Abstract
The θ subunit (holE gene product) of Escherichia coli DNA polymerase (Pol) III holoenzyme is a tightly bound component of the polymerase core. Within the core (α-ɛ-θ), the α and ɛ subunits carry the DNA polymerase and 3′ proofreading functions, respectively, while the precise function of θ is unclear. holE homologs are present in genomes of other enterobacteriae, suggestive of a conserved function. Putative homologs have also been found in the genomes of bacteriophage P1 and of certain conjugative plasmids. The presence of these homologs is of interest, because these genomes are fully dependent on the host replication machinery and contribute few, if any, replication factors themselves. To study the role of these θ homologs, we have constructed an E. coli strain in which holE is replaced by the P1 homolog, hot. We show that hot is capable of substituting for holE when it is assayed for its antimutagenic action on the proofreading-impaired dnaQ49 mutator, which carries a temperature-sensitive ɛ subunit. The ability of hot to substitute for holE was also observed with other, although not all, dnaQ mutator alleles tested. The data suggest that the P1 hot gene product can substitute for the θ subunit and is likely incorporated in the Pol III complex. We also show that overexpression of either θ or Hot further suppresses the dnaQ49 mutator phenotype. This suggests that the complexing of dnaQ49-ɛ with θ is rate limiting for its ability to proofread DNA replication errors. The possible role of hot for bacteriophage P1 is discussed.
Escherichia coli DNA polymerase III holoenzyme (HE) is the enzyme responsible for the faithful duplication of the bacterial chromosome. HE is a multisubunit, dimeric complex that replicates simultaneously the leading and lagging strands at the replication fork (for a review, see reference 31). Each half of the dimeric complex contains a polymerase (Pol) III core unit consisting of three subunits, α, ɛ, and θ, which are tightly bound in the linear order α-ɛ-θ. Of these, α (135 kDa) is the DNA polymerase (dnaE gene product), ɛ (28 kDa) is the exonucleotidic proofreader responsible for editing polymerase insertion errors (dnaQ gene product), and θ (8 kDa) is a small subunit whose function is not well defined (holE gene product). The HE (17 subunits total, 10 distinct) further consists of the two β-clamps (one for each core) that serve to tether the polymerase to the DNA and the DnaX complex (τ2γδδ′χψ), which is responsible for connecting the two polymerases and for loading and unloading the β-clamp in the discontinuously synthesized lagging strand. The precise mechanisms by which HE is capable of replicating the bacterial chromosome with high speed and high accuracy are the subject of active investigation (21, 31, 33, 52).
Our laboratory has been interested in the roles and mechanisms of the ɛ and θ subunits within the Pol III core, particularly with respect to their contributions to the fidelity of the replication process (37, 48-50). ɛ is a critical subunit with at least dual functions. It is a fidelity subunit whose loss leads to strongly increased mutation rates (7, 13, 42, 48, 49). Second, it provides an important structural role within the core, most likely by its stabilizing effect on the α subunit (18, 23, 49). Consequently, a deletion mutant of the dnaQ gene is essentially nonviable unless compensated for by suppressor mutations in the polymerase subunit. ɛ has been investigated by genetic and physical methods. It consists of two separate domains, an N-terminal catalytic domain, which also contains the θ-binding site, and a C-terminal domain essential for binding to the α subunit. The structure of the N-terminal catalytic domain has been determined (5, 12), and the θ interaction domain on ɛ has been defined (3).
Much less is known about the θ subunit. θ binds to ɛ (but not to α) (2, 47), and this binding to ɛ was shown to increase the activity of the 3′ exonuclease of the ɛ subunit on a terminal G · T mismatch in vitro by about 2.5-fold (47), supporting the idea of a possible fidelity role for θ. In vivo experiments showed that a ΔholE strain is normally viable (45), indicating that the subunit is not essential. A possible role for θ in controlling the fidelity of DNA replication was further suggested by measurements of mutant frequencies in mismatch repair-defective strains, in which ΔholE derivatives showed two- to five-fold-higher mutator activities for selected mutational markers (50). Based on these results, it was suggested that θ exerts a positive effect on replication fidelity, presumably indirectly by promoting the proofreading ability of ɛ. These experiments also revealed θ to play a particularly important role in stabilizing the temperature-sensitive dnaQ49 allele (50), which produces a temperature-sensitive ɛ subunit (7, 13, 41, 42). At low temperature (28°C), a dnaQ49 strain displays only a modest mutator phenotype. However, a dnaQ49 ΔholE strain shows, at this temperature, a very high mutator activity (1,000-fold enhanced over that of the single-dnaQ49 level) (50). A similar effect of θ was observed with several other dnaQ mutator alleles, particularly those that appeared to carry a structurally impaired ɛ subunit (50). Thus, it was proposed that one function of θ is to stabilize ɛ. Such a stabilizing function is likely to be beneficial, as ɛ appears to be intrinsically unstable (8) or poorly folded (5, 10).
The studies reported here focus on the holE homolog, named hot (homolog of theta), present in the genome of bacteriophage P1 (25). P1 phage relies entirely on the E. coli replication machinery for its replication during both the lytic and lysogenic stages (25). The phage encodes only a few other potential replication proteins, such as ban (a dnaB helicase homolog), ssb (an ssb homolog), and humD (a umuD′ homolog), which have been shown to complement corresponding host defects, but it does not contain any other putative components of the Pol III HE (25). Thus, the presence of this putative homolog of the Pol III core is of interest. θ homologs have been found, in addition, in certain large conjugational plasmids of enterobacteriae, Salmonella pSLT (30) and Proteus vulgaris Rts1 (34), further supporting a meaningful function for this protein. Indirectly, study of these proteins may help elucidate the function of θ in E. coli. For example, the greater in vitro stability of Hot than that of θ has permitted determination of the solution structure of Hot by nuclear magnetic resonance methods (4).
Here, we have investigated whether Hot, the P1 θ homolog, can substitute for θ in stabilizing certain dnaQ alleles characterized by structurally impaired ɛ subunits. We show that the presence of the hot gene stabilizes dnaQ49 and similar dnaQ alleles, suggesting that Hot protein is indeed capable of substituting for θ. We also observed that at least one dnaQ allele can be stabilized by θ but not by Hot, indicating that the interactions of the two proteins with ɛ are not completely identical. We discuss the possible reasons why the bacteriophage P1 may have a θ homolog.
MATERIALS AND METHODS
Strains and media.
The E. coli strains used, along with information on their construction, are listed in Table 1. P1 transductions were performed using P1virA. P1 used for the cloning of the hot gene was from a P1 c1-100 Tn9 lysogen (25) obtained from P. Schendel (Genetics Institute, Cambridge, MA). The construction of MG1655 derivatives NR13104 (ΔholE203) and NR16351 (ΔholE204::hot) is detailed below. The various dnaQ alleles (Table 1) were transduced using linkage (∼40%) with transposon zae-502::Tn10, followed by testing for the associated dnaQ mutator phenotype (scoring for rifampin-resistant [Rifr] mutants). The donor strains NR11569, NR11572, NR11573, and NR11641 were dnaQ derivatives of NR9501 or NR9601 as described previously (48). Plasmid pKO3 (24) was obtained from G. Church (Harvard Medical School). LB broth was the standard recipe (39). Unless indicated otherwise, antibiotics were added as follows: ampicillin at 100 μg/ml, chloramphenicol at 20 μg/ml, tetracycline at 15 μg/ml, and rifampin at 100 μg/ml. Solid media contained 1.5% agar (Difco).
