Abstract
Shigella dysenteriae serotype 1, a major cause of bacillary dysentery in humans, can use heme as a source of iron. Genes for the transport of heme into the bacterial cell have been identified, but little is known about proteins that control the fate of the heme molecule after it has entered the cell. The shuS gene is located within the heme transport locus, downstream of the heme receptor gene shuA. ShuS is a heme binding protein, but its role in heme utilization is poorly understood. In this work, we report the construction of a chromosomal shuS mutant. The shuS mutant was defective in utilizing heme as an iron source. At low heme concentrations, the shuS mutant grew slowly and its growth was stimulated by either increasing the heme concentration or by providing extra copies of the heme receptor shuA on a plasmid. At intermediate heme concentrations, the growth of the shuS mutant was moderately impaired, and at high heme concentrations, shuS was required for growth on heme. The shuS mutant did not show increased sensitivity to hydrogen peroxide, even at high heme concentrations. ShuS was also required for optimal utilization of heme under microaerobic and anaerobic conditions. These data are consistent with the model in which ShuS binds heme in a soluble, nontoxic form and potentially transfers the heme from the transport proteins in the membrane to either heme-containing or heme-degrading proteins. ShuS did not appear to store heme for future use.
The Shigella spp. are the primary cause of bacillary dysentery in humans. Following ingestion, this pathogen transits the digestive system to the colon, where it invades the cells of the colonic epithelium. It then replicates inside the epithelial cells and spreads to adjacent cells. The resulting inflammation and tissue damage are responsible for the bloody diarrhea characteristic of dysentery.
Iron is often a limiting nutrient for microbial growth, especially in the context of the host. The shigellae have an absolute requirement for iron, and multiple iron acquisition systems have been identified in Shigella dysenteriae serotype 1. These include genes for the synthesis and utilization of enterobactin (13), a common siderophore also produced by Escherichia coli, Salmonella, and several other species of enteric bacteria. The S. dysenteriae genome contains the iroA locus (23), which contains genes for the synthesis and transport of the recently discovered siderophore salmochelin (7). Also present are the ferrous transporters Feo (10, 26) and Sit (26). Although Sit has been reported to transport primarily manganese in Salmonella enterica serovar Typhimurium (11), genetic studies of Shigella indicate that it can transport iron (26).
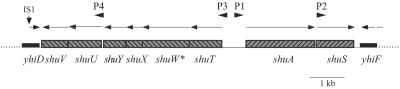
Heme is the most abundant source of iron in the mammalian host (for reviews, see references 5 and 33). We previously demonstrated that S. dysenteriae transports heme (12) and have cloned and characterized the heme transport locus (16, 17, 38). This locus contains eight genes that are flanked by genes with near identity to E. coli K-12 yhiD and yhiF (Fig. 1), and other than the presence of the heme transport genes, the overall map of this region is similar to that of E. coli K-12 (38). None of the genes in the heme transport locus have homologues anywhere in the genome of E. coli K-12; however, heme transport loci with the same organization and nearly identical DNA sequences are present in many heme-utilizing pathogenic E. coli strains, including O157:H7 and uropathogenic strains (8, 17, 21, 31, 35, 38).
FIG. 1.
Map of the S. dysenteriae heme transport locus. The bars indicate the individual genes, and the arrows indicate the direction of transcription. The arrowheads labeled P1, P2, P3, and P4 show the locations of the four predicted promoters. The asterisk on shuW indicates that this is a defective gene in S. dysenteriae.
The heme transport locus open reading frames have been described in various levels of detail. The best-characterized gene, shuA, encodes a receptor that transports the intact heme moiety across the outer membrane (17). The energy for heme transport is transduced to ShuA in a process that requires TonB, ExbB, and ExbD (22), which are not encoded within the heme transport locus (16, 17). shuTUV likely encode a periplasmic binding protein-dependent ABC transport system that shuttles heme through the periplasm and across the inner membrane. ShuT, ShuU, and ShuV have homology with periplasmic binding proteins, inner membrane permeases, and ATPases, respectively. The locus also contains the poorly characterized shuWXY genes, which appear to be cotranscribed with shuT (Fig. 1). These genes are conserved in several other heme transport loci (38) but are not required for heme utilization, and their functions remain unknown. In S. dysenteriae, shuW contains a premature stop codon, but in pathogenic E. coli strains, the corresponding open reading frame, chuW, is predicted to encode a full-length protein.
The other gene in the locus is shuS, which is located immediately downstream of shuA (Fig. 1). This gene was required for optimal heme utilization when portions of the S. dysenteriae heme transport locus were expressed in S. enterica serovar Typhimurium (38). In this assay, the disruption of shuS had approximately a twofold effect on colony size, and this defect was most pronounced when both shuS and the ABC transporter genes were deleted. Many of the characterized heme transport loci contain shuS homologues, including those of Yersinia enterocolitica (28), Yersinia pestis (30), Pseudomonas aeruginosa (20), Bordetella spp. (19, 32), Enterