Abstract
FliG is a component of the switch complex on the rotor of the bacterial flagellum. Each flagellar motor contains about 25 FliG molecules. The protein of Escherichia coli has 331 amino acid residues and comprises at least two discrete domains. A C-terminal domain of about 100 residues functions in rotation and includes charged residues that interact with the stator protein MotA. Other parts of the FliG protein are essential for flagellar assembly and interact with the MS ring protein FliF and the switch complex protein FliM. The crystal structure of the middle and C-terminal parts of FliG shows two globular domains joined by an α-helix and a short extended segment that contains two well-conserved glycine residues. Here, we describe targeted cross-linking studies of FliG that reveal features of its organization in the flagellum. Cys residues were introduced at various positions, singly or in pairs, and cross-linking by a maleimide or disulfide-inducing oxidant was examined. FliG molecules with pairs of Cys residues at certain positions in the middle domain formed disulfide-linked dimers and larger multimers with a high yield, showing that the middle domains of adjacent subunits are in fairly close proximity and putting constraints on the relative orientation of the domains. Certain proteins with single Cys replacements in the C-terminal domain formed dimers with moderate yields but not larger multimers. On the basis of the cross-linking results and the data available from mutational and electron microscopic studies, we propose a model for the organization of FliG subunits in the flagellum.
Bacterial flagella are turned by rotary motors that obtain energy from the membrane gradient of protons or sodium ions (2, 17, 25). In many species, the motor can rotate either counterclockwise (CCW) or clockwise (CW), and cells regulate their swimming behavior by controlling reversals in motor direction (26). In the enteric species Escherichia coli and Salmonella enterica serovar Typhimurium, CCW flagellar rotation allows cells to swim in relatively straight paths, whereas CW rotation causes cells to tumble (i.e., rapidly change orientation). When a cell moves up a gradient of a chemical attractant or other positive stimulus, the probability of CW rotation is decreased so that the run in a favorable direction is extended. Other modes of motor control are found in some other species; the motors of Rhodobacter sphaeroides alternately start and stop, while those of Sinorhizobium meliloti rotate at variable speeds (1). Whatever the mode of swimming, the swimming is regulated by sensory cues to bias cell movement toward attractant stimuli and away from repellents.
The molecular mechanisms of motor rotation and direction switching are not understood. The proteins that function in rotation have been identified, mainly through genetic studies of E. coli and Salmonella. The stator (nonrotating part) is formed from the proteins MotA and MotB (10-12, 33, 36). The motor contains several independently functioning stator complexes (5, 6), each with the subunit composition MotA4MotB2 (24, 32). The stator complexes are located in the cytoplasmic membrane surrounding the basal body (21), where they function to conduct either protons or sodium ions (depending on the motor) and couple ion flow to rotation (4, 15, 37). MotB contains a conserved, functionally critical aspartate residue (Asp32 in E. coli MotB) that appears to have a direct role in ion conduction (50). Charge-neutralizing mutations of this aspartate induce a conformational change in the stator complex, as evidenced by the altered protease susceptibility of MotA (23). On the basis of these findings, it was proposed that proton (or Na+) association/dissociation at Asp32 might drive conformational changes in the stator that act on the rotor to drive rotation. A power stroke mechanism such as this is consistent with the torque-speed characteristic of the motor as measured by Berry and Berg (3). The motor torque was found to be nearly constant over a wide range of speeds, which is predicted for a mechanism in which energy from the ion gradient is used to apply force to the rotor, but not for a Brownian ratchet mechanism in which the gradient is used to rectify thermally driven movements.
The rotor proteins that function to support rotation are FliG, FliM, and FliN. Together, these proteins form the “switch complex” that is essential for assembly, rotation, and CW/CCW switching of the flagellum (45). FliG is involved most directly in motor rotation (19, 28). Its C-terminal domain (FliGC), in particular, is essential for rotation but dispensable for flagellar assembly (28). FliM is closely involved in CW/CCW switching (34) and interacts with the chemotaxis signaling molecule phospho-CheY (7). The precise role of FliN is unknown, but it is essential for flagellar assembly and has been implicated in the process of flagellum-specific export that delivers the proteins forming the exterior structures (the rod, hook, and filament) to their sites of installation (39, 44). FliN also appears to be involved in switching, but its specific role in this process is unknown (19).
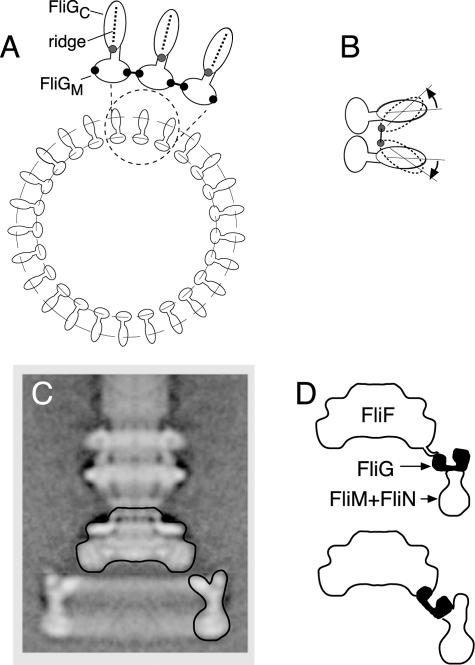
The rotor contains many molecules of FliG, probably about 25 (38, 47). FliG binds to the MS ring protein FliF (13, 16, 22, 31), and electron microscopic studies have suggested that a portion of FliG might be located on the cytoplasmic face of the MS ring (14, 38, 40) (Fig. 1). It has been difficult to assign specific features in micrographs to FliG, however, and the precise location of FliG in the rotor remains uncertain. The crystal structure of the middle and C-terminal domains of FliG (FliGMC) from Thermotoga maritima revealed two compact, globular domains connected by an α-helix and a short segment with extended structure (8). The extended segment includes two well-conserved Gly residues and might therefore be flexible. The C-terminal domain of FliG contains well-conserved charged residues that are important for rotation (27) and that interact with charged residues of the stator protein MotA (48, 49). The structure shows that these charged residues are clustered on a prominent ridge in the C-terminal domain (8, 29). In the flagellum, FliG is presumably positioned with this ridge near the stator. FliG interacts with FliM (22, 30, 42), and sequence alignments have identified conserved surface features in both the middle and C-terminal domains that might function in binding FliM or other proteins in the rotor (8).