TABLE 1.
E. coli strains used in this study
| Strain | Relevant genotype | Reference or construction |
|---|---|---|
| MG1655 | Wild-type | 44 |
| NR13104 | ΔholE203 | This work |
| NR16315 | ΔholE204::hot | This work |
| NR9695 | dnaQ49 zae-502::Tn10 | 38 |
| NR11569 | dnaQ920 zae-502::Tn10 | 48 |
| NR11572 | dnaQ923 zae-502::Tn10 | 48 |
| NR11573 | dnaQ924 zae-502::Tn10 | 48 |
| NR11641 | dnaQ928 zae-502::Tn10 | 48 |
| NR16316 | dnaQ920 | MG1655 × P1/NR11569 |
| NR16317 | ΔholE203 dnaQ920 | NR13104 × P1/NR11569 |
| NR16318 | ΔholE204::hot dnaQ920 | NR16315 × P1/NR11569 |
| NR16319 | dnaQ923 | MG1655 × P1/NR11572 |
| NR16320 | ΔholE203 dnaQ923 | NR13104 × P1/NR11572 |
| NR16321 | ΔholE204::hot dnaQ923 | NR16315 × P1/NR11572 |
| NR16322 | dnaQ924 | MG1655 × P1/NR11573 |
| NR16323 | ΔholE203 dnaQ924 | NR13104 × P1/NR11573 |
| NR16324 | ΔholE204::hot dnaQ924 | NR16315 × P1/NR11573 |
| NR16325 | dnaQ928 | MG1655 × P1/NR11641 |
| NR16326 | ΔholE203 dnaQ928 | NR13104 × P1/NR11641 |
| NR16327 | ΔholE204::hot dnaQ928 | NR16315 × P1/NR11641 |
DNA isolation, PCR, and DNA sequencing.
E. coli genomic DNA was prepared using the DNeasy tissue kit (QIAGEN Sciences). Bacteriophage P1 DNA was purified from a P1 c1-100 Tn9 lysogen using the QIAGEN large construction kit (QIAGEN Sciences). Plasmid DNAs were purified with a Qiaprep spin miniprep kit (QIAGEN Sciences). All preparative PCRs were performed using the Expand high-fidelity PCR system (Roche). Analytic PCRs were performed using Taq DNA polymerase (Invitrogen). Conditions for PCRs were as recommended by the manufacturer. Oligo 6.8 software (Molecular Biology Insights, Inc., Cascade, CO) was used to design oligonucleotide primers (Table 2) and to determine annealing temperatures. DNA sequencing of plasmids pUC18hot and pKO3hot or the PCR product containing the chromosomal ΔholE::hot insert (strain NR16315) (Table 1) was performed using the Big Dye Terminator v1.1 cycle sequencing kit (Applied Biosystems, Foster City, CA) and Perkin-Elmer ABI model 377 and model 3100 sequencers. Both DNA strands were sequenced.
TABLE 2.
Oligonucleotide primers used in PCRs
| Name | Sequencea |
|---|---|
| GR-1b | 5′-CACGCAATAACCTTCACACTCCAAATTTATAACCATTCTTAATCTCCTCATCATTCGCGG-3′ |
| GR-2 | 5′-AAGGAAAAAAGCGGCCGCTGTAGTGTCCTTTCGTTTTATGCCC-3′ |
| GR-3b | 5′-GTTATAAATTTGGAGTGTGAAGGTTATTGCGTGTAAAACTTATACAGAGTTACACTTTCTTACATAACG-3′ |
| GR-4 | 5′-CGCACGCATGTCGACCGGTTGTTCATCGACCACC-3′ |
| Pr1 | 5′-GAGAAATGCGGCCGCTGTAGTGTCCTTTCGTTTTATGCCC-3′ |
| Pr2c | 5′-CATTCTTAATCTCCTCATCATTCGC-3′ |
| Pr3c | 5′-GCGAATGATGAGGAGATTAAGAATGTACGATTGGAATATTGCAGC-3′ |
| Pr4d | 5′-CTCAATAGGATCCGAGAAGTTCAAAACGGTTAACTACC-3′ |
| Pr5d | 5′-ACTTCTCGGATCCTATTGAGCAGAGTTACACTTTCTTACATAACGC-3′ |
| Pr6 | 5′-CAAATCAGTCGACGCCAGCAGGTCGGGTTCTCC-3′ |
Underlines represent NotI (GR-2, Pr1) or SalI (GR-4, Pr6) sequences. Double underlines indicate holE or hot start (GR-1, Pr3) or stop (GR-3, Pr5) codons. P1 hot sequences (in Pr2, Pr4, and Pr5) are italicized.
The first 33 bases of GR-1 and GR-3 are complementary (boldface).
The first 25 bases of Pr2 and Pr3 are complementary (boldface) and include the hot ATG start codon.
The first 20 bases of Pr4 and Pr5 are complementary (boldface).
Construction of NR13104 containing a precise holE deletion (ΔholE203).
Strains containing partial deletions of the holE coding sequence have been described previously (45). In the present study, we used a strain, NR13104, containing a complete deletion of the holE gene, created by the method of Link et al. (24), as described below. NR13104 contains a precise chromosomal deletion of the 226 bp between the holE start and stop codons, replaced by a 36-bp stuffer fragment encoding the 12-mer peptide MVINLVCEGYCV. The primers used are listed in Table 2. Outside primers were GR-2, positioned 496 bp upstream of the holE ATG codon (and containing a NotI restriction site in its 5′ tail), and GR-4, positioned 494 bp downstream of the holE stop codon (and containing a SalI restriction site). Inside primers were GR-1 and GR-3, which contain a 33-base overlapping sequence at their 5′ ends encoding the 12-residue stuffer fragment. Genomic DNA of strain MG1655 was prepared and amplified by PCR in two separate reactions using primer pairs GR-1-GR-2 and GR-3-GR-4. The resulting two products were then mixed together in a second round of PCR amplification using the outside primers GR-2 and GR-4. Due to the complementarity of the GR1 and GR3 sequences, this yielded an ∼1,000-bp PCR product containing the holE deletion. The fragment was purified and inserted between the NotI and SalI sites of plasmid pKO3 (24), yielding plasmid pSTBΔholE. This plasmid was used for integration into the MG1655 chromosome at the holE sequence, leading to replacement of the holE+ gene by the ΔholE (ΔholE203) fragment by the method of Link et al. (24).
Cloning of the P1 hot gene under the control of the holE promoter.
