Abstract
FpvR is a presumed cytoplasmic membrane-associated anti-sigma factor that controls the activities of extracytoplasmic function sigma factors PvdS and FpvI responsible for transcription of pyoverdine biosynthetic genes and the ferric pyoverdine receptor gene, fpvA, respectively. Using deletion analysis and an in vivo bacterial two-hybrid system, FpvR interaction with these σ factors was confirmed and shown to involve the cytoplasmic N-terminal 67 amino acid resides of FpvR. FpvR bound specifically to a C-terminal region of FpvI corresponding to region 4 of the σ70 family of sigma factors. FpvR and FpvI mutant proteins compromised for this interaction were generated by random and site-directed PCR mutagenesis and invariably contained secondary structure-altering proline substitution in predicted α-helices within the FpvR N terminus or FpvI region 4. PvdS was shown to bind to the same N-terminal region of FpvR, and FpvR mutations compromising FpvI binding also compromised PvdS binding, although some mutations had a markedly greater impact on PvdS binding. Apparently, these two σ factors bind to FpvR in a substantially similar but not identical fashion. Intriguingly, defects in FpvR binding correlated with a substantial drop in yields of the FpvI and to a lesser extent PvdS σ factors, suggesting that FpvR-bound FpvI and PvdS are stable while free and active sigma factor is prone to turnover.
Iron is an essential nutrient whose acquisition by Pseudomonas aeruginosa is often facilitated by high-affinity iron-chelating molecules termed siderophores that, together with cell surface receptors specific for the iron-siderophore complexes, serve to provide the organism with iron under the most nutritionally dilute conditions (35). A major siderophore produced by P. aeruginosa and, indeed, all fluorescent pseudomonads is pyoverdine, a mixed catecholate-hydroxamate siderophore characterized by a conserved dihydroxyquinoline chromophore to which is attached a peptide chain of variable length and composition (5, 30). This variation likely explains the noted specificity vis-à-vis pyoverdine utilization by Pseudomonas spp., where a given strain will often use only its own pyoverdine and not that of other Pseudomonas strains (6, 14), and suggests that the peptide moiety is involved in receptor recognition and binding. A number of genes for the biosynthesis of pyoverdine have been identified to date in P. aeruginosa (19, 20, 26-28, 32, 48-50) and cluster within a region of the chromosome referred to as the pvd locus (47), although an operon implicated in synthesis of the chromophore, pvcABCD (45, 46), occurs elsewhere. Still, a pvd gene (pvdL) has also been reported to function in chromophore synthesis (31), raising questions about the necessity of the pvc genes in this.
The ferric pyoverdine receptor of P. aeruginosa PAO1 (FpvA or FpvAI) is a ca. 90-kDa outer membrane protein inducible under conditions of iron limitation (29, 36) and encoded by the fpvA gene (37) that is also found in the pvd locus (32). FpvA exhibits features typical of receptors whose activities are dependent upon the energy-coupling TonB protein (reviewed in reference 39), and a tonB gene (tonB1) has been reported in P. aeruginosa and is required for FpvA-mediated iron-pyoverdine uptake (38). In addition to its role in transporting ferric pyoverdine, FpvA plays a critical role in controlling expression of the fpvA gene (3, 40) as well as genes of pyoverdine biosynthesis (18, 43), in both instances mediating pyoverdine stimulation of FpvA (12) and pyoverdine synthesis (18). Pyoverdine-dependent, FpvA-mediated stimulation of fpvA and pvd gene expression involves extracytoplasmic-function (ECF) sigma factor/anti-sigma factor pairs FpvI/FpvR (3, 40) and PvdS/FpvR (18), respectively, whereby siderophore interaction with FpvA initiates a signal transduction cascade that triggers the release of the sigma factors by FpvR, freeing them to activate fpvA (FpvI) or pvd (PvdS) gene expression. This is reminiscent of PupIR control of the PupB ferric pseudobactin BN7/8 receptor in Pseudomonas putida WCS358 (16, 17) and FecIR control of the FecA ferric dicitrate receptor in Escherichia coli (4). FecR is a cytoplasmic membrane-spanning protein whose N terminus occurs in the cytoplasm (51), where it interacts with the C-terminal region 4 of FecI (24) and thus controls the activity of this ECF sigma factor. To understand the nature of FpvR's dual control of the FpvI and PvdS sigma factors in P. aeruginosa, we assessed the interactions between FpvR and its sigma factors. Here we confirm the interaction of the FpvR N terminus with the C-terminal regions of PvdS and FpvI and identify key features in FpvR and FpvI important for these interactions.
MATERIALS AND METHODS
Bacterial strains and growth media.
Bacterial strains and plasmids used in this study are listed in Table 1. Routine growth was performed in Luria-Bertani medium (Luria broth base; Difco). Ampicillin (100 μg/ml) and tetracycline (10 μg/ml) were added to the medium as required.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| K767 | PAO1 prototroph | 25a |
| E. coli | ||
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 1a |
| SU202 | lexA71::Tn5 sulA211 sulA::lacZ Δ(lacIPOZYA)169 F′ lacIqlacZΔM15::Tn9 | 9 |
| Plasmids | ||
| pDP804 | LexA1-87408-Jun zipper fusion | 9 |
| pMS604 | LexA1-87WT-Fos zipper fusion | 9 |
| pAR011 | pDP804::FpvI | This study |
| pAR012 | pMS604::FpvR1-92 | This study |
| pAR013 | pDP804::PvdS | This study |
| pAR014 | pDP804::FpvI95-159 | This study |
| pAR015 | pDP804::FpvI1-92 | This study |
| pAR016 | pMS604::FpvR1-74 | This study |
| pAR017 | pMS604::FpvR24-92 | This study |
| pAR018 | pMS604::FpvR24-74 | This study |
| pAR019 | pMS604::FpvR1-52 | This study |
| pAR020 | pDP804::FpvI95-159 (L103P) | This study |
| pAR021 | pDP804::FpvI95-159 (L103P, L115P) | This study |
| pAR022 | pDP804::FpvI95-159 (L103P, N144S, M146V, C149G) | This study |
| pAR023 | pDP804::FpvI95-159 (L106S, I127T, R135G) | This study |
| pAR024 | pDP804::FpvI95-159 (L119, R152P, M153I) | This study |
| pAR025 | pDP804::FpvI95-159 (L103A) | This study |
| pAR026 | pDP804::FpvI95-159 (L106S) | This study |
| pAR027 | pDP804::FpvI95-159 (I127T) | This study |
| pAR028 | pDP804::FpvI95-159 (L115P) | This study |
| pAR029 | pDP804::FpvI95-159 (L119P) | This study |
| pAR030 | pDP804::FpvI95-159 (L131P) | This study |
| pAR031 | pDP804::FpvI95-159 (R152P) | This study |
| pAR032 | pMS604::FpvR1-74 (L23P) | This study |
| pAR033 | pMS604::FpvR1-74 (E31stop) | This study |
| pAR034 | pMS604::FpvR1-74 (W39stop) | This study |
| pAR035 | pMS604::FpvR1-74 (W57stop) | This study |
| pAR036 | pMS604::FpvR1-74 (H18Y, L59P, L63P) | This study |
| pAR037 | pMS604::FpvR1-74 (C20Y, W57L) | This study |
| pAR038 | pMS604::FpvR1-74 (H18Y) | This study |
| pAR039 | pMS604::FpvR1-74 (C20Y) | This study |
| pAR040 | pMS604::FpvR1-74 (W57L) | This study |
| pAR041 | pMS604::FpvR1-74 (L59P) | This study |
| pAR042 | pMS604::FpvR1-74 (L63P) | This study |
| pAR043 | pMS604::FpvR1-67 | This study |
Amino acid changes in the FpvI and FpvR proteins encoded by the indicated plasmids are indicated in parentheses. “Stop” indicates a nonsense mutation in the codon for the indicated amino acid, leading to truncation of FpvR.
DNA techniques.
Standard protocols were used for restriction endonuclease digestions, ligations, transformation of electrocompetent E. coli, and agarose gel electrophoresis, as described by Sambrook and Russell (42). Plasmid isolation was performed using the alkaline lysis method (42) or using a plasmid Midi kit (QIAGEN, Mississauga, Ontario, Canada). Genomic DNA was extracted from P. aeruginosa following the protocol of Barcak et al. (2). Oligonucleotide synthesis and nucleotide sequencing were carried out by Cortec DNA Services Inc., Kingston, Ontario, Canada.
Construction of LexA fusion plasmids.
Full-length fpvI was amplified by PCR using the primer pair lexA-fpvIf (5′-AGTCCTCGAGATGGAAAACCATTATCGGGA-3′; XhoI site underlined) and lexA-fpvIr2 (5′-AGTCAGATCTTCAGTCGGCTTCCCAT-3′; BglII site underlined) and an annealing temperature of 60.5°C but otherwise exactly as described previously (40). The resulting amplicon was digested with XhoI and BamHI and cloned into XhoI-BamHI-restricted plasmid pDP804, creating plasmid pAR011, expressing an FpvI protein which is fused, at its N terminus, to the DNA-binding domain (residues 1 to 87) of wild-type LexA. Full-length pvdS was amplified by PCR as above using an annealing temperature of 58.8°C and the primer pair lexA-pvdSf (5′-AGTCCTCGAGATGTCGGAACAACTGTCTAC-3′; XhoI site underlined) and lexA-pvdSr (5′-AGTCAGATCTCGGCGCTGAGGAATGCTC-3′; BglII site underlined) and cloned into pDP804, yielding pAR013, producing a LexA1-87-PvdS fusion. Plasmids pAR015 and pAR014, pDP804 derivatives carrying portions of the fpvI gene encoding residues 1 to 92 and 95 to 159, respectively, fused to LexA1-87, were generated by cloning PCR products obtained following amplification with primer pairs lexA-fpvI95-159f (5′-AGTCCTCGAGCGCCTGGACAACCTGCAG-3′; XhoI site underlined) and lexA-fpvIr2 (above), and lexA-fpvIf (above) and lexA-fpvI1-92r (5′-AGTCAGATCTACTGGTCGACCACCGTCTG-3′; BglII site underlined), respectively. Reaction conditions were as described above except that annealing temperatures of 58.4°C (FpvI1-92; pAR015) and 61.8°C (FpvI95-159; pAR014) were employed. The region of fpvR encoding the cytoplasmic portion of FpvR (amino acid residues 1 to 92) was PCR amplified using an annealing temperature of 62.2°C and the primer pair lexA-fpvRf (5′-AGTCACCGGTGATGAAGACACCCTCTCC-3′; AgeI site underlined) and lexA-fpvR1-92r2 (5′-AGTCCTCGAGTCAGCGTGCCTGGCTCTTC-3′; XhoI site underlined) and cloned into AgeI-XhoI-restricted pMS604, creating plasmid pAR012, in which FpvR1-92 was fused, at its N terminus, to the DNA-binding domain (residues 1 to 87) of an altered LexA protein (LexA-408) that recognizes a different operator sequence than does wild-type LexA (9). Plasmids pAR016 to pAR019 and pAR043, pMS604 derivatives encoding additional truncated versions of FpvR (pAR016, FpvR1-74; pAR017, FpvR24-92; pAR018, FpvR24-74; pAR019, FpvR1-52; pAR043, FpvR1-67) fused to LexA-408 were constructed by cloning PCR products generated from reactions carried out with primer pairs lexA-fpvRf (above) and lexA-fpvRΔ75-92r (5′-AGTCCTCGAGTCAGTGGTTGCGAAGCTCG-3′; XhoI site underlined), lexA-fpvRΔ1-23f (5′-AGTCACCGGTGCACGCCGAGGACTTC-3′; AgeI site underlined) and lexA-fpvR1-92r2 (above), lexA-fpvRΔ1-23f (above) and lexA-fpvRΔ75-92r (above), lexA-fpvRf (above) and lexA-fpvR1-52r (5′-AGTCCTCGAGTCATTCTGCGTATTCGGCGGCG-3′; XhoI site underlined), and lexA-fpvRf (above) and lexA-fpvR1-67r (5′-AGTCCTCGAGTCACGGCGTTCGCGGCAGCAG-3′; XhoI site underlined), respectively. Again, reaction conditions were as described above except that annealing temperatures of 58.8°C (FpvR1-74), 64°C (FpvR24-74 and FpvR24-92), 58.4°C (FpvR1-52), and 59.4°C (FpvR1-67) were employed.
PCR-based random mutagenesis of fpvI and fpvR.
The fpvI gene of pDP804 derivative pAR014, encoding the FpvI95-159 protein, and the fpvR gene of pMS604 derivative pAR016, encoding the FpvR1-74 protein, were individually subjected to PCR-based random mutagenesis using primer pairs lexA-fpvI95-159f (above) and lexA-fpvIr2 (above), and lexA-fpvRf (above) and lexA-fpvRΔ75-92r (above), respectively, as described previously (22) but with modifications. These modifications included the use of ThermoPol (1×) buffer (New England Biolabs) as the reaction buffer, template plasmid DNA that was not linearized before the PCR, and dCTP (not dATP) at a final concentration of 40 μM. All other deoxynucleoside triphosphates were included at a final concentration of 200 μM. The reaction mixture was heated at 98°C for 3 min, followed by 30 cycles of 98°C for 45 s, 60°C (for fpvR) or 62°C (for fpvI) for 45 s, and 72°C for 1 min, followed by 72°C for 5 min. PCR amplicons were digested with XhoI and BglII (fpvI) or AgeI and XhoI (fpvR) and cloned into appropriately restricted pAR014 and pAR016, respectively, which had been gel purified (Prep-a-gene; Bio-Rad) to be free from the wild-type fpvI and fpvR sequences present on these vectors. This effectively permitted replacement of the wild-type FpvI95-159- and FpvR1-74-encoding fpvI and fpvR genes of pAR014 and pAR106, respectively, with PCR-mutagenized versions thereof.
PCR-based site-directed mutagenesis of fpvI and fpvR.
Selected mutations were introduced into the fpvI and fpvR genes of plasmids pAR014 and pAR016, respectively, using a protocol provided with the QuickChange Site-Directed mutagenesis kit from Stratagene. Using the Primer Generator (http://www.med.jhu.edu/medcenter/primer/primer.cgi), primer pairs were designed to introduce L103A (5′-TGCAGCGCGCGGCGGCCGAGTTGCC-3′ and its reverse complement; mutated bases responsible for the amino acid substitution and the introduction of an EagI site [boldfaced] are underlined and italicized, respectively), L106S (5′-CGCTGGCCGAGTCGCCGGCCATCTG-3′ and its reverse complement; mutated base responsible for the amino acid substitution is underlined), L115P (5′-CGCAGTTCCTTCCCATTGCGCAAGCTG-3′ and its reverse complement; the mutated base responsible for the amino acid substitution is underlined), L119P (5′-CTGTTGCGCAAGCCGGACGGCCTGTC-3′ and its reverse complement; the mutated base responsible for the amino acid substitution and introduction of HpaII/MspI site [boldfaced] is underlined), I127T (5′-TCCCATTCGCAGACCGCGGAgCATCTC-3′ and its reverse complement; mutated bases responsible for the amino acid substitution and the introduction of a SacII site [boldfaced] are underlined and italicized, respectively; the lowercase base was changed to eliminate a Sau3A site), L131P (5′-ATCGCCGAACATCCCAATATTTCCCGCA-3′ and its reverse complement; mutated base responsible for the amino acid substitution and introduction of FokI site [boldfaced] is underlined), and R152P (5′-CACTGTCGGGTACCCATGCGCGAATGG-3′ and its reverse complement; mutated bases responsible for the amino acid substitution and the introduction of a KpnI site [boldfaced] are underlined and italicized, respectively) mutations into FpvI95-159 and H18Y (5′-CAGGACGCCGCATATTGGTGCATGCG-3′ and its reverse complement; mutated bases responsible for the amino acid substitution and the introduction of an MslI site [boldfaced] are underlined and italicized, respectively), C20Y (5′-CAGGACGCCGCGCACTGGTACATGCGCCTGCACG-3′ and its reverse complement; mutated bases responsible for the amino acid substitution and the loss of a SphI site are underlined and boldfaced, respectively), W57L (5′-GAAATGGAGGAGATCCTGGCGCTCAGCGAAC-3′ and its reverse complement; mutated bases responsible for the amino acid substitution and loss of a BglII site are underlined and boldfaced, respectively), L59P (5′-GAGATCTGGGCGCCCAGCGAACTGCTG-3′ and its reverse complement; mutated base responsible for the amino acid substitution and loss of a BlpI site is underlined), and L63P (5′-CTCAGCGAACTGCCGCCGCGGACGC-3′ and its reverse complement; mutated bases responsible for the amino acid substitution and the introduction of a SacII site [boldfaced] are underlined and italicized, respectively) mutations into FpvR1-74. In most instances mutant primers contained base changes needed to introduce the desired amino acid substitutions in FpvR as well as loss or introduction of a restriction site (silent mutations) that could be used to readily verify the mutant sequence in the final plasmid constructs. Reaction mixtures contained 2.5 U of Pfu DNA polymerase (native; Fermentas), 1× Mg2+ Pfu reaction buffer, 200 μM of each deoxyribonucleoside triphosphate, 80 ng of template plasmid (either pAR014 or pAR016), and 30 pmol of each primer in a final volume of 50 μl. Mixtures were first heated at 98°C for 3 min prior to Pfu polymerase addition, followed by 30 cycles of 98°C for 45 s, 55°C for 45 s, and 72°C for 8 to 9 min, before finishing with 72°C for 5 min. Upon completion of the reaction, samples were digested with DpnI (New England Biolabs) at 37°C overnight in order to eliminate template (i.e., methylated) DNA. Digests were then dialyzed for 20 min against double-distilled water and electroporated into E. coli DH5α. Mutated pAR014 or pAR016 was recovered from individual colonies selected on L agar containing ampicillin (pAR014) or tetracycline (pAR016) and sequenced to ensure that the intended mutation had been introduced into the fpvI and fpvR genes present on these vectors.
Bacterial two-hybrid system.
To assess an interaction between FpvI and FpvR in vivo and the impact of truncations and mutations on this interaction, fpvI-carrying pDP804 (or its truncated or mutated derivatives) and fpvR-carrying pMS604 (or its truncated or mutated derivatives) were electroporated into E. coli strain SU202 and plated onto 1% lactose-MacConkey agar containing ampicillin and tetracycline. E. coli strain SU202 harbors a chromosomal lacZ gene engineered to contain a hybrid lexA operator sequence in the promoter region to which a heterodimer only of the LexAWT-LexA408 DNA-binding domains encoded by pDP804 and pMS604, respectively, can bind. pDP804- and pMS604-encoded LexA proteins lack the natural dimerization domains of this protein, but fusion of the individual LexA DNA-binding domains encoded by these vectors to proteins that do interact in vivo can promote their dimerization and, ultimately, binding to the lexA hybrid operator upstream of lacZ in SU202, effectively repressing lacZ expression. Thus, any interaction between FpvI and FpvR sequences encoded by the various fpvI- and fpvR-containing pDP804 and pMS604 derivatives, respectively, should promote dimerization of the LexA DNA-binding domains of these vectors and repression of lacZ, observable as lack of or reduction in β-galactosidase activity (i.e., pale pink to white colonies on lactose-MacConkey agar; in control experiments, E. coli SU202 carrying pDP804 or pMS604 produced only red colonies [β-galactosidase positive] on lactose-MacConkey agar, consistent with the failure to form the LexAWT-LexA408 heterodimers needed to repress lacZ in this strain). So, E. coli SU202 carrying pDB804 and pMS604 vectors with various truncated and site-directed mutant fpvI and fpvR sequences was cultured overnight with antibiotics, plated onto lactose-MacConkey agar, and screened for the absence (pink or white colonies) or presence (red colonies) of β-galactosidase, as an indication of relevant FpvI and FpvR sequences interacting (pink or white colonies) or not (red colonies). Results were then confirmed using a more quantitative β-galactosidase assay as described below. In screening randomly mutagenized fpvI and fpvR sequences for defects responsible for compromised FpvI-FpvR interaction, the ligation mixtures producing the pAR104 and pAR106 derivatives that carry randomly PCR mutagenized fpvI and fpvR, respectively (see above), were electroporated directly into E. coli SU202, which was then plated onto 1% lactose-MacConkey agar containing ampicillin and tetracycline and screened as above. Finally, to assess an interaction between PvdS and FpvR, the pvdS-carrying pDP804 derivative pAR013 was introduced into E. coli SU202 together with the fpvR-carrying pAR012 or pAR106 (and mutant versions thereof) and screened as above. In all instances, whole-cell protein extracts were prepared and screened (using immunoblotting with anti-LexA antibodies, see below) for the production of LexA-FpvI, LexA-FpvR, and LexA-PvdS fusions, as appropriate, to ensure production of truncated and mutant versions of these proteins.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting.
Whole-cell extracts were prepared as described previously (40), electrophoresed on 15% (wt/vol) sodium dodecyl sulfate-polyacrylamide gels, and transferred to Immobilon-P polyvinylidene difluoride membranes (Millipore) (54). Equal loading of protein in all wells was confirmed by rapid Coomassie blue staining of duplicated gels (11). Membranes were probed with monoclonal anti-LexA antibodies (Invitrogen) as described previously (54).
β-Galactosidase assay.
E. coli SU202 containing pDP804 (or its derivatives) and pMS604 (or its derivatives) was grown in Luria-Bertani medium supplemented with ampicillin (100 μg/ml) and tetracycline (10 μg/ml) for approximately 18 h at 37°C. Cultures were then diluted 1:49 into fresh antibiotic-supplemented medium and incubated at 37°C to an optical density at 600 nm of 0.8 to 1.0 before being assayed for β-galactosidase activity as described previously (40).
RESULTS
FpvR and FpvI interact in vivo.
FpvR negatively influences FpvI-mediated expression of fpvA (3), and given that previously described FpvR homologues control the activity of their cognate ECF sigma factors via a direct interaction (e.g., FecR binds FecI [10]), it seemed likely that FpvR would interact directly with FpvI. To assess this, a bacterial two-hybrid system (9) was employed whereby the fpvI and fpvR genes were cloned in frame to coding sequences for the DNA-binding domain of LexA on plasmids pDP804 and pMS604, respectively, and introduced into E. coli SU202 carrying a chromosomal lacZ gene under the control of a LexA operator. LexA binding to its operator and subsequent repression of lacZ in this strain requires prior dimerization of the LexA-binding domains encoded by pDP804 and pMS604, necessitating interaction of the FpvI and FpvR sequences fused to the LexA DNA-binding domains of these vectors. As such, lack of β-galactosidase activity is a measure of FpvI-FpvR interaction. The two-hybrid vectors also contain sequences encoding Jun and Fos zipper motifs (known to interact) fused to lexA, such that E. coli SU202 carrying these vectors demonstrates substantial repression of lacZ (Fig. 1). The unaltered vectors thus provide a positive control for the system, although the Jun and Fos zipper-encoding sequences will be disrupted upon cloning fpvI and/or fpvR sequences, making lacZ repression dependent upon FpvI-FpvR interaction.
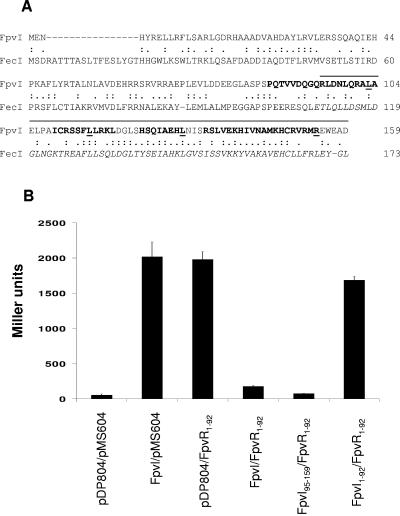
FIG. 1.
Interaction of the FpvI C-terminal region with FpvR1-92. (A) Alignment of FpvI and FecI highlighting the previously defined region 4 of FecI (italicized) and the corresponding region of FpvI (residues 95 to 159; overlined) that was assessed for an ability to interact with FpvR1-92. Residues in FpvI whose mutation in this study compromised FpvR95-159 stability and/or its interaction with FpvR are underlined. Four putative α-helices in the FpvI C terminus (Fig. 3) are in bold. (B) β-Galactosidase activity of E. coli SU202 carrying pDP804 or derivatives thereof expressing the indicated FpvI proteins and pMS604 or its derivative expressing FpvR1-92.
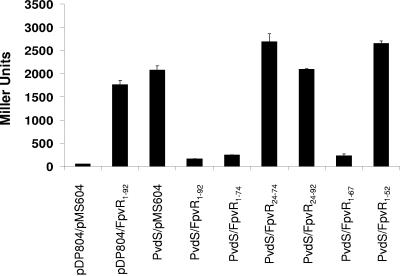
Because FpvR is predicted to possess a single cytoplasmic membrane-spanning region with the N terminus of the protein exposed to the cytoplasm (see Fig. 4A), only the 5′ end of the fpvR gene (encoding residues 1 to 92) was cloned into pMS604 for use in these studies. E. coli SU202 carrying only one of the fpvI or fpvR vectors and an “empty” second vector demonstrated high levels of β-galactosidase activity (Fig. 1B), consistent with the absence of a suitable binding partner to facilitate LexA dimerization and, thus, repression of lacZ in this reporter strain. In contrast, E. coli SU202 expressing full-length FpvI fused to LexA (in pDP804 derivative pAR011) and FpvR1-92 fused to LexA (in pMS604 derivative pAR012) expressed very low levels of β-galactosidase activity (Fig. 1B), consistent with lacZ repression and thus FpvI-FpvR1-92 interaction in this strain. Previous studies with the FpvI homologue FecI demonstrated that the C-terminal region 4 of this ECF sigma factor (Fig. 1A) was sufficient for its interaction with its cognate regulatory partner, FecR. Thus, a deletion derivative of fpvI encoding the corresponding region of FpvI (FpvI95-159; overlined in Fig. 1A) was engineered, and its interaction with FpvR1-92 assessed. As seen in Fig. 1B, E. coli SU202 expressing FpvI95-159 and FpvR1-92 fused to LexA in pAR014 and pAR012, respectively, again exhibited low levels of β-galactosidase activity, consistent with the C-terminal, putative region 4 of FpvI interacting with the N-terminal cytoplasmic extension of FpvR. As expected, the N-terminal region of FpvI (FpvI1-92) did not interact with FpvR1-92 (Fig. 1B).
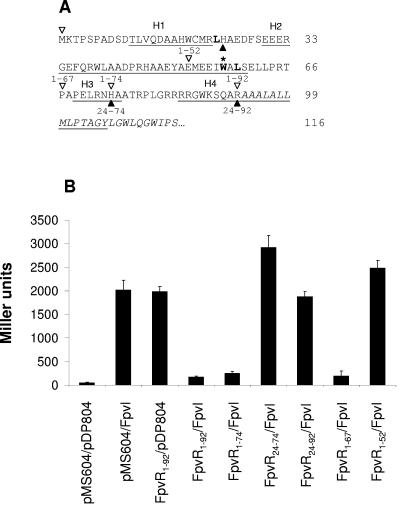
FIG. 4.
Interaction of FpvR deletion derivatives with FpvI. (A) N-terminal, proposed cytoplasmic portion of FpvR upstream of a putative membrane-spanning domain (italicized) highlighting four putative α-helical domains (underlined and labeled H1 to -4). Regions of FpvR encoded on pMS604 and assessed for an interaction with FpvI are marked by open arrowheads above the sequence (for FpvR constructs beginning at residue 1 and extending to one of four C-terminal endpoints at residues 52, 67, 74, and 92, dubbed 1-52, 1-67, 1-74, and 1-92, respectively) or filled arrowheads below the sequence (for those beginning at residue 24 and extending to one of two C-terminal endpoints at residues 74 and 92, dubbed 24-74 and 24-92, respectively). Two leucine residues whose mutation in this study compromised FpvR stability and/or interaction with FpvI are boldfaced. The tryptophan residue at which a nonsense mutant version of FpvR truncates (Fig. 5A, lane 5) is boldfaced and highlighted with an asterisk. (B) β-Galactosidase activity of E. coli SU202 carrying pMS604 or derivatives thereof expressing the indicated FpvR proteins and pDP804 or its derivative expressing FpvI. Immunoblotting and β-galactosidase assays were performed on samples prepared from the same culture in all instances, and the data shown are representative of three replicates.
Mutations in FpvI impacting FpvR binding.
Having localized the FpvR-binding domain of FpvI to the C-terminal half of the protein, corresponding to region 4 of this putative ECF sigma factor, it was of interest to identify residues and/or features (i.e., secondary structures) of this region important for binding. To this end, plasmid pAR014 encoding LexA-FpvI95-159 was randomly mutagenized via PCR and introduced into E. coli SU202 carrying the LexA-FpvR1-92 vector pAR012, and mutations in FpvI95-159 compromising its interaction with FpvR1-92 were recovered from β-galactosidase-positive (i.e., red) colonies on MacConkey agar. Of 50,000 colonies screened only five β-galactosidase-positive colonies were recovered that subsequently produced detectable FpvI95-159 (Fig. 2A, lanes 2 to 6) and carried only missense mutations in fpvI95-159 (many frameshift and nonsense mutations were recovered among several dozen β-galactosidase-positive colonies originally identified, but these were not studied further).
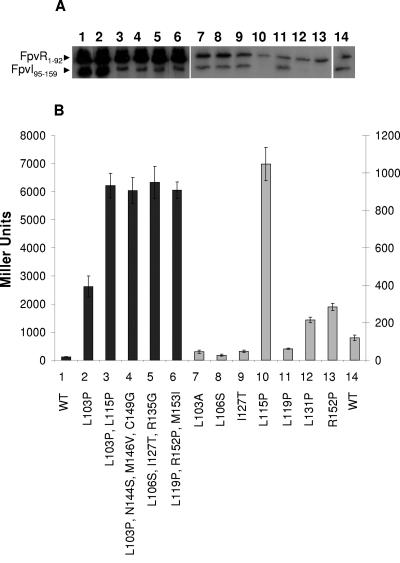
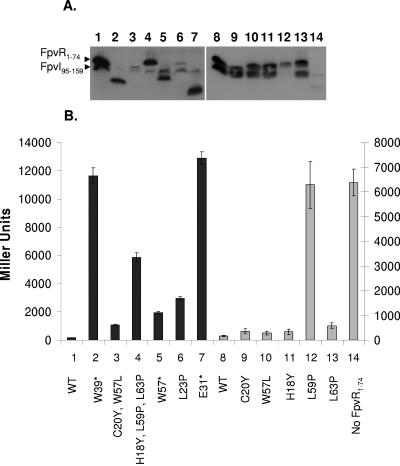
FIG. 2.
Impact of mutations in FpvI95-159 on its interaction with FpvR1-92. (A) Western immunoblot of whole-cell extracts of E. coli SU202 carrying pAR012 (pMS604::fpvR1-92) and pDP804 derivatives expressing mutated (lanes 2 to 13) or wild-type (lanes 1 and 14) FpvI95-159 developed with antibodies to LexA. Specific mutations are indicated and numbered in panel B with the numbers used to match the mutation and β-galactosidase data to the corresponding immunoblot lane in panel A. (B) β-Galactosidase activity of E. coli SU202 carrying pDP804 or derivatives thereof expressing the indicated mutant FpvI95-159 proteins and pAR012 expressing FpvR1-92. WT, wild-type FpvI95-159. The scale to the left applies to the darkly shaded bars (1 to 6) while the scale to the right applies to the lightly shaded bars (7 to 14). Variability in protein yields (A) and β-galactosidase activity (B) for FpvI and FpvR derivatives shown in lanes 1 to 6 versus lanes 7 to 14 reflects the fact that these data were obtained from experiments carried out independently on different days. Immunoblotting and β-galactosidase assays were performed on samples prepared from the same culture in all instances, and the data shown are representative of three replicates.
Most of the recovered mutant fpvI genes carried multiple missense mutations (Fig. 2B, bars 3 to 6), although a single L103P mutation was recovered that had no impact on FpvI95-159 yields (Fig. 2A, lane 2, cf. lane 1) but markedly compromised its interaction with FpvR1-92 (Fig. 2B). The presence of multiple mutations in FpvI correlated with substantially reduced yields of the mutant FpvI95-159 protein (Fig. 2A, lanes 3 to 6), possibly reflecting protein instability. To determine if a specific mutation of the multiple mutations in mutant FpvI95-159 proteins was responsible for the loss of interaction with FpvR1-92 (and/or apparent instability of the mutant proteins), attempts were made to construct mutant versions of FpvI95-159 carrying these mutations individually and to assess the impact on protein yields and interaction with FpvR1-92. Several of these were constructed, and most yielded wild-type levels of FpvI95-159 (e.g., Fig. 2A, lanes 7 to 9 and 11, cf. lane 14) and were proficient in interacting with FpvR1-92 (Fig. 2B, L103A, L106S, I127T, and L119P). The L115P mutation, which in concert with the L103P mutation yielded substantially reduced FpvI95-159 levels (Fig. 2A, lane 3) and wholly abrogated FpvR1-92 binding (Fig. 2B, bar 3), alone produced an unstable FpvI95-159 protein (Fig. 2A, lane 10) unable to interact with FpvR1-92 (Fig. 2B, bar 10). Attempts to construct an R135G mutant of FpvI95-159 failed, although given that the L106S I127T R135G triple mutant was unstable (Fig. 2A, lane 5) and unable to interact with FpvR1-92 (Fig. 2B, bar 5) and that individual L106S (Fig. 2, lane/bar 8) and I127T (Fig. 2, lane/bar 9) mutations yielded wild-type levels of FpvI95-159 (cf. lane 14) that interacted with FpvR1-92, either all three mutations were needed to compromise binding to FpvR1-92 and/or FpvI95-159 production or the R135G mutation alone was predominantly responsible for the defect. The R152P mutation found first together with L119P and M153I in a mutant showing reduced FpvI95-159 production (Fig. 2A, lane 6) and loss of FpvR1-92 interaction (Fig. 2B, bar 6) alone seemed to compromise FpvI95-159 production (Fig. 2A, lane 13), and while it did reduce FpvI95-159 interaction with FpvR1-92 (Fig. 2B, bar 13), it did not compromise this to the same extent as did other mutations (e.g., L115P; Fig. 2B, bar 10).
Modeling the C-terminal region of FpvI on the available crystal structure of region 4 of σ70 (PDB ID 1KU7) revealed the presence of four α-helices (Fig. 3) with the L103P, L115P, and R152P mutations shown (above) to compromise FpvI95-159 production and interaction with FpvR1-92 mapping to the C-terminal ends of three of these (Fig. 1A and 3). Proline substitutions within α-helices will have a major impact on secondary structure, highlighting the importance of these α-helices for FpvR1-74 interaction and/or protein stability. To assess the importance of the fourth α-helix (actually third from the N terminus of the C-terminal domain but the only one unaltered in hitherto-described mutants) for FpvR binding and possibly stability, a proline substitution was engineered at residue L131 of FpvI95-159, at the C terminus of this α-helix (Fig. 1A and 3). As seen in Fig. 2A (lane 12), this substitution compromised FpvI95-159 production or stability and had a modest but measurable negative impact on FpvR1-92 binding (Fig. 2B, bar 12).
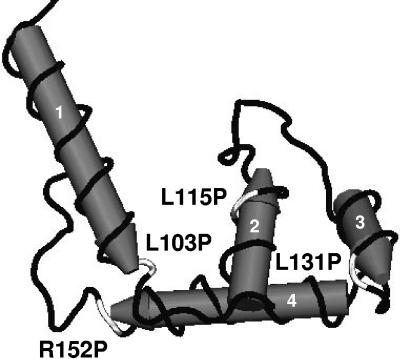
FIG. 3.
Three-dimensional model of the C-terminal region of FpvI implicated in FpvR binding. The C-terminal portion of FpvI was modeled on the structure of region 4 of the related σ70 sigma factor (structure coordinates available from the Protein Data Bank, ID 1KU7 [7]) and reveals the presence of four α-helices (labeled 1 to 4; highlighted also in Fig. 1A) on which are mapped (in white) mutations that compromise FpvI stability and/or interaction with FpvR. The N-terminal-most proximal helix exists for the most part upstream of the putative region 4 of FpvI and outside residues encompassed by FpvI95-159 (Fig. 1A) and is included here only to highlight the position of the L103P mutation that occurs at the end of this helix.
Identification of the FpvI-binding region of FpvR.
While the first 92 residues of FpvR are predicted to be cytoplasmic and are sufficient for FpvR interaction with FpvI, it was of interest to see whether the entirety of the N-terminal cytoplasmic domain was necessary for this interaction. A variety of deletion derivatives of FpvR were constructed, and as seen in Fig. 4B the presence of residues 1 to 67, which contain the first two of four α-helices predicted to occur in the cytoplasmic domain of FpvR (Fig. 4A), was sufficient for an interaction with FpvI95-159. The FpvR1-52 construct, which is disrupted in the second α-helix (i.e., carries only the first helix), and any construct lacking the first 24 amino acids of FpvR (and so lacking the first predicted α-helix) (Fig. 4A) failed to interact with FpvI95-159 (Fig. 4B).
Mutations in FpvR impacting FpvI binding.
Using a strategy outlined above, mutagenesis of fpvR1-74 on plasmid pAR016 was undertaken to identify key residues or features of the FpvR N terminus important for its interaction with FpvI95-159. Of several thousand potential mutants screened for lost FpvI95-159 binding, only six β-galactosidase-positive (i.e., red on MacConkey) colonies were recovered, and three of these carried nonsense mutations in fpvR codons corresponding to W39, W57, and E31. All of these yielded truncated FpvR proteins of the appropriate size (Fig. 5A, lanes 2, 5, and 7) with, intriguingly, the W57stop mutant retaining some ability to interact with FpvI95-159 (Fig. 5B, bar 5). While β-galactosidase activity increases from 166 Miller units for wild-type FpvR1-74 (Fig. 5B, bar 1) to 1,950 Miller units for this mutant (Fig. 5B, bar 5), consistent with a defect in FpvI95-159 binding, this is still less than what is seen for the W39stop and E31stop mutants (ca. 12,000 Miller units; Fig. 5B, bars 2 and 7), which appear to be wholly defective in FpvI95-159 binding. W57 occurs near the C-terminal end of predicted α-helix 2 of the FpvR N terminus (Fig. 4A), and so the resultant FpvR1-56 retains the bulk of this α-helix as well as α-helix 1, which appears to be sufficient for FpvR interaction with FpvI. The other FpvR nonsense mutants lack substantial portions of α-helix 2, which may explain their inability to interact with FpvI95-159 (Fig. 5B, bars 2 and 7) despite production of substantial quantities of FpvR1-74 (Fig. 4B, lanes 2 and 7). The yields of FpvR1-74 with mutations C20Y and W57L (Fig. 5A, lane 3) and L23P (Fig. 5A, lane 6) are poor, and while there is a substantial defect in FpvI95-159 binding (β-galactosidase activity increases to 1,068 [Fig. 5B, bar 3] and 2,963 [Fig. 5, bar 6] Miller units, respectively), some interaction is clearly still occurring. Neither of the C20Y or W57L mutations alone, however, had any impact on FpvR1-74 levels (Fig. 5A, lanes 9 and 10) or interaction with FpvI95-159 (Fig. 5B, bars 9 and 10), indicating that both are necessary for the defect. An intriguing FpvR1-74 mutant carried three mutations, H18Y, L59P, and L63P, which together had a substantial negative impact on FpvI95-159 binding (Fig. 5B, bar 4), although levels of the mutant FpvR1-74 protein were not adversely affected (Fig. 5A, lane 4). Still, the protein ran anomalously, with an apparent molecular weight that was larger than that of wild-type FpvA1-74 (more easily seen in Fig. 7A, lane 4). Of these three mutations, only the L59P substitution was responsible for the defect in FpvI95-159 interaction (Fig. 5B, bar 12).
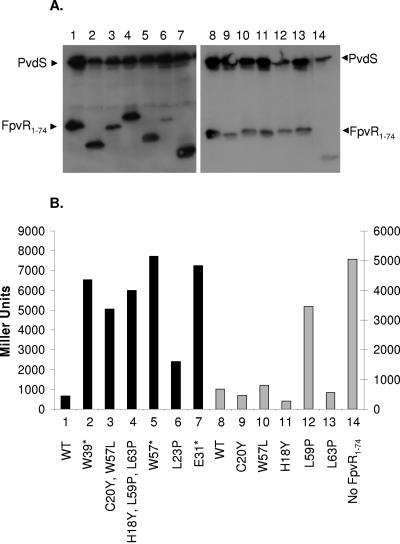
FIG. 5.
Impact of mutations in FpvR1-74 on its interaction with FpvI95-159. Western immunoblot of whole-cell extracts of E. coli SU202 carrying pAR014 (pDP804::fpvI95-159) and pMS604 (lane 14) or its derivatives expressing wild-type (lanes 1 and 8) or mutated (lanes 2 to 7 and 9 to 13) FpvR1-74 developed with antibodies to LexA. Specific mutations are indicated and numbered in panel B with the numbers used to match the mutation and β-galactosidase data to the corresponding immunoblot lane in panel A. (B) β-Galactosidase activity of E. coli SU202 carrying pMS604 or derivatives thereof expressing the indicated mutant FpvR1-74 proteins and pAR014 expressing FpvI95-159. Asterisks indicate the presence of a nonsense mutation in the corresponding fpvR codon, leading to truncation of the protein. WT, wild-type FpvR1-74. The scale to the left applies to the darkly shaded bars (1 to 7), while the scale to the right applies to the lightly shaded bars (8 to 14). Variability in protein yields (A) and β-galactosidase activity (B) for FpvI and FpvR derivatives shown in lanes 1 to 6 versus 7 to 14 reflects the fact that these data were obtained from experiments carried out independently on different days. Immunoblotting and β-galactosidase assays were performed on samples prepared from the same culture in all instances, and the data shown are representative of three replicates.
FIG. 7.
Impact of mutations in FpvR1-74 on its interaction with PvdS. Western immunoblot of whole-cell extracts of E. coli SU202 carrying pAR013 (pDP804::pvdS) and pMS604 (lane 14) or its derivatives expressing wild-type (lanes 1 and 8) or mutated (lanes 2 to 7 and 9 to 13) FpvR1-74 developed with antibodies to LexA. Specific mutations are indicated and numbered in panel B with the numbers used to match the mutation and β-galactosidase data to the corresponding immunoblot lane in panel A. (B) β-Galactosidase activity of E. coli SU202 carrying pMS604 or derivatives thereof expressing the indicated mutant FpvR1-74 proteins and pAR013 expressing PvdS. Asterisks indicate the presence of a nonsense mutation in the corresponding fpvR codon, leading to truncation of the protein. WT, wild-type FpvR1-74. Immunoblotting and β-galactosidase assays were performed on samples prepared from the same culture in all instances, and the data shown are representative of three replicates. The scale to the left applies to the darkly shaded bars (1 to 7), while the scale to the right applies to the lightly shaded bars (8 to 14). Variability in protein yields (A) and β-galactosidase activity (B) for FpvI and FpvR derivatives shown in lanes 1 to 6 versus 7 to 14 reflects the fact that these data were obtained from experiments carried out independently on different days. Immunoblotting and β-galactosidase assays were performed on samples prepared from the same culture in all instances, and the data shown are representative of three replicates.