FIG. 1.
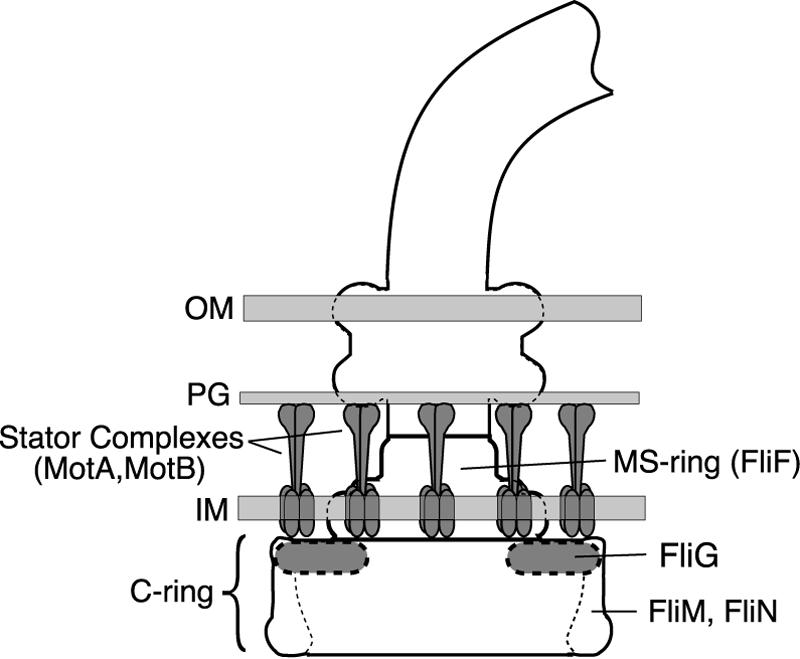
Schematic diagram of the flagellar basal body. FliG is believed to be in the upper (membrane-proximal) part of the C ring, where it can interact with the MS ring protein FliF and the stator protein MotA. Its exact location is unknown. OM, outer membrane; PG, peptidoglycan; IM, inner membrane.
While the crystal structure of FliGMC provides a useful starting point, to understand the functions of FliG in the flagellum, we must determine how the ∼25 FliG subunits are organized. Here, we used a targeted cross-linking approach to deduce features of FliG subunit organization. Several positions on the surface of FliGMC were mutated to cysteine, singly and in pairs, and cysteine-specific cross-linking was examined using either maleimide or disulfide-inducing oxidants. The results indicate that the middle domains (FliGM) are adjacent to each other in the flagellum, with their long axes pointing toward each other. The FliGC domains do not appear to be in close contact, but they are able to undergo domain rotations or other movements that allow cross-linking between proteins with single Cys replacements in certain positions. We propose a model for FliG organization in which the FliGC domains are at an outer radius, where they interact with the stator, while the FliGM domains are nearer the MS ring.
MATERIALS AND METHODS
Strains, plasmids, and mutagenesis.
The FliG-null strain DFB225 (28) was used for assays of mutant FliG function and for cross-linking experiments with cysteine-containing mutant proteins. To control for cross-linking that might occur independent of flagellar assembly, parallel experiments were carried out with strain RP3098, a flhDC mutant that does not assemble flagella (a gift from J. S. Parkinson).
Cysteine replacements were made by site-directed mutagenesis of the fliG gene in pSL27, using the Altered Sites procedure (Promega). Plasmid pSL27 is a derivative of pAlter-1 (Promega). It expresses FliG at about twice the wild-type level and complements a fliG-null strain to wild-type function in a soft-agar swarming assay (28). Mutagenesis reactions were carried out by using the manufacturer's protocol, except that the concentration of mutagenic oligonucleotide was increased two- to fivefold in some reactions. Mutations were confirmed by dideoxy sequencing, and then mutant plasmids were transformed into the fliG-null strain DFB225 and the nonflagellated control strain RP3098.
The function of the mutant FliG proteins with Cys replacements was tested in soft-agar swarming assays. Cells of strain DFB225 transformed with pSL27 variants encoding the mutant proteins were cultured to saturation in TB (1% tryptone, 0.5% NaCl) and diluted 1:500 or 1:1,000 in fresh medium, and then 1-μl portions of the diluted cultures were spotted onto soft-agar tryptone plates (1% tryptone, 0.5% NaCl, 0.30% Bacto agar [Difco]). The plates were incubated at 32°C, and once swarming commenced, the colony diameters were measured at regular (approximately hourly) intervals. Rates were computed from plots of swarm diameter versus time and compared to the rates for wild-type controls (strain DFB225 transformed with wild-type pSL27) included on the same plates.
Cross-linking.
Freezer stocks of DFB225 cells containing the Cys mutant pSL27 plasmids were used to inoculate 1- to 2-ml cultures, which were grown to saturation in LB (1% tryptone, 0.5% yeast extract, 0.5% NaCl) containing 125 μg/ml ampicillin at 37°C with shaking. Cultures were diluted 100-fold in TB containing ampicillin (typically 4 ml) buffered at pH 7.5 with 43 mM sodium phosphate and incubated overnight at 32°C with shaking in culture tubes (17 by 100 mm). The optical density at 600 nm was measured for each culture, and then the cells were pelleted by centrifugation (≥2,000 × g, 15 to 30 min) and resuspended in a volume of XL buffer (20 mM sodium phosphate, pH 7.4, 150 mM NaCl) calculated to give an optical density at 600 nm of 10. Samples (100 μl) were dispensed and chilled on ice for at least 10 min before cross-linking.