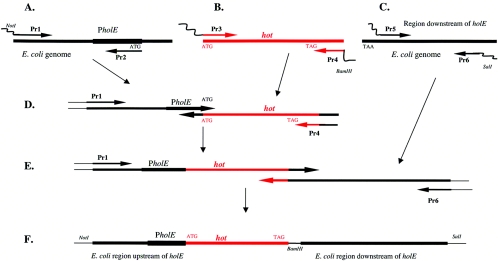
To clone the P1 hot gene under the control of the holE promoter, three steps of PCR were used as diagrammed in Fig. 1. The primers used are listed in Table 2. In the first step, we created three separate PCR products (A, B, and C in Fig. 1). In reaction A, a 499-bp fragment of DNA upstream of the E. coli holE gene was amplified using primers Pr1 and Pr2 on E. coli MG1655 chromosomal DNA, yielding a product that included the holE promoter but ended at the ATG start codon. In reaction B, primers Pr3 and Pr4 were used on bacteriophage P1 DNA to create a 313-bp fragment containing the entire P1 hot coding sequence starting at the ATG start codon and ending 27 nucleotides past the TAG stop codon. In reaction C, 564 bp of E. coli chromosomal DNA downstream of the holE gene was amplified using primers Pr5 and Pr6 on E. coli chromosomal DNA. The “outside” primers Pr1 and Pr6 contained tails carrying NotI and SalI restriction sequences, respectively, to facilitate the eventual cloning of the desired construct. “Inside” primer pairs Pr2-Pr3 and Pr4-Pr5 contained complementary tails and sequences (Table 2) that permitted assembly of these three fragments by overlap PCR into one large, 1,399-bp fragment, as described below. In the second step (Fig. 1D), fragments A and B were joined using Pr1 and Pr4 based on the overlap provided by Pr2 and Pr3. The resulting fragment contained the P1 hot coding sequence fused to the holE promoter, while at the same time holE was deleted starting from its ATG codon. In the final step (Fig. 1E), the fragment generated in step 2 was joined to fragment C using primers Pr1 and Pr6, facilitated by the overlap of primers Pr4 and Pr5. This joined the hot gene to the E. coli chromosomal sequence downstream of the holE gene. In the final product, the hot TAG stop codon was followed by 27 nucleotides of downstream hot sequence and then by 12 unrelated nucleotides containing a BamHI site (not relevant to the present study), which replaced 8 nucleotides that normally follow the holE TAA stop codon. The final 1,399-bp product contained the P1 hot open reading frame surrounded on either side by about 500 nucleotides of E. coli chromosomal DNA (ΔholE::hot). This product was blunt ended into the SmaI site of plasmid pUC18, yielding pUC18hot. The plasmid was sequenced to confirm the correctness of the construct.
FIG. 1.
Three steps of PCR to replace the holE coding sequence by the P1 hot open reading frame. Red lines indicate P1 genomic DNA, and the black lines indicate E. coli genomic DNA. PholE denotes the holE promoter. Arrows indicate the 5′→3′ direction of DNA primers. See Materials and Methods for further details and Table 2 for primer sequences.
Plasmid constructions.
In addition to pUC18hot described above, two additional pUC18 plasmids were made, as well as corresponding derivatives of low-copy-number plasmid pKO3 (24). Primers Pr1 and Pr6 (Table 2) were used to obtain a PCR fragment from genomic DNA of strains MG1655 (holE+) and NR13104 (ΔholE). These fragments contained the holE gene and the ΔholE deletion, respectively, flanked by approximately 500 base pairs of genomic sequence on either side of holE. The fragments were inserted directly into the SmaI restriction site of pUC18, yielding pUC18holE and pUC18ΔholE, respectively. Plasmid pKO3 is a low-copy-number (three to five copies) temperature-sensitive vector that can be used for creating gene deletions or genomic replacements (24). The PCR inserts from the three pUC18 plasmids were transferred into pKO3 using the NotI-SalI restriction sites contained in their ends, yielding pKO3holE, pKO3hot, and pKO3ΔholE, respectively.
Insertion of the P1 hot gene into the E. coli chromosome.
The pKO3hot plasmid containing the P1 hot gene under the control of the holE promoter (see above) was used to insert the hot construct into the E. coli chromosome by the method of Link et al. (24). NR13104 (ΔholE203) cells were transformed by pKO3hot, and transformants were selected at 40°C on LB plates containing chloramphenicol. Several chloramphenicol-resistant colonies were diluted in LB broth and plated at 30°C on LB broth containing 5% sucrose. Single colonies were tested for loss of chloramphenicol resistance as well as the presence of the hot gene by PCR using primers Pr3 and Pr4 (Table 2). A PCR product containing the recombinant gene was sequenced to confirm the correctness of the construct. The new chromosomal allele was designated ΔholE204::hot (Table 1)
Protein expression.
Expression of θ and Hot was assayed by Western blotting using a rabbit anti-θ polyclonal antibody (prepared by Biocon, Inc., Rockville, MD). Strains containing various pUC18 plasmids (see above) were grown overnight at 37°C in 1 ml LB broth plus ampicillin (500 μg/ml). Control strains without plasmid were grown in LB broth. Cells were centrifuged and washed with Tris-buffered saline; the pellet was sonicated and lysed in 100 μl of Tricine-sodium dodecyl sulfate loading buffer (Invitrogen) containing 5% β-mercaptoethanol. Protein samples (8 μl) were electrophoresed through a 16% polyacrylamide-Tricine-sodium dodecyl sulfate gel (Invitrogen) and transferred to a nitrocellulose membrane using a MilliBlot graphite electroblotter apparatus (Millipore). To remove nonspecific binding activities, the anti-θ antibody was pretreated (3 h at room temperature) with a cell lysate of strain NR13104 (ΔholE) (prepared by sonication) in I-block solution (Applied Biosystems), and the precipitate was removed by centrifugation. Goat anti-rabbit antibody labeled with alkaline phosphatase (Bio-Rad) was used as secondary antibody. The Western-star kit (Applied Biosystems) was used for membrane development, as recommended by the manufacturer. Purified θ, used as a protein marker, was as described previously (22).
Mutant frequencies.
To measure mutant frequencies for each strain, the frequency of Rifr mutants in overnight cultures was determined. Ten to 15 cultures for each strain started from individual colonies were grown overnight in 1 ml LB broth at 37°C. For each culture, a 50- to 100-μl aliquot of a 10−6 dilution was spread on an LB plate to determine the total number of viable cells, and 100 μl of the undiluted or appropriately diluted culture was spread on LB broth-Rif plates to determine the number of rifampin-resistant mutants. The mutant frequency for each culture was calculated by dividing the total number of mutants by the total number of viable cells. The data were analyzed using the statistical analysis software Prism (GraphPad).
Database searches and protein sequences comparison.
BLAST (BLASTP) searches for homologs of E. coli θ protein were performed with the GenBank (http://www.ncbi.nlm.nih.gov/) and coliBASE (http://colibase.bham.ac.uk) databases, using the sequence of E. coli θ as a query. The Klebsiella pneumoniae sequence was found by translated BLAST (TBLASTN) on the NCBI microbial genome database. A ClustalW multiple-amino-acid sequence alignment was created using BioEdit software with BLOSUM62 as the similarity matrix (11). This alignment was used in conjunction with Tree Top analysis tools of the GeneBee Molecular Biology server (http://www.genebee.msu.su/genebee.html) to create a phylogenetic tree of θ proteins (by topological algorithm).
RESULTS
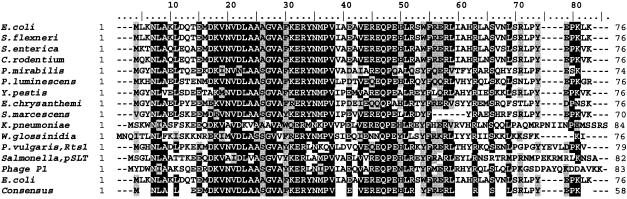
Alignment of θ with its homologs. In Fig. 2, we present an amino acid alignment of E. coli θ with several homologs as found by BLAST searches in GenBank and coliBASE (see Materials and Methods). The bacteria in this group (Shigella flexneri, Salmonella enterica, Citrobacter rodentium, Proteus mirabilis, Photorhabdus luminescens, Yersinia pestis, Erwinia chrysanthemi, Serratia marcescens, Klebsiella pneumoniae, and Wigglesworthia glossinidia) are all gram-negative enterobacteriae with relatively close relatedness to E. coli. The homology with θ is generally good, as expected for these related organisms. The homology is strongest in the N-terminal part (residues 1 to 56 of θ) and is more diverse in the remaining C-terminal part (residues 57 to 76). The alignment also includes two homologs residing on large conjugative plasmids: plasmid Rts1, originally isolated from Proteus vulgaris (34), and plasmid pSLT from Salmonella enterica serovar Typhimurium LT2 (30). Rts1 is a large (2.1-Mb) conjugative plasmid, while pSLT (0.94 Mb) is the conjugative virulence plasmid of S. enterica serovar Typhimurium LT2. Their homology with θ is lower but still generally good (44 to 54% identical, 57 to 62% similar).
FIG. 2.
Alignment of θ homologs. A ClustalW multiple-amino-acid sequence alignment was created using BioEdit software (11) and appears with the indicated consensus sequence. The sequences were obtained by BLAST searches of the coliBASE database (P. mirabilis, C. rodentium, S. marcescens) or the NCBI database (others) as described in Materials and Methods. Multiple, but identical, entries were found for most of the θ homologs, except for Y. pestis and Y. pestis KIM. The θ homolog from the latter contained several extra amino acids at its N terminus. These are not shown here. The E. coli θ sequence is presented on both the top and bottom to facilitate comparisons. Identical amino acids (70%) are shown with a black background, and similar residues (70%) are shown with a gray background. The indicated species are in the order Escherichia coli, Shigella flexneri, Salmonella enterica, Citrobacter rodentium, Proteus mirabilis, Photobacter luminescens, Yersinia pestis, Erwinia chrysanthemi, Serratia marcescens, Klebsiella pneumoniae, Wigglesworthia glossinidia, Proteus vulgaris, Salmonella sp., bacteriophage P1, and Escherichia coli.
The alignment includes the bacteriophage P1 Hot protein, the product of the phage hot gene (the mnemonic hot derives from homolog of theta) (25). Hot also has good homology to θ (47% identity, 61% similarity), particularly in the N-terminal part. In the following, we describe experiments aimed at finding out whether the P1 hot gene product can act as a functional replacement for θ in its role of Pol III accessory factor.
hot expression in E. coli.
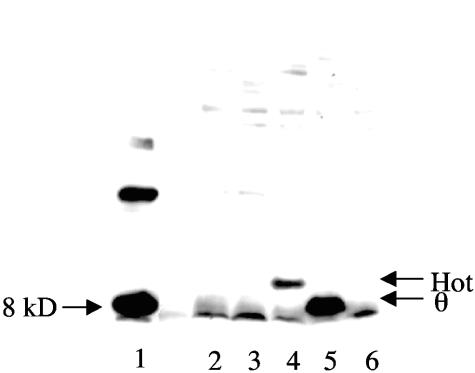
The hot gene on P1 has been reported to be expressed from a “late” promoter (19, 20, 25). Late P1 promoters regulate the expression of proteins needed at the end of the phage cycle, including the coat proteins needed for phage packaging (25). The late promoters require the P1 gene 10 (lpa) product, a transcription factor specific for late phage gene expression (19, 20, 25). Expression of this transcription factor is reportedly toxic to E. coli (19), and therefore we decided to not use the P1 native expression system. Instead, we used PCR methods to create a construct in which the hot gene is expressed from the E. coli holE promoter. Specifically, we amplified the hot coding sequence and inserted it in front of the holE promoter with its ATG start codon precisely in place of the holE ATG codon, while holE itself was deleted in the process. The procedure is diagrammed in Fig. 1 and described in more detail in the Materials and Methods. Initially the new construct was obtained in multicopy plasmid pUC18, yielding pUC18hot. Western blots on extracts from a ΔholE strain containing pUC18hot showed that the recombinant cells produced a significant amount of Hot protein (Fig. 3, lane 4). The size of Hot is consistent with its molecular size (83 amino acids). It is clearly distinct from θ (76 amino acids), either as purified θ or in extracts of pUC18holE containing strains (lanes 1 and 5). pUC18holE is identical to pUC18hot except that it contains holE instead of hot (see Materials and Methods). The antibody used was a polyclonal antibody generated against θ, and the recognition of Hot by this antibody is consistent with presumed similarities between θ and Hot. The antibody was not capable of detecting either θ (Fig. 3, lane 6) or Hot (not shown) in extracts when expressed from a single chromosomal gene copy.
FIG. 3.
Expression of θ and Hot from pUC18 plasmids detected by Western blot analysis using anti-θ antibody. Strain NR13104 (ΔholE) containing various pUC plasmids was grown overnight at 37°C in LB broth plus ampicillin (500 μg/ml). The control strains NR13104 (ΔholE) and MG1655 (hol+) were grown in LB broth. See Materials and Methods for details on lysate preparation, gel electrophoresis, and the Western blot procedure. Lane 1, purified θ protein; lane 2, ΔholE strain, no plasmid; lane 3, ΔholE strain containing pUC18; lane 4, ΔholE strain containing pUC18hot; lane 5, ΔholE strain containing pUC18holE; lane 6, MG1655 (wild type). The higher-molecular-mass band in lane 1 may represent a θ dimer, but this was not further investigated.
Single-copy replacement of holE by hot.
Cells containing pUC18hot and pUC18holE produced small and heterogeneously sized colonies. Also, the plasmids were rapidly lost unless high levels of ampicillin were included in the medium. These results suggested that excessive amounts of θ of Hot are deleterious. When the inserts were transferred to the low-copy-number vector pKO3 (three to five copies) (see Materials and Methods), colonies were of normal size and were capable of stable plasmid maintenance. However, for our study to investigate possible replacement of θ by Hot under normal single-copy conditions, we decided to generate a strain in which hot is integrated, as a single copy, in the E. coli chromosome, replacing holE. This replacement was readily possible using the pKO3hot plasmid by the method of Link et al. (24). This yielded strain NR16315 (ΔholE204::hot) (Table 1 and Materials and Methods), in which hot is expressed chromosomally from the native holE promoter.
