Abstract
Mycoplasma mobile glides on a glass surface in the direction of its tapered end by an unknown mechanism. Two large proteins, Gli349 and Gli521, were recently reported to be involved in glass binding and force generation/transmission, respectively, in M. mobile gliding. These proteins are coded tandemly with two other open reading frames (ORFs) in the order p123-gli349-gli521-p42 on the genome. In the present study, reverse transcriptase PCR analysis suggested that these four ORFs are transcribed cistronically. To characterize the p123 gene coding a 123-kDa protein (Gli123) of 1,128 amino acids, we raised polyclonal antibody against the Gli123 protein. Immunoblotting for Gli123 revealed that Gli123 was missing in a mutant strain, m12, which was previously isolated and characterized by a deficiency in glass binding. Sequencing analysis showed a nonsense mutation at the 523rd amino acid of the protein in the m12 mutant. Immunofluorescence microscopy with the polyclonal antibody showed that Gli123 is localized at the head-like protrusion's base, the cell neck, which is specialized for gliding, as observed for Gli349 and Gli521. Localization of the gliding proteins, Gli349 and Gli521, was disturbed in the m12 mutant, suggesting that Gli123 is essential for the positioning of gliding proteins in the cell neck.
Mycoplasmas are commensal and occasionally parasitic bacteria with small genome sizes that lack a peptidoglycan layer (27). Several mycoplasma species form membrane protrusions such as the head-like structure in Mycoplasma mobile and the attachment organelle in M. pneumoniae. These species exhibit gliding motility, the movement of cells over surfaces in the direction of the protrusion, which is believed to be involved in the pathogenicity of mycoplasmas (4, 14, 21). Mycoplasmas have no flagella or pili on their surface and no genes related to known bacterial motility in the genome (29, 36). In addition, no homologs of motor proteins that are common in eukaryotic motility have been found (6-8, 12, 26, 31).
M. mobile, isolated from the gills of a freshwater fish in the early 1980s, is the fastest-gliding mycoplasma (15, 28). It glides smoothly and continuously on glass at an average speed of 2.0 to 4.5 μm/s or 3 to 7 times the length of the cell per s, exerting a force up to 27 piconewtons (pN) (9, 23, 28). We have previously identified two large proteins, Gli349 (a 349-kDa protein) (37) and Gli521 (a 521-kDa protein) (35), involved in the gliding mechanism of M. mobile as determined on the basis of analyses of gliding-defective mutants (25) and inhibitory antibodies (18, 35). Analysis of the inhibitory effects of the anti-Gli349 and anti-Gli521 antibodies on gliding mycoplasmas has suggested that Gli349 and Gli521 are responsible for hemadsorption and glass binding and for force generation and/or transmission, respectively (35, 37). Gli349 and Gli521 are localized exclusively at the base of a head-like structure, designated the “neck,” specialized in gliding and binding (18, 24, 37). Rapid-freeze and freeze fracture rotary-shadow electron microscopy visualization has shown that many spike-like structures 50 nm in length stick out around the neck and are bound to the glass surface with their distal ends (22). The spike seems to be composed of a Gli349 molecule and to function as a “leg” in the gliding mechanism, as its subcellular localization and apparent volume agree with those of Gli349, and the spike has not been found in a nonbinding mutant (22). Depletion of cellular ATP by the addition of arsenate reduces both ATP concentrations and gliding speed, suggesting that the gliding is driven by the energy of ATP (11). These observations lead us to the assumption that cells are propelled by spikes composed of Gli349 repeatedly binding to and releasing from the glass, driven by the force exerted from or through the Gli521 molecule based on the energy of ATP hydrolysis (21). This assumption is likely applicable to the gliding motility of other mycoplasma species with slower gliding speeds, as they appear to glide in a similar manner (5, 13), and the antibodies against the binding proteins show common effects in gliding (33).
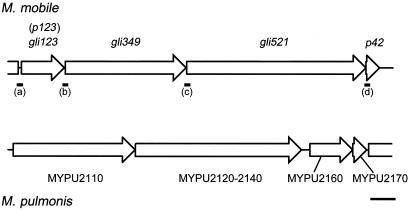
In the genome of M. mobile, the gli349 and gli521 genes are ordered tandemly with two other open reading frames (ORFs) (Fig. 1) as p123-gli349-gli521-p42, suggesting that the four ORFs are working as an operon (12, 35, 37). This gene arrangement is mostly conserved in another gliding mycoplasma, M. pulmonis, a mouse pathogen. In M. pulmonis, MYPU-2110, 2120 to 2140, and 2160 form a cluster in this order, corresponding to gli349, gli521, and p123, respectively (Fig. 1). Our recent sequencing analyses of M. pulmonis showed that gap regions between ORFs 2120 and 2130 and ORFs 2130 and 2140 are actually fused into one large ORF in the standard strain of M. pulmonis (ATCC19612) (35). p123 of M. mobile codes for a polypeptide of 1,128 amino acids with a predicted molecular mass of 123,316 Da and an isoelectric point of 5.06. The primary structure of the gene product is in part similar to that of the ORF MYPU-2160, i.e., 24% identity and 40% similarity were found in the region spanning amino acid residues 857 to 1126 of the p123 gene of M. mobile. The P123 protein has no cysteine residues, no significant similarity to other protein sequences, and no motifs, as observed for Gli349 and Gli521 (19, 35, 37). Using SOSUI, an algorithm to predict a membrane-spanning segment (10), a transmembrane segment preceded by a positively charged region can be predicted near the N terminus, as observed in Gli349 and Gli521 (35, 37). These observations suggest that P123 is also related to the gliding mechanism.
FIG. 1.
Organization of genes encoding gliding proteins in the M. mobile genome and the corresponding region of M. pulmonis. DNA regions analyzed by RT-PCR are indicated by solid bars marked (a) to (d). Scale bar, 2 kb.
In this study, we examined the p123 gene products and a nonbinding mutant truncated for this protein and concluded that this protein participates in the gliding mechanism of M. mobile.
MATERIALS AND METHODS
Strains and culture conditions.
M. mobile strain 163K (ATCC 43663) and its mutants (25) were grown at 25°C in Aluotto medium (2, 25) to an optical density at 600 nm of approximately 0.07, which corresponds to 7 × 108 CFU/ml.
RT-PCR.
The total RNA was obtained with a RNeasy Mini kit (QIAGEN, Hilden, Germany) according to the manufacturer's instruction, except for the lysozyme treatment. Reverse transcriptase PCRs (RT-PCRs) were performed with Ready-To-Go RT-PCR beads (GE Healthcare) after the total RNA was treated by DNaseI. A random primer of 6 nucleotides was used as the first-strand primer, and subsequent PCR was carried out using the following primer sets for the corresponding gap regions presented in Fig. 1: (a), 5′-GTGGTACCAATGTACATTCC-3′ and 5′-GCTTGATTCTTCTGAATTTGC-3′; (b), 5′-GACACCAACATCTGGAACTG-3′ and 5′-GGTAAAGCATTTGGACTTAG-3′; (c), 5′-TCAATCAATTCATTCCCTCC-3′ and 5′-TTTCTCATAACTTCCTGCTG-3′; and (d), 5′-GAACAATCAAATCCTGGAGC-3′ and 5′-GAGCAATGTCATTGGATTTTG-3′. The control reactions were performed with reverse transcriptase inactivated at 95°C for 10 min to ensure that there was no contaminating DNA in the RNA samples assayed. The reaction products were analyzed by 1% acrylamide gel electrophoresis and sequenced by using the former primer of each set, except for gap region (c), for which another primer, 5′-AATGTTAGTGGCGATCTATC-3′, neighboring the former primer on the genome was used.
Immunoblot analysis.
Three primer sets (N-terminal, 5′-CCCCCCATGGGACCAATTATTGCAGTAGGA-3′ and 5′-CCCCCTCGAGTCTCACAGATTCATTAGCAC-3′; middle, 5′-CCCCCCATGGGTGCTAATGAATCTGTGAGA-3′ and 5′-CCCCCTCGAGCCCTGTTAAATATCTTGCTC-3′; and C-terminal, 5′-CCCCCCATGGTTAAGCTACCTGGTCTTATT-3′ and 5′-CCCCCTCGAGTTCTACAGTATTGCCTTTTT-3′) were used to amplify parts of the gli123 gene, comprising 12 to 421, 415 to 744, and 717 to 1,116 amino acids, respectively (see Fig. 3). The chromosomal DNA of M. mobile was obtained by use of a Genomic-tip system (QIAGEN). The amplified DNA fragments were inserted into an expression vector, pET-30a (EMD Biosciences, Darmstadt, Germany). Their protein products were expressed in Escherichia coli BL21(DE3) and purified under denaturing conditions with an Ni-nitrilotriacetic acid (NTA) column. The polyclonal antibody was produced as described previously (18, 30) by immunizing female DDY mice with a 40-μg antigen for each animal.
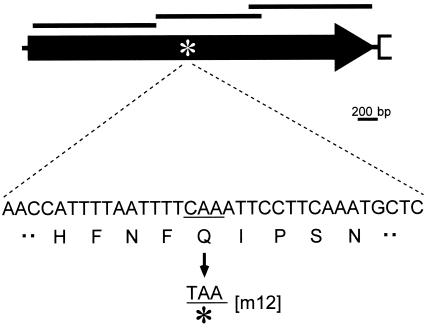
FIG. 3.
Schematic and mutation site of gli123 (p123). The gli123 gene is represented by a solid arrow. The regions corresponding to the recombinants are presented by the top solid bars. The mutation site in the m12 mutant is indicated by an asterisk. A partial sequence around the mutation site is indicated under the asterisk. A triplet of CAA coding glutamic acid is mutated to a TAA nonsense codon in the m12 mutant.
Quantification of gliding proteins.
The gliding proteins were quantified from a Coomassie brilliant blue (CBB)-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel. Mycoplasma cells of the wild-type and mutant strains were collected by centrifugation at 12,000 × g for 10 min at 4°C. The cells were washed twice with phosphate-buffered saline consisting of 75 mM Na-phosphate (pH 7.3) and 68 mM NaCl, lysed, and subjected to SDS-PAGE. The protein amount of the lysate was adjusted from the optical density at 600 nm of the culture. The band intensity was measured by NIH Image for gel images scanned by a transparent scanner (GT9800F; Epson, Nagano, Japan). The linear relationship between the band intensity and protein amount was confirmed by measuring the intensities of the bands of different amounts of lysate.
DNA sequencing of the mutated gene.
A 3.6-kb DNA fragment containing the gli123 gene was amplified by PCR with primers 5′-GACCAATTATTGCAGTAGGA-3′ and 5′-GTGTATGTACTTTTGATTGC-3′ and then sequenced using internal primer positioning at approximately 500-nucleotide intervals.
Immunofluorescence microscopy.
Cultured cells were bound to cleaned glass and stained for the gliding proteins as previously described (18, 35, 37). Amounts of 100- and 10-fold dilutions were used, respectively, for the anti-Gli123 antiserum and the hybridoma medium containing monoclonal antibody against Gli349 or Gli521. The remaining procedures were carried out as previously described (32, 34, 37).
RESULTS
Four ORFs in the gene cluster were transcribed cistronically.
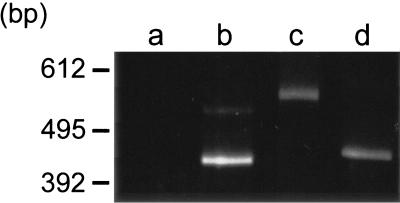
To determine the continuity of transcripts of the four ORFs, we conducted RT-PCR analysis for the four ORF gaps presented in Fig. 1. In the gel electrophoresis, RT-PCR products were found near the positions of expected sizes for gaps b to d but not for gap a (Fig. 2). The mobility of the DNA product for gap c in the gel was slower than expected. However, sequencing analysis showed that the expected sequences were amplified for gap c, as observed for gaps b and d. Presumably, the product for gap c tends to form special three-dimensional structures. These results indicate that at least some transcripts continue over gaps b to d, suggesting that these ORFs are working in an operon.
FIG. 2.
RT-PCR analysis of transcripts of gene clusters including gliding genes. The DNA regions covering the ORF gaps a to d were analyzed by RT-PCR. The expected lengths of the PCR products are 484, 455, 475, and 457 bp for regions a, b, c, and d, respectively. The DNA lengths shown by a marker are indicated on the left.
Gli123 is missing in the nongliding mutant m12.
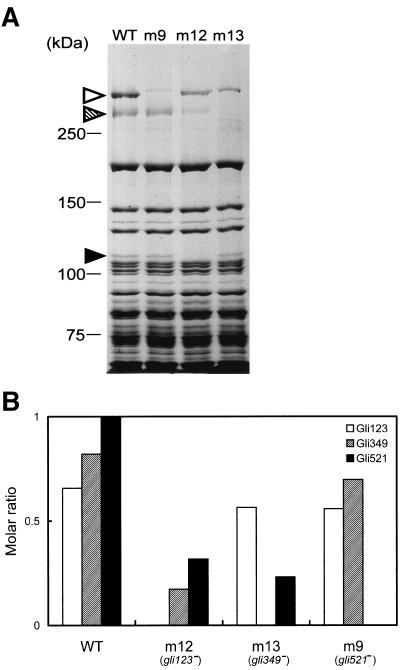
To characterize the protein product of p123 gene, we raised antisera against recombinant proteins of N-terminal, middle, and C-terminal portions of this ORF, comprising amino acids 12 to 421, 415 to 744, and 717 to 1116, respectively (Fig. 3). All of the antibodies recognized a single protein band at approximately the 125-kDa position, the expected size, in the whole-cell lysate of the wild-type strain in immunoblot analysis, as presented for an antibody against the middle part in Fig. 4. These results indicate that these antibodies recognize the product of p123. The existence of P123 was examined for the previously isolated ten gliding mutants, m6, m9, m12, m13, m14, m23, m26, m27, m29, and m34 (25). P123 was not detected only in the m12 mutant, which is characterized by a deficiency in glass binding and consequently in gliding (Fig. 4). In a CBB-stained SDS-PAGE gel image, a protein band was found in the wild-type strain at the corresponding position but not in mutant m12 (Fig. 5A), suggesting that this band is derived from p123. DNA sequencing of the genome from mutant m12 showed a nonsense mutation in the p123 gene at the position of the 523rd amino acid residue (Fig. 3); more specifically, a CAA codon in the wild-type strain is mutated to a TAA nonsense codon in mutant m12. We therefore renamed this ORF gli123. The antibodies raised against the N-terminal and middle portions were expected to react with the truncated peptide of Gli123 band in the m12 mutant. However, the truncated peptide band was not detected, although the antisera against the N-terminal and middle portions of Gli123 detected this protein in the wild-type strain, suggesting that the truncated peptide is unstable or untranslated.
