Abstract
The pnhA gene of Pasteurella multocida encodes PnhA, which is a member of the Nudix hydrolase subfamily of dinucleoside oligophosphate pyrophosphatases. PnhA hydrolyzes diadenosine tetra-, penta-, and hexaphosphates with a preference for diadenosine pentaphosphate, from which it forms ATP and ADP. PnhA requires a divalent metal cation, Mg2+ or Mn2+, and prefers an alkaline pH of 8 for optimal activity. A P. multocida strain that lacked a functional pnhA gene, ACP13, was constructed to further characterize the function of PnhA. The cellular size of ACP13 was found to be 60% less than that of wild-type P. multocida, but the growth rate of ACP13 and its sensitivity to heat shock conditions were similar to those of the wild type, and the wild-type cell size was restored in the presence of a functional pnhA gene. Wild-type and ACP13 strains were tested for virulence by using the chicken embryo lethality model, and ACP13 was found to be up to 1,000-fold less virulent than the wild-type strain. This is the first study to use an animal model in assessing the virulence of a bacterial strain that lacked a dinucleoside oligophosphate pyrophosphatase and suggests that the pyrophosphatase PnhA, catalyzing the hydrolysis of diadenosine pentaphosphates, may also play a role in facilitating P. multocida pathogenicity in the host.
The Nudix (nucleoside diphosphate linked to another moiety, X) hydrolase family of proteins consists of approximately 800 members found in more than 200 prokaryotic and eukaryotic species (6, 49). The hydrolases may be divided into subfamilies based on the substrate specificity: dinucleoside oligophosphate pyrophosphatases, NADH, ADP-ribose, nucleotide sugars, or ribo- and deoxyribonucleoside triphosphates (19). Catalytic activity is located within the Nudix motif, which has a consensus sequence of GX5EX7REUXEEXGU, where X represents any amino acid and U represents Ile, Leu, or Val.
Enzymatic activity of Nudix hydrolases has been well established (7, 14, 16); however, how this activity relates to cellular function is less understood. Recently, much attention has been given to the dinucleoside oligophosphate pyrophosphatase subfamily of Nudix hydrolase, whose members catalyze the hydrolysis of dinucleoside oligophosphates (tetra-, penta-, and hexaphosphates). Cellular dinucleoside oligophosphates are by-products of the reactions of aminoacyl-tRNA synthetases (12, 45) and are usually present in small amounts in both prokaryotic and eukaryotic cells (36). Many suggestions have been put forth as to the function of these molecules within the cell. Reports indicate that the dinucleoside oligophosphates are signaling molecules that can increase more than 100-fold when cells are exposed to conditions such as heat shock and oxidative stress, and this increase can lead to cellular toxicity (8, 32). The role of the Nudix hydrolase is to remove the excess amounts of these molecules to restore cellular balance. In Escherichia coli, an increased concentration of dinucleoside oligophosphates, such as adenosine (5′)-tetraphospho-(5′)-adenosine (Ap4A), is proposed to be a signal for cell division (38, 39) and to be involved in the stress responses by modulating protein refolding by chaperone proteins (21, 30).
The dinucleoside oligophosphate pyrophosphatases of some bacteria have been associated with invasion of host cells. The pyrophosphatase of Bartonella bacilliformis, IalA, has been linked to this organism's ability to invade human erythrocytes; expression of IalA within noninvasive E. coli renders it invasive (37). The orthologous YgdP protein of E. coli K1 has been strongly associated with this organism's ability to invade human brain microvascular endothelial cells (5). Recent studies of the pyrophosphatase YgdP of Salmonella enterica serovar Typhimurium have demonstrated this protein's involvement in adherence and invasion of mammalian cells (28). In addition, the Rickettsia prowazekii dinucleoside oligophosphate pyrophosphatase gene, invA, was shown to have increased expression during the early stages of host cell infection (23). How these pyrophosphatases are associated with bacterial invasion is not yet clear. However, they may play a role in enhancing the intracellular survival of the invading bacteria by controlling the toxic levels of dinucleoside oligophosphates, which can accumulate during invasion of the host cell (7, 14, 16, 36, 27). In this study we have identified the gene pnhA, which encodes the dinucleoside oligophosphate pyrophosphatase of the bacterial pathogen Pasteurella multocida.
Pasteurella multocida is a facultative gram-negative pathogen that is the causative agent of a wide range of diseases in both wild and domestic animals. These include atrophic rhinitis in swine, hemorrhagic septicemia and pneumonic pasteurellosis in cattle and buffalo, and fowl cholera in most avian species (13, 18, 26, 40). Fowl cholera is a highly infectious disease which affects both wild and domesticated birds and results in a heavy environmental toll as well as significant economic losses to the poultry industry (26, 41). Investigation of P. multocida pathogenesis has been challenging for researchers. However, a number of P. multocida virulence factors have been identified, including PMT toxin in strains causing atrophic rhinitis (20), capsule in serogroups A and B (11, 15), several iron acquisition proteins, such as hemoglobin-binding protein, TonB, ExbD, and ExbB (9, 10, 22), and outer membrane proteins, such as PmOmpA (17). In this study we describe the expression of pnhA, as well as the purification and the enzymatic properties of the PnhA protein. We also present for the first time the use of an animal model to study the possible role of dinucleoside oligophosphate pyrophosphatase in bacterial pathogens.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. All E. coli strains were cultured on LB medium at 35°C, and all P. multocida strains were grown on heart infusion (HI) medium at 37°C. Medium was supplemented with the appropriate antibiotics when necessary. Standard growth conditions were used to generate a growth curve of the P. multocida wild-type and mutant strains. Bacterial strains were made electrocompetent (29) and were transformed with plasmids via electroporation using the Gene Pulser apparatus (Bio-Rad, Hercules, CA) set at 1.8 kV, 200 Ω, and 25 μF.
TABLE 1.
Strains and plasmids used in this study
| Strain | Description | Source or reference |
|---|---|---|
| E. coli strains | ||
| BL21 (DE3) | F−ompT hsdSB (rB− mB−) gal dcm (DE3) (Cmr) | Novagen |
| JM109 | endA1 recA1 gyrA96 thi hsdR17 (rK− mK+) relA1 supE44 Δ(lac-proAB) [F′ traD36 proAB laqIqZΔM15] | Promega |
| ACP1008 | JM109 containing pACP1005 | This study |
| ACP1009 | BL21(DE3) containing pACP1005 and pGroESL | This study |
| P. multocida strains | ||
| X-73 | Fowl cholera-causing, wild-type strain; serogroup A, serotype 1 | 42 |
| ACP13 | ΔpnhA::tet(M) (Tetr) | This study |
| ACP17 | ΔpnhA::tet(M) containing pACP1007 | This study |
| Plasmids | ||
| pAL99 | P. multocida-E. coli shuttle vector containing promoter region of P. multocida gene tpiA (Knr) | B. Adlera |
| pET30b | Expression vector (Knr) | Novagen |
| pGroESL | Plasmid containing groEL and groES genes and the T7 lac promoter (Cmr) | 16 |
| pVB101 | pBR322 containing tet(M) gene from Tn916 (Apr Tetr) | 11 |
| pACP1000 | pWSK29 containing entire coding region of pnhA and promoter region (Apr) | This study |
| pACP1002 | pACP1000 containing pnhA::tet(M) mutagenesis cassette (Apr Tetr) | This study |
| pACP1005 | pET30b containing promoterless pnhA gene (Knr) | This study |
| pACP1007 | pAL99 containing pnhA gene from pACP1005 (Knr) | This study |
Kind gift from Ben Adler, Department of Microbiology, Monash University, Victoria, Australia.