For cross-linking with 1,6-bismaleimidohexane (BMH), 100-μl samples of cells were mixed with 2 μl of 50 mM BMH (dissolved in dimethyl sulfoxide and stored at −20°C), which resulted in a final BMH concentration of approximately 1 mM. Cells were incubated on ice for 30 min, and then the reaction was quenched by adding 2.5 μl of 2-mercaptoethanol (BME). Control samples received 2 μl of dimethyl sulfoxide and then 2.5 μl of BME. In a small number of experiments we used the longer bifunctional cross-linker 1,4-di-(2-pyridyldithio-propionamido)butane (DPDPB). The protocols were similar to those used with BMH, except that the DPDPB concentration was 2 mM and cross-linking was done at room temperature for 90 min.
In some cross-linking experiments we used samples solubilized in Triton X-100. Cells were cultured as described above, except that in the second culturing step we used 25 ml of buffered TB in 125-ml flasks. Cells were collected by centrifugation, frozen at −70°C, thawed, and then resuspended in 0.5 ml of 0.5 M sucrose, 10 mM Tris, pH 8.0, on ice. Lysozyme was added from a 20-mg/ml stock solution to a final concentration of 1 mg/ml, and then EDTA was added from a 0.5 M stock solution to a final concentration of 120 mM. Samples were stirred for 30 min on ice and then solubilized by addition of 650 μl of a cocktail containing 127 mM MgSO4, 6.4% Triton X-100, 840 μM 4-amidino-phenylmethanesulfonyl fluoride, 8 μg/ml pepstatin, 17 μg/ml leupeptin, and 170 μg/ml DNase I. Samples were stirred for 3 h on ice and then centrifuged (5,600 × g, 20 min) to remove unsolubilized material. Aliquots (100 μl) of the supernatant were used for cross-linking experiments by using the protocols described above, except that the final sodium dodecyl sulfate (SDS) concentration was 2% (rather than 7%) in the samples loaded on gels.
Disulfide cross-linking was induced by iodine. Cells were washed and resuspended in XL buffer as described above and then treated with I2 at a concentration of 0.2 mM, added from a 20 mM working stock solution in ethanol. Following 10 min of incubation on ice, unreacted Cys residues were blocked with 20 mM N-ethylmaleimide for 5 min on ice. In experiments to monitor disulfide rereduction, dithiothreitol was added from a 1 M stock solution (in water) to a final concentration of 20 mM.
In some of the iodine-induced cross-linking experiments (those with Cys at positions 297, 298, and 299 in FliGC) we used 50 mM Tris, pH 8.0, 50 mM NaCl, and the experiments were done at room temperature. The other steps were the same as those described above.
Following cross-linking, cells were collected by centrifugation and resuspended in 100 μl of a loading buffer containing 62.5 mM Tris, pH 6.8, 7% (wt/vol) SDS, 10% glycerol, and ∼0.01% bromophenol blue. For samples cross-linked with BMH, the loading buffer also included 5% BME. Samples were heated for 10 min at >95°C, refrigerated briefly, and either sonicated for 15 to 20 s (Branson model 450 with a microtip; power, 1; duty cycle, 50%) and loaded on SDS-polyacrylamide gels (typically 7.5%) (10 μl/well) or frozen at −70°C for later use. Following electrophoresis, the gels were soaked in transfer buffer (20% [vol/vol] methanol, 48 mM Tris base, 39 mM glycine, 0.0375% [wt/vol] SDS) at room temperature with slow shaking for at least 10 min. Proteins were transferred to nitrocellulose membranes by using a Bio-Rad semidry blotting apparatus at 13 V for 48 min. After transfer, the membranes were soaked for at least 30 min in 4% dry nonfat milk in 20 mM Tris, pH 8.0, 500 mM NaCl at room temperature with slow shaking. Immunoblotting was carried out using anti-FliG (dilution factor, between 1:2,500 and1:5,000) and horseradish peroxidase-conjugated goat anti-rabbit secondary antibody. Blots were visualized using WestPico SuperSignal reagents (Pierce) and X-ray film. Bands were quantified by using video densitometry and the public domain NIH Image program (developed at the National Institutes of Health and available at http://rsb.info.nih.gov/nih-image/)
RESULTS
Cys replacement mutations.
Site-directed mutagenesis was used to make cysteine replacements at 22 positions on the surface of FliGMC, singly and in various pairwise combinations. The positions of the replacements on the structure are shown in Fig. 2, which also shows the double Cys replacements that were studied. In FliGM, Cys replacements were made at positions that sampled most of the surface of the domain. In FliGC, the substitutions were made on the faces around the active site ridge, encircling the domain at approximately its widest part. This choice of positions was based on mutational data which indicated that the active site ridge interacts with the stator protein MotA (49) and is therefore not likely to face toward adjacent FliG subunits.
FIG. 2.
Locations of Cys replacements in FliG. The diagrams show the crystal structure of residues 115 to 327 of T. maritima FliG (PDB accession no. 1lkv). The two views are related by 180° rotation, as indicated. The residue numbers are the numbers for the E. coli protein. The dashed line in FliGC indicates the charge-bearing ridge (27, 29) that interacts with MotA (49). Double Cys replacements made for the cross-linking study are listed on the right.
Cross-linking in FliGM.
