selection. After at least 24 h of growth, spotted colonies were screened by fluorescence microscopy using a low-power (2.5×) objective. We observed a variety of colony fluorescence phenotypes, such as uniform fluorescence, concentric rings, and radial spokes, which were clearly distinct from the wild-type ring pattern shown in Fig. 1 (see figures in the supplemental material). These colonies were selected for further characterization. After transducing the transposon insertions back into MDG147 and verifying that the mutant phenotypes were stable, we determined the locations of the transposon insertions by DNA sequencing. This revealed insertions in 14 different genes (see the supplemental material).
To determine whether any of the isolated mutants showed altered porin expression under conditions other than growth on solid medium A agar, we measured the CFP and YFP fluorescence of liquid cultures in glucose minimal medium. Of the 14 mutants, 1 exhibited a particularly dramatic change in porin expression, with a roughly 17-fold decrease in ompF transcription and a 2-fold increase in ompC transcription (Fig. 2a). This mutant, strain EPB54-012, contained an insertion in cpxA, which encodes a histidine kinase. The insertion was just after bp 277 of the coding sequence for cpxA, which is the second gene in an operon with the gene for the cognate response regulator, cpxR. Colonies of the cpxA::kan mutant on medium A agar differed only subtly from the colonies with the original ring phenotype (see the supplemental material). However, due to the dramatic difference in porin transcription between the mutant and wild-type strains for growth in glucose minimal medium, we decided to further characterize this mutant.
FIG. 2.
Deletion of cpxA results in activation of ompC and repression of ompF in the MG1655 (a) and MC4100 (b) strain backgrounds. Porin expression was measured as CFP fluorescence (corresponding to ompC) (open bars) and YFP fluorescence (corresponding to ompF) (gray bars) in arbitrary units (AU), normalized by OD600, for strains grown in glucose minimal medium to the mid-log phase (see Materials and Methods). (a) Strains MDG147 (wild type), EPB54-012 (cpxA::kan), EPB62 (cpxR::spc, polar on cpxA), EPB28 [Δ(ackA-pta)::cat], and EPB81 [cpxA::kan Δ(ackA-pta)::cat]. (b) Strains MDG131 (wild type), EPB55-012 (cpxA::kan), EPB80 (cpxR::spc, polar on cpxA), EPB10 [Δ(ackA-pta)::cat], and EPB83 [cpxA::kan Δ(ackA-pta)::cat]. The bars indicate the means of at least three independent experiments, and the error bars indicate the standard deviations.
Altered porin expression in the cpxA::kan mutant occurs under conditions associated with high levels of CpxR-P.
To verify that the porin phenotype associated with EPB54-012 (cpxA::kan) was due to the loss of CpxA expression, we constructed an independent deletion of cpxA in MDG147, strain EPB134. This strain showed the same repression of ompF and activation of ompC that were observed for EPB54-012 (data not shown). In addition, transformation of EPB54-012 with a plasmid that expressed cpxA from the trc promoter (pEB50) restored wild-type transcription of ompF and ompC (data not shown), indicating that the mutant phenotype was strictly due to loss of CpxA. We then disrupted both cpxA and cpxR in MDG147 by introducing a cpxR::spc mutation (which exerts a polar effect on the downstream cpxA gene [T. J. Silhavy, personal communication]). Elimination of CpxR and CpxA together had no effect on porin expression (Fig. 2a). The simplest explanation for the results described above is that the change in porin expression in the cpxA strains was due to high levels of CpxR-P as a result of phosphorylation by acetyl phosphate. For wild-type cells in unstressed conditions, CpxR-P levels are low because of the phosphatase activity of CpxA (58). However in cpxA strains, conditions associated with high levels of acetyl phosphate, such as growth on glucose (43, 67, 68), apparently lead to high levels of CpxR-P (11, 12, 17).
To test this hypothesis, we eliminated production of acetyl phosphate in our fluorescent reporter strains by deleting ackA and pta (43, 67, 68). Fluorescence measurements for cultures grown in glucose minimal medium revealed that, indeed, deletion of ackA-pta in a cpxA strain restored porin expression to wild-type levels (Fig. 2a). In addition, when the cpxA strain was grown in minimal medium with glycerol as the carbon source, conditions that result in low levels of acetyl phosphate (43, 67, 68), the porin expression was the same as that in the cpxA+ strain (data not shown). When the same experiment was performed using minimal glycerol medium supplemented with 30 mM acetate, which results in high levels of acetyl phosphate (43, 67, 68), we observed elevated ompC transcription and greatly reduced ompF transcription for the cpxA strain (data not shown). Again, the porin levels returned to the wild-type levels when either cpxR or ackA-pta was deleted from the cpxA strain (data not shown).