DNA manipulation, analysis, and RT-PCR.
Standard PCR, DNA ligation, restriction analysis, and gel electrophoresis were carried out as described by Ausubel et al. (4). Standard automated DNA sequencing was performed at the University of Wisconsin—Madison Biotechnology Center. Expression of both the pnhA and pnhB genes in wild-type and mutant strains was determined via RT-PCR under standard conditions (4).
Cloning of the pnhA gene.
The coding region of the pnhA gene, which contained a translational stop, but without the promoter region, was amplified via PCR using P. multocida genomic DNA, the 5′ primer (CRp064: 5′-GATTAAAGGACATATGATGATCGATTTCGATGGC-3′) with an engineered NdeI site and the 3′ primer (CRp065: 5′-GAAAAAAGGTGAAGCTTCTCAACCCCTTG-3′) with a HindIII engineered site. The PCR-amplified product was digested with NdeI and HindIII and ligated into the corresponding sites of the pET30b vector (Novagen, San Diego, CA). This permitted the cloning of the pnhA gene upstream of the T7 promoter and downstream of the T7 transcriptional terminator. The resultant construct, pACP1005, was used to transform E. coli JM109 cells (ACP1008) for storage and E. coli BL21(DE3) cells containing the pGroESL plasmid (16) for expression (ACP1009). Purified pACP1005 DNA was sequenced using a T7 primer and primer CRp065 to confirm that the cloned PCR product was the pnhA gene.
Expression of pnhA and purification of PnhA.
One colony of the PnhA expression strain, ACP1009, was inoculated into 40 ml of LB medium containing 30 μg/ml of kanamycin and 50 μg/ml of chloramphenicol and cultured overnight at 37°C. This culture was then used to inoculate 2 liters of LB containing 30 μg/ml of kanamycin and 50 μg/ml of chloramphenicol. After reaching an optical density at 600 nm of 0.3, the culture was transferred to a 22°C incubator and grown to an A600 of 0.8. The cells were then induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside, incubated for an additional 12 h, harvested, and washed in buffered saline solution. The washed cell pellet was resuspended in 2.5 volumes of buffer A (50 mM Tris-Cl, pH 7.5, 1 mM EDTA, 0.1 mM dithiothreitol), disrupted with a French pressure cell (16,000 lb/in2), and centrifuged to remove cellular debris. The supernatant, containing the expressed PnhA protein (fraction I), was adjusted to approximately 10 mg/ml with buffer A, and 10% streptomycin solution was added slowly to a final concentration of 1.5% to precipitate the protein. The precipitate, containing PnhA (fraction II), was collected by centrifugation, resuspended in buffer A containing 1 M ammonium sulfate, applied to a DEAE-Sepharose column, and eluted with buffer A. Fractions containing the PnhA protein were brought to 70% saturation with ammonium sulfate, and the precipitate (fraction III) was collected and dissolved in a minimal volume of buffer A. Fraction III was applied to a Sephadex G-100 gel filtration column equilibrated with buffer A containing 200 mM sodium chloride and eluted with the same buffer. In the early purification steps, enzyme assays and sodium dodecyl sulfate-polyacrylamide gel electrophoresis were used to identify the PnhA protein within the fractions. Final fractions identified spectrometrically, A280, were visualized by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and those containing the purified PnhA protein were pooled (fraction V) and stored under −80°C or used in the enzyme assay.
Enzyme assay.
A standard reaction mixture contained, in a 50-μl volume, 50 mM Tris-Cl, pH 8.0, 5 mM Mg2+, 2 mM substrate, 0.5 unit of yeast inorganic pyrophosphatase for phosphatase-sensitive substrates, such as nucleoside triphosphates, or 4 units of alkaline phosphatase for phosphatase-resistant substrates, and 0.1 to 2 milliunits of enzyme. After 15 min at 37°C, the reaction was terminated by adding 250 μl 4 mM EDTA solution and the liberated inorganic orthophosphate was assayed by the colorimetric procedure of Ames and Dubin (3). One unit of enzyme catalyzes the hydrolysis of one μmol of substrate per min under these conditions.
Construction of pnhA mutagenesis cassettes.
Construct pACP1000 (Table 1), which contained the entire coding region of the pnhA gene, was used to construct the pnhA mutagenesis cassette. A 3.2-kb BamHI fragment, which contained the tetracycline M gene, tet(M), was excised from the plasmid pVB101 and was blunt ended using DNA polymerase I large (Klenow) fragment (Promega). The modified fragment was then ligated to a ScaI site of pACP1000, which was located within the coding region of the pnhA gene to generate pACP1002. pACP1002 contained the mutagenesis cassette, which comprised the pnhA gene, which was insertionally inactivated by the tet(M) gene.
Construction of P. multocida pnhA mutant.
The cloned mutagenesis cassette was further amplified by PCR, using pACP1002 as a template and using a 5′ primer, CRp012 (5′-GGCTACCGTCCGAATGTAGG-3′), and a 3′ primer, CRp009 (5′-GACAAGACCGCCACCGATACC-3′). The resulting PCR product was a 3.8-kb fragment that contained the tet(M) gene and 156 bp of the pnhA gene at the 5′ end of tet(M) and 445 bp of the pnhA gene at the at the 3′ end of tet(M). The product was end filled using DNA polymerase I large (Klenow) fragment (Promega), and the product was then ligated to create circularized molecules. Approximately 3 μg of the circularized mutagenesis cassette was used to transform wild-type P. multocida by electroporation (29), and the transformants were selected on HI solid medium containing 12.5 μg/ml of tetracycline. This allelic exchange method generated the pnhA mutant ACP13. The presence of the correct double-crossover event within the ACP13 chromosome was confirmed by PCR and Southern blot analysis.
Complementation of the P. multocida pnhA mutant, ACP13.
For complementation of the ACP13 mutant strain, the pnhA gene from pACP1005 was excised and inserted into the P. multocida-E. coli shuttle vector pAL99. This ensured that the gene was under to control of the P. multocida tpiA promoter. The resultant construct, pACP1007, was used to transform ACP13 cells. Transformants were selected on HI medium containing 50 μg/ml kanamycin and 12.5 μg/ml tetracycline. A kanamycin- and tetracycline-resistant transformant, which contained pACP1007, was then designated ACP17.
Metabolic phenotype and heat shock stress response.
The abilities of the wild-type P. multocida, ACP13, and ACP17 strains to metabolize a variety of different carbohydrates were determined by using the API 50 CH system (bioMérieux, France). This system tests assimilation, oxidation, and fermentation of 49 carbohydrate sources. In addition, 19 different enzyme activities of wild-type P. multocida, ACP13, and ACP17 were also examined using the API ZYM kit (bioMérieux). To assess the affect of heat shock conditions, 1 ml of the wild-type P. multocida, ACP13, or ACP17 strain was transferred from 37°C to 42°C and 45°C and 50°C, and cell samples were taken at different time points from 0 to 30 min. Viable counts were performed before and after exposure.
Embryo lethality assay.
The test was performed on embryonated specific-pathogen-free chicken eggs (Charles River Laboratories, Wilmington, MA), and the method followed was based on the method described by Wooley et al. (48). The eggs were incubated at 37.7°C for 11 days and were turned each day. When the eggs were 12 days old, they were candled and marked in an area that was relatively free of large blood vessels for inoculation. One ml of overnight culture (108 CFU) of either the P. multocida wild-type strain, ACP13, or ACP17 was washed in 0.85% saline, centrifuged, and then resuspended in 1 ml 0.85% saline. Groups of four eggs were used per bacterial strain per dilution. The site at which inoculation occurred was swabbed with iodine, a 1-mm hole was then punched into the shell, and the area was swabbed with iodine again. Using a syringe and a 25G 5/8-inch needle, a 200-μl sample of bacteria was inoculated directly into the allantoic sac of the egg. The hole was sealed with Elmer's glue, and the inoculated eggs were incubated at 37.7°C and candled daily to assess viability over 72 h. Eggs deemed to be on the onset of morbidity were euthanized. Onset of morbidity was marked by the loss of vascular structure within the egg and hemorrhaging of the embryo (visualized via candling). Upon completion of the experiment, autopsies were performed and tissue samples from various parts of the embryo were taken, suspended in 0.85% saline, and plated onto HI media with or without tetracycline to confirm the presence or absence of bacterial cells. DNA from bacterial cells recovered from the embryo lethality experiments was used as a template in PCRs, with appropriate primers to confirm the presence or absence of the tet(M) or pnhA gene.
Electron microscopy.
Bacterial strains grown overnight at 37°C in liquid medium were washed in phosphate-buffered saline, centrifuged, and resuspended again in phosphate-buffered saline. Washed cells were allowed to adhere to a formvar-coated copper grid (300 mesh) for 10 s and then negatively stained with 0.5% phosphotungstic acid solution for 10 s. A Zeiss model EM 10 transmission electron microscope (TEM) was used to visualize the cells.
Nucleotide sequence accession number.
The GenBank accession number for pnhA is AAR14276.
RESULTS
Characterization of the pnhA gene.
Plasmid pACP1000 contained a 596-bp open reading frame whose deduced amino acid sequence shared strong similarity to those of the dinucleoside oligophosphate pyrophosphatase subfamily of Nudix hydrolases, including YgdP and IalA (7, 14, 16). The open reading frame was subsequently named pnhA (P. multocida Nudix hydrolase) and was identical to the published PM0082 sequence (35). The pnhA gene was found to be located within an operon along with only one other gene, pnhB, and RT-PCR revealed that both genes are cotranscribed in X-73 (data not shown). The protein product of the pnhB gene was predicted by the PSORT program (version WWW; Human Genome Center, University of Tokyo [http://psort.nibb.ac.jp/form.html]) to contain eight transmembrane domains and was shown to have similarity to hypothetical bacterial permeases. The putative protein encoded by pnhA, PnhA, had a predicted size of 23 kDa and contained a Nudix motif located near the N terminus of the protein sequence (Fig. 1), which is characteristic of all dinucleoside oligophosphate pyrophosphatases. Bessman and colleagues (7) observed that in addition to the Nudix motif, pyrophosphatases, such as YgdP of E. coli and IalA, contained amino acid residues adjacent to and within the motif that are highly conserved. PnhA, along with other pyrophosphatases from bacterial pathogens, also contained the conserved residues within the Nudix motif region (Fig. 1, shaded sequence).
FIG. 1.
Alignment of dinucleoside oligophosphate pyrophosphatases from various bacterial pathogens. Only the Nudix motif (shown in bold) and the highly conserved sequence (shaded) are shown for each of the dinucleoside oligophosphate pyrophosphatases. Asterisks indicate identical amino acid residues, and ≫ indicates proteins that have been identified as dinucleoside oligophosphate pyrophosphatases. All of the above sequences were obtained via BLASTP (2) and by using the PnhA amino acid sequence as the query sequence.
Expression and purification of PnhA.
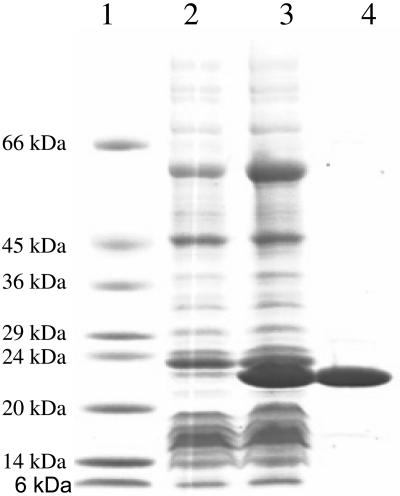
The PnhA protein expressed well but was insoluble in the E. coli hosts (ACP1008) under standard conditions and was thus unsuitable for purification and enzymatic studies. Accordingly, pnhA was expressed at a reduced temperature in ACP1009, which coproduced the chaperonins, GroEL and GroES. As was observed previously with the invasion-associated enzyme from Bartonella bacilliformis (16), the PnhA protein expressed well, as evidenced by the appearance of a major 23-kDa band in the crude extract of induced cells (Fig. 2, lane 3), and was readily soluble. Figure 2 shows the appearance of a protein banding at approximately 23 kDa in the induced culture and also the purified protein (fraction V) used for the subsequent enzymatic studies.
FIG. 2.
Expression and purification of the PnhA Nudix hydrolase protein. Shown is a gradient polyacrylamide gel (4 to 20%) containing 1% sodium dodecyl sulfate, stained with Coomassie brilliant blue. Lane 1 is protein standards with the indicated molecular masses; lane 2 contains approximately 10 μg of crude extract of uninduced cells of ACP1009; lane 3 contains approximately 20 μg of crude extract of isopropyl-β-d-thiogalactopyranoside-induced ACP1009; lane 4 contains approximately the same number of enzyme units of the purified PnhA protein (from fraction V) as in lane 3.