All of the spontaneous FpvR1-74 mutants (Fig. 5A, lanes 2 to 7) and the specifically constructed L59P mutant (Fig. 5A, lane 12) produced detectable FpvR1-74 protein (although yields of the C20Y W57L double [Fig. 5A, lane 3] and L23P single [Fig. 5A, lane 6] mutants were markedly reduced) that was to some extent compromised for interaction with FpvI95-159 (Fig. 5B). Intriguingly, in all of these instances FpvI95-159 levels were drastically reduced (Fig. 5A, lanes 2 to 7, cf. lane 1; Fig. 5A, lane 12, cf. lane 8). Moreover, in the absence of FpvR1-74, FpvI95-159 is barely detectable (Fig. 5A, lane 14), suggesting that in the absence of FpvR (or FpvR binding) FpvI is unstable.
Interaction of FpvR with ECF sigma factor PvdS.
FpvR is known to control the activity of PvdS, an ECF sigma factor responsible for expression of pyoverdine biosynthetic genes and thus pyoverdine under iron-limiting conditions (21, 52). Its ability to interact with FpvR was assessed as above for FpvI, and regions of FpvR necessary for this were elucidated. As seen in Fig. 6, E. coli SU202 coexpressing PvdS (from pDP804 derivative pAR013) and the cytoplasmic domain of FpvR (i.e., FpvR1-92; from pMS604 derivative pAR012) produced low levels of β-galactosidase activity, consistent with these proteins interacting in vivo. As with its interaction with FpvI, FpvR derivatives encompassing residues 1 to 67 but not 1 to 52 were proficient in interacting with PvdS (Fig. 6), highlighting the likely significance of helices 1 and 2 of the FpvR N-terminal region for interaction with PvdS as well. All of the previously described randomly generated FpvR1-74 mutants were defective in PvdS binding, as they were for FpvI binding, though to a generally greater extent (Fig. 7B, bars 2 to 7). This was particularly true of the C20Y W57L double (Fig. 7B, bar 3) and W57stop nonsense (Fig. 7B, bar 5) mutants, which previously demonstrated some evidence of FpvI binding but appear to be wholly compromised for PvdS binding. As with FpvI binding, too, the FpvR1-74 L23P mutant protein that was produced at very low levels (Fig. 7A, lane 6) showed reduced but measurable PvdS binding (Fig. 7B, bar 6). Finally, the L59P mutant FpvR1-74 protein was substantially defective in PvdS binding (Fig. 7B, bar 12). While PvdS levels tended to decrease in cells expressing mutant FpvR1-74 proteins compromised for PvdS binding (Fig. 7A, lanes 2 to 7, cf. lane 1; lane 12, cf. lane 8) and in cells lacking FpvR1-74 (Fig. 7A, lane 14), the effect was much less striking than for FpvI.
FIG. 6.
Interaction of FpvR deletion derivatives with PvdS. β-Galactosidase activity of E. coli SU202 carrying pMS604 or derivatives thereof expressing the indicated FpvR proteins and pDP804 or its derivative expressing PvdS. Data are representative of three replicates.
DISCUSSION
ECF σ factors are typically sequestered in an inactive complex by anti-sigma factors that often span the cytoplasmic membrane, binding being promoted by sequences present within the N-terminal, cytoplasmic domain of the anti-sigma factor (e.g., σE and its anti-σ factor RseA in E. coli [8], FecI and its anti-σ factor FecR also in E. coli [10], and possibly AlgU and its anti-σ factor MucA in P. aeruginosa [41]). In a recent study, several putative ECF sigma factors in Bacillus subtilis were shown to interact with putative membrane-spanning anti-sigma factors via the latter's N-terminal, presumed cytoplasmic domains (53). In the current study, the first 67 amino acid residues of FpvR, encompassing two putative α-helices, are sufficient for FpvI interaction. Interestingly, truncation of the second helix at W57 (the putative helix extends to S60) reduces but does not wholly abrogate FpvI binding while a proline substitution at L59 completely eliminates FpvI binding, presumably because the latter more markedly alters the structure of the N-terminal FpvI-binding domain. A proline substitution at residue L23 of FpvR, immediately following the first putative α-helix of the FpvR N terminus, also shows some negative impact on FpvI binding, though unlike the W57stop and L59P mutations which did not adversely impact FpvR1-74 levels, this substitution drastically reduced FpvR1-74 yields. Thus, it is not entirely clear whether this mutation solely impacted protein stability, and only indirectly FpvI binding, or whether it impacted both. Clearly, however, only very small amounts of the FpvI and FpvR binding partners are needed to dimerize LexA and so repress lacZ expression in E. coli SU202 (e.g., the C20Y W57L FpvA1-74 mutant produces levels of FpvA1-74 comparable to the L23P mutant [Fig. 5A, cf. lanes 3 and 6] and yet E. coli SU202 expressing this mutant protein [and FpvI95-159] fused to LexA produces markedly less β-galactosidase activity [1,068 Miller units, cf. 2,968 Miller units; Fig. 5B, bars 3 and 6]). The greater defect in FpvI binding (as indicated by less lacZ repression) for the L23P mutant cannot wholly be explained by reduced FpvR1-74 levels and must be attributable, at least in part, to a direct negative impact on binding. That proline substitutions, noted disruptors of protein secondary structure, were alone among single missense mutations identified in FpvR as compromising FpvI binding strongly suggests that the overall structure of the two-helix N-terminal domain of FpvR and not specific residues are important for FpvI binding.
The observation that levels of the FpvR1-74 binding partner FpvI95-159 markedly declined in E. coli SU202 expressing FpvR mutant proteins compromised for FpvI interaction and in E. coli SU202 lacking FpvR clearly indicates that FpvI95-159 is unstable when it is free of FpvR, possibly reflecting the natural situation in vivo with full-length FpvI. Indeed, in initial two-hybrid studies with full-length FpvI, FpvI levels were noticeably higher when the protein was coexpressed with FpvR1-92 in E. coli SU202 than when it was expressed alone (data not shown). Similarly, levels of full-length PvdS declined, albeit less markedly, in E. coli SU202 carrying mutant FpvR1-74 or not expressing this protein relative to E. coli SU202 expressing wild-type FpvR1-74. A similar reduction in FecI yields was also noted for FecI mutant proteins compromised for binding to its cognate anti-sigma factor FecR (24). Whether this reflects the natural situation whereby free and active ECF sigma factors are readily turned over, perhaps to ensure that continued activation of target genes is dependent upon ongoing synthesis of the sigma factors (an indicator that they are still required) and allowing the system to quickly respond to absence of inducing conditions, is unclear. Certainly, proteolysis and proteolytic control of sigma factor activity have been reported in bacteria (15, 25, 34). Still, it cannot be ruled out that the observed instability of FpvI, PvdS, and FecI in these two-hybrid studies is an artifact of their expression as fusions with LexA. In light, however, of the apparent instability of FpvI95-159 when it can't interact with FpvR1-74 and the fact that FpvI mutations compromised for but retaining some FpvI binding ability show undetectable levels of FpvI95-159 (e.g., the L131P and R152 mutant proteins), it is likely that apparent defects in FpvR1-74 binding attributable to proline substitutions in FpvI95-159 are not due to protein instability and thus insufficient FpvI95-159 to partner with FpvR1-74. Rather, these substitutions probably impact FpvR binding, and unbound FpvI95-159 is susceptible, e.g., to cellular proteases.
Region 4 of σ70 family sigma factors, including ECF σ factors, is implicated in recognition of −35 promoter sequences but also appears to participate in core RNA polymerase (RNAP) binding (33). Thus, FpvR binding to this region of FpvI might block core polymerase recruitment or promoter binding. Certainly, a number of soluble anti-sigma factors have been shown to interfere with ECF sigma factor binding to core RNAP (e.g., ChrR and its ECF sigma factor σE in Rhodobacter sphaeroides [1]; RsrA and its ECF sigma factor σR in Streptomyces coelicolor [23]) though generally via interaction with region 2 of these σ factors (1, 23). The AsiA anti-sigma factor of phage T4 that controls σ70 activity in E. coli is known to bind region 4 and to interfere with promoter binding (44), though AsiA is a soluble protein and it's not clear that a membrane-bound anti-sigma factor binding to the same region of a sigma factor would act in the same fashion. Indications are that FecR does not impede FecI binding to its cognate −35 region (24), though, given that FecR does not function solely, if at all, as an anti-sigma factor and is, in fact, required for FecI activity (under inducing, i.e., citrate+ conditions) (4), it is far from clear that FpvR control of FpvI would mimic FecR. Certainly, FpvR mutants show greatly enhanced FpvI-dependent expression of fpvA (3) (and greatly enhanced expression of pyoverdine biosynthetic genes dependent upon another ECF sigma factor controlled by FpvR, PvdS [18]) consistent with it functioning as a bona fide anti-sigma factor. Given the known involvement of region 4 sequences in σ70 binding to core RNAP (33), it may be that FpvI holoenzyme assembly is impeded by FpvR binding to region 4 of FpvI. Still, binding, and thus sequestration, of FpvI at the cytoplasmic membrane by FpvR may itself be sufficient to prevent FpvI-mediated transcription of fpvA, since it is not at all clear that a membrane-bound holoenzyme would be capable of transcription. The MucA anti-sigma factor of P. aeruginosa, for example, does not interfere with core RNAP binding to the AlgU ECF sigma factor but sequesters the latter and indeed assembled AlgU-RNAP holoenzyme at the membrane, preventing transcription of target genes (41).