It was possible that the results described above were somehow unique to the MG1655 strain background used to construct MDG147. To verify that this was not the case, we also looked at the effect of cpx mutations in the fluorescent reporter strain MDG131 (4), which was derived from MC4100. Due to strain differences, the CFP fluorescence was lower and the YFP fluorescence was higher in MDG131 than in MDG147. However, the relative changes in the CFP and YFP signals for the cpxA strains relative to the cpxA+ strains in the MDG131 background were comparable to what we observed for MDG147 (Fig. 2).
To further test whether porin expression was mediated by CpxR-P, we used the allele cpxRD51A, which encodes a CpxR variant with an alanine at position 51 in place of aspartate (16). Strains with this mutation behave like cpxR null strains (16). A deletion of cpxA was constructed in a strain with the cpxRD51A mutation and grown on glucose minimal media. As shown in Fig. 3, porin expression in the cpxA cpxRD51A strain was comparable to porin expression in the wild-type strain.
FIG. 3.
Elimination of the site of phosphorylation on CpxR eliminates the dependence of porin expression on CpxA. The strains used were MDG147 (wild type), EPB54-012 (cpxA::kan), EPB128 (cpxRD51A), and EPB218 (cpxRD51A cpxA::cat). Porin expression was measured as CFP fluorescence (corresponding to ompC) (open bars) and YFP fluorescence (corresponding to ompF) (gray bars) in arbitrary units (AU), normalized by OD600, for strains grown in glucose minimal medium to the mid-log phase (see Materials and Methods). The bars indicate the means of at least three independent experiments, and the error bars indicate the standard deviations.
Taken together, the results described above indicate that in the absence of CpxA and in the presence of a high concentration of acetyl phosphate, high levels of CpxR-P lead to strong repression of ompF and activation of ompC transcription.
Activation of CpxA decreases ompF expression and increases ompC expression.
If the interpretation described above is correct, then activation of CpxA, which results in high levels of CpxR-P, should also lead to repression of ompF and activation of ompC. To test this hypothesis, we stimulated CpxA by overexpressing the periplasmic protein NlpE (64). Introduction of plasmid pLD404 (64), which constitutively overexpresses NlpE, into MDG147 resulted in a ∼threefold increase in ompC transcription and an ∼11-fold decrease in ompF transcription compared with the fluorescence levels for a strain containing an empty control vector (Fig. 4). Overexpression of NlpE in EPB62 (cpxR cpxA) did not affect ompF or ompC transcription (Fig. 4).
FIG. 4.
Overexpression of NlpE, which activates the Cpx system, decreases ompF expression and increases ompC expression. The strains used were MDG147/pBR322, MDG147/pLD404, EPB62/pBR322, and EPB62/pLD404. Plasmid pLD404 was used to overexpress NlpE from a constitutive promoter, and pBR322 was used as a control. Porin expression was measured as CFP fluorescence (corresponding to ompC) (open bars) and YFP fluorescence (corresponding to ompF) (gray bars) in arbitrary units (AU), normalized by OD600. Strains were grown in glucose minimal medium with 50 μg/ml ampicillin to the mid-log phase (see Materials and Methods). The bars indicate the means of at least three independent experiments, and the error bars indicate the standard deviations.
We also tested a constitutively active variant of CpxA, designated CpxA*, which lacks phosphatase activity (58). As shown in Fig. 5, the CpxA* strain exhibited a ∼threefold increase in ompC transcription and a ∼10-fold decrease in ompF transcription. Expression of CpxA* in the absence of CpxR resulted in no change in porin expression (Fig. 5).
FIG. 5.
CpxA* strain exhibits altered porin expression. The strains used were MDG147/pTrc99a (wild type), EPB54-012/pTrc99a (cpxA), EPB62/pTrc99a (cpxR cpxA), EPB54-012/pAS09b (cpxA*), and EPB62/pAS09b (cpxR cpxA*). Plasmid pAS09b was used to express CpxA*, and plasmid pTrc99a was used as a control. Porin expression was measured as CFP fluorescence (corresponding to ompC) (open bars) and YFP fluorescence (corresponding to ompF) (gray bars) in arbitrary units (AU), normalized by OD600. Strains were grown in glycerol minimal medium with 50 μg/ml ampicillin to the mid-log phase (see Materials and Methods). The bars indicate the means of at least three independent experiments, and the error bars indicate the standard deviations.
High CpxR-P decreases OmpF protein levels and increases OmpC protein levels.