Properties of the PnhA protein.
The YgdP (7) and IalA (16) enzymes catalyze the hydrolysis of diadenosine oligophosphates with a preference for Ap5A and Ap4A, respectively. Accordingly, a number of diadenosine oligophosphates were tested, and as with E. coli YgdP (7) and R. prowazekii InvA (24), PnhA preferred Ap5A, although Ap4A and Ap6A were readily hydrolyzed (Table 2). As also seen with IalA, YgdP, and InvA, PnhA showed little activity toward Ap3A. Furthermore, little or no activity was observed toward other Nudix hydrolase substrates, such as ADP-ribose, NADH, UDP-glucose, GDP-mannose, FAD, or (deoxy)nucleoside triphosphates.
TABLE 2.
Relative specificities of Nudix hydrolases
| Substrated | Relative specificitya (%) of:
|
||||
|---|---|---|---|---|---|
| PnhA | YgdPc | IalAc | InvAc | NudAc | |
| Ap5A | 100b | 100 | 77 | 100 | 34 |
| Ap6A | 43.5 | 92 | 67 | 66 | 45 |
| Ap4A | 13.5 | 14 | 100 | 5 | 100 |
| Ap3A | <1 | <1 | <1 | <1 | 7 |
| GDP-mannose | <0.5 | <1 | <1 | <1 | <1 |
| ADP-ribose | <0.5 | <1 | <1 | 4 | 0 |
| UDP-glucose | <0.5 | NDe | <1 | ND | ND |
| NADH | <0.5 | <1 | <1 | <1 | 0 |
| FAD | <0.5 | ND | ND | ND | ND |
Relative specificities were expressed as the percentage of activity of each enzyme, with the preferred substrate set at 100.
Substrates were present at a concentration of 1 mM or 2 mM in the standard assay.
The data for E. coli YgdP, B. bacilliformis IalA, R. prowazekii InvA, and H. pylori NudA were taken from references 7, 16, 24, and 34, respectively, and included for comparison.
Ap5A, adenosine (5′)-pentaphospho-(5′)-adenosine; Ap6A, adenosine (5′)-hexaphospho-(5′)-adenosine, Ap4A, adenosine (5′)-tetraphospho-(5′)-adenosine; Ap3A, adenosine (5′)-triphospho-(5′)-adenosine.
ND, not determined.
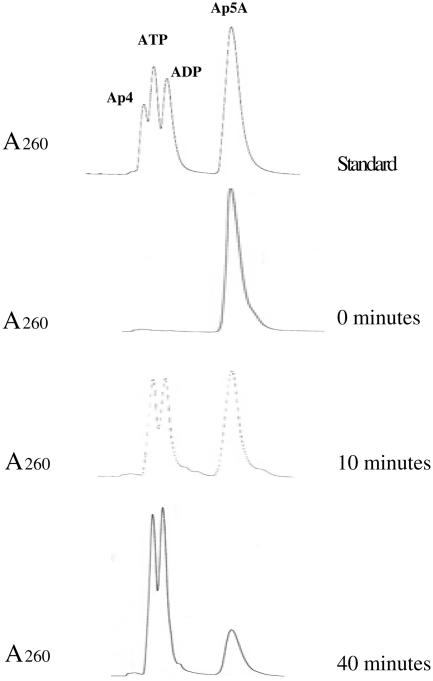
To identify the products of Ap5A hydrolysis, a scaled-up reaction, omitting alkaline phosphatase, was incubated under standard conditions, and at 0, 10, and 40 min, aliquots were analyzed by high-performance liquid chromatography. As the chromatograph indicates, there was a decrease in the Ap5A and the concomitant formation of ATP and ADP during the incubation (Fig. 3). No fraction of Ap4 was observed throughout the course of the reaction. Thus, the reaction may be written as Ap5A + H2O → ATP + ADP.
FIG. 3.
Reaction products from hydrolysis of Ap5A by PnhA. High-performance liquid chromatography was used to identify the reaction products as described previously (16). Samples were collected at 0, 10, and 40 min of reaction, respectively.
Kinetic studies on the hydrolysis of Ap5A yielded the following: Km, 0.5 mM; kcat, 2.1 s−1; Vmax, 5.3 units mg−1; kcat/Km, 4.1 × 103 s−1 M−1 (data not shown).
In accordance with other Nudix hydrolases, PnhA had an absolute requirement for a divalent cation, was maximally functional at 5 mM Mg2+ (100%) and Mn2+ (96%), and was optimally active at pH 8.0.
Construction and characterization of the ACP13 mutant strain.
The pnhA mutant was generated via allelic exchange using a mutagenesis cassette which contained a tetracycline resistance gene, tet(M), within the pnhA gene. The cassette was generated via PCR, circularized, and introduced into wild-type P. multocida. Five putative mutants, screened by PCR and confirmed by Southern hybridization, contained within the chromosome the mutagenesis cassette, and one of these colonies was designated ACP13. Using specific primers and total ACP13 RNA, RT-PCR experiments confirmed the transcription of the tet(M) gene and the absence of an intact pnhA and pnhB transcript (data not shown). Within P. multocida, the lgt gene is found downstream of the pnhA-B operon. The lgt gene, which encodes the prelipoprotein diacylglycerol transferase enzyme, is an essential gene in gram-negative bacteria (44). Consequently, it is unlikely that the insertion of the mutagenesis cassette within pnhA had any effect on the expression of the lgt gene, since it would have resulted in a nonviable phenotype.
The phenotype of ACP13 compared to that of the P. multocida wild-type strain was investigated by observing growth rate, response to heat shock, cellular morphology, and metabolic activities (Table 3). Wild-type and ACP13 strains were grown in culture medium, and their growth rates were compared based on cell turbidity and cell viability. There was very little difference in growth rate between the two isogenic strains, since doubling times for both strains were approximately 30 to 32 min and both entered the lag, log, and stationary phases at similar times. Similarly, no difference was seen between the wild-type P. multocida and ACP13 when exposed to heat shock conditions. At temperatures of 42°C, 45°C, and 50°C, the ACP13 strain died at the same rate as the wild-type strain. The starting cell culture was 1010 CFU/ml, and the final viable count for cells at 42°C and 45°C was 109 CFU/ml; for those at 50°C it was 107 CFU/ml.
TABLE 3.
Characterization of ACP13 strain
| Phenotypic characteristic | Comparison of ACP13 and WT |
|---|---|
| Growth rate | Similar to WT,e with generation time of 30-32 min |
| Response to heat shocka | Decrease in viable cell count was similar to WT |
| Cell sizeb | 60% smaller than WT |
| Carbohydrate utilizationc | Similar to WT |
| Enzymatic activitiesd | ACP13 showed lower activity for alkaline phosphatase, esterase (C4), esterase lipase (C8), acid phosphatase, and napthol-phosphohydrolase |
Cells were exposed to temperatures of 42°C, 45°C, and 50°C.