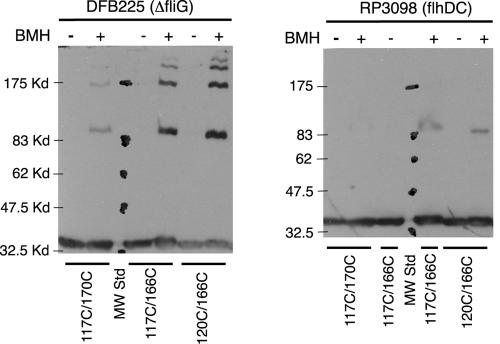
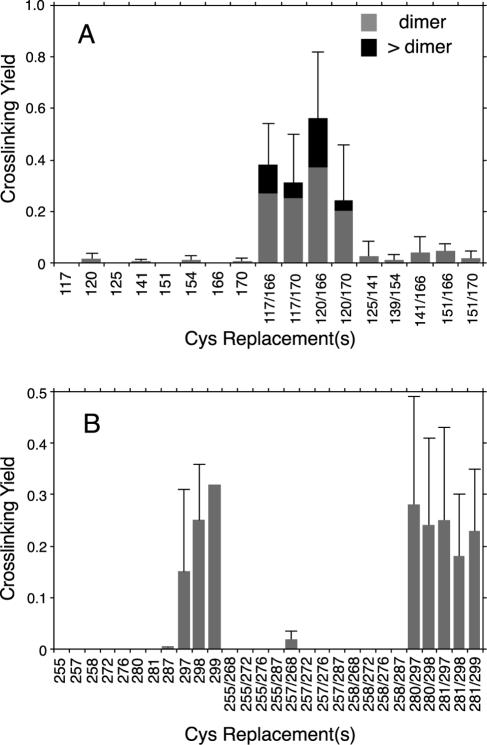
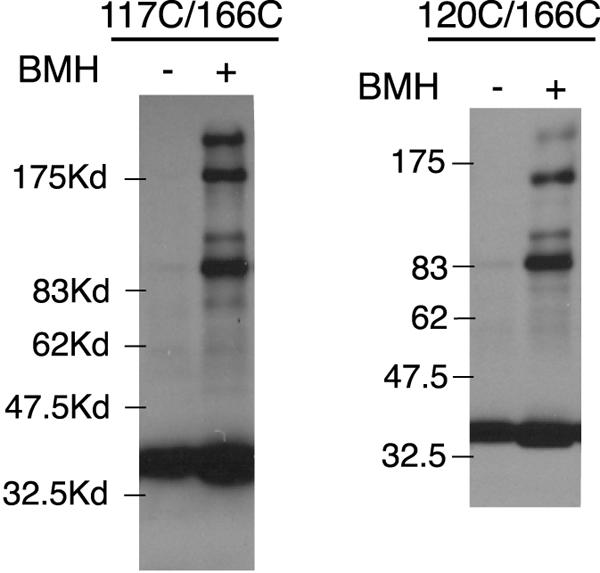
Proteins with single and double Cys replacements in FliGM were expressed from plasmid pSL27 in cells of the fliG-null strain DFB225, and cross-linking was examined using the bifunctional, sulfhydryl-specific cross-linker BMH. Products of cross-linking were characterized by using FliG immunoblots. The majority of the proteins with single Cys residues in FliGM showed little or no cross-linking with BMH. Most of the doubly substituted proteins also did not show significant cross-linking, but four (117/166, 117/170, 120/166, and 120/170) gave a product at about the position expected for a FliG dimer and also a series of larger products with fairly high yields. Representative results are shown in Fig. 3. The product yields for BMH cross-linking of proteins with Cys residues in FliGM were quantified by densitometry and are summarized in Fig. 4A.
FIG. 3.
Examples of BMH cross-linking of proteins with two Cys replacements in FliGM. The positions of the Cys replacements are indicated at the bottom. DFB225 is a fliG-null strain, which is flagellated and motile when the fliG gene is provided on the plasmid. Strain RP3098 contains a deletion of the flagellar regulatory genes flhDC and does not assemble flagella even when the fliG plasmid is present. Kd, kilodaltons.
FIG. 4.
Yields of BMH-induced cross-linking in FliG proteins with one or two Cys replacements. (A) Summary of yields for Cys at positions in the middle domain (FliGM). The shaded bars indicate the yields of cross-linked dimer, and the solid bars indicate the combined yields of all multimers larger than dimers. The error bars indicate the standard deviations of two or more determinations. (B) Summary of yields of cross-linked dimers for Cys at positions in the C-terminal domain. Larger multimers were not observed for any of the Cys replacements in FliGC.
To verify that cross-linking occurred within flagella rather than by a collisional mechanism, the same experiments were conducted with mutant strain RP3098, which does not express any chromosomal flagellar genes and does not assemble flagella. The Cys replacement proteins were expressed from the same plasmid (pSL27), and the levels of the FliG proteins in the nonflagellated strain were similar to those observed in the flagellated strain. The cross-linking yields were greatly reduced in the nonflagellated strain. Only a small amount of dimer and none of the higher-molecular-weight products were observed (Fig. 3).
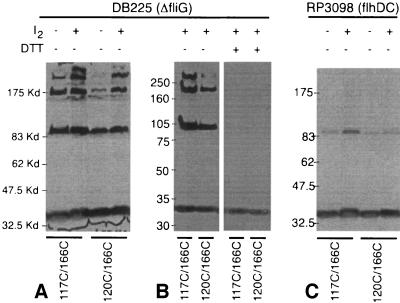
BMH is fairly long and might cross-link between Cys residues that are separated by more than 10 Å. To put tighter constraints on residue proximity, we examined disulfide cross-linking in the 117/166, 117/170, 120/166, and 120/170 double mutants and in the corresponding single-Cys controls. Cells expressing the Cys mutant proteins were treated with iodine to induce disulfide bond formation, and products of the reaction were examined as described above, using anti-FliG immunoblots. The results for the 117/166 and 120/166 double mutants are shown in Fig. 5. Iodine treatment of these double mutants yielded several cross-linked products, in a pattern similar to that seen with BMH. High-yield cross-linking was not observed in the corresponding single-Cys mutants (Fig. 4), and the dimer and higher-molecular-weight bands were essentially eliminated when samples were treated with DTT before electrophoresis. Thus, they were the products of disulfide cross-linking. Experiments with the nonflagellated flhDC strain resulted in none of the higher-molecular-weight products and only a small amount of dimer, indicating that the cross-linking occurred within flagella and not through collision.
FIG. 5.
(A) Iodine-induced cross-linking of proteins with double Cys replacements in FliGM. Some preexisting disulfide cross-linking was seen in both of the mutants, and cross-linking was increased by treatment with iodine. (B) Effect of treatment with DTT. (C) Cross-linking experiment with the nonflagellated flhDC strain. Kd, kilodaltons.
While the high-yield cross-linking was observed only in cells that assembled flagella, this cross-linking could nevertheless have involved a protein that was not in the flagellum, such as a protein nearby in the membrane. To rule out cross-linking to nearby proteins in the membrane, we solubilized the flagella with the detergent Triton X-100 (final concentration, 1%) before cross-linking with iodine. Triton X-100 releases the basal bodies from the membrane but leaves the C rings intact (14). For both the 117C/166C and 120C/166C proteins, the yields and distribution of cross-linked products were similar to those seen in the experiments with whole cells (Fig. 6).