To verify that the observed regulation of ompF and ompC transcription by the Cpx system results in regulation of porin protein levels, we examined the porin content in the cell envelope. Wild-type, cpxA, and cpxRA strains were grown in glucose minimal medium, and cell envelopes were analyzed by urea SDS-PAGE. The OmpC levels were ∼2.2-fold higher in the cpxA strain than in the wild-type strain (Fig. 6). The OmpF levels in the cpxA strain were reduced to such an extent that they were below the detection level of the Coomassie blue-stained gel (Fig. 6). Disruption of both cpxA and cpxR, on the other hand, restored the outer membrane porin content to wild-type levels (Fig. 6). The results described above complement previous work in which a cpxA mutant was found to have reduced levels of OmpF in the membrane (44). In that study, the cause for this reduction in OmpF levels was not identified, but it was hypothesized to be due to an inability of CpxA− cells to properly incorporate OmpF into the outer membrane.
FIG. 6.
Porin protein levels are altered by Cpx activation. (a) SDS-PAGE analysis of cell envelopes of wild-type (MDG147), cpxA (EPB54-012), and cpxR cpxA (EPB62) cells grown in glucose minimal medium. (b) Quantification of protein levels. The open bars indicate the OmpC concentration, and the gray bars indicate the OmpF concentration, measured in arbitrary units (AU). The OmpF levels of EPB54-012 were below the level of detection of the assay (indicated by an asterisk). The bars indicate the means from three independent gels, and the error bars indicate the standard deviations. The gel contained 6 M urea and was stained with Coomassie blue.
Repression of ompF by CpxR-P does not require MicF.
Since OmpF expression is almost completely repressed in the presence of high CpxR-P levels, we wanted to determine whether CpxR-P could act through MicF, a small RNA that binds to and induces degradation of ompF mRNA (reviewed in reference 14). We therefore introduced a deletion of micF into wild-type and cpxA strains. The micF deletion that we used also eliminates ompC. (The micF gene lies upstream of and is transcribed in the opposite direction from ompC.) SDS-PAGE analysis of porin protein levels revealed that for cultures growing in glucose minimal medium, disruption of cpxA still resulted in reduced levels of OmpF in the ΔmicF strain (Fig. 7). Thus, CpxR-P does not require MicF to repress OmpF expression.
FIG. 7.
Repression of OmpF by the Cpx system is independent of MicF: SDS-PAGE analysis of cell envelopes of EPB84 [Δ(micF-ompC)] and EPB89 [Δ(micF-ompC) cpxA::kan]. The gel contained 6 M urea and was stained with Coomassie blue.
CpxR-P represses ompF in the absence of EnvZ.
One difficulty in separating the role of OmpR from the roles of other cellular components in regulating porin expression is that OmpR is required for transcription of ompF and ompC (63). Indeed, fluorescence analysis showed that a cpxA ompR double mutant failed to express either porin when it was grown in glucose minimal medium (data not shown), indicating that in the absence of OmpR, CpxR-P cannot act as a transcriptional activator of ompC or ompF.
OmpC expression increases with increasing OmpR-P levels, whereas OmpF is maximally expressed at intermediate levels of OmpR-P (61). It was thus possible that CpxR-P represses ompF and activates ompC by increasing OmpR-P levels. To test this possibility, we took advantage of the fact that in the envZ strain EPB91, phosphorylation of OmpR by acetyl phosphate leads to an OmpF+/− OmpC− phenotype (27; data not shown). Thus, in this background, a small increase in the OmpR-P level should result in an increase in ompF transcription, and a large increase in the OmpR-P level should result in an increase in ompC transcription. However, we found that for growth in glucose minimal medium, disruption of cpxA in EPB91 further reduced ompF transcription (Fig. 8). The level of transcription of ompC remained below the level of detection in the envZ cpxA strain (data not shown). Therefore, we concluded that the repression of ompF by CpxR-P does not require EnvZ or an increase in OmpR-P levels. Activation of ompC transcription by CpxR-P, on the other hand, is still dependent on increased levels of OmpR-P.
FIG. 8.

CpxR represses ompF in an EnvZ− strain. EPB91 (envZ) and EPB97 (envZ cpxA) were grown in glucose minimal medium to the mid-log phase. The expression of ompF was measured as YFP fluorescence normalized by OD600. The bars indicate the means of at least three independent experiments, and the error bars indicate the standard deviations.
Inactivation of the Cpx system does not affect the osmoregulation of ompF and ompC transcription.