In a previous study assessing FecI-FecR interaction, a limited number of FecI mutants compromised for FecR binding were obtained, and these invariably carried multiple amino acid substitutions or proline substitutions in residues predicted to occur in α-helices of FecI region 4 (24), reminiscent of results presented here for FpvI. Intriguingly, three of the proline substitutions in FpvI that compromised FpvR binding occurred in leucine residues (L103, L115, and L131) that are conserved in FecI (Fig. 1A). Presumably, this reflects a need to drastically alter the secondary structure of the C termini of these ECF sigma factors in order to impede anti-sigma factor binding and emphasizes, therefore, the importance of secondary structure in region 4 for anti-sigma factor binding. The proline substitutions reported here to negatively impact FpvR binding occurred within (L115P and L131P, in α-helices 2 and 3, respectively) or just outside (L103P, at the end of a helix that occurs for the most part outside region 4, where it likely impacts the disposition of downstream helices, and R152P, which occurs at the end of helix 4, where it might be expected to influence disposition of upstream helices) a region of FpvI predicted to include the three α-helices implicated as the major structural (and functional) determinants of region 4 of σ70 (7). Apparently, FpvI structures required for anti-sigma factor binding are the same as those necessary for promoter binding (i.e., sigma factor activity), as appears to be the case for FecI—proline substitutions in FecI region 4 that impeded FecR binding also compromised FecI sigma factor activity (24).
FpvR is unique in its natural interaction, in vivo, with two different ECF sigma factors, PvdS and FpvI. Anti-ECF sigma factor proteins tend to be very selective, partnering only with a single sigma factor, usually encoded by a linked gene (13). Indeed, a previous study of two putative ECF sigma factor-anti-sigma factor pairs in P. aeruginosa confirmed the expected interaction of cognate binding partners and demonstrated the specificity of that interaction (24). In the current study, FpvR was shown to interact with both of its ECF sigma factors, and this interaction involved the same N-terminal region of FpvR and was similarly negatively impacted by proline substitutions at L23 and especially L59 that were expected to disrupt secondary structure of the N-terminal domain. Still, subtle differences were noted, with truncation of FpvR owing to a W57stop mutation having a markedly greater impact on PvdS than FpvI95-159 binding, suggesting that PvdS may interact slightly differently with FpvR than FpvI does. Consistent with this, the C20Y W57L double mutant of FpvR1-74 also had a substantially greater impact on binding of PvdS than did FpvI. Still, it is possible that these differences reflect the use of full-length PvdS versus N-terminally truncated FpvI, with the latter perhaps being more proficient at partnering with FpvR1-74 and so being less affected by FpvR1-74 mutations. Certainly, the N-terminally truncated FpvI95-159 showed improved FpvR1-74 binding relative to the full-length protein (Fig. 1). Still, other mutations in FpvR1-74 (e.g., L23P and L59P) do not impact PvdS binding to a greater extent, suggesting that the differential effect of some FpvR1-74 mutations on PvdS versus FpvI binding truly reflects differences in their binding to this anti-sigma factor.
Acknowledgments
This work was supported by an operating grant from the Canadian Institutes of Health Research to K.P. G.A.R. was the recipient of a CCFF graduate student scholarship.
REFERENCES
- 1.Anthony, J. R., J. D. Newman, and T. J. Donohue. 2004. Interactions between the Rhodobacter sphaeroides ECF sigma factor, σE, and its anti-sigma factor, ChrR. J. Mol. Biol. 341:345-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1992. Short protocols in molecular biology, 2nd ed. John Wiley & Sons, Inc., New York, N.Y.
- 2.Barcak, G. J., M. S. Chandler, R. J. Redfield, and J. F. Tomb. 1991. Genetic systems in Haemophilus influenzae. Methods Enzymol. 204:321-342. [DOI] [PubMed] [Google Scholar]
- 3.Beare, P. A., R. J. For, L. W. Martin, and I. L. Lamont. 2003. Siderophore-mediated cell signalling in Pseudomonas aeruginosa: divergent pathways regulate virulence factor production and siderophore receptor synthesis. Mol. Microbiol. 47:195-207. [DOI] [PubMed] [Google Scholar]
- 4.Braun, V., S. Mahren, and M. Ogierman. 2003. Regulation of the FecI-type ECF sigma factor by transmembrane signalling. Curr. Opin. Microbiol. 6:173-180. [DOI] [PubMed] [Google Scholar]
- 5.Budzikiewicz, H. 1997. Siderophores of fluorescent pseudomonads. Z. Naturforsch. Sect. C 52:713-720. [PubMed] [Google Scholar]
- 6.Buyer, J. S., and J. Leong. 1986. Iron transport-mediated antagonism between plant growth-promoting and plant-deleterious Pseudomonas strains. J. Biol. Chem. 261:791-794. [PubMed] [Google Scholar]
- 7.Campbell, E. A., O. Muzzin, M. Chlenov, J. L. Sun, C. A. Olson, O. Weinman, M. L. Trester-Zedlitz, and S. A. Darst. 2002. Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol. Cell 9:527-539. [DOI] [PubMed] [Google Scholar]
- 8.Campbell, E. A., J. L. Tupy, T. M. Gruber, S. Wang, M. M. Sharp, C. A. Gross, and S. A. Darst. 2003. Crystal structure of Escherichia coli σE with the cytoplasmic domain of its anti-sigma RseA. Mol. Cell 11:1067-1078. [DOI] [PubMed] [Google Scholar]
- 9.Dmitrova, M., G. Younes-Cauet, P. Oertel-Buchheit, D. Porte, M. Schnarr, and M. Granger-Schnarr. 1998. A new LexA-based genetic system for monitoring and analyzing protein heterodimerization in Escherichia coli. Mol. Gen. Genet. 257:205-212. [DOI] [PubMed] [Google Scholar]
- 10.Enz, S., S. Mahren, U. H. Stroeher, and V. Braun. 2000. Surface signaling in ferric citrate transport gene induction: interaction of the FecA, FecR, and FecI regulatory proteins. J. Bacteriol. 182:637-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faguy, D. M., D. P. Bayley, A. S. Kostyukova, N. A. Thomas, and K. F. Jarrell. 1996. Isolation and characterization of flagella and flagellin proteins from the thermoacidophilic archaea Thermoplasma volcanium and Sulfolobus shibatae. J. Bacteriol. 178:902-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gensberg, K., K. Hughes, and A. W. Smith. 1992. Siderophore-specific induction of iron uptake in Pseudomonas aeruginosa. J. Gen. Microbiol. 138:2381-2387. [DOI] [PubMed] [Google Scholar]
- 13.Helmann, J. D. 2002. The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 46:47-110. [DOI] [PubMed] [Google Scholar]
- 14.Hohnadel, D., and J.-M. Meyer. 1988. Specificity of pyoverdine-mediated iron uptake among fluorescent Pseudomonas strains. J. Bacteriol. 170:4865-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horikoshi, M., T. Yura, S. Tsuchimoto, Y. Fukumori, and M. Kanemori. 2004. Conserved region 2.1 of Escherichia coli heat shock transcription factor σ32 is required for modulating both metabolic stability and transcriptional activity. J. Bacteriol. 186:7474-7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koster, M., W. van Klompenburg, W. Bitter, J. Leong, and P. Weisbeek. 1994. Role for the outer membrane ferric siderophore receptor PupB in signal transduction across the bacterial cell envelope. EMBO J. 13:2805-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koster, M., J. van de Vossenberg, J. Leong, and P. J. Weisbeek. 1993. Identification and characterization of the pupB gene encoding an inducible ferric-pseudobactin receptor of Pseudomonas putida WCS358. Mol. Microbiol. 8:591-601. [DOI] [PubMed] [Google Scholar]
- 18.Lamont, I. L., P. A. Beare, U. Ochsner, A. I. Vasil, and M. L. Vasil. 2002. Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 99:7072-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamont, I. L., and L. W. Martin. 2003. Identification and characterization of novel pyoverdine synthesis genes in Pseudomonas aeruginosa. Microbiology 149:833-842. [DOI] [PubMed] [Google Scholar]
- 20.Lehoux, D. E., F. Sanschagrin, and R. C. Levesque. 2000. Genomics of the 35-kb pvd locus and analysis of novel pvdIJK genes implicated in pyoverdine biosynthesis in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 190:141-146. [DOI] [PubMed] [Google Scholar]
- 21.Leoni, L., N. Orsi, V. de Lorenzo, and P. Visca. 2000. Functional analysis of PvdS, an iron starvation sigma factor of Pseudomonas aeruginosa. J. Bacteriol. 182:1481-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leung, D. L., E. Chen, and D. V. Goeddel. 1989. A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique 1:11-15. [Google Scholar]
- 23.Li, W., C. E. Stevenson, N. Burton, P. Jakimowicz, M. S. Paget, M. J. Buttner, D. M. Lawson, and C. Kleanthous. 2002. Identification and structure of the anti-sigma factor-binding domain of the disulphide-stress regulated sigma factor σR from Streptomyces coelicolor. J. Mol. Biol. 323:225-236. [DOI] [PubMed] [Google Scholar]
- 24.Mahren, S., S. Enz, and V. Braun. 2002. Functional interaction of region 4 of the extracytoplasmic function sigma factor FecI with the cytoplasmic portion of the FecR transmembrane protein of the Escherichia coli ferric citrate transport system. J. Bacteriol. 184:3704-3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mandel, M. J., and T. J. Silhavy. 2005. Starvation for different nutrients in Escherichia coli results in differential modulation of RpoS levels and stability. J. Bacteriol. 187:434-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25a.Masuda, N., and S. Ohya. 1992. Cross-resistance to meropenem, cephems, and quinolones in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 36:1847-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McMorran, B. J., H. M. Kumara, K. Sullivan, and I. L. Lamont. 2001. Involvement of a transformylase enzyme in siderophore synthesis in Pseudomonas aeruginosa. Microbiology 147:1517-1524. [DOI] [PubMed] [Google Scholar]
- 27.McMorran, B. J., M. E. Merriman, I. T. Rombel, and I. L. Lamont. 1996. Characterization of the pvdE gene which is required for pyoverdine synthesis in Pseudomonas aeruginosa. Gene 176:55-59. [DOI] [PubMed] [Google Scholar]
- 28.Merriman, T. R., M. E. Merriman, and I. L. Lamont. 1995. Nucleotide sequence of pvdD, a pyoverdine biosynthetic gene from Pseudomonas aeruginosa: PvdD has similarity to peptide synthetases. J. Bacteriol. 177:252-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer, J.-M., D. Hohnadel, A. Khan, and P. Cornelis. 1990. Pyoverdin-facilitated iron uptake in Pseudomonas aeruginosa: immunological characterization of the ferripyoverdin receptor. Mol. Microbiol. 4:1401-1405. [DOI] [PubMed] [Google Scholar]
- 30.Meyer, J. M. 2000. Pyoverdines: pigments, siderophores and potential taxonomic markers of fluorescent Pseudomonas species. Arch. Microbiol. 174:135-142. [DOI] [PubMed] [Google Scholar]
- 31.Mossialos, D., U. Ochsner, C. Baysse, P. Chablain, J.-P. Pirnay, N. Koedam, H. Budzikiewicz, D. U. Fernàndez, M. Schäfer, J. Ravel, and P. Cornelis. 2002. Identification of new, conserved, non-ribosomal peptide synthetases from fluorescent pseudomonads involved in the biosynthesis of the siderophore pyoverdine. Mol. Microbiol. 45:1673-1685. [DOI] [PubMed] [Google Scholar]
- 32.Ochsner, U. A., P. J. Wilderman, A. I. Vasil, and M. L. Vasil. 2002. GeneChip expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthetic genes. Mol. Microbiol. 45:1277-1287. [DOI] [PubMed] [Google Scholar]
- 33.Paget, M. S., and J. D. Helmann. 2003. The σ70 family of sigma factors. Genome Biol. 4:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peterson, C. N., N. Ruiz, and T. J. Silhavy. 2004. RpoS proteolysis is regulated by a mechanism that does not require the SprE (RssB) response regulator phosphorylation site. J. Bacteriol. 186:7403-7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poole, K., and G. A. McKay. 2002. Iron acquisition and its control in Pseudomonas aeruginosa: many roads lead to Rome. Front. Biosci. 8:d661-d686. [DOI] [PubMed] [Google Scholar]
- 36.Poole, K., S. Neshat, and D. Heinrichs. 1991. Pyoverdine-mediated iron transport in Pseudomonas aeruginosa: involvement of a high-molecular-mass outer membrane protein. FEMS Microbiol. Lett. 78:1-5. [PubMed] [Google Scholar]
- 37.Poole, K., S. Neshat, K. Krebes, and D. E. Heinrichs. 1993. Cloning and nucleotide sequence analysis of the ferripyoverdine receptor gene fpvA of Pseudomonas aeruginosa. J. Bacteriol. 175:4597-4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poole, K., Q. Zhao, S. Neshat, D. E. Heinrichs, and C. R. Dean. 1996. The tonB gene of Pseudomonas aeruginosa encodes a novel TonB protein. Microbiology 142:1449-1458. [DOI] [PubMed] [Google Scholar]
- 39.Postle, K., and R. J. Kadner. 2003. Touch and go: tying TonB to transport. Mol. Microbiol. 49:869-882. [DOI] [PubMed] [Google Scholar]
- 40.Redly, A., and K. Poole. 2003. Pyoverdine-mediated regulation of FpvA synthesis in Pseudomonas aeruginosa: involvement of a probable ECF sigma factor, FpvI. J. Bacteriol. 185:1261-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rowen, D. W., and V. Deretic. 2000. Membrane-to-cytosol redistribution of ECF sigma factor AlgU and conversion to mucoidy in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Mol. Microbiol. 36:314-327. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Shen, J.-S., A. Meldrum, and K. Poole. 2002. FpvA receptor involvement in pyoverdine biosynthesis in Pseudomonas aeruginosa. J. Bacteriol. 184:3268-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simeonov, M. F., R. J. Bieber Urbauer, J. M. Gilmore, K. Adelman, E. N. Brody, A. Niedziela-Majka, L. Minakhin, T. Heyduk, and J. L. Urbauer. 2003. Characterization of the interactions between the bacteriophage T4 AsiA protein and RNA polymerase. Biochemistry 42:7717-7726. [DOI] [PubMed] [Google Scholar]
- 45.Stintzi, A., P. Cornelis, D. Hohnadel, J.-M. Meyer, C. Dean, K. Poole, S. Kourambas, and V. Krishnapillai. 1996. Novel pyoverdine biosynthesis gene(s) of Pseudomonas aeruginosa PAO. Microbiology 142:1181-1190. [DOI] [PubMed] [Google Scholar]
- 46.Stintzi, A., Z. Johnson, M. Stonehouse, U. Ochsner, J. M. Meyer, M. L. Vasil, and K. Poole. 1999. The pvc gene cluster of Pseudomonas aeruginosa: role in synthesis of the pyoverdine chromophore and regulation by PtxR and PvdS. J. Bacteriol. 181:4118-4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsuda, M., H. Miyazaki, and T. Nakazawa. 1995. Genetic and physical mapping of genes involved in pyoverdin production in Pseudomonas aeruginosa PAO. J. Bacteriol. 177:423-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vandenende, C. S., M. Vlasschaert, and S. Y. Seah. 2004. Functional characterization of an aminotransferase required for pyoverdine siderophore biosynthesis in Pseudomonas aeruginosa PAO1. J. Bacteriol. 186:5596-5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vasil, M. L., and U. A. Ochsner. 1999. The response of Pseudomonas aeruginosa to iron: genetics, biochemistry and virulence. Mol. Microbiol. 34:399-413. [DOI] [PubMed] [Google Scholar]
- 50.Visca, P., A. Ciervo, and N. Orsi. 1994. Cloning and nucleotide sequence of the pvdA gene encoding the pyoverdin biosynthetic enzyme l-ornithine N5-oxygenase in Pseudomonas aeruginosa. J. Bacteriol. 176:1128-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Welz, D., and V. Braun. 1998. Ferric citrate transport of Escherichia coli: functional regions of the FecR transmembrane regulatory protein. J. Bacteriol. 180:2387-2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson, M. J., and I. L. Lamont. 2000. Characterization of an ECF sigma factor protein from Pseudomonas aeruginosa. Biochem. Biophys. Res. Commun. 273:578-583. [DOI] [PubMed] [Google Scholar]
- 53.Yoshimura, M., K. Asai, Y. Sadaie, and H. Yoshikawa. 2004. Interaction of Bacillus subtilis extracytoplasmic function (ECF) sigma factors with the N-terminal regions of their potential anti-sigma factors. Microbiology 150:591-599. [DOI] [PubMed] [Google Scholar]
- 54.Zhao, Q., X.-Z. Li, A. Mistry, R. Srikumar, L. Zhang, O. Lomovskaya, and K. Poole. 1998. Influence of the TonB energy-coupling protein on efflux-mediated multidrug resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 42:2225-2231. [DOI] [PMC free article] [PubMed] [Google Scholar]