Abstract
In a previous work, we demonstrated that the Mycobacterium tuberculosis Rv2358-furB operon is induced by zinc. In this study, the orthologous genes from Mycobacterium smegmatis mc2155 were inactivated and mutants analyzed. Rv2358 protein was purified and found to bind upstream of the Rv2358 gene. Binding was inhibited by Zn2+ ions.
The long-recognized phenomenon of nutritional immunity, in which the sequestration of iron and possibly other metals occurs as a nonspecific host response to infection (1), hints in general terms at the possibility of a keen competition between host and parasite for essential metal ions. Two major mechanisms play the prominent roles in governing metal ion homeostasis and resistance. The first one involves the uptake or efflux of specific heavy metal ions across biomembranes (15), while the second involves the specific chelation of metals by intracellular chaperons, e.g., metallothioneins (12). In prokaryotes, the expression of these genes is tightly controlled by specific “metal-sensing” transcriptional regulators (3, 5) clustered in four distinct families: Fur (4), DtxR (10), MerR (2), and SmtB/ArsR (3). In each family, representative members are dimers, with each subunit containing an amino-terminal helix-turn-helix DNA-binding domain and an adjacent metal-binding domain.
Mycobacterium tuberculosis genome encodes two regulators of the Fur family (FurA and FurB), two regulators of the DtxR family (IdeR and SirR), three regulators of the MerR family (Rv1674c, Rv1994c, and Rv3334), and seven regulators of the SmtB/ArsR family (Rv0324, Rv0576, Rv2034, Rv2358, Rv2640c, Rv2642, and Rv3744) (http://genolist.pasteur.fr/TubercuList/). IdeR is the only one whose role has been well characterized. Like Fur and DtxR, IdeR binds iron and then interacts with a specific sequence in the operator region of iron-regulated genes to control their transcription (11, 13).
The genetic linkage of furA and katG in all species of mycobacteria has suggested that FurA may control katG expression. Indeed, recent studies have demonstrated that it is a negative regulator of katG (18). Further, we showed that transcription of both genes is induced upon oxidative stress and that FurA negatively controls transcription of its own gene (7, 14).
In mycobacteria, the furB gene is located immediately downstream of the Rv2358 gene, encoding a putative regulator of the SmtB/ArsR family. Using Mycobacterium smegmatis as a model system, we found that the M. tuberculosis Rv2358 and furB genes are cotranscribed by a common promoter which is induced by zinc (8), thus suggesting that this operon is involved in zinc homeostasis. In this study, we report results on inactivation of Ms2358 (the Rv2358 ortholog) and furB genes in M. smegmatis and the in vitro interaction between the Rv2358 protein and a palindromic sequence located immediately upstream of the transcriptional start site of the Rv2358-furB operon.
Ms2358 and furB knockout strains and zinc tolerance.
To investigate the physiological role of these genes, we produced two M. smegmatis mutants in which Ms2358 and furB were inactivated by partial gene deletion and insertion of a hygromycin resistance cassette, according to the protocol described by Jacobs et al. (6), and the two strains were named mcJA8 and mcJF3, respectively.
In order to confirm the involvement of Ms2358 and FurB in metal homeostasis, we tested the relative resistance of the wild type (wt) and the two mutants to ZnSO4 and to the chelating agent EDTA by using an agar disk diffusion assay. Filter paper disks soaked with 10 μl of 100 mM ZnSO4 or 0.5 M EDTA were put onto 20-ml 7H11 plates that were inoculated with 0.5 ml of M. smegmatis cultures, grown in 7H9 medium up to an optical density at 600 nm of 0.6. Plates were incubated at 37°C for 48 h and inhibitory boundary measured. While no difference in zinc tolerance could be detected among wild-type and mutant strains, tolerance to EDTA was decreased in mcJA8 but not in mcJF3. The increased sensitivity to chelating agents, shown by the Ms2358 knockout strain, was partially reversed after complementation with pMVA2, a pMV261 (16) derivative containing a wt copy of Ms2358 under the control of the hsp60 promoter (Table 1).
TABLE 1.
Sensitivities of M. smegmatis strains to EDTA
| M. smegmatis strain | Diameter (cm)a |
|---|---|
| mc2155 | 2.2 ± 0.1 |
| mcJF3 | 2.1 ± 0.1 |
| mcJA8 | 3.3 ± 0.1 |
| mcJA8(pMVA2) | 2.6 ± 0.1 |
The values represent the averages ± the standard deviations of the diameters of the inhibition zones in cm. The experiments, performed in triplicate, were repeated two times with independent bacterial cultures.
The regulation of the Rv2358-furB operon is mediated by Rv2358.