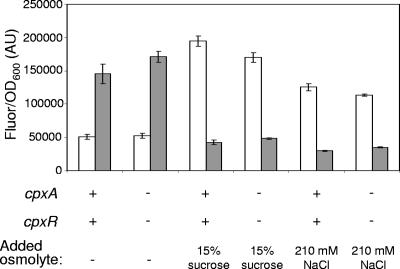
High osmolarity has been shown to increase transcription of several Cpx-regulated genes and to increase the level of CpxR-P (32, 56). To determine whether the Cpx system plays a role in the osmoregulation of ompF and ompC, we compared the levels of YFP and CFP fluorescence of wild-type and cpxR::spc strains grown in minimal glucose media with and without 15% sucrose or 210 mM NaCl. We found that elimination of CpxR and CpxA had no effect on the activation of ompC and repression of ompF in response to a high sucrose or NaCl concentration (Fig. 9).
FIG. 9.
Porin osmoregulation in cpxRA+ and cpxRA strains. The strains used were MDG147 (wild type) and EPB62 (cpxR::spc). Porin expression was measured as CFP fluorescence (corresponding to ompC) (open bars) and YFP fluorescence (corresponding to ompF) (gray bars) in arbitrary units (AU), normalized by OD600. Strains were grown in glucose minimal medium or glucose minimal medium supplemented with 15% sucrose or 210 mM NaCl to the mid-log phase (see Materials and Methods).
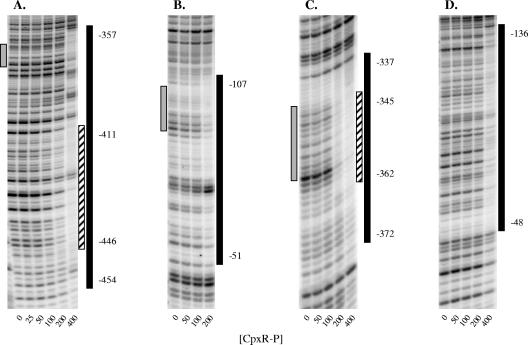
DNase I footprinting indicates that CpxR binds to sites upstream of ompF and ompC.
To determine if CpxR binds to sites near ompF and ompC, we performed a DNase I footprinting analysis (Fig. 10). The results are summarized in Fig. 11. For the region upstream of ompF, CpxR-P protected regions from position −454 to position −357 and from position −107 to position −51 upstream of the ompF transcriptional start (with a region of higher affinity from position −446 to position −411). Within these intervals were sites identified in a genome-wide search for CpxR binding sites (Fig. 10A and B) (15). However, the extended patterns of protection in both of these regions suggest there are multiple CpxR binding sites that were not previously predicted. The distal region of protection overlaps the F4 site (positions −384 to −351) protected by OmpR, and the proximal site overlaps the F1 site (positions −98 to −80), the F2 site (positions −77 to −60), and a portion of the F3 site (positions −57 to −40) protected by OmpR (28, 29, 48, 66). For ompC, we observed protection in regions extending from position −372 to position −337 and from position −136 to position −48 upstream of the ompC transcription start (Fig. 10C and D). The distal region includes the region from position −362 to position −345 (Fig. 10C), which was predicted to be a CpxR binding site (15). The region from position −136 to position −48, however, was not predicted to contain a CpxR binding site. This region overlaps the C1 binding site (positions −95 to −78), the C2 binding site (positions −74 to −57), and part of the C3 binding site (positions −54 to −37) for OmpR at the ompC promoter (41, 66). Together, the results of our DNase I footprinting analysis strongly suggest that CpxR acts directly at the ompC and ompF promoters to regulate expression of these genes.
FIG. 10.
DNase I footprinting of CpxR-P at ompF and ompC. CpxR-P protects two distinct regions upstream of ompF (A and B) and of ompC (C and D). The concentration of CpxR-P (in nM) used in each lane is indicated at the bottom. The coordinates indicate positions relative to the ompF or ompC transcription start sites. Predicted CpxR binding sites (15) are indicated on the left by gray bars. Regions of protection are indicated by black bars and regions of higher affinity are indicated by striped bars on the right.
FIG. 11.
Summary of the results of DNase I footprinting of CpxR-P at ompF (a) and ompC (b). Regions of protection are indicated by black bars, and regions of higher affinity are indicated by striped bars. OmpR binding sites C1, C2, and C3 at ompC and F1, F2, F3, and F4 at ompF are also indicated. The numbers indicate the positions relative to the ompC and ompF transcription start sites.
DISCUSSION
The Cpx and σE pathways respond to cell envelope stress. These two pathways are largely distinct, both in terms of their activating signals and in terms of the genes that they regulate. However, there is at least some overlap; the degP and skp genes are members of both the Cpx and σE regulons, and many other Cpx and σE-regulated genes have similar functional roles (1, 2, 18, 57). Based on the results presented here and recent work on σE-regulated genes (60), ompC and ompF are two additional members of the Cpx and σE regulons. However, porin regulation seems to highlight the differences rather than the similarities between the Cpx and σE systems. In particular, while σE downregulates expression of both porins (60), Cpx downregulates OmpF but upregulates OmpC.