FIG. 6.
Cross-linking in samples solubilized in Triton X-100. Cys replacements are indicated at the top. Kd, kilodaltons.
FliM contains a single native Cys residue and is thought to be near FliG in the flagellum (30, 42, 45). To see if any of the cross-linked products contained FliM, we cross-linked the 117C/166C and 120C/166C double-mutant proteins with BMH and used anti-FliM immunoblots to examine the products. The blots showed a single strong band corresponding to the FliM monomer but no bands at higher molecular weight (data not shown). FliM is therefore not a component of the cross-linked products.
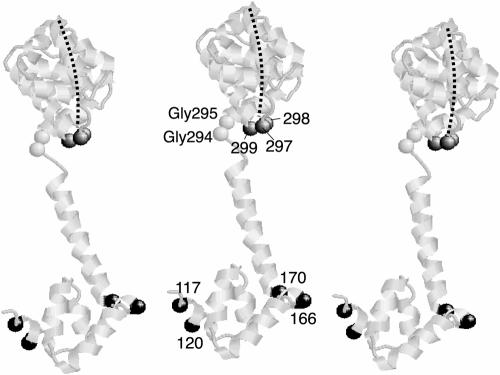
Because the high-yield cross-linking required assembled flagella and two Cys residues in FliGM, still occurred in flagella solubilized in detergent, and did not involve FliM, we conclude that the products are dimers and larger multimers of FliG formed by cross-linking between adjacent subunits within the flagellum. The positions in FliGM that gave the highest yields of FliG dimers and multimers (positions 117, 120, 166, and 170) are at opposite ends of the FliGM domain (cf. Fig. 8).
FIG. 8.
Model for the arrangement of FliG subunits. Three copies of the FliGMC protein from T. maritima are shown. The Gly-Gly motif and the positions giving high-yield cross-linking are indicated by space-filled alpha carbons and E. coli residue numbers. The FliGM domain is oriented with residues 117 and 120 of one subunit near residues 166 and 170 of the adjacent subunit. The FliGC domain is positioned with the active site ridge (indicated by the dashed line) toward the viewer and oriented in an approximately radial direction relative to the axis of the flagellum. The relative orientation of FliGM and FliGC domains is somewhat different from that in the crystal structure and was adjusted by making sterically allowed changes in torsion angles of the Gly-Gly linker. The three positions in FliGC that gave a cross-linked dimer with a moderate yield are also indicated. One of the residues in FliGC that gave a cross-linked dimer (residue 297 in E. coli; residue 299 in T. maritima) is at the end of the charge-bearing ridge and is among the charged residues important for function.
Cross-linking in FliGC.
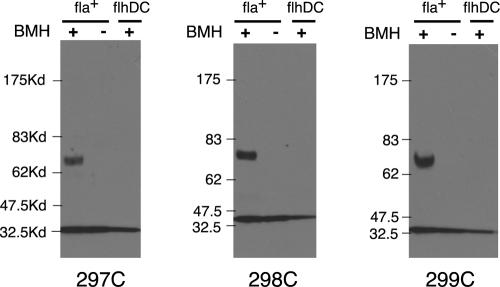
We next examined cross-linking of proteins with single or double Cys replacements in the C-terminal domain. The product yields for BMH cross-linking experiments with FliGC are summarized in Fig. 4B. Most of the Cys-containing proteins did not show measurable cross-linking with BMH. A moderate level of cross-linking was observed for the proteins with single Cys replacements at position 297, 298, or 299 and also for double mutants containing Cys at any of these positions. Residues 297, 298, and 299 are grouped at one end of the active site ridge. Cross-linking gave a single product, at the position expected for a FliG dimer (Fig. 7), and was again much stronger in the strain that assembles flagella than in the nonflagellated control strain. We conclude that the product is a FliG dimer formed by cross-linking between adjacent FliGC domains in the flagellum.
FIG. 7.
BMH-induced cross-linking of proteins with single Cys residues in FliGC. The positions of the Cys replacements are indicated at the bottom; double Cys mutants containing any of these replacements showed similar yields of cross-linking (see Fig. 4). The 297C, 298C, and 299C proteins gave similar dimer yields when cross-linking was induced by iodine (not shown). Kd, kilodaltons.
The results were similar when cross-linking was induced by iodine. Upon oxidation, the proteins with Cys residues at position 297, 298, or 299 cross-linked to form a FliG dimer with a moderate yield (data not shown). Other positions in FliGC gave little or no cross-linking. A small number of experiments were done using the bifunctional cross-linker DPDPB, which is longer than BMH. Even under conditions expected to maximize cross-linking (high concentration, long incubation time, room temperature) we observed only dimer formation, and only with Cys at position 297, 298, or 299 (data not shown).
Function of the proteins showing strong cross-linking.
To confirm that the proteins that showed high-yield cross-linking were not altered by the Cys replacements, we measured the swarming rates of fliG-null cells expressing the mutant proteins. All of the mutant proteins that exhibited cross-linking (117/166, 117/170, 120/166, 120/170, 297, 298, and 299) supported swarming at normal rates (data not shown).
DISCUSSION
Zhao and coworkers measured levels of FliG in purified flagellar basal bodies of Salmonella and estimated that the basal body contains around 40 molecules of FliG and a similar number of FliM molecules (47). En face electron micrographs of C rings showed a subunit structure with rotational symmetry that was most often 34-fold but varied somewhat (±2) between specimens (41, 46). Because FliM is thought to be in the part of the structure with ∼34-fold symmetry, the number of FliM molecules is probably 34 per basal body, which is close to the estimate of Zhao et al. (47). The MS ring/proximal rod is formed from the protein FliF (18, 43). The number of FliF subunits was estimated to be approximately 26, using both scanning transmission electron microscopy (35) and 35S labeling of basal body proteins (20). A recent electron microscopic reconstruction of the MS ring showed 26-fold symmetry (38). FliG is believed to be present at the same number of copies as FliF, because when FliF and FliG are fused genetically into a single protein, flagella are still assembled and can function well (13). The 26-fold symmetry of MS rings is retained and becomes somewhat more pronounced when the rings become associated with FliG (38). Thus, while the subunit number might vary slightly among specimens (46), a typical motor should have close to 26 copies of FliG.