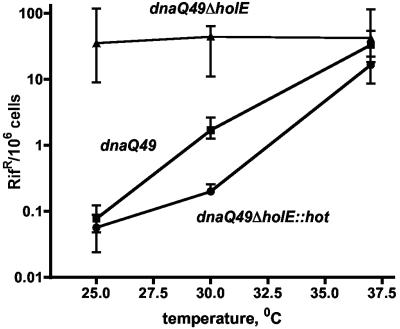
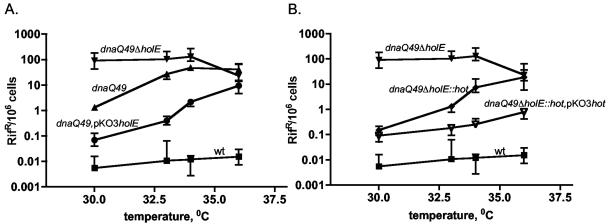
One conveniently assayable phenotype associated with the lack of θ (ΔholE strain) is reduced stability of the dnaQ49 mutator mutant. The dnaQ49 mutant (containing the V96G mutation) carries a defective polymerase III ɛ subunit whose stability is greatly dependent on the θ subunit (50). The dnaQ49 mutant is only a modest mutator at low temperatures (25°C), but it becomes a very strong mutator (up to 1,000-fold enhanced) at 37°C due to collapse of its proofreading ability (13, 17, 36, 40). In contrast, the dnaQ49 ΔholE strain is an exceptionally strong mutator even at the lowest temperature (50).
To assay the effect of hot on the dnaQ49 mutator, we compared the mutabilities of three dnaQ49 strains: the dnaQ49 holE+, dnaQ49 ΔholE, and dnaQ49 ΔholE::hot strains. The results in Fig. 4 show that, as before (50), the dnaQ49 ΔholE strain is an extremely strong mutator regardless of the temperature compared to the dnaQ49 holE+ strain, which shows greatly reduced mutator activity at the lower temperature and reaches high levels of mutagenesis only at 37°C. Quantitatively, the presence of θ reduces the mutant frequency at 25°C by about 1,000-fold. Importantly, the results also show that the dnaQ49 ΔholE::hot strain—containing hot instead of holE—behaves like the dnaQ49 strain; i.e., it displays low mutability at low temperature. This indicates that Hot, like θ, can stabilize the dnaQ49 ɛ subunit. Interestingly, the stabilizing effect of Hot appears actually greater than that of θ. In this experiment, the mutant frequency of the dnaQ49 ΔholE::hot strain is 3- to 10-fold lower at the three temperatures than that of the dnaQ49 holE+ strain. In four independent experiments, the average reductive effect of Hot was 2.8-, 15-, and 5.7-fold greater than the reductive effect of θ at 25, 30, and 37°C, respectively. Based on these experiments, we conclude that P1 Hot can replace the θ subunit and must be considered a functional homolog of θ.
FIG. 4.
Mutability of dnaQ49 strains containing a chromosomal holE+, ΔholE, or ΔholE::hot allele. Twelve to 18 independent cultures for each strain were incubated overnight at the indicated temperatures; plates were incubated at 30°C. The median mutant frequencies along with the indicated interquartile ranges were calculated using Prism (GraphPad) (see Materials and Methods). Note the log scale on the y axis.
Multiple copies of holE or hot further suppress the dnaQ49 mutator effect.
The above-described experiments were also performed with the holE and hot genes on the low-copy-number plasmid pKO3, as shown in Fig. 5. Panel A shows that expression of θ from pKO3holE at temperatures between 30 and 34°C reduces the mutant frequency of the dnaQ49 strain by 1 order of magnitude compared to that of a strain with a single chromosomal holE copy (note that pKO3 has a temperature-sensitive replication origin and cannot be grown above 36°C). A similar effect is seen in the case of Hot protein expressed from pKO3hot (Fig. 5B). At 30°C, a single copy of hot exerts a maximal effect, although at a higher temperature, multiple copies are clearly more effective than the single copy. Overall, the mutability of the dnaQ49 strain with multiple hot copies shows only weak temperature dependence. These results, showing that multiple copies of holE and hot further stabilize the dnaQ49 mutant, have implications for the precise mechanism underlying the dnaQ49 mutator effect. In addition, they reinforce the greater efficacy of Hot in suppressing the dnaQ49 mutator effect.
FIG. 5.
Reduction of the dnaQ49 mutator activity by multiple copies of the holE (A) or hot (B) gene. In panel A, the mutant frequency of the dnaQ49 ΔholE strain is compared to those of its counterparts containing either one copy (dnaQ49) or multiple copies (dnaQ49 pKO3holE) of the holE gene. The MG1655 (wild-type [wt]) strain is shown as a control. In panel B, the dnaQ49 ΔholE strain is compared to its counterparts containing either one copy (dnaQ49 ΔholE::hot) or multiple copies (dnaQ49 ΔholE::hot pKO3hot) of the P1 hot gene. All strains not containing either pKO3holE or pKO3hot carried pKO3ΔholE. Cultures were grown overnight at 30, 33, 34, or 36°C in LB broth plus chloramphenicol; plates were incubated at 30°C. Mutant frequencies are based on 12 independent cultures for each strain (see Materials and Methods). The median values and interquartile ranges (indicated) were calculated using Prism software (GraphPad).
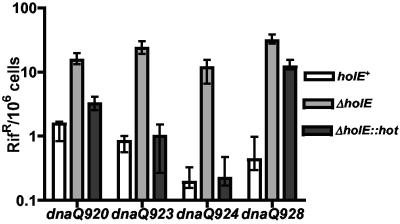
Effect of hot on other dnaQ mutator alleles.
In the previous study on the θ subunit (50), we also investigated a series of dnaQ alleles other than dnaQ49 (V96G). We found several additional alleles whose mutability was further elevated in the ΔholE background. We concluded that these other dnaQ alleles, like dnaQ49, contain certain structural defects that are exacerbated by the lack of a supporting θ subunit. In the experiment whose results are shown in Fig. 6, we tested the relative efficiency of θ and Hot in stabilizing these other dnaQ mutators. Hot, like θ, was capable of restoring low mutability to the ΔholE dnaQ920 (R56W), ΔholE dnaQ923 (H66Y), and ΔholE dnaQ924 (L171F) strains, further corroborating the ability of Hot to substitute for θ. Interestingly, little or no effect of Hot was observed for the dnaQ928 (G17S) allele, although this allele is readily stabilized by θ (Fig. 6). These results suggest that while both θ and Hot interact with and presumably stabilize ɛ, there are additional aspects to this interaction that are not identical for θ and Hot.
FIG. 6.
Effect of θ and Hot on dnaQ920, dnaQ923, dnaQ924, and dnaQ928 mutants. Mutant frequencies were calculated for 12 independent cultures for each strain (see Materials and Methods). Cultures were grown overnight at 37°C in LB broth; plates were also incubated at 37°C. (Note that the mutant frequencies for the mutator strains are not comparable to those reported previously [50] because the latter were determined in the mismatch repair-deficient mutL background). Data were analyzed using Prism software (GraphPad). The graph shows median values and the interquartile ranges for the frequency of rifampin-resistant mutants. The x axis indicates, for each dnaQ allele, the three strains containing the holE+, ΔholE, or ΔholE::hot chromosomal configuration.
DISCUSSION
The present data indicate that the phage P1 Hot protein, a homolog of the E. coli Pol III θ subunit, can compensate for the lack of θ, as observed through the stabilizing effect of the hot gene in several dnaQ strains. This compensation most logically involves a direct substitution for θ in its interaction with the ɛ subunit and, likely, the incorporation of Hot into the Pol III core. The presence of this functional homolog of θ in the bacteriophage raises the question of why the phage contains this homolog. The answer is likely intertwined with the precise function of θ in E. coli.
The role of θ.