Although the structure of OmpC has never been determined, permeability measurements suggest that it has a smaller pore than OmpF (52). In particular, toxins, such as bile salts and cephalosporins, pass through OmpF more readily than they pass through OmpC. Based on these observations, it has been suggested that reciprocal regulation of the two porins provides a means for E. coli to balance the need for nutrients and the need for protection from toxins (52). In environments such as freshwater supplies, which contain low levels of nutrients and toxins, the higher permeability OmpF would provide an advantage. In animal intestinal tracts, on the other hand, which are rich in nutrients and toxins, OmpC would provide greater protection. In light of our results, it is possible that in the natural environments for E. coli, toxins that induce the Cpx system are primarily permeable through OmpF. The Cpx regulation of porin expression would then be a simple way to provide protection from these compounds while avoiding the more drastic measure used by σE 60) of downregulating expression of both OmpF and OmpC.
A previous genome-wide analysis identified possible CpxR-P binding sites in the ompC and ompF regulatory regions. However, the consensus in the ompF promoter region was determined to be weak and was not characterized further (15). Our DNase I footprinting analysis provided evidence that CpxR-P acts directly at the ompF and ompC promoters. Repression of ompF by CpxR could result from prevention of either OmpR or RNA polymerase binding. Alternatively, CpxR could stimulate DNA loop formation and interaction with the upstream repressive site F4 (reviewed in reference 34). We identified a CpxR binding site >300 bp upstream of ompC. Although this site is at the terminus of micF, we found that a deletion of micF did not prevent CpxR-mediated repression of ompF. This site, however, may be important for ompC transcription. We found that high levels of CpxR-P failed to activate transcription of ompC in the absence of OmpR. This suggests that CpxR binding upstream of ompC acts in conjunction with OmpR binding to activate transcription.
Our results indicate that porin regulation by the CpxA-CpxR system and by the EnvZ-OmpR system converges at the porin promoters. Interestingly, this is not the first example of convergence of these two regulatory systems. Recent studies have shown that expression of curli fimbriae is also regulated by both CpxR and OmpR (17, 32). Other recent work with Chlamydia has identified interactions between OmpR and a CpxR homologue, ChxR (35). Unfortunately, without a better understanding of the stimulus for EnvZ and the mechanism of transcriptional control by OmpR and CpxR, it is difficult to determine the regulatory logic for the porin promoters. Two-component regulatory systems have generally been regarded as separate signaling cascades. However, increasingly, examples are emerging in which these systems are interconnected at various steps in their signaling pathways (for example, see references 6, 20, 24, 32, 47, 49, 65). Given the large regulons controlled by many of these systems, it seems likely that many more examples of complex promoters regulated by multiple two-component systems will emerge.
Supplementary Material
Acknowledgments
We thank T. Silhavy and members of the Silhavy lab for helpful discussions.
This work was supported by grants NSF MCB0212925 (to M.G.), NSF MCB 0243085 (to L.J.K.), NIH GM58746 (to L.J.K.), and NRSA F32-GM68364 (to D.W.) and by a predoctoral fellowship from the American Heart Association (to E.B.).
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ades, S. E. 2004. Control of the alternative sigma factor sigmaE in Escherichia coli. Curr. Opin. Microbiol. 7:157-162. [DOI] [PubMed] [Google Scholar]
- 2.Alba, B. M., and C. A. Gross. 2004. Regulation of the Escherichia coli sigma-dependent envelope stress response. Mol. Microbiol. 52:613-619. [DOI] [PubMed] [Google Scholar]
- 3.Amann, E., B. Ochs, and K. J. Abel. 1988. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69:301-315. [DOI] [PubMed] [Google Scholar]