Determined by TEM.
Carbohydrate utilization was determined by using the API 50 CH system.
Different enzyme activities were examined using the API ZYM kit.
WT, wild type.
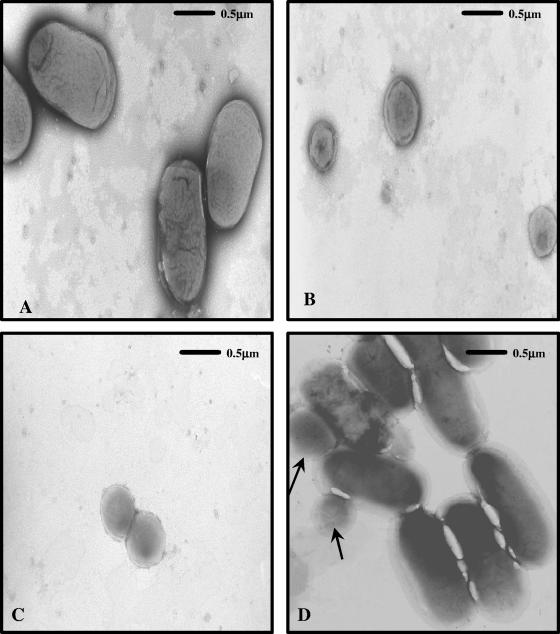
However, when the strains were observed via TEM, there were marked differences in cellular morphology between the wild type (Fig. 4 A) and ACP13 (Fig. 4B and C). The cell size of ACP13 was approximately 60% smaller than that of the wild-type strain. Figure 4C shows an ACP13 cell undergoing cell division, which suggests that the mutant cells do not reach normal wild-type size during their life cycle. A pnhA-complemented mutant was generated to determine if the difference in cell morphology between the wild-type strain and ACP13 was due to the mutation within the pnhA gene. A fragment containing the promoterless pnhA gene was cloned into the E. coli-P. multocida shuttle vector, pAL99, directly downstream of the tpiA promoter to generate pACP1007. ACP13 was transformed with pACP1007, resulting in the strain ACP17. Electron microscopy revealed that the cell morphology of ACP17 was restored to wild-type size and consistently shown to be indistinguishable from that of the wild type (Fig. 4D). The growth rate of ACP17 was also found to be indistinguishable from the growth rates of the wild-type P. multocida strain in HI broth. Complementation of ACP13 and ACP17 with an intact pnhB gene was not possible, since repeatedly the gene could not be successfully cloned. It was assumed that the gene product, PnhB (eight-transmembrane protein), may have been lethal to both E. coli and P. multocida when present in more than one copy within the cell.
FIG. 4.
Morphological characterization of wild-type P. multocida, ACP13, and ACP17. Electron micrographs of negatively stained (A) wild-type P. multocida, (B) ACP13 mutant, (C) ACP13 cell undergoing replication, and (D) ACP17 complemented mutant. The arrows (D) indicate small cells that are assumed to be those that have lost the pACP1007 plasmid with the functional pnhA and are thus exhibiting the mutant phenotype (B, C). The TEM micrographs were taken at magnification ×20,000.
In order to assess the presence of metabolic differences among wild-type P. multocida, ACP13, and ACP17, a number of metabolic enzyme activities using the API 50 CH and API ZYM systems were tested. No significant, reproducible (n = 4) differences were observed among the strains in regard to carbohydrate utilization. However, the enzymatic activities of alkaline phosphatase, esterase (C4), esterase lipase (C8), acid phosphatase, and naphthol-phosphohydrolase were consistently (n = 4) relatively lower for ACP13 and ACP17 than for the wild-type strain. No difference in activity was observed for the remaining enzymatic activities (C14 lipase, leucine arylamidase, valine arylamidase, cystine arylamidase, trypsin, α-chymotrypsin, α-galactosidase, β-galactosidase, β-glucuronidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, and α-fucosidase).
The ACP13 mutant was attenuated for virulence.
The ability of ACP13 to cause disease within the host was assessed using the chicken embryo lethality assay (Table 4). Avian species are the natural host for P. multocida, and most species are infected with serogroup A P. multocida strains. Thus, the use of chicken embryos was a relevant model for assessing P. multocida virulence. Eggs were inoculated via the allantoic sac with various doses of either wild-type, ACP13, or ACP17 strains and were assessed for viability over a 72-h period. After the eggs were deemed morbid, they were euthanized and autopsied, and tissue samples were taken from the embryo. Wild-type P. multocida had a 50% infective dose of 10 CFU 24 h postinfection. In contrast, 24 h postinfection, no deaths were recorded for eggs infected with 104 CFU or less of ACP13. After 48 h postinfection, all eggs inoculated with wild-type P. multocida were deemed morbid; however, no eggs inoculated with <102 CFU of ACP13 were found dead, and only half of the eggs inoculated with 103 and 104 CFU were dead after 48 h (Table 4). The chicken embryo lethality model shows that after 48 h postinfection, the ability of ACP13 to infect chicken embryos was dramatically reduced, approximately 1,000-fold, from that of the wild type, which can successfully infect embryos at only 10 CFU. This attenuation of virulence was expected, since other bacterial dinucleoside oligophosphate pyrophosphatase mutants also display reduced virulence when tested in their respective virulence assays (5, 28).
TABLE 4.
Assessment of P. multocida wild-type, ACP13, and ACP17 virulence using the chicken embryo lethality model
| Injected dose (CFU/ml) | % Survival of embryonated eggsa
|
|||||
|---|---|---|---|---|---|---|
| WT
|
ACP13
|
ACP17b
|
||||
| 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | |
| 108 | 0 | 0 | 50 | 0 | ND | ND |
| 107 | 0 | 0 | 50 | 0 | ND | ND |
| 106 | 0 | 0 | 50 | 0 | ND | ND |
| 105 | 0 | 0 | 75 | 25 | 66 | 33 |
| 104 | 25 | 0 | 100 | 50 | 66 | 33 |
| 103 | 25 | 0 | 100 | 50 | 66 | 33 |
| 102 | 50 | 0 | 100 | 100 | 100 | 33 |
| 10 | 50 | 0 | 100 | 100 | ND | ND |
These results are an average for four separate experiments. WT, wild type; ND, not determined.
Six eggs were used for each group that received ACP17.
Unexpectedly, the complemented mutant, ACP17, did not infect the embryos in a manner similar to that of the wild-type strain. No eggs infected with the wild-type strain survived 48 h postinfection; in contrast, 33% of the eggs infected with ACP17, at dosages from 102 to 105 CFU/ml, were able to survive 48 h postinfection. Thus, it appeared that ACP17 was not fully restored to wild-type virulence. ACP17 cells recovered from the embryos exhibited wild-type, full cell size, morphology when examined via TEM, and PCR confirmed the presence of an intact pnhA gene on pACP1007.