FIG. 4.
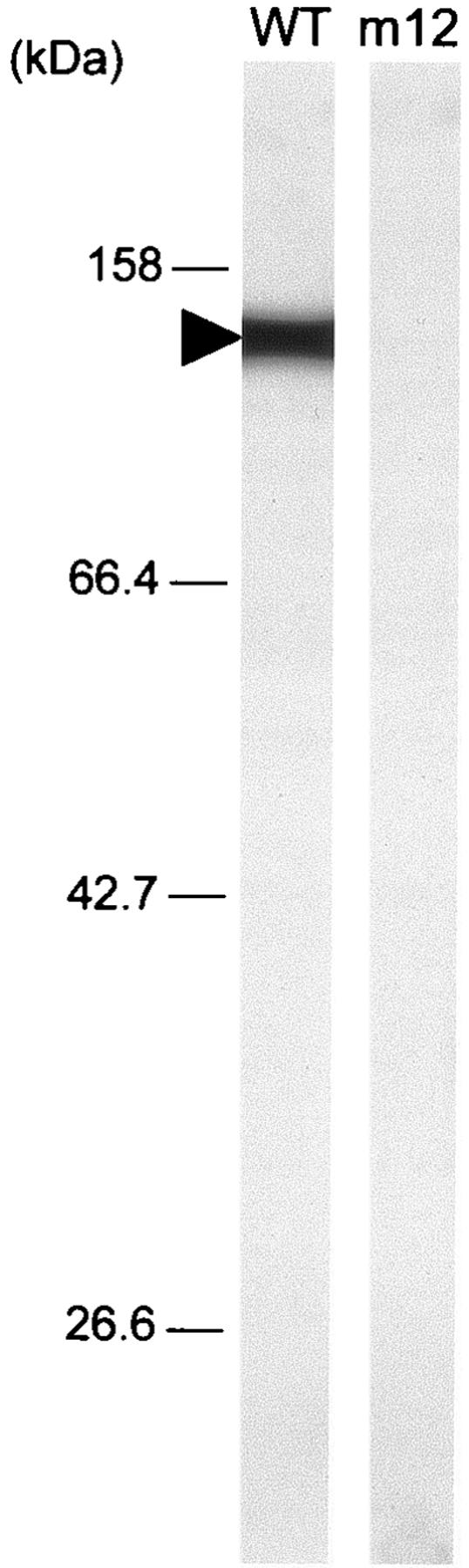
Immunoblotting of wild-type and m12 mutant strains for Gli123. Whole-cell lysate of each strain was subjected to immunoblotting following SDS-10% PAGE with an anti-Gli123 polyclonal antiserum against the middle portion. The molecular mass is indicated on the left. The blotting signal is indicated by a solid triangle.
FIG. 5.
Expression of gliding proteins in wild-type and gliding mutant strains. (A) Protein profiles of the wild-type and gliding mutants. Whole-cell lysates were subjected to SDS-5.5% PAGE and stained by CBB. The mutant strains m9, m12, and m13 have a nonsense mutation in the gli521, gli123, and gli349 genes, respectively. The protein bands of Gli521, Gli349, and Gli123 are indicated by open, hatched, and solid triangles, respectively. The molecular mass is indicated on the left. (B) The molar ratio of gliding proteins in the wild-type and mutant strains. The intensity of each gliding protein band of CBB-stained SDS-PAGE gel was measured, divided by the molecular mass of each protein, and normalized by the value of Gli521 in the wild-type strain. The analyzed strains are indicated on the bottom.
Relative amounts of gliding proteins in the gliding mutants.
The molar ratios of gliding proteins were estimated for the wild-type and mutant strains by dividing the band intensity of the CBB-stained gel by the molecular weights (Fig. 5B). The ratios of Gli123, Gli349, and Gli521 in the wild-type strain were estimated as 0.65, 0.82, and 1, respectively. The ratio of Gli123 was reduced only slightly to 86% and 85% of that of the wild-type level in m13 and m9 mutants, respectively. The ratio of Gli521 was significantly reduced to 32% and 23% of that of the wild-type level in m12 and m13 mutants, respectively. The ratio of Gli349 was reduced to 85% and 21% of that of the wild-type level in m9 and m12 mutants, respectively. In the lane of m9 of the gel image (Fig. 5A), another band can be seen at a similar position with Gli521. This band did not react with the anti-Gli521 antibodies, indicating that it was derived from another ORF (35). Thus, the band intensity of Gli521 was estimated after subtracting the intensity of the band observed in m9 from that of Gli521 in the other strains.
Subcellular localization of Gli123.