Because the C ring has the appearance of an axially symmetric structure, we might expect its subunits to be arranged with axial symmetry. More complex arrangements involving local symmetry axes can be imagined, however, and are probably relevant in the case of FliN, which can form a stable tetramer in solution (9). The cross-linking results obtained with FliGM are most simply interpreted in terms of an axially symmetric array of subunits, oriented so that residues 117 and 120 of one subunit are near residues 166 and 170 of the adjacent subunit (Fig. 8). Within the FliGM domain, the residue 117/120 pair is separated from the 166/170 pair by about 2.8 nm (average of the four Cα-Cα distances). The alpha carbons of Cys residues in a disulfide bond are separated by about 0.5 nm, and so if disulfide cross-linking occurred with no movement of the subunits, the intersubunit spacing would be approximately 3.3 nm. For 26 subunits and a spacing of 3.3 nm, the subunits would fall on a ring with a diameter of 27 nm (3.3 × 26/π). This represents a lower boundary for the subunit spacing and ring diameter, because the subunits might have some freedom to move and cross-linking could trap them in positions away from equilibrium.
It has been difficult to locate FliG precisely in electron micrographs of flagellar basal bodies. The MS rings appear to be thicker in samples that contain FliG than in samples depleted of FliG (14, 40), which might indicate that part of the protein lies on the underside of the MS ring. The increased thickness would account for only a small part of the mass of FliG, however, so the bulk of FliG might be located somewhat apart from the MS ring. FliF has two transmembrane segments, a large periplasmic domain that forms much of the MS ring and the proximal rod, and a C-terminal domain consisting of about 90 residues located in the cytoplasm. FliG attaches to a fairly well conserved segment of about 15 residues near the C terminus of FliF (16). Other parts of the FliF C-terminal domain are less conserved and are predicted to be largely helical and possibly nonglobular (i.e., extended in conformation) in E. coli and its close relatives. This segment of FliF could form a linker that reaches out from the ordered part of the MS ring that is visible in micrographs to contact FliG at a greater radius. If the linker were relatively flexible, then FliG would remain largely disordered and invisible in micrographs until FliM and FliN joined the assembly and completed the C ring.
In an effort to locate FliG, Thomas et al. (40) examined the structures of basal bodies containing FliF-FliG fusion proteins, including a fusion protein that lacks a substantial portion of the FliG amino-terminal domain. Careful comparison of wild-type and fusion mutant basal bodies indicated that the amino-terminal domain of FliG might lie at a radius of 16.5 nm. This would require an intersubunit spacing of 4 nm for the FliGM domains if the motor contains 26 FliG molecules and the FliGM domains are evenly spaced. This is somewhat greater than the estimate given above based on immobile subunits, but it is within the range of the cross-linking results if we allow for some movement.
Because FliGC interacts with MotA (49) and MotA is located in the membrane surrounding the MS ring (21), we think that FliGC is likely to lie at a greater radius than FliGM. Electron microscopic reconstructions revealed two concentrations of electron density in the upper (membrane-proximal) part of the C ring, at radii of about 17 nm and 22 nm (14, 40). These two features might correspond to the FliGM and FliGC domains, respectively. Alternatively, FliGC might correspond to the inner feature (at a radius of 17 nm), with FliGM attached to the MS ring at an even smaller radius. These alternative arrangements for FliG are illustrated in Fig. 9C and D.
FIG. 9.
Model for the arrangement of FliG subunits in the flagellum. (A) Overall arrangement of 25 FliG subunits, viewed along the axis of rotation. The FliGM domains are at an inner position to allow interaction with FliF, while the FliGC domains are at a larger radius to allow interaction with the stator complexes. The ridge bearing the conserved charged residues is oriented radially. The positions in FliGM that yielded cross-linked dimers and multimers are indicated in the enlarged view by black circles, and the region in FliGC where Cys residues gave cross-linked dimers is indicated by gray circles. (B) Hypothesized small rotations of FliGC that could bring together positions 297, 298, and 299 in adjacent subunits. (C) Image of the flagellar basal body obtained from electron microscopic reconstructions (40). The outlines indicate the features sketched in panel D. (D) Two possibilities for the location of FliG relative to features seen in the electron microscopic reconstruction.
Previously, we suggested that the ridge on FliGC that bears the functionally important charged residues might be oriented parallel to the edge of the rotor, so that charged residues on the ridge could interact successively with charged groups on the stator as the rotor turns. If the ridge were oriented in this way, then adjacent subunits would approach each other most closely through positions at opposite ends of the ridge, and we would expect the highest yield of cross-linking in mutants with Cys residues at both ends of the ridge. This was not observed. Instead, cross-linking occurred in fair yield for proteins with single Cys replacements at one end of the ridge and was not significantly greater in proteins with Cys at the other end also. Cross-linking between the single-Cys proteins implies that equivalent positions in adjacent subunits can approach close enough to form a disulfide cross-link. This would require some movement of the domains or a more drastic rearrangement such as local unfolding. The required movement would be especially large if the ridge were oriented parallel to the edge of the rotor, as it would then involve the rotation of one or both domains by more than 90°. An approximately radial orientation of the ridge appears more likely, because small rotations of the domain would then be sufficient to bring together the positions that showed cross-linking, particularly if the residue 297/298/299 end of the ridge were pointed inward (toward the axis of the flagellum [Fig. 9]).
If the charged ridge is oriented radially, then the shorter dimension of the domain would be directed toward the adjacent subunit, and we would predict a sizable gap between the domains. The gap between adjacent FliGC domains could be anywhere in the range from 2 nm to 3.3 nm, depending upon whether FliG is in the outer or inner location discussed above and what symmetry FliGC adopts (even if 26 copies of FliGC are present, it might locally adopt the 34-fold symmetry of the C ring). This relatively large spacing could account for the absence of any strong cross-linking between proteins with Cys replacements on the sides of the domain, even with the long reagents BMH and DPDPB. Thus, while the FliGC domains appear to be able to undergo rotational movements that bring the 297/298/299 ends of adjacent domains into proximity, they are constrained from bringing their sides together. Motor switching seems likely to involve movements of FliGC, and we are presently examining switch mutants to determine whether cross-linking yields are affected by mutations that alter motor bias or rates of switching.