- 4.Batchelor, E., and M. Goulian. 2003. Robustness and the cycle of phosphorylation and dephosphorylation in a two-component regulatory system. Proc. Natl. Acad. Sci. USA 100:691-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Batchelor, E., T. J. Silhavy, and M. Goulian. 2004. Continuous control in bacterial regulatory circuits. J. Bacteriol. 186:7618-7625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Birkey, S. M., W. Liu, X. Zhang, M. F. Duggan, and F. M. Hulett. 1998. Pho signal transduction network reveals direct transcriptional regulation of one two-component system by another two-component regulator: Bacillus subtilis PhoP directly regulates production of ResD. Mol. Microbiol. 30:943-953. [DOI] [PubMed] [Google Scholar]
- 7.Bolivar, F. 1978. Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique EcoRI sites for selection of EcoRI generated recombinant DNA molecules. Gene 4:121-136. [DOI] [PubMed] [Google Scholar]
- 8.Casadaban, M. J. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104:541-555. [DOI] [PubMed] [Google Scholar]
- 9.Chen, S., A. Zhang, L. B. Blyn, and G. Storz. 2004. MicC, a second small-RNA regulator of Omp protein expression in Escherichia coli. J. Bacteriol. 186:6689-6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Danese, P. N., G. R. Oliver, K. Barr, G. D. Bowman, P. D. Rick, and T. J. Silhavy. 1998. Accumulation of the enterobacterial common antigen lipid II biosynthetic intermediate stimulates degP transcription in Escherichia coli. J. Bacteriol. 180:5875-5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danese, P. N., and T. J. Silhavy. 1998. CpxP, a stress-combative member of the Cpx regulon. J. Bacteriol. 180:831-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Danese, P. N., W. B. Snyder, C. L. Cosma, L. J. Davis, and T. J. Silhavy. 1995. The Cpx two-component signal transduction pathway of Escherichia coli regulates transcription of the gene specifying the stress-inducible periplasmic protease, DegP. Genes Dev. 9:387-398. [DOI] [PubMed] [Google Scholar]
- 13.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delihas, N., and S. Forst. 2001. MicF: an antisense RNA gene involved in response of Escherichia coli to global stress factors. J. Mol. Biol. 313:1-12. [DOI] [PubMed] [Google Scholar]
- 15.De Wulf, P., A. M. McGuire, X. Liu, and E. C. Lin. 2002. Genome-wide profiling of promoter recognition by the two-component response regulator CpxR-P in Escherichia coli. J. Biol. Chem. 277:26652-26661. [DOI] [PubMed] [Google Scholar]
- 16.DiGiuseppe, P. A., and T. J. Silhavy. 2003. Signal detection and target gene induction by the CpxRA two-component system. J. Bacteriol. 185:2432-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dorel, C., O. Vidal, C. Prigent-Combaret, I. Vallet, and P. Lejeune. 1999. Involvement of the Cpx signal transduction pathway of E. coli in biofilm formation. FEMS Microbiol. Lett. 178:169-175. [DOI] [PubMed] [Google Scholar]
- 18.Duguay, A. R., and T. J. Silhavy. 2004. Quality control in the bacterial periplasm. Biochim. Biophys. Acta 1694:121-134. [DOI] [PubMed] [Google Scholar]
- 19.Egger, L. A., H. Park, and M. Inouye. 1997. Signal transduction via the histidyl-aspartyl phosphorelay. Genes Cells 2:167-184. [DOI] [PubMed] [Google Scholar]
- 20.Eguchi, Y., and R. Utsumi. 2005. A novel mechanism for connecting bacterial two-component signal-transduction systems. Trends Biochem. Sci. 30:70-72. [DOI] [PubMed] [Google Scholar]
- 21.Ferrario, M., B. R. Ernsting, D. W. Borst, D. E. Wiese, 2nd, R. M. Blumenthal, and R. G. Matthews. 1995. The leucine-responsive regulatory protein of Escherichia coli negatively regulates transcription of ompC and micF and positively regulates translation of ompF. J. Bacteriol. 177:103-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forst, S., J. Delgado, A. Rampersaud, and M. Inouye. 1990. In vivo phosphorylation of OmpR, the transcription activator of the ompF and ompC genes in Escherichia coli. J. Bacteriol. 172:3473-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forst, S., and M. Inouye. 1988. Environmentally regulated gene expression for membrane proteins in Escherichia coli. Annu. Rev. Cell Biol. 4:21-42. [DOI] [PubMed] [Google Scholar]
- 24.Gunn, J. S., and S. I. Miller. 1996. PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J. Bacteriol. 178:6857-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Head, C. G., A. Tardy, and L. J. Kenney. 1998. Relative binding affinities of OmpR and OmpR-phosphate at the ompF and ompC regulatory sites. J. Mol. Biol. 281:857-870. [DOI] [PubMed] [Google Scholar]
- 26.Hernday, A. D., B. A. Braaten, G. Broitman-Maduro, P. Engelberts, and D. A. Low. 2004. Regulation of the pap epigenetic switch by CpxAR: phosphorylated CpxR inhibits transition to the phase ON state by competition with Lrp. Mol. Cell 16:537-547. [DOI] [PubMed] [Google Scholar]