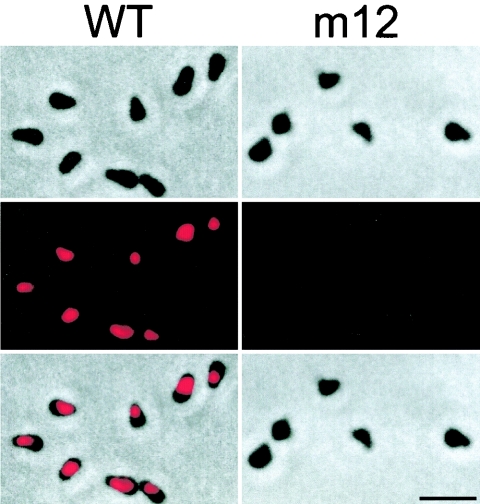
The localization of Gli123 in the wild-type cells was examined by immunofluorescence microscopy with antisera against a middle portion of Gli123 (Fig. 6, left panels). The results indicate that Gli123 is located at the cell neck, which is specialized for gliding (18, 24, 37). No signal was detected for Gli123 when the m12 mutant was probed (Fig. 6, right panels). Gli123 was not detected by using antisera against N- or C-terminal portions of this protein (data not shown).
FIG. 6.
Subcellular localization of Gli123 as examined by immunofluorescence microscopy. The upper, middle, and lower panels show phase-contrast, fluorescent, and merged images, respectively. The left and right panels show the images of the wild-type and mutant m12 strains, respectively. Bar, 2 μm.
Subcellular localization of gliding proteins in gliding mutants.
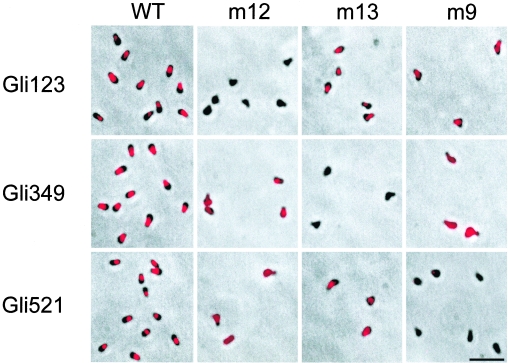
Gli123 localizes at the cell neck, a position similar to that seen in Gli349 and Gli521 (35, 37), with a comparable protein amount in the wild-type strain. These observations suggest relationships among these three proteins, possibly including physical interactions. To address this possibility, the localization dependency of these proteins was examined by immunofluorescence microscopy of the gliding mutants truncated for these proteins (Fig. 7) (Table 1). In the wild-type cells, Gli123, Gli349, and Gli521 localized similarly at the cell neck. In mutant m12 with truncated Gli123, no signal was detected for Gli123, as shown in Fig. 6. The signals of Gli349 and Gli521 in this mutant were intense enough for detection, but their positions were significantly disturbed. In mutant m13, with truncated Gli349, both Gli123 and Gli521 were focused at the cell neck, while no signals were detected for Gli349, as previously observed (37). Mutant m9, with a truncated Gli521 protein, showed no signal for Gli521, as previously reported, consistent with the finding that the monoclonal antibody recognizes the C-terminal side region of the nonsense mutation (35). In this mutant, the signals of Gli123 were found as foci at the cell neck, while those of Gli349 were distributed over the cell.
FIG. 7.
Subcellular localization of gliding proteins in gliding mutants. Fluorescence signal merged with phase-contrast images are shown. The strain is indicated on the top of each column. The mutant strains m12, m13, and m9 have a mutation in the gli123, gli349, and gli521 genes, respectively. The gliding protein examined for localization is indicated on the left of each panel. Bar, 2 μm.
TABLE 1.
Summary of fluorescent signals for gliding proteins in the mutantsa
| Protein | Result for strainb:
|
|||
|---|---|---|---|---|
| WT | m12 (gli123 mutant) | m13 (gli349 mutant) | m9 (gli521 mutant) | |
| Gli123 | ||||
| Signal intensity | ++ | − | ++ | ++ |
| Ordinal signal localization | ++ | ND | + | + |
| Gli349 | ||||
| Signal intensity | ++ | + | − | + |
| Ordinal signal localization | ++ | ± | ND | − |
| Gli521 | ||||
| Signal intensity | ++ | + | ++ | − |
| Ordinal signal localization | ++ | − | + | ND |
Results for indicated strains and proteins correspond to those shown in Fig. 7.
Signal intensities: ++, highest; +, high; ±, moderate; −, lowest. Ordinal signal localization: ++, well focused into neck; +, localized to neck; ±, partially localized to neck; −, distributed through whole cell body. ND, not determined. WT, wild-type.
DISCUSSION
We focused on a 123-kDa protein of M. mobile, which is encoded upstream of two gliding genes, gli349 and gli521, (35, 37), and concluded, based on the following reasons, that this protein, named Gli123, also plays a role in the gliding mechanism: (i) the gli123 gene is likely to be coded in the same operon with those previously identified genes involved in the gliding mechanism; (ii) Gli123 is missing in the nonbinding and nongliding mutant, m12, due to a nonsense mutation; (iii) Gli123 is localized at the cell neck specialized for the gliding mechanism; (iv) Gli123 exists in cells at levels comparable to those of other gliding proteins; (v) the subcellular localization of other gliding proteins is disturbed in the m12 mutant; (vi) the levels of the other gliding proteins, Gli349 and Gli521, are reduced in the m12 mutant.
Immunofluorescence microscopy using the antibodies against three different portions of Gli123 showed that only antibodies against the middle portion can bind to Gli123 molecules in a cell, although the other antibodies can also detect the Gli123 protein in immunoblotting (Fig. 6). This observation suggests that the N- and C-terminal portions of Gli123 are occluded in the gliding machinery embedded in a cell.
The antibody against the middle portion, unlike the antibodies against Gli349 and Gli521, does not appear to affect gliding (35, 37), although the anti-Gli123 antibody also can bind to the molecule from the outside of a cell. This observation may suggest that the participation of Gli123 in the gliding mechanism does not involve a conformational change during gliding, because inhibition by the antibody is thought to be caused by blocking the conformational change. Alternatively, Gli123 executes such a conformational change in a portion not accessible to the antibodies of the antisera used.
We examined the localization of gliding proteins in the wild-type and mutant strains (Fig. 7). All gliding proteins, Gli123, Gli349, and Gli521, were localized at the cell neck in the wild-type cells. The localization of Gli349 and Gli521 was found to be disturbed in mutant m12, which is defective in Gli123. This is a contrast to the localization of Gli123 and Gli521, which is not disturbed in m13, a mutant defective in Gli349. Similarly, the Gli123 localization was not disturbed by the depletion of Gli349 or Gli521, as shown by the results obtained with mutants m13 or m9, respectively, although the localization of Gli349 was much disturbed in mutants m9 and m12. These observations suggest that Gli123 is dominant in the localization to the other gliding proteins. Then which of the other two proteins is dominant in these two? Gli349 localization to the cell neck was lost in mutant m9, but that of Gli521 was slightly affected in mutant m13. This finding suggests that the positioning of Gli349 depends on Gli521, while that of Gli521 does not depend on Gli349. This positioning hierarchy, Gli123-Gli521-Gli349, may suggest physical interactions among these three proteins and may explain why all gliding mutants exhibit similar phenotypes, i.e., nonbinding and nongliding. The molar ratio of gliding proteins in the cells was estimated at near 1:1:1 for Gli123, Gli349, and Gli521 (Fig. 5), suggesting that these proteins form a complex with this molar ratio. The gliding proteins are predicted to have a transmembrane segment at a terminal position and to primarily be exposed outside of the cells, as predicted from the accessibility of antibodies (18, 35, 37). These common predicted topologies for these three proteins may also support the presence of interactions among them.
Quantification of gliding proteins in the wild-type and mutant strains suggested a hierarchy of the amounts of these proteins similar to that of localization (Fig. 5). The amounts of Gli349 and Gli521 were reduced in mutant m12, which is defective in Gli123. This may be explained by the instability of these proteins caused by the failure in their localization to the proper sites. However, the relationship between Gli349 and Gli521 may be more complicated. The amount of Gli521 was reduced in mutant m13, which is defective in Gli349, but that of Gli349 was not reduced in m9, which is defective in Gli521, inconsistent with the hierarchy of protein localization, Gli123-Gli521-Gli349. Another possible explanation for the hierarchy in the amount of gliding protein is the translational coupling, where the 30S subunit of the ribosome just after the first translation can easily find the Shine-Dalgarno site of the following ORF (1). This assumption can explain the results showing that the amount of Gli521 was affected by the truncation of Gli123 or Gli349 and that of Gli349 was affected by the truncation of Gli123.
The truncated peptide of Gli123 failed to be detected, suggesting the instability of this peptide (Fig. 4). Such instability was also observed for the truncated peptide of Gli349 in mutant m13 (37). This may be explained by the unfolded structure or the improper localization of these peptides.
The hierarchy of protein components has been reported for the gliding and adhesion machinery, the so-called “attachment organelle” of M. pneumoniae (16, 17, 20, 21). HMW1 and HMW2 proteins included in this organelle should function from the initial stage of organelle formation, because the loss of either protein results in the failure of formation, causing irregular localization of the other six components and their instability (3, 32, 34, 38). Although no sequence similarities have been found between Gli123 and HMW1 or HMW2, these proteins may have common roles in the formation of gliding machinery.
In the present study, we identified a novel protein involved in the gliding machinery of M. mobile, which would be another clue to understanding the unknown mechanism of mycoplasma gliding.
Acknowledgments
This work was supported in part by Grants-in-Aid for Scientific Research (C) and Science Research on Priority Areas (“Genome Science” and “Infection and host response”) from the Ministry of Education, Science, Sports, Culture, and Technology to M.M.