We have previously suggested (50) that θ might act as a chaperonin for the ɛ subunit. This suggestion was based on the stabilizing effect of θ on several impaired dnaQ alleles, as well as on Saccharomyces cerevisiae tri-hybrid assays showing a strongly improved α-ɛ interaction in the presence of θ (50). The proposal is consistent with other observations of θ, such as the tight interaction between ɛ and θ (47), the intrinsic instability of ɛ in vivo (8), and the improved in vitro behavior of purified ɛ in the presence of θ (5, 10, 12). We also noted that θ is found among organisms with a 3′ proofreading activity that is present on a separate subunit (rather than being part of the polymerase polypeptide), suggesting that the need for θ may be related to the peculiarities of a free ɛ subunit. On the other hand, based on recent GenBank searches, we note that θ is not found among several other gram-negative bacteria that also contain an isolated proofreading activity, such as Vibrio cholerae, Haemophilus influenzae, Pseudomonas aeruginosa, or Photobacterium profundum. Therefore, the relevance of this connection remains unclear. We cannot exclude the possibility that in those organisms, θ orthologs exist but are not detected by BLAST search due to sequence diversion. Whether the stability of ɛ will be a limiting factor for efficient or faithful replication in a given organism will likely depend on multiple factors, such as the intrinsic stability of the particular ɛ, the nature of its interactions with the α subunit, and the levels of chaperone/heat shock proteins (8), which may all differ in different taxa of gram-negative bacteria.
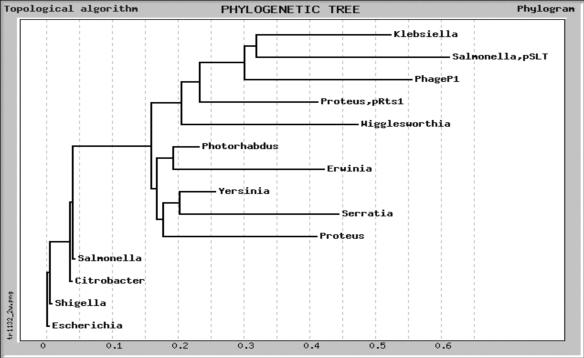
The relatedness of θ proteins.
The homology of the various θ proteins as presented in Fig. 2 is extensive, particularly in the N-terminal part. Additional insight into the relatedness of the proteins may be gleaned from Fig. 7, where we show a phylogenetic tree constructed based on the relatedness to θ. It is clear that, within this scheme, E. coli θ, along with the proteins from the closely related Salmonella, Shigella, and Citrobacter species, is more distantly related to P1 Hot. Hot is part of a separate grouping that contains the two plasmid-encoded homologs (Salmonella pSLT and Proteus Rst1) as well as the proteins from Klebsiella and Wigglesworthia. Thus, it is unlikely that hot is a recent acquisition from its current host, E. coli. Instead, it may have its origin in a different species. The same argument applies to the homologs encoded by the Salmonella pSLT and Proteus Rts1 plasmids, which also differ significantly from their host homologs. Thus, P1 Hot may be optimized for interaction with a phylogenetically different ɛ subunit. We note that Wigglesworthia glossinidia is an endosymbiont (of tsetse flies) with a greatly reduced genome (698 kb, ∼15% of the E. coli chromosome) (1). That the organism has retained the θ homolog despite the severe size reduction of its genome is a further suggestion that the θ homolog fulfills a relevant function. On the other hand, two other symbionts from the enterobacterial group whose genomes have been sequenced and which also have a reduced genome, Buchnera aphidicola (43, 51) and Blochmannia floridanus (9), do not possess a θ homolog.
FIG. 7.
Phylogenetic tree of θ protein homologs. The θ homologs are as shown in the alignment of Fig. 2. The tree was obtained using Tree Top sequence analysis tools on the GeneBee Molecular Biology Server (http://www.genebee.msu.su/genebee.html) and a topological algorithm.
Relative effects of θ and Hot in suppressing dnaQ mutator alleles.
Our results with the dnaQ49 mutant show that Hot protein is more effective than θ in stabilizing this allele (Fig. 4). This observation may indicate that Hot interacts more effectively with ɛ, leading to additional stabilization of this subunit. Alternatively, Hot protein may be intrinsically more stable, leading to a higher effective protein concentration available for interaction with ɛ. As seen from Fig. 5, increased θ or Hot lead to additional reduction in the dnaQ49 mutator activity. Further support for this hypothesis comes from structural studies performed on θ and Hot. Nuclear magnetic resonance analysis of the θ solution structure showed it to be a rather unstructured protein (15, 22). Hot behaved significantly better in these experiments, permitting determination of its solution structure (4). Circular dichroism studies also indicated that the stability of Hot was greater than that of θ (4). On the other hand, experiments with the dnaQ920, dnaQ923, and dnaQ924 alleles showed θ and Hot to be similarly effective in stabilizing these alleles, while dnaQ928 could be stabilized only by θ (Fig. 6). These results are suggestive of differences in the precise ɛ-θ and ɛ-Hot interactions. In a subsequent report (A. K. Chikova and R. M. Schaaper, unpublished data), we describe certain circumstances in which the substitution of Hot for θ actually increases mutagenesis. Analysis of the ɛ-θ and ɛ-Hot complexes by structural methods may shed further light on these questions.
Effects of multicopy holE or hot on the dnaQ49 mutant.
The data of Fig. 5 indicate that the dnaQ49 mutator effect can be further reduced when these proteins are expressed from plasmid pKO3, when protein levels may be expected to be increased three- to fivefold based on the plasmid copy number. This observation has implications for the nature of the dnaQ49 defect. The dnaQ49 mutation is genetically recessive (13, 29). This has been interpreted to indicate that the encoded ɛ subunit is defective in its binding to the Pol III α subunit. Indeed, yeast two-hybrid analyses have shown the α-ɛ interaction to be severely diminished for this mutant (14, 49, 50). On the other hand, it is likely that part of the dnaQ49 defect reflects a catalytic deficiency. The amino acid substitution responsible for the dnaQ49 defect, V96G, resides in the N-terminal catalytic domain, while the primary determinant for interaction with the α subunit resides in the C-terminal domain (residues 187 to 243) (35, 49). Also, the V96G mutation resides near the exonuclease II motif (48), which provides a component of the ɛ catalytic site.
The observation that θ and Hot overexpression further reduces the dnaQ49 mutant frequency suggests that in dnaQ49 strains the chromosome is replicated, at least part of the time, by a form of HE that lacks θ. In fact, this form of HE may also lack ɛ, as yeast two- and three-hybrid assays have shown the DnaQ49-α interaction to be dramatically reduced in the absence of θ (14, 49, 50). HE containing a core consisting of only the α subunit has been prepared in vitro and shown to synthesize DNA effectively, albeit with reduced processivity (16, 28, 46). Chromosomal DNA synthesis by this kind of HE would naturally be highly mutagenic due to the lack of any proofreading. The existence of such a proofreading-defective form of HE in vivo has been speculated upon (6), but our current results may provide a first experimental indication of their existence.
What is the function of Hot in P1?