DISCUSSION
Relatively few P. multocida enzymes have been characterized. This study characterizes the PnhA protein from P. multocida, which belongs to the dinucleoside oligophosphate pyrophosphatase subfamily of Nudix hydrolase. PnhA has enzymatic characteristics similar to those of three other bacterial pyrophosphatases, YgdP of E. coli (7), InvA of R. prowazekii (24), and YgdP of Salmonella (28), all of which can hydrolyze a number of dinucleoside oligophosphates but preferentially hydrolyze Ap5A. PnhA hydrolyzes Ap5A at a kcat of 2.1 s−1 and a Km of 0.5 mM, always resulting in the production of one ATP molecule, and for optimal activity at pH 8.0, PnhA requires a divalent cation.
During cellular stress, the levels of dinucleoside oligophosphates are high within the cell (21, 30), and it is predicted that the Nudix hydrolases are involved in hydrolyzing these molecules to avoid cellular toxicity (8). However, recent studies of the dinucleoside oligophosphate pyrophosphatase YgdP of Salmonella indicate that the role of this enzyme may not be linked to the oxidative stress response (28). In addition, the pyrophosphatase NudA of H. pylori was constitutively expressed and did not appear to be involved in the cellular heat shock response (34). Similarly, the lack of a functional pnhA gene did not have a significant impact on the growth rate of the mutant ACP13 or its ability to survive under heat shock stress conditions. However, microscopy analysis revealed that the cell size for ACP13 was 60% smaller than that for the wild-type P. multocida strain (Fig. 4A to C). Based on the work of Nishimura and colleagues (38, 39), the difference in cell size between wild-type P. multocida and ACP13 may be due to an increase in the concentration of dinucleoside oligophosphates within the cells, resulting from the lack of a functional PnhA protein. Small-cell morphology was observed in an E. coli Nudix hydrolase mutant that contained a 100-fold increase of Ap4A, which in turn was thought to trigger the initiation of early cell division and subsequently result in a small cell size (38). High concentrations of dinucleoside oligophosphates in both prokaryotic and eukaryotic cells have been linked to a number of physiological effects, such as inhibition of ATP-sensitive K+ channels (31), induction of cell apoptosis and cell differentiation (46), and inhibition of adenylate kinase activity (33). Dinucleoside oligophosphates are thought to be the signaling molecules that couple cell division with DNA replication, and an increase in these molecules does not allow this coupling to occur, resulting in premature cell division (39). Thus, in the absence of PnhA, the amount of Ap5A was presumably high, triggering early cell division, and giving rise to the smaller cell morphology. Further support of this hypothesis was observed when a functional pnhA gene was introduced into ACP13, which resulted in the restoration of wild-type morphology (Fig. 4D). Therefore, although the internal concentration of Ap5A was not measured in this study, functional PnhA in ACP17 presumably eliminated the excess amounts of the dinucleoside oligophosphates from the cell to maintain normal cell division and restore wild-type cell size. These data further support the role of dinucleoside oligophosphates as signaling molecules within bacterial cells.
Since initial studies showed that IalA of B. bacilliformis not only was a dinucleoside oligophosphate pyrophosphatase but was also involved in this pathogen's ability to invade human erythrocytes (14, 37), three other pyrophosphatases have been identified with the same characteristics: the YgdP proteins of both E. coli K1 and Salmonella (5, 28) and the InvA protein of R. prowazekii (23). In addition, a DNA fragment from Actinobacillus actinomycetemcomitans that contained a gene with a high similarity to apaH enables noninvasive E. coli to invade KB cells (43). To determine if PnhA belongs to this set of dinucleoside oligophosphate pyrophosphatases, we investigated the abilities of ACP13 and ACP17 to infect chicken embryos. Given that the P. multocida wild-type strain used in this study is highly virulent for chickens, the chicken embryo lethality model was a relevant model for assessing the virulence of the strains. As predicted, within this in vivo model ACP13 had a marked decrease in virulence, and its ability to infect and cause morbidity was delayed and reduced from that of the wild-type strain (Table 4). The virulence of the pnhA-complemented mutant, ACP17, compared to that of the wild-type strain was not fully restored. This incomplete restoration of virulence in P. multocida was also observed for complemented acapsular mutants by Chung et al. (15). An acapsular mutant was complemented with capsular genes, and although capsular production was restored, the complemented mutant was unable to infect chickens. It is of interest to note that the P. multocida-E. coli shuttle vector pAL99 used in this study to complement ACP13 was a derivative of pPBA1101, which was used by Chung and colleagues to complement their acapsular mutant. Thus, the use of this P. multocida-E. coli shuttle vector may be a factor that impacts on the restoration of wild-type virulence in the complemented mutants. Alternatively, due to ACP17 lacking a functional pnhB gene, it is possible that PnhB as well as PnhA may play a role in the ability of P. multocida to cause infection within the host.
The pnhA and pnhB genes were found to be within a single operon. Database analysis revealed that Haemophilus influenzae, Actinobacillus pleuropneumoniae, Vibrio cholerae, Pseudomonas aeruginosa, and Neisseria meningitidis contain genes similar to pnhB; interestingly, none of the enteric bacteria contain a pnhB-like gene. Only the pnhB-like genes of H. influenzae and A. pleuropneumoniae were found within an operon along with a gene encoding a protein that shares high similarity to PnhA. Further bioinformatic analysis using PSORT indicates that PnhB is most likely located within the bacterial membrane, and the presence of eight transmembrane regions supports this location. A number of diadenosine oligophosphate hydrolases have been identified as ecto-enzymes that are located on the cell surface within eukaryotes (1, 47); in contrast, the InvA protein of rickettsial species has been localized to the cytoplasm (25). Rickettsial species do not contain a PnhB orthologue, and it may be that within P. multocida, PnhB is required to export PnhA to the cell surface, where it, along with other virulence factors, plays a role in the disease process. Consequently, in the absence of functional PnhB, PnhA would not be found on the cell surface, decreasing the ability of P. multocida to cause disease. Detailed analyses of the location of PnhA within P. multocida and further characterization of PnhB have been undertaken.
At present, PnhA is the fourth member of the dinucleoside oligophosphate pyrophosphatase subfamily that is also associated with bacterial infectivity. How these pyrophosphatases contribute to a pathogen's ability to cause disease remains unclear. Within Salmonella, increased amounts of dinucleoside oligophosphates result in the loss of host cell invasion (28). This, in conjunction with our observations, suggests that an increase in the level of dinucleoside oligophosphates needs to be prevented during invasion so as not to disrupt or trigger cellular processes like cell division. It is plausible that the induction of cell division due to a high level of dinucleoside oligophosphates could inhibit host cell invasion. The use of an animal model in this study to assess the loss of PnhA is significant, since it extends our knowledge beyond the cultured cell line and indicates that dinucleoside oligophosphate pyrophosphatases are involved in the entire infection process.
Acknowledgments
We thank Patrick Pfaffle and Janet Huie from the Biology Department at Carthage College, WI, for their help and use of the TEM. We also thank Priscilla Johanesen, Medical College of Wisconsin, for helpful comments.
This work was supported by grant no. 463 from the U.S. Poultry and Egg Association (to C.G.R.) and grant no. GM 18649 from the National Institutes of Health (to M.J.B.).
REFERENCES
- 1.Aguilar, J. S., R. Reyes, A. C. Asensio, S. Oakin, P. Rotllan, and R. Miledi. 2001. Ectoenzymatic breakdown of diadenosine polyphosphates by Xenopus laevis oocytes. Eur. J. Biochem. 268:1289-1297. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ames, B. N., and D. T. Dubin. 1960. The role of polyamines in the neutralization of bacteriophage deoxyribonucleic acid. J. Biol. Chem. 235:769-775. [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1989. Current protocols in molecular biology. Wiley, New York, N.Y.
- 5.Badger, J. L., C. A. Wass, and K. S. Kim. 2000. Identification of Escherichia coli K1 genes contributing to human brain microvascular endothelial cell invasion by differential fluorescence induction. Mol. Microbiol. 36:174-182. [DOI] [PubMed] [Google Scholar]
- 6.Bessman, M. J., D. N. Frick, and S. F. O'Handley. 1996. The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J. Biol. Chem. 271:25059-25062. [DOI] [PubMed] [Google Scholar]
- 7.Bessman, M. J., J. D. Walsh, C. A. Dunn, J. Swaminathan, J. E. Weldon, and J. Shen. 2001. The gene ygdP, associated with the invasiveness of Escherichia coli K1, designates a Nudix hydrolase, Orf176, active on adenosine (5′)-pentaphospho-(5′)-adenosine (Ap5A). J. Biol. Chem. 276:37834-37838. [DOI] [PubMed] [Google Scholar]
- 8.Bochner, B. R., P. C. Lee, S. W. Wilson, C. W. Cutler, and B. N. Ames. 1984. AppppA and related adenylylated nucleotides are synthesized as a consequence of oxidation stress. Cell 37:225-232. [DOI] [PubMed] [Google Scholar]
- 9.Bosch, M., M. E. Garrido, M. Llagostera, A. M. P. de Rozas, I. Badiola, and J. Barbé. 2002. Characterization of the Pasteurella multocida hgbA gene encoding a hemoglobin-binding protein. Infect. Immun. 70:5955-5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bosch, M., E. Garrido, M. Llagostera, A. M. P. de Rozas, I. Badiola, and J. Barbe. 2002. Pasteurella multocida exbB, exbD and tonB genes are physically linked but independently transcribed. FEMS Microbiol. Lett. 210:201-208. [DOI] [PubMed] [Google Scholar]
- 11.Boyce, J. D., and B. Adler. 2000. The capsule is a virulence determinant in the pathogenesis of Pasteurella multocida M1404 (B:2). Infect. Immun. 68:3463-3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brevet, A., J. Chen, F. Leveque, P. Plateau, and S. Blanquet, S. 1989. In vivo synthesis of adenylylated bis(5′-nucleosidyl) tetraphosphates (Ap4N) by Escherichia coli aminoacyl-tRNA synthetases. Proc. Natl. Acad. Sci. USA 86:8275-8279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carter, G. R. 1967. Pasteurellosis: Pasteurella multocida and Pasteurella haemolytica. Adv. Vet. Sci. Comp. Med. 11:321-379. [PubMed] [Google Scholar]
- 14.Cartwright, J. L., P. Britton, M. F. Minnick, and A. G. McLennan. 1999. The IalA invasion gene of Bartonella bacilliformis encodes a (de)nucleoside polyphosphate hydrolase of the MutT motif family and has homologs in other invasive bacteria. Biochem. Biophys. Res. Commun. 256:474-479. [DOI] [PubMed] [Google Scholar]
- 15.Chung, J. Y., I. Wilkie, J. D. Boyce, K. M. Townsend, A. J. Frost, M. Ghoddusi, and B. Adler. 2001. Role of capsule in the pathogenesis of fowl cholera caused by Pasteurella multocida serogroup A. Infect. Immun. 69:2487-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conyers, G. B., and M. J. Bessman. 1999. The gene, ialA, associated with the invasion of human erythrocytes by Bartonella bacilliformis, designates a nudix hydrolase active on dinucleoside 5′-polyphosphates. J. Biol. Chem. 274:1203-1206. [DOI] [PubMed] [Google Scholar]
- 17.Dabo, S. M., A. W. Confer, and R. A. Quijano-Blas. 2003. Molecular and immunological characterization of Pasteurella multocida serotype A:3 OmpA: evidence of its role in P. multocida interaction with extracellular matrix molecules. Microb. Pathog. 35:147-157. [DOI] [PubMed] [Google Scholar]
- 18.De Alwis, M. C. L. 1992. Pasteurellosis in production animals: a review, p. 11-22. In B. E. Patten, T. L. Spencer, R. B. Johnson, D. Hoffmann and L. Lehane, (ed.), Pasteurellosis in production animals. ACIAR Publishing, Canberra, Australia.
- 19.Dunn, C. A., S. F. O'Handley, D. N. Frick, and M. J. Bessman. 1999. Studies on the ADP-ribose pyrophosphatase subfamily of the nudix hydrolases and tentative identification of trgB, a gene associated with tellurite resistance. J. Biol. Chem. 274:32318-32324. [DOI] [PubMed] [Google Scholar]
- 20.Foged, N. T., J. P. Nielsen, and S. E. Jorsal. 1989. Protection against progressive atrophic rhinitis by vaccination with Pasteurella multocida toxin purified by monoclonal antibodies. Vet. Rec. 125:7-11. [DOI] [PubMed] [Google Scholar]
- 21.Fuge, E. K., and S. B. Farr. 1993. AppppA-binding protein E89 is the Escherichia coli heat shock protein ClpB. J. Bacteriol. 175:2321-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuller, T. E., M. J. Kennedy, and D. E. Lowery. 2000. Identification of Pasteurella multocida virulence genes in a septicemic mouse model using signature-tagged mutagenesis. Microb. Pathog. 29:25-38. [DOI] [PubMed] [Google Scholar]
- 23.Gaywee, J., S. Radulovic, J. A. Higgins, and A. F. Azad. 2002. Transcriptional analysis of Rickettsia prowazekii invasion gene homolog (invA) during host cell infection. Infect. Immun. 70:6346-6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaywee, J., W. Xu, S. Radulovic, M. J. Bessman, and A. F. Azad. 2002. The Rickettsia prowazekii invasion gene homolog (invA) encodes a Nudix hydrolase active on adenosine (5′)-pentaphospho-(5′)-adenosine. Mol. Cell Proteomics 1:179-185. [DOI] [PubMed] [Google Scholar]
- 25.Gaywee, J., J. B. Sacci, Jr., S. Radulovic, M. S. Beier, and A. F. Azad. 2003. Subcellular localization of rickettsial invasion protein, InvA. Am. J. Trop. Med. Hyg. 68:92-96. [PubMed] [Google Scholar]
- 26.Glisson, J. R. 1998. Bacterial respiratory diseases of poultry. Poult. Sci. 77:1139-1142. [DOI] [PubMed] [Google Scholar]
- 27.Guranowski, A. 2000. Specific and nonspecific enzymes involved in the catabolism of mononucleoside and dinucleoside polyphosphates. Pharmacol. Ther. 87:117-139. [DOI] [PubMed] [Google Scholar]
- 28.Ismail, T. M., C. A. Hart, and A. G. McLennan. 2003. Regulation of dinucleoside polyphosphate pools by the YgdP and ApaH hydrolases is essential for the ability of Salmonella enterica serovar typhimurium to invade cultured mammalian cells. J. Biol. Chem. 278:32602-32607. [DOI] [PubMed] [Google Scholar]
- 29.Jablonski, L., N. Sriranganathan, S. M. Boyle, and G. R. Carter. 1992. Conditions for transformation of Pasteurella multocida by electroporation. Microb. Pathog. 12:63-68. [DOI] [PubMed] [Google Scholar]
- 30.Johnstone, D. B., and S. B. Farr. 1991. AppppA binds to several proteins in Escherichia coli, including the heat shock and oxidative stress proteins DnaK, GroEL, E89, C45 and C40. EMBO J. 10:3897-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jovanovic, A., S. Zhang, A. E. Alekseev, and A. Terzic. 1996. Diadenosine polyphosphate-induced inhibition of cardiac KATP channels: operative state-dependent regulation by a nucleoside diphosphate. Pflugers Arch. 431:800-802. [DOI] [PubMed] [Google Scholar]
- 32.Lee, P. C., B. R. Bochner, and B. N. Ames. 1983. AppppA, heat-shock stress, and cell oxidation. Proc. Natl. Acad. Sci. USA 80:7496-7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lienhard, G. E., and I. I. Secemski. 1973. P1,P5-Di(adenosine-5′)pentaphosphate, a potent multisubstrate inhibitor of adenylate kinase. J. Biol. Chem. 248:1121-1123. [PubMed] [Google Scholar]
- 34.Lundin, A., C. Nilsson, M. Gerhard, D. I. Andersson, M. Krabbe, and L. Engstrand. The NudA protein in the gastric pathogen Helicobacter pylori is an ubiquitous and constitutively expressed dinucleoside polyphosphate hydrolase. 2003. J. Biol. Chem. 278:12574-12578. [DOI] [PubMed] [Google Scholar]
- 35.May, B. J., Q. Zhang, L. L. Li, M. L. Paustian, T. S. Whittam, and V. Kapur. 2001. Complete genomic sequence of Pasteurella multocida, Pm70. Proc. Natl. Acad. Sci. USA 98:3460-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McLennan, A. G. 2000. Dinucleoside polyphosphates—friend or foe? Pharmacol. Ther. 87:73-89. [DOI] [PubMed] [Google Scholar]
- 37.Mitchell, S. J., and M. F. Minnick. 1995. Characterization of a two-gene locus from Bartonella bacilliformis associated with the ability to invade human erythrocytes. Infect. Immun. 63:1552-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishimura, A., S. Moriya, H. Ukai, K. Nagai, M. Wachi, and Y. Yamada. 1997. Diadenosine 5′,5‴-P1,P4-tetraphosphate (Ap4A) controls the timing of cell division in Escherichia coli. Genes Cells 2:401-413. [DOI] [PubMed] [Google Scholar]
- 39.Nishimura, A. 1998. The timing of cell division: Ap4A as a signal. Trends Biochem. Sci. 23:157-159. [DOI] [PubMed] [Google Scholar]
- 40.Pedersen, K. B., and F. Elling. 1984. The pathogenesis of atrophic rhinitis in pigs induced by toxigenic Pasteurella multocida. J. Comp. Pathol. 94:203-214. [DOI] [PubMed] [Google Scholar]
- 41.Rhoades, K. R., and R. B. Rimler. 1989. Fowl cholera, p. 95-113. In C. Adlam, and J. M. Rutter (ed.), Pasteurella and Pasteurellosis. Academic Press Limited, London, United Kingdom.
- 42.Rimler, R. B. 1990. Comparisons of Pasteurella multocida lipopolysaccharides by sodium dodecyl sulfate-polyacrylamide gel electrophoresis to determine relationship between group B and E hemorrhagic septicemia strains and serologically related group A strains. J. Clin. Microbiol. 28:654-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saarela, M., S. Asikainen, S. Alaluusua, and P. Fives-Taylor. 1998. apaH polymorphism in clinical Actinobacillus actinomycetemcomitans isolates. Anaerobe 4:139-144. [DOI] [PubMed] [Google Scholar]
- 44.Sankaran, K., and H. C. Wu. 1994. Lipid modification of bacterial prolipoprotein: transfer of diacylglyceryl moiety from phosphatidylglycerol. J. Biol. Chem. 269:19701-19706. [PubMed] [Google Scholar]
- 45.Sillero, A., and M. A. Sillero. 2000. Synthesis of dinucleoside polyphosphates catalyzed by firefly luciferase and several ligases. Pharmacol. Ther. 87:91-102. [DOI] [PubMed] [Google Scholar]
- 46.Vartanian, A., I. Prudovsky, H. Suzuki, I. Dal Pra, and L. Kisselev. 1997. Opposite effects of cell differentiation and apoptosis on Ap3A/Ap4A ratio in human cell cultures. FEBS Lett. 415:160-162. [DOI] [PubMed] [Google Scholar]
- 47.Von Drygalski, A., and A. Ogilvie. 2000. Ecto-diadenosine 5′,5‴-P1,P4-tetraphosphate (Ap4A)-hydrolase is expressed as an ectoenzyme in a variety of mammalian and human cells and adds new aspects to the turnover of Ap4A biofactors. 11:179-187. [DOI] [PubMed]
- 48.Wooley, R. E., P. S. Gibbs, T. P. Brown, and J. J. Maurer. 2000. Chicken embryo lethality assay for determining the virulence of avian Escherichia coli isolates. Avian Dis. 44:318-324. [PubMed] [Google Scholar]
- 49.Xu, W., P. Gauss, J. Shen, C. A. Dunn, and M. J. Bessman. 2002. The gene e.1 (nudE.1) of T4 bacteriophage designates a new member of the Nudix hydrolase superfamily active on flavin adenine dinucleotide, adenosine 5′-triphospho-5′-adenosine, and ADP-ribose. J. Biol. Chem. 277:23181-23185. [DOI] [PubMed] [Google Scholar]