REFERENCES
- 1.Adhin, M. R., and J. van Duin. 1990. Scanning model for translational reinitiation in eubacteria. J. Mol. Biol. 213:811-818. [DOI] [PubMed] [Google Scholar]
- 2.Aluotto, B. B., R. G. Wittler, C. O. Williams, and J. E. Faber. 1970. Standardized bacteriologic techniques for the characterization of mycoplasma species. Int. J. Syst. Bacteriol. 20:35-58. [Google Scholar]
- 3.Balish, M. F., S. M. Ross, M. Fisseha, and D. C. Krause. 2003. Deletion analysis identifies key functional domains of the cytadherence-associated protein HMW2 of Mycoplasma pneumoniae. Mol. Microbiol. 50:1507-1516. [DOI] [PubMed] [Google Scholar]
- 4.Bredt, W. 1979. Motility, p. 141-145. In M. F. Barile, S. Razin, J. G. Tully, and R. F. Whitcomb (ed.), The mycoplasmas, vol. 1. Academic Press, New York, N.Y. [Google Scholar]
- 5.Bredt, W. 1968. Motility and multiplication of Mycoplasma pneumoniae. A phase contrast study. Pathol. Microbiol. (Basel) 32:321-326. [DOI] [PubMed] [Google Scholar]
- 6.Chambaud, I., R. Heilig, S. Ferris, V. Barbe, D. Samson, F. Galisson, I. Moszer, K. Dybvig, H. Wroblewski, A. Viari, E. P. Rocha, and A. Blanchard. 2001. The complete genome sequence of the murine respiratory pathogen Mycoplasma pulmonis. Nucleic. Acids Res. 29:2145-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fraser, C. M., J. D. Gocayne, O. White, M. D. Adams, R. A. Clayton, R. D. Fleischmann, C. J. Bult, A. R. Kerlavage, G. Sutton, J. M. Kelley, R. D. Fritchman, J. F. Weidman, K. V. Small, M. Sandusky, J. Fuhrmann, D. Nguyen, R. Utterback, D. M. Saudek, C. A. Phillips, J. M. Merrick, J.-F. Tomb, B. A. Dougherty, K. F. Bott, P.-C. Hu, T. S. Lucier, S. N. Peterson, H. O. Smith, C. A. Hutchison III, and J. C. Venter. 1995. The minimal gene complement of Mycoplasma genitalium. Science. 270:397-403. [DOI] [PubMed] [Google Scholar]
- 8.Himmelreich, R., H. Hilbert, H. Plagens, E. Pirkl, B.-C. Li, and R. Herrmann. 1996. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 24:4420-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiratsuka, Y., M. Miyata, and T. Q. P. Uyeda. 2005. Living microtransporter by uni-directional gliding of Mycoplasma along microtracks. Biochem. Biophys. Res. Commun. 331:318-324. [DOI] [PubMed] [Google Scholar]
- 10.Hirokawa, T., B. C. Seah, and S. Mitaku. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378-379. [DOI] [PubMed] [Google Scholar]
- 11.Jaffe, J. D., M. Miyata, and H. C. Berg. 2004. Energetics of gliding motility in Mycoplasma mobile. J. Bacteriol. 186:4254-4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaffe, J. D., N. Stange-Thomann, C. Smith, D. DeCaprio, S. Fisher, J. Butler, S. Calvo, T. Elkins, M. G. FitzGerald, N. Hafez, C. D. Kodira, J. Major, S. Wang, J. Wilkinson, R. Nicol, C. Nusbaum, B. Birren, H. C. Berg, and G. M. Church. 2004. The complete genome and proteome of Mycoplasma mobile. Genome Res. 14:1447-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kenri, T., S. Seto, A. Horino, Y. Sasaki, T. Sasaki, and M. Miyata. 2004. Use of fluorescent-protein tagging to determine the subcellular localization of Mycoplasma pneumoniae proteins encoded by the cytadherence regulatory locus. J. Bacteriol. 186:6944-6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirchhoff, H. 1992. Motility, p. 289-306. In J. Maniloff, R. N. McElhaney, L. R. Finch, and J. B. Baseman (ed.), Mycoplasmas—molecular biology and pathogenesis. American Society for Microbiology, Washington, D.C.
- 15.Kirchhoff, H., R. Rosengarten, W. Lotz, M. Fischer, and D. Lopatta. 1984. Flask-shaped mycoplasmas: properties and pathogenicity for man and animals. Israel J. Med. Sci. 20:848-853. [PubMed] [Google Scholar]
- 16.Krause, D. C., and M. F. Balish. 2004. Cellular engineering in a minimal microbe: structure and assembly of the terminal organelle of Mycoplasma pneumoniae. Mol. Microbiol. 51:917-924. [DOI] [PubMed] [Google Scholar]
- 17.Krause, D. C., and M. F. Balish. 2001. Structure, function, and assembly of the terminal organelle of Mycoplasma pneumoniae. FEMS Microbiol. Lett. 198:1-7. [DOI] [PubMed] [Google Scholar]
- 18.Kusumoto, A., S. Seto, J. D. Jaffe, and M. Miyata. 2004. Cell surface differentiation of Mycoplasma mobile visualized by surface protein localization. Microbiology 150:4001-4008. [DOI] [PubMed] [Google Scholar]
- 19.Metsugi, S., A. Uenoyama, J. Adan-Kubo, M. Miyata, K. Yura, H. Kono, and N. Go. 2005. Sequence analysis of the gliding protein Gli349 in Mycoplasma mobile. Biophysics 1:33-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyata, M. 2002. Cell division, p. 117-130. In R. Herrmann and S. Razin (ed.), Molecular biology and pathogenicity of mycoplasmas. Kluwer Academic/Plenum Publishers, London, United Kingdom.