Acknowledgments
This work was supported by grant 8R01-EB2041 from the National Institutes of Biomedical Imaging and Bioengineering. B.J.L. received partial support from NIH training grant 5T32-GM08537. M.D. was supported by a grant from the Biosciences Undergraduate Research Program at the University of Utah. The Protein-DNA core facility at the University of Utah receives support from the National Cancer Institute (grant 5P30 CA42014).
We thank Matthew Borrowman, Karen Jordan, Monica Longtin, Kate Sadler, Akiko Tsunoda, and Christina M. Yong for assistance with cross-linking experiments and swarming assays.
REFERENCES
- 1.Armitage, J. P., and R. Schmitt. 1997. Bacterial chemotaxis: Rhodobacter sphaeroides and Sinorhizobium meliloti—variations on a theme? Microbiology 143:3671-3682. [DOI] [PubMed] [Google Scholar]
- 2.Berg, H. C., and R. A. Anderson. 1973. Bacteria swim by rotating their flagellar filaments. Nature 245:380-382. [DOI] [PubMed] [Google Scholar]
- 3.Berry, R. M., and H. C. Berg. 1999. Torque generated by the flagellar motor of Escherichia coli while driven backward. Biophys. J. 76:580-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blair, D. F., and H. C. Berg. 1990. The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell 60:439-449. [DOI] [PubMed] [Google Scholar]
- 5.Blair, D. F., and H. C. Berg. 1988. Restoration of torque in defective flagellar motors. Science 242:1678-1681. [DOI] [PubMed] [Google Scholar]
- 6.Block, S. M., and H. C. Berg. 1984. Successive incorporation of force-generating units in the bacterial rotary motor. Nature 309:470-472. [DOI] [PubMed] [Google Scholar]
- 7.Bren, A., and M. Eisenbach. 1998. The N terminus of the flagellar switch protein, FliM, is the binding domain for the chemotactic response regulator, CheY. J. Mol. Biol. 278:507-514. [DOI] [PubMed] [Google Scholar]
- 8.Brown, P. N., C. P. Hill, and D. F. Blair. 2002. Crystal structure of the middle and C-terminal domains of the flagellar rotor protein FliG. EMBO J. 21:3225-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, P. N., M. A. A. Mathews, L. A. Joss, C. P. Hill, and D. F. Blair. 2005. Crystal structure of the flagellar rotor protein FliN from Thermotoga maritima. J. Bacteriol. 187:2890-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chun, S. Y., and J. S. Parkinson. 1988. Bacterial motility: membrane topology of the Escherichia coli MotB protein. Science 239:276-278. [DOI] [PubMed] [Google Scholar]
- 11.Dean, G. E., R. M. Macnab, J. Stader, P. Matsumura, and C. Burke. 1984. Gene sequence and predicted amino-acid sequence of the MotA protein, a membrane-associated protein required for flagellar rotation in Escherichia coli. J. Bacteriol. 143:991-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeMot, R., and J. Vanderleyden. 1994. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both Gram-positive and Gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol. Microbiol. 12:333-334. [DOI] [PubMed] [Google Scholar]
- 13.Francis, N. R., V. M. Irikura, S. Yamaguchi, D. J. DeRosier, and R. M. Macnab. 1992. Localization of the Salmonella typhimurium flagellar switch protein FliG to the cytoplasmic M-ring face of the basal body. Proc. Natl. Acad. Sci. USA 89:6304-6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Francis, N. R., G. E. Sosinsky, D. Thomas, and D. J. DeRosier. 1994. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J. Mol. Biol. 235:1261-1270. [DOI] [PubMed] [Google Scholar]
- 15.Garza, A. G., L. W. Harris-Haller, R. A. Stoebner, and M. D. Manson. 1995. Motility protein interactions in the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA 92:1970-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grunenfelder, B., S. Gehrig, and U. Jenal. 2003. Role of the cytoplasmic C terminus of the FliF motor protein in flagellar assembly and rotation. J. Bacteriol. 185:1624-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirota, N., M. Kitada, and Y. Imae. 1981. Flagellar motors of alkalophilic Bacillus are powered by an electrochemical potential gradient of Na+. FEBS Lett. 132:278-280. [Google Scholar]
- 18.Homma, M., Y. Komeda, T. Iino, and R. M. Macnab. 1987. The flaFIX gene product of Salmonella typhimurium is a flagellar basal body component with a signal peptide for export. J. Bacteriol. 169:1493-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Irikura, V. M., M. Kihara, S. Yamaguchi, H. Sockett, and R. M. Macnab. 1993. Salmonella typhimurium fliG and fliN mutations causing defects in assembly, rotation, and switching of the flagellar motor. J. Bacteriol. 175:802-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones, C. J., R. M. Macnab, H. Okino, and S.-I. Aizawa. 1990. Stoichiometric analysis of the flagellar hook-(basal body) complex of Salmonella typhimurium. J. Mol. Biol. 212:377-387. [DOI] [PubMed] [Google Scholar]
- 21.Khan, S., M. Dapice, and T. S. Reese. 1988. Effects of mot gene expression on the structure of the flagellar motor. J. Mol. Biol. 202:575-584. [DOI] [PubMed] [Google Scholar]
- 22.Kihara, M., G. U. Miller, and R. M. Macnab. 2000. Deletion analysis of the flagellar switch protein FliG of Salmonella. J. Bacteriol. 182:3022-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kojima, S., and D. F. Blair. 2001. Conformational change in the stator of the bacterial flagellar motor. Biochemistry 40:13041-13050. [DOI] [PubMed] [Google Scholar]
- 24.Kojima, S., and D. F. Blair. 2004. Solubilization and purification of the MotA/MotB complex of Escherichia coli. Biochemistry 43:26-34. [DOI] [PubMed] [Google Scholar]
- 25.Larsen, S. H., J. Adler, J. J. Gargus, and R. W. Hogg. 1974. Chemomechanical coupling without ATP: the source of energy for motility and chemotaxis in bacteria. Proc. Natl. Acad. Sci. USA 71:1239-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larsen, S. H., R. W. Reader, E. N. Kort, W.-W. Tso, and J. Adler. 1974. Change in direction of flagellar rotation is the basis of the chemotactic response in E. coli. Nature 249:74-77. [DOI] [PubMed] [Google Scholar]