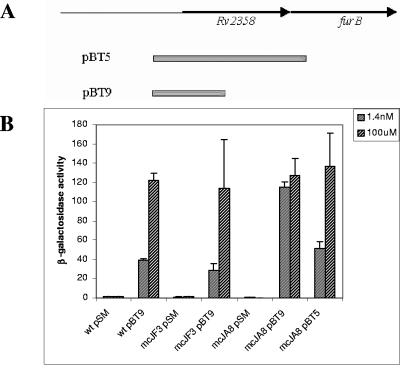
To verify whether zinc-dependent expression of the Rv2358-furB operon is mediated by Rv2358 or FurB, we analyzed its promoter activity in the wt and in the mutant strains. M. smegmatis mc2155, mcJA8, and mcJF3 were transformed with pBT9 and pBT5. These plasmids are pSM128 derivatives carrying different transcriptional fusions of M. tuberculosis Rv2358-furB promoter region to a promoterless lacZ gene (Fig. 1A) (8): pBT9 carries the promoter region of the operon plus a partial Rv2358 sequence, while pBT5 contains the promoter region and the complete Rv2358 sequence. After growth under conditions in which metal ions were limited, zinc was added to the cultures at physiological noninducing (1.4 nM) or inducing (100 μM) concentrations, as previously described (8). Cells were disrupted by sonication, and levels of β-galactosidase activity of the extracts were measured as described by Miller (9). The enzyme activity was expressed as nanomoles of o-nitrophenol-β-d-galactopyranoside converted to o-nitrophenol min−1 mg−1 of total proteins. β-Galactosidase activity of strains transformed with pBT9 revealed that the zinc-dependent regulation of the Rv2358-furB promoter was lost in mcJA8 but maintained in mcJF3 (Fig. 1B). Promoter activity was restored in mcJA8 mutant transformed with pBT5 plasmid, thus suggesting a direct involvement of Rv2358 protein in the transcriptional regulation of the operon.
FIG. 1.
β-Galactosidase assays of M. smegmatis mc2155 wild-type and mcJA8 and mcJF3 mutant strains, assayed at physiological (1.4 nM) and inducing (100 μM) zinc concentrations. (A) pSM derivatives pBT5 and pBT9 containing the promoter fragments of the Rv2358-furB operon. (B) β-Galactosidase activity results.
Rv2358 binds specifically to the Rv2358-furB promoter region.
The Rv2358 coding region was cloned into pET-15b (Invitrogen), and the recombinant protein was overproduced in Escherichia coli BL21(DE3)plysE (17) and purified. M. tuberculosis FurB protein was overproduced with the pGEX-6P-I system (Pharmacia-Amersham) in E. coli XL1-Blue and purified as previously described (8).
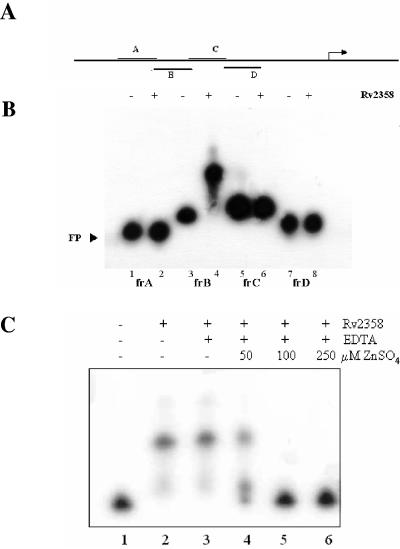
Rv2358 was used in electrophoretic mobility shift assays (EMSA) to determine its ability to bind the Rv2358-furB promoter region. Overlapping fragments of about 100 bp, mapping on the promoter region contained in pBT9, were amplified by PCR with specific primers from M. tuberculosis genomic DNA, end labeled with [γ-32P]dATP (6,000 Ci/mmol) by using T4 kinase, and used as probes (fragments A, B, C, and D [Fig. 2A ]). Binding reaction mixtures [20 mM Tris-HCl, pH 7.8, 1 mM EDTA, 0.1 μg of poly(dI-dC)/ml, 0.5 mM spermidine, 3% glycerol] containing 0.27 pmol of the labeled probe were incubated with purified Rv2358 protein (5.5 pmol) for 20 min at room temperature. Reaction mixtures were loaded onto a nondenaturing 8% polyacrylamide gel containing 40 mM Tris-acetate, pH 8, and 1 mM EDTA.
FIG. 2.