It seems most logical to analyze this question in the context of the function of θ. Assuming that the function of θ is to support the intrinsically unstable ɛ subunit, one possibility is that the function of Hot may be to help sequester and stabilize ɛ for the benefit of the phage. As P1 is dependent on Pol III HE for its replication (25), establishing sufficient amounts of ɛ for incorporation in the Pol III core may be one way to ensure the availability of a sufficient number of Pol III HE molecules for phage replication. There are only limited numbers of Pol III core and Pol III HE molecules per cell (26, 27, 32), making this an issue of relevance to the phage. The need for additional HE would be most acute in the lytic cycle but could also be felt during P1 plasmid maintenance. One interesting concern with this model is that hot has been classified as a late gene (19, 20, 25), and Hot protein would be expected to be produced primarily during the phage packaging stage. One (speculative) possibility is that Hot, after being produced late in infection, is encapsulated in the virion and released upon infection (25). On the other hand, the hot promoter does not conform exactly to the late promoter consensus, because its −10 and −22 sequences, characteristic for the P1 late genes, are spaced 9 nucleotides apart instead of the canonical 4 (25). Thus, it will be important to investigate the precise timing of Hot expression in the phage life cycle, including the prophage stage, to address these questions. It will also be of interest to investigate the viability or properties of a P1 mutant lacking hot.
Acknowledgments
We thank M. Lobocka for many helpful discussions on the subject of P1 Hot, Sharon Taft-Benz for constructing the holE chromosomal deletion, and J. Fortune and M. Graziewicz of the NIEHS for their critical review of the manuscript for this paper.
REFERENCES
- 1.Akman, L., A. Yamashita, H. Watanabe, K. Oshima, T. Shiba, M. Hattori, and S. Aksoy. 2002. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat. Genet. 32:402-407. [DOI] [PubMed] [Google Scholar]
- 2.Carter, J. R., M. A. Franden, R. Aebersold, D. R. Kim, and C. S. McHenry. 1993. Isolation, sequencing and overexpression of the gene encoding the θ subunit of DNA polymerase III holoenzyme. Nucleic Acids Res. 21:3281-3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeRose, E. F., T. Darden, S. Harvey, S. Gabel, F. W. Perrino, R. M. Schaaper, and R. E. London. 2003. Elucidation of the ɛ-θ interface of Escherichia coli DNA polymerase III by NMR spectroscopy. Biochemistry 42:3635-3644. [DOI] [PubMed] [Google Scholar]
- 4.DeRose, E. F., T. W. Kirby, G. A. Mueller, A. K. Chikova, R. M. Schaaper, and R. E. London. 2004. Phage like it HOT: solution structure of the bacteriophage P1-encoded HOT protein, a homolog of the θ subunit of E. coli DNA polymerase III. Structure 12:2221-2231. [DOI] [PubMed] [Google Scholar]
- 5.DeRose, E. F., D. Li, T. Darden, S. Harvey, F. W. Perrino, R. M. Schaaper, and R. E. London. 2002. Model for the catalytic domain of the proofreading epsilon subunit of Escherichia coli DNA polymerase III based on NMR structural data. Biochemistry 41:94-110. [DOI] [PubMed] [Google Scholar]
- 6.Dorazi, R. 2003. Can tRNAs act as antisense RNA? The case of mutA and dnaQ. J. Theor. Biol. 225:383-388. [DOI] [PubMed] [Google Scholar]
- 7.Echols, H., C. Lu, and P. M. J. Burgers. 1983. Mutator strains of Escherichia coli, mutD and dnaQ, with defective exonucleolytic editing by DNA polymerase III holoenzyme. Proc. Natl. Acad. Sci. USA 80:2189-2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foster, P. L., and M. G. Marinus. 1992. Levels of epsilon, an essential replication subunit of Escherichia coli DNA polymerase III, are controlled by heat shock proteins. J. Bacteriol. 174:7509-7516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gil, R., F. J. Silva, E. Zientz, F. Delmotte, F. González-Candelas, A. Latorre, C. Rausell, J. Kamerbeek, J. Gadau, B. Holldobler, R. C. H. J. van Ham, R. Gross, and A. Moya. 2003. The genome sequence of Blochmannia floridanus: comparative analysis of reduced genomes. Proc. Natl. Acad. Sci. USA 100:9388-9393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta, R., S. M. Hamdan, N. E. Dixon, M. M. Sheil, and J. L. Beck. 2004. Application of electrospray ionization mass spectrometry to study the hydrophobic interaction between the ɛ and θ subunits of DNA polymerase III. Protein Sci. 13:2878-2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 12.Hamdan, S., P. D. Carr, S. E. Brown, D. L. Ollis, and N. E. Dixon. 2002. Structural basis of proofreading during replication of the Escherichia coli chromosome. Structure 10:535-546. [DOI] [PubMed] [Google Scholar]
- 13.Horiuchi, T., H. Maki, and M. Sekiguchi. 1978. A new conditional lethal mutator (dnaQ49) in Escherichia coli K12. Mol. Gen. Genet. 163:277-283. [DOI] [PubMed] [Google Scholar]
- 14.Jonczyk, P., A. Nowicka, I. J. Fijalkowska, R. M. Schaaper, and Z. Ciesla. 1998. In vivo protein interactions within the Escherichia coli DNA polymerase III core. J. Bacteriol. 180:1563-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keniry, M. A., H. A. Berthon, J. Y. Yang, C. S. Miles, and N. E. Dixon. 2000. NMR solution structure of the θ subunit of DNA polymerase III from Escherichia coli. Protein Sci. 9:721-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, D. R., and C. S. McHenry. 1996. In vivo assembly of overproduced DNA polymerase III. Overproduction, purification, and characterization of the α, α-ɛ, and α-ɛ-θ subunits. J. Biol. Chem. 271:20681-20689. [DOI] [PubMed] [Google Scholar]
- 17.Krishnaswamy, S., J. A. Rogers, R. J. Isbell, and R. G. Fowler. 1993. The high mutator activity of the dnaQ49 allele of Escherichia coli is medium-dependent and results form both defective 3′→5′ proofreading and methyl-directed mismatch repair. Mutat. Res. 288:311-319. [DOI] [PubMed] [Google Scholar]
- 18.Lancy, E. D., M. R. Lifsics, D. G. Kehres, and R. Maurer. 1989. Isolation and characterization of mutants with deletions in dnaQ, the gene for the editing subunit of DNA polymerase III in Salmonella typhimurium. J. Bacteriol. 171:5572-5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lehnherr, H., A. Guidolin, and W. Arber. 1991. Bacteriophage P1 gene 10 encodes a trans-activating factor required for late expression. J. Bacteriol. 173:6438-6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lehnherr, H., A. Guidolin, and W. Arber. 1992. Mutational analysis of the bacteriophage P1 late promoter sequence Ps. J. Mol. Biol. 228:101-107. [DOI] [PubMed] [Google Scholar]
- 21.Leu, F. P., R. Georgescu, and M. O'Donnell. 2003. Mechanism of the E. coli τ processivity switch during lagging-strand synthesis. Mol. Cell 11:315-327. [DOI] [PubMed] [Google Scholar]