- 21.Miyata, M. 2005. Gliding motility of mycoplasmas—the mechanism cannot be explained by current biology, p. 137-163. In A. Blanchard and G. Browning (ed.), Mycoplasmas: pathogenesis, molecular biology, and emerging strategies for control. Horizon Scientific Press, Norwich, United Kingdom.
- 22.Miyata, M., and J. Petersen. 2004. Spike structure at interface between gliding Mycoplasma mobile cell and glass surface visualized by rapid-freeze and fracture electron microscopy. J. Bacteriol. 186:4382-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyata, M., W. S. Ryu, and H. C. Berg. 2002. Force and velocity of Mycoplasma mobile gliding. J. Bacteriol. 184:1827-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyata, M., and A. Uenoyama. 2002. Movement on the cell surface of gliding bacterium, Mycoplasma mobile, is limited to its head-like structure. FEMS Microbiol. Lett. 215:285-289. [DOI] [PubMed] [Google Scholar]
- 25.Miyata, M., H. Yamamoto, T. Shimizu, A. Uenoyama, C. Citti, and R. Rosengarten. 2000. Gliding mutants of Mycoplasma mobile: relationships between motility and cell morphology, cell adhesion and microcolony formation. Microbiology 146:1311-1320. [DOI] [PubMed] [Google Scholar]
- 26.Papazisi, L., T. S. Gorton, G. Kutish, P. F. Markham, G. F. Browning, D. K. Nguyen, S. Swartzell, A. Madan, G. Mahairas, and S. J. Geary. 2003. The complete genome sequence of the avian pathogen Mycoplasma gallisepticum strain R(low). Microbiology 149:2307-2316. [DOI] [PubMed] [Google Scholar]
- 27.Razin, S., D. Yogev, and Y. Naot. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62:1094-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosengarten, R., and H. Kirchhoff. 1987. Gliding motility of Mycoplasma sp. nov. strain 163K. J. Bacteriol. 169:1891-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosengarten, R., and H. Kirchhoff. 1989. Growth morphology of Mycoplasma mobile 163K on solid surfaces: reproduction, aggregation, and microcolony formation. Curr. Microbiol. 18:15-22. [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Sasaki, Y., J. Ishikawa, A. Yamashita, K. Oshima, T. Kenri, K. Furuya, C. Yoshino, A. Horino, T. Shiba, T. Sasaki, and M. Hattori. 2002. The complete genomic sequence of Mycoplasma penetrans, an intracellular bacterial pathogen in humans. Nucleic Acids Res. 30:5293-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seto, S., G. Layh-Schmitt, T. Kenri, and M. Miyata. 2001. Visualization of the attachment organelle and cytadherence proteins of Mycoplasma pneumoniae by immunofluorescence microscopy. J. Bacteriol. 183:1621-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seto, S., T. Kenri, T. Tomiyama, and M. Miyata. 2005. Involvement of P1 adhesin in gliding motility of Mycoplasma pneumoniae as revealed by the inhibitory effects of antibody under optimized gliding conditions. J. Bacteriol. 187:1875-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seto, S., and M. Miyata. 2003. Attachment organelle formation represented by localization of cytadherence protein and formation of electron-dense core in the wild-type and mutant strains of Mycoplasma pneumoniae. J. Bacteriol. 185:1082-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seto, S., A. Uenoyama, and M. Miyata. 2005. Identification of 521-kilodalton protein (Gli521) involved in force generation or force transmission for Mycoplasma mobile gliding. J. Bacteriol. 187:3502-3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimizu, T., and M. Miyata. 2002. Electron microscopic studies of three gliding mycoplasmas, Mycoplasma mobile, M. pneumoniae, and M. gallisepticum, by using the freeze-substitution technique. Curr. Microbiol. 44:431-434. [DOI] [PubMed] [Google Scholar]
- 37.Uenoyama, A., A. Kusumoto, and M. Miyata. 2004. Identification of a 349-kilodalton protein (Gli349) responsible for cytadherence and glass binding during gliding of Mycoplasma mobile. J. Bacteriol. 186:1537-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willby, M. J., M. F. Balish, S. M. Ross, K. K. Lee, J. L. Jordan, and D. C. Krause. 2004. HMW1 is required for stability and localization of HMW2 to the attachment organelle of Mycoplasma pneumoniae. J. Bacteriol. 186:8221-8228. [DOI] [PMC free article] [PubMed] [Google Scholar]