EMSA experiments and probes used. (A) Fragments A, B, C, and D, mapping in the Rv2358 promoter region, are shown. (B) EMSA results with fragment A (lanes 1 and 2), fragment B (lanes 3 and 4), fragment C (lanes 5 and 6), and fragment D (lanes 7 and 8) as probes. Odd lanes represent negative controls without Rv2358 protein. (C) Rv2358 binding on fragment B is specifically inhibited by the addition of zinc.
Rv2358 was able to retard specifically the migration of fragment B (Fig. 2B, lanes 3 and 4). The specificity of binding was further investigated in competition experiments: gel shift was partially inhibited by a 200-fold excess of unlabeled fragment B but not by a similar amount of an unrelated DNA fragment (data not shown).
As shown in Fig. 2C, Zn2+ inhibits the Rv2358-fragment B complex formation. The shift bands, corresponding to the complex, partially disappeared when the Zn2+ concentration was 50 μM and completely disappeared when the concentrations of Zn2+ were raised to 100 and to 250 μM, respectively.
In similar experiments, we showed that other divalent metal ions, such as Cd2+, Cu2+, Fe2+, Ni2+, Mn2+, and Ni2+, are unable to inhibit Rv2358-fragment B complex formation (data not shown). Taken together, these findings demonstrate that Rv2358 DNA binding activity is exclusively regulated by Zn2+.
Two different Rv2358-DNA complexes could be detected at intermediate protein concentrations, but only the fast-migrating one was stable when protein-DNA stoichiometry was increased up to 200-fold. The presence of a reducing agent, such as dithiothreitol, also caused the loss of the slower-migrating band (data not shown). These evidences would suggest the labile formation of higher-molecular-weight complexes because of multimerization of Rv2358 on the DNA probe, the stability of which is clearly dependent on the redox state of the protein.
Binding experiments performed under the same conditions with the purified FurB protein were unsuccessful (data not shown), confirming the exclusive role of Rv2358 in the operon regulation.
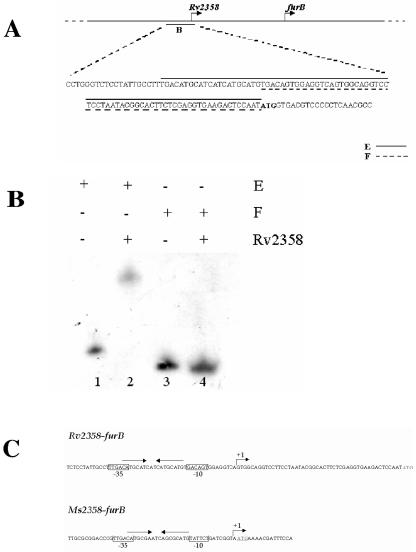
In order to identify more precisely the Rv2358 binding site, fragment B (126 bp) was divided in two smaller fragments, E and F (83 and 62 bp, respectively) (Fig. 3A). When these two fragments were used in EMSA, only fragment E was found to be still retained by Rv2358 (Fig. 3B). Sequence analysis of this fragment showed the presence of a palindromic sequence, probably the Rv2358 recognition site, with homology to 12-2-12 inverted repeats, described as binding sites of members of ArsR/SmtB family regulators (3).
FIG. 3.
Identification of the Rv2358 binding site. Fragments E and F, internal to fragment B (A), were used in EMSA experiments, but only fragment E was retained by Rv2358 protein (B). (C) Promoter/operator sequence organization in M. tuberculosis and M. smegmatis. Palindrome localization between −35 and −10 sites is significantly conserved in M. tuberculosis and M. smegmatis. The consensus promoter is boxed; transcriptional start point, as mapped by experiments with rapid amplification of cDNA ends, is shown by the curved arrow, and palindrome orientation is shown by straight arrows.
Experiments with rapid amplification of cDNA ends mapped the 5′ end of M. tuberculosis Rv2358-furB transcript 12 bp downstream of the previously identified inverted repeats which overlaps the putative −35 (TTGACA) and −10 (GACAGT) promoter region. Interestingly, we found a similar palindromic sequence in the Ms2358-furB upstream region in M. smegmatis (Fig. 3C).
Conclusions.
From these results, it appears that the Rv(Ms)2358 protein is responsible for the zinc-dependent repression of the Rv(Ms)2358-furB operon. This finding is consistent with a simple model of derepression, in which metal binding by the sensor protein weakens the DNA binding affinity significantly, such that RNA polymerase can load and initiate transcription of the operon.
Until now, no regulatory function could be attributed to FurB protein; mycobacterial furB could be the first member of the fur family not to be autoregulated. Our hypothesis is that Rv2358 could act as the “metal sensor” which controls the expression of FurB in response to zinc availability. On the other hand, FurB could be involved in the regulation of still-unidentified metal transport systems. However, we cannot rule out a further role of Rv2358 in zinc homeostasis. Usually, SmtB/ArsR regulators are involved in control of genes responsible for metal-specific efflux pumps, membrane-bound transporters, metal reductases, or metal-sequestering proteins (3). Its zinc-dependent DNA binding activity is in agreement with its hypothetical role in regulation of efflux systems or other resistance determinants, able to remove metal ions at high toxic concentrations.