- 27.Hsing, W., and T. J. Silhavy. 1997. Function of conserved histidine-243 in phosphatase activity of EnvZ, the sensor for porin osmoregulation in Escherichia coli. J. Bacteriol. 179:3729-3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang, K.-J., and M. M. Igo. 1996. Identification of the bases in the ompF regulatory region which interact with the transcription factor OmpR. J. Mol. Biol. 262:615-628. [DOI] [PubMed] [Google Scholar]
- 29.Huang, K.-J., J. L. Schieberl, and M. M. Igo. 1994. A distant upstream site involved in the negative regulation of the Escherichia coli ompF gene. J. Bacteriol. 176:1309-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang, L., P. Tsui, and M. Freundlich. 1990. Integration host factor is a negative effector of in vivo and in vitro expression of ompC in Escherichia coli. J. Bacteriol. 172:5293-5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hung, D. L., T. L. Raivio, C. H. Jones, T. J. Silhavy, and S. J. Hultgren. 2001. Cpx signaling pathway monitors biogenesis and affects assembly and expression of P pili. EMBO J. 20:1508-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jubelin, G., A. Vianney, C. Beloin, J. M. Ghigo, J. C. Lazzaroni, P. Lejeune, and C. Dorel. 2005. CpxR/OmpR interplay regulates curli gene expression in response to osmolarity in Escherichia coli. J. Bacteriol. 187:2038-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawaji, H., T. Mizuno, and S. Mizushima. 1979. Influence of molecular size and osmolarity of sugars and dextrans on the synthesis of outer membrane proteins O-8 and O-9 of Escherichia coli K-12. J. Bacteriol. 140:843-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kenney, L. J. 2002. Structure/function relationships in OmpR and other winged-helix transcription factors. Curr. Opin. Microbiol. 5:135-141. [DOI] [PubMed] [Google Scholar]
- 35.Koo, I. C., D. Walthers, T. L. Nicholson, L. J. Kenney, and R. S. Stephens. Submitted for publication.
- 36.Lan, C. Y., and M. M. Igo. 1998. Differential expression of the OmpF and OmpC porin proteins in Escherichia coli K-12 depends upon the level of active OmpR. J. Bacteriol. 180:171-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langen, G. R., J. R. Harper, T. J. Silhavy, and S. P. Howard. 2001. Absence of the outer membrane phospholipase A suppresses the temperature-sensitive phenotype of Escherichia coli degP mutants and induces the Cpx and σE extracytoplasmic stress responses. J. Bacteriol. 183:5230-5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larsen, R. A., M. M. Wilson, A. M. Guss, and W. W. Metcalf. 2002. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch. Microbiol. 178:193-201. [DOI] [PubMed] [Google Scholar]
- 39.Lee, Y. M., P. A. DiGiuseppe, T. J. Silhavy, and S. J. Hultgren. 2004. P pilus assembly motif necessary for activation of the CpxRA pathway by PapE in Escherichia coli. J. Bacteriol. 186:4326-4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu, X., and T. Ferenci. 2001. An analysis of multifactorial influences on the transcriptional control of ompF and ompC porin expression under nutrient limitation. Microbiology 147:2981-2989. [DOI] [PubMed] [Google Scholar]
- 41.Maeda, S., and T. Mizuno. 1990. Evidence for multiple OmpR-binding sites in the upstream activation sequence of the ompC promoter in Escherichia coli: a single OmpR-binding site is capable of activating the promoter. J. Bacteriol. 172:501-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mattison, K., R. Oropeza, N. Byers, and L. J. Kenney. 2002. A phosphorylation site mutant of OmpR reveals different binding conformations at ompF and ompC. J. Mol. Biol. 315:497-511. [DOI] [PubMed] [Google Scholar]
- 43.McCleary, W. R., and J. B. Stock. 1994. Acetyl phosphate and the activation of two-component response regulators. J. Biol. Chem. 269:31567-31572. [PubMed] [Google Scholar]
- 44.McEwen, J., L. Sambucetti, and P. M. Silverman. 1983. Synthesis of outer membrane proteins in cpxA cpxB mutants of Escherichia coli K-12. J. Bacteriol. 154:375-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mecsas, J., P. E. Rouviere, J. W. Erickson, T. J. Donohue, and C. A. Gross. 1993. The activity of sigma E, an Escherichia coli heat-inducible sigma factor, is modulated by expression of outer membrane proteins. Genes Dev. 7:2618-2628. [DOI] [PubMed] [Google Scholar]
- 46.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. CSHL Press, Plainview, N.Y.