- 27.Lloyd, S. A., and D. F. Blair. 1997. Charged residues of the rotor protein FliG essential for torque generation in the flagellar motor of Escherichia coli. J. Mol. Biol. 266:733-744. [DOI] [PubMed] [Google Scholar]
- 28.Lloyd, S. A., H. Tang, X. Wang, S. Billings, and D. F. Blair. 1996. Torque generation in the flagellar motor of Escherichia coli: evidence of a direct role for FliG but not for FliM or FliN. J. Bacteriol. 178:223-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lloyd, S. A., F. G. Whitby, D. F. Blair, and C. P. Hill. 1999. Structure of the C-terminal domain of FliG, a component of the rotor in the bacterial flagellar motor. Nature 400:472-475. [DOI] [PubMed] [Google Scholar]
- 30.Mathews, M. A. A., H. L. Tang, and D. F. Blair. 1998. Domain analysis of the FliM protein of Escherichia coli. J. Bacteriol. 180:5580-5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oosawa, K., T. Ueno, and S.-I. Aizawa. 1994. Overproduction of the bacterial flagellar switch proteins and their interactions with the MS ring complex in vitro. J. Bacteriol. 176:3683-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sato, K., and M. Homma. 2000. Functional reconstitution of the Na+-driven polar flagellar motor component of Vibrio alginolyticus. J. Biol. Chem. 275:5718-5722. [DOI] [PubMed] [Google Scholar]
- 33.Silverman, M., and M. Simon. 1976. Operon controlling motility and chemotaxis in E. coli. Nature 264:577-580. [DOI] [PubMed] [Google Scholar]
- 34.Sockett, H., S. Yamaguchi, M. Kihara, V. M. Irikura, and R. M. Macnab. 1992. Molecular analysis of the flagellar switch protein FliM of Salmonella typhimurium. J. Bacteriol. 174:793-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sosinsky, G. E., N. R. Francis, D. J. DeRosier, J. S. Wall, M. N. Simon, and J. Hainfeld. 1992. Mass determination and estimation of subunit stoichiometry of the bacterial hook-basal body flagellar complex of Salmonella typhimurium by scanning transmission electron microscopy. Proc. Natl. Acad. Sci. USA 89:4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stader, J., P. Matsumura, D. Vacante, G. E. Dean, and R. M. Macnab. 1986. Nucleotide sequence of the Escherichia coli motB gene and site-limited incorporation of its product into the cytoplasmic membrane. J. Bacteriol. 166:244-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stolz, B., and H. C. Berg. 1991. Evidence for interactions between MotA and MotB, torque-generating elements of the flagellar motor of Escherichia coli. J. Bacteriol. 173:7033-7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki, H., K. Yonekura, and K. Namba. 2004. Structure of the rotor of the bacterial flagellar motor revealed by electron cryo-microscopy and single-particle image analysis. J. Mol. Biol. 337:105-113. [DOI] [PubMed] [Google Scholar]
- 39.Tang, H., S. Billings, X. Wang, L. Sharp, and D. F. Blair. 1995. Regulated underexpression and overexpression of the FliN protein of Escherichia coli and evidence for an interaction between FliN and FliM in the flagellar motor. J. Bacteriol. 177:3496-3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas, D., D. G. Morgan, and D. J. DeRosier. 2001. Structures of bacterial flagellar motors from two FliF-FliG gene fusion mutants. J. Bacteriol. 183:6404-6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas, D. R., D. G. Morgan, and D. J. DeRosier. 1999. Rotational symmetry of the C ring and a mechanism for the flagellar rotary motor. Proc. Natl. Acad. Sci. USA 96:10134-10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toker, A. S., and R. M. Macnab. 1997. Distinct regions of bacterial flagellar switch protein FliM interact with FliG, FliN and CheY. J. Mol. Biol. 273:623-634. [DOI] [PubMed] [Google Scholar]
- 43.Ueno, T., K. Oosawa, and S.-I. Aizawa. 1992. M ring, S ring and proximal rod of the flagellar basal body of Salmonella typhimurium are composed of subunits of a single protein, FliF. J. Mol. Biol. 227:672-677. [DOI] [PubMed] [Google Scholar]
- 44.Vogler, A. P., M. Homma, V. M. Irikura, and R. M. Macnab. 1991. Salmonella typhimurium mutants defective in flagellar filament regrowth and sequence similarity of FliI to F0F1, vacuolar, and archaebacterial ATPase subunits. J. Bacteriol. 173:3564-3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamaguchi, S., S.-I. Aizawa, M. Kihara, M. Isomura, C. J. Jones, and R. M. Macnab. 1986. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J. Bacteriol. 168:1172-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Young, H. S., H. Dang, Y. Lai, D. J. DeRosier, and S. Khan. 2003. Variable symmetry in Salmonella typhimurium flagellar motors. Biophys. J. 84:571-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao, R., N. Pathak, H. Jaffe, T. S. Reese, and S. Khan. 1996. FliN is a major structural protein of the C-ring in the Salmonella typhimurium flagellar basal body. J. Mol. Biol. 261:195-208. [DOI] [PubMed] [Google Scholar]
- 48.Zhou, J., and D. F. Blair. 1997. Residues of the cytoplasmic domain of MotA essential for torque generation in the bacterial flagellar motor. J. Mol. Biol. 273:428-439. [DOI] [PubMed] [Google Scholar]
- 49.Zhou, J., S. A. Lloyd, and D. F. Blair. 1998. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA 95:6436-6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou, J., L. L. Sharp, H. L. Tang, S. A. Lloyd, S. Billings, T. F. Braun, and D. F. Blair. 1998. Function of protonatable residues in the flagellar motor of Escherichia coli: a critical role for Asp 32 of MotB. J. Bacteriol. 180:2729-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]