- 22.Li, D., D. L. Allen, S. Harvey, F. W. Perrino, R. M. Schaaper, and R. E. London. 1999. A preliminary CD and NMR study of the Escherichia coli DNA polymerase III θ subunit. Proteins 36:111-116. [DOI] [PubMed] [Google Scholar]
- 23.Lifsics, M. R., E. D. Lancy, Jr., and R. Maurer. 1992. DNA replication defect in Salmonella typhimurium mutants lacking the editing (ɛ) subunit of DNA polymerase III. J. Bacteriol. 174:6965-6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Link, A. J., D. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lobocka, M. B., D. J. Rose, G. Plunkett III, M. Rusin, A. Samojedny, H. Lehnherr, M. B. Yarmolinsky, and F. R. Blattner. 2004. Genome of bacteriophage P1. J. Bacteriol. 186:7032-7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maki, H., and A. Kornberg. 1985. The polymerase subunit of DNA polymerase III of Escherichia coli. II. Purification of the α subunit, devoid of nuclease activities. J. Biol. Chem. 260:12987-12992. [PubMed] [Google Scholar]
- 27.Maki, H., S. Maki, and A. Kornberg. 1988. DNA polymerase III holoenzyme of Escherichia coli. IV. The holoenzyme is an asymmetric dimer with twin active sites. J. Biol. Chem. 263:6570-6578. [PubMed] [Google Scholar]
- 28.Marians, K. J., H. Hiasa, D. R. Kim, and C. S. McHenry. 1998. Role of the core DNA polymerase III subunits at the replication fork. α is the only subunit required for processive replication. J. Biol. Chem. 273:2452-2457. [DOI] [PubMed] [Google Scholar]
- 29.Maruyama, M., T. Horiuchi, H. Maki, and M. Sekiguchi. 1983. A dominant (mutD5) and a recessive (dnaQ49) mutator of Escherichia coli. J. Mol. Biol. 167:757-771. [DOI] [PubMed] [Google Scholar]
- 30.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik. J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature (London) 413:852-856. [DOI] [PubMed] [Google Scholar]
- 31.McHenry, C. S. 2003. Chromosomal replicases as asymmetric dimers: studies of subunit arrangement and functional consequences. Mol. Microbiol. 29:1157-1165. [DOI] [PubMed] [Google Scholar]
- 32.McHenry, C. S., and A. Kornberg. 1977. DNA polymerase III holoenzyme of Escherichia coli. J. Biol. Chem. 252:6478-6484. [PubMed] [Google Scholar]
- 33.McInerney, P., and M. O'Donnell. 2004. Functional uncoupling of twin polymerases. Mechanism of polymerase dissociation from a lagging-strand block. J. Biol. Chem. 279:21543-21551. [DOI] [PubMed] [Google Scholar]
- 34.Murata, T., M. Ohnishi, T. Ara, J. Kaneko, C.-G. Han, Y. F. Li, K. Takashima, H. Nojima, K. Nakayama, A. Kaji, Y. Kamio, T. Miki, H. Mori, E. Ohtsubo, Y. Terawaki, and T. Hayashi. 2002. Complete nucleotide sequence of plasmid Rts1: implications for evolution of large plasmid genomes. J. Bacteriol. 184:3194-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perrino, F. W., S. Harvey, and S. M. McNeill. 1999. Two functional domains of the ɛ subunit of DNA polymerase III. Biochemistry 38:16001-16009. [DOI] [PubMed] [Google Scholar]
- 36.Schaaper, R. M. 1988. Mechanisms of mutagenesis in the Escherichia coli mutator mutD5: role of DNA mismatch repair. Proc. Natl. Acad. Sci. USA 85:8126-8130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schaaper, R. M. 1993. Base selection, proofreading, and mismatch repair during DNA replication in Escherichia coli. J. Biol. Chem. 268:23762-23765. [PubMed] [Google Scholar]
- 38.Schaaper, R. M., and R. Cornacchio. 1992. An Escherichia coli dnaE mutation with suppressor activity toward mutD5. J. Bacteriol. 174:1974-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schaaper, R. M., B. N. Danforth, and B. W. Glickman. 1985. Rapid repeated cloning of mutant lac repressor genes. Gene 39:181-189. [DOI] [PubMed] [Google Scholar]
- 40.Schaaper, R. M., and M. Radman. 1989. The extreme mutator effect of Escherichia coli mutD5 results from saturation of mismatch repair by excessive DNA replication errors. EMBO J. 8:3511-3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheuermann, R., S. Tam, P. M. J. Burgers, C. Lu, and H. Echols. 1983. Identification of the ɛ-subunit of Escherichia coli DNA polymerase III holoenzyme as the dnaQ gene product: a fidelity subunit for DNA replication. Proc. Natl. Acad. Sci. USA 80:7085-7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheuermann, R. H., and H. Echols. 1984. A separate editing exonuclease for DNA replication: the ɛ subunit of Escherichia coli DNA polymerase III holoenzyme. Proc. Natl. Acad. Sci. USA 81:7747-7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shigenobu, S., H. Watanabe, M. Hattori, Y. Sakai, and H. Ishikawa. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature (London) 407:81-86. [DOI] [PubMed] [Google Scholar]
- 44.Singer, M., T. A. Baker, G. Schnitzler, S. M. Deischel, M. Goel, W. Dove, K. J. Jaacks, A. D. Grossman, J. W. Erickson, and C. A. Gross. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol. Rev. 53:1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Slater, S. C., M. R. Lifsics, M. O'Donnell, and R. Maurer. 1994. holE, the gene coding for the θ subunit of DNA polymerase III of Escherichia coli: characterization of a holE mutant and comparison with a dnaQ (ɛ-subunit) mutant. J. Bacteriol. 176:815-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Studwell, P. S., and M. O'Donnell. 1990. Processive replication is contingent on the exonuclease subunit of DNA polymerase III holoenzyme. J. Biol. Chem. 265:1171-1178. [PubMed] [Google Scholar]
- 47.Studwell-Vaughan, P. S., and M. O'Donnell. 1993. DNA polymerase III accessory proteins: θ encoded by holE. J. Biol. Chem. 268:11785-11791. [PubMed] [Google Scholar]
- 48.Taft-Benz, S. A., and R. M. Schaaper. 1998. Mutational analysis of the 3′→5′ proofreading exonuclease of Escherichia coli DNA polymerase III. Nucleic Acids Res. 26:4005-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taft-Benz, S. A., and R. M. Schaaper. 1999. The C-terminal domain of DnaQ contains the polymerase binding site. J. Bacteriol. 181:2963-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taft-Benz, S. A., and R. M. Schaaper. 2004. The θ subunit of Escherichia coli DNA polymerase III: a role in stabilizing the ɛ proofreading subunit. J. Bacteriol. 186:2774-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Ham, R. C. H. J., J. Kamerbeek, C. Palacios, C. Rausell, F. Abascal, U. Bastolla, J. M. Fernandez, L. Jiménez, M. Postigo, F. J. Silva, J. Tamames, E. Viguera, A. Latorre, A. Valencia, F. Morán, and A. Moya. 2003. Reductive genome evolution in Buchnera aphidicola. Proc. Natl. Acad. Sci. USA 100:581-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Williams, C. R., A. K. Snyder, P. Kuzmic, and M. O'Donnell. 2004. Mechanism of loading the Escherichia coli DNA polymerase III sliding clamp. I. Two distinct activities for individual ATP sites in the γ complex. J. Biol. Chem. 279:4376-4385. [DOI] [PubMed] [Google Scholar]