- 47.Minagawa, S., H. Ogasawara, A. Kato, K. Yamamoto, Y. Eguchi, T. Oshima, H. Mori, A. Ishihama, and R. Utsumi. 2003. Identification and molecular characterization of the Mg2+ stimulon of Escherichia coli. J. Bacteriol. 185:3696-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mizuno, T., M. Kato, Y. L. Jo, and S. Mizushima. 1988. Interaction of OmpR, a positive regulator, with the osmoregulated ompC and ompF genes of Escherichia coli. Studies with wild-type and mutant OmpR proteins. J. Biol. Chem. 263:1008-1012. [PubMed] [Google Scholar]
- 49.Mouslim, C., and E. A. Groisman. 2003. Control of the Salmonella ugd gene by three two-component regulatory systems. Mol. Microbiol. 47:335-344. [DOI] [PubMed] [Google Scholar]
- 50.Nakayama, S., and H. Watanabe. 1998. Identification of cpxR as a positive regulator essential for expression of the Shigella sonnei virF gene. J. Bacteriol. 180:3522-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nevesinjac, A. Z., and T. L. Raivio. 2005. The Cpx envelope stress response affects expression of the type IV bundle-forming pili of enteropathogenic Escherichia coli. J. Bacteriol. 187:672-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nikaido, H. 1996. Outer membrane, p. 29-47. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaecter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. ASM Press, Washington, D.C.
- 53.Otto, K., and T. J. Silhavy. 2002. Surface sensing and adhesion of Escherichia coli controlled by the Cpx-signaling pathway. Proc. Natl. Acad. Sci. USA 99:2287-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pratt, L. A., W. Hsing, K. E. Gibson, and T. J. Silhavy. 1996. From acids to osmZ: multiple factors influence synthesis of the OmpF and OmpC porins in Escherichia coli. Mol. Microbiol. 20:911-917. [DOI] [PubMed] [Google Scholar]
- 55.Pratt, L. A., and T. J. Silhavy. 1996. The response regulator SprE controls the stability of RpoS. Proc. Natl. Acad. Sci. USA 93:2488-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prigent-Combaret, C., E. Brombacher, O. Vidal, A. Ambert, P. Lejeune, P. Landini, and C. Dorel. 2001. Complex regulatory network controls initial adhesion and biofilm formation in Escherichia coli via regulation of the csgD gene. J. Bacteriol. 183:7213-7223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Raivio, T. L., and T. J. Silhavy. 2001. Periplasmic stress and ECF sigma factors. Annu. Rev. Microbiol. 55:591-624. [DOI] [PubMed] [Google Scholar]
- 58.Raivio, T. L., and T. J. Silhavy. 1997. Transduction of envelope stress in Escherichia coli by the Cpx two-component system. J. Bacteriol. 179:7724-7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ramani, N., L. Huang, and M. Freundlich. 1992. In vitro interactions of integration host factor with the ompF promoter-regulatory region of Escherichia coli. Mol. Gen. Genet. 231:248-255. [DOI] [PubMed] [Google Scholar]
- 60.Rhodius, V., and C. A. Gross. Personal communication.
- 61.Russo, F. D., and T. J. Silhavy. 1991. EnvZ controls the concentration of phosphorylated OmpR to mediate osmoregulation of the porin genes. J. Mol. Biol. 222:567-580. [DOI] [PubMed] [Google Scholar]
- 62.Slauch, J. M., and T. J. Silhavy. 1989. Genetic analysis of the switch that controls porin gene expression in Escherichia coli K-12. J. Mol. Biol. 210:281-292. [DOI] [PubMed] [Google Scholar]
- 63.Slauch, J. M., and T. J. Silhavy. 1996. The porin regulon: a paradigm for the two-component regulatory systems, p. 383-417. In E. C. C. Lin and A. S. Lynch (ed.), Regulation of gene expression in Escherichia coli. Chapman & Hall, New York, N.Y.
- 64.Snyder, W. B., L. J. Davis, P. N. Danese, C. L. Cosma, and T. J. Silhavy. 1995. Overproduction of NlpE, a new outer membrane lipoprotein, suppresses the toxicity of periplasmic LacZ by activation of the Cpx signal transduction pathway. J. Bacteriol. 177:4216-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Soncini, F. C., and E. A. Groisman. 1996. Two-component regulatory systems can interact to process multiple environmental signals. J. Bacteriol. 178:6796-6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsung, K., R. E. Brissette, and M. Inouye. 1989. Identification of the DNA-binding domain of the OmpR protein required for transcriptional activation of the ompF and ompC genes of Escherichia coli by in vivo DNA footprinting. J. Biol. Chem. 264:10104-10109. [PubMed] [Google Scholar]
- 67.Wanner, B. L., and M. R. Wilmes-Riesenberg. 1992. Involvement of phosphotransacetylase, acetate kinase, and acetyl phosphate synthesis in control of the phosphate regulon in Escherichia coli. J. Bacteriol. 174:2124-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wolfe, A. J. 2005. The acetate switch. Microbiol. Mol. Biol Rev. 69:12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.