Our results are in favor of the hypothesis that cotranscription of Rv(Ms)2358 and furB genes could represent the key element for the compensation between import and export of zinc.
Acknowledgments
We thank Riccardo Manganelli for his helpful discussion and critical reading of the manuscript.
This work was supported by grants COFIN-2003 from MIUR and FAR-2004 from University of Pavia.
REFERENCES
- 1.Beisel, W. R. 1977. Magnitude of the host nutritional responses to infection. Am. J. Clin. Nutr. 28:1236-1247. [DOI] [PubMed] [Google Scholar]
- 2.Brown, N. L., J. V. Stoyanov, S. P. Kidd, and J. L. Hobman. 2003. The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 27:145-163. [DOI] [PubMed] [Google Scholar]
- 3.Busenlehner, L. S., M. A. Pennella, and D. P. Giedroc. 2003. The SmtB/ArsR family of metalloregulatory transcriptional repressors: structural insights into prokaryotic metal resistance. FEMS Microbiol. Rev. 27:131-143. [DOI] [PubMed] [Google Scholar]
- 4.Escolar, L., J. Perez-Martin, and V. De Lorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finney, L. A., and T. V. O'Halloran. 2003. Transition metal speciation in the cell: insights from the chemistry of metal ion receptors. Science 300:931-936. [DOI] [PubMed] [Google Scholar]
- 6.Jacobs, W. R., Jr., S. B. Snapper, M. Tuckman, and B. R. Bloom. 1989. Mycobacteriophage vector systems. Rev. Infect. Dis. 11:S404-S410. [DOI] [PubMed] [Google Scholar]
- 7.Milano, A., F. Forti, C. Sala, G. Riccardi, and D. Ghisotti. 2001. Transcriptional regulation of furA and katG upon oxidative stress in Mycobacterium smegmatis. J. Bacteriol. 183:6801-6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Milano, A., M. Branzoni, F. Canneva, A. Profumo, and G. Riccardi. 2004. The Mycobacterium tuberculosis Rv2358-furB is a zinc-induced operon. Res. Microbiol. 155:192-200. [DOI] [PubMed] [Google Scholar]
- 9.Miller, J. M. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 10.Oram, D. M., A. Avdalovic, and R. K. Holmes. 2004. Analysis of genes that encode DtxR-like transcriptional regulators in pathogenic and saprophytic corynebacterial species. Infect. Immun. 72:1885-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pohl, E., R. K. Holmes, and W. G. J. Hol. 1999. Crystal structure of the iron-dependent regulator (IdeR) from Mycobacterium tuberculosis shows both metal binding site fully occupied. J. Mol. Biol. 285:1145-1156. [DOI] [PubMed] [Google Scholar]
- 12.Robinson, N. J., S. K. Whitehall, and J. S. Cavet. 2001. Microbial metallothioneins. Adv. Microb. Physiol. 44:183-213. [DOI] [PubMed] [Google Scholar]
- 13.Rodriguez, G. M., M. I. Voskuil, B. Gold, G. K. Schoolnik, and I. Smith. 2002. ideR, an essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect. Immun. 70:3371-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sala, C., F. Forti, E. Di Florio, F. Canneva, A. Milano, G. Riccardi, and D. Ghisotti. 2003. Mycobacterium tuberculosis FurA autoregulates its own expression. J. Bacteriol. 185:5357-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Solioz, M., and C. Vulpe. 1996. CPx-type ATPases: a class of P-type ATPases that pump heavy metals. Trends Biochem. Sci. 21:237-241. [PubMed] [Google Scholar]
- 16.Stover, C. K., V. F. de la Cruz, T. R. Fuerst, J. E. Burlein, L. A. Benson, L. T. Bennet, G. P. Bansall, J. F. Young, M. H. Lee, G. F. Hatfull, S. B. Snapper, R. G. Barletta, W. R Jacobs, Jr., and B. R. Bloom. 1991. New use of BCG for recombinant vaccines. Nature 251:456-460. [DOI] [PubMed] [Google Scholar]
- 17.Studier, Y. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 18.Zahrt, T. C., J. Song, J. Siple, and V. Deretic. 2001. Mycobacterial FurA is a negative regulator of catalase-peroxidase gene katG. Mol. Microbiol. 39:1174-1185. [DOI] [PubMed] [Google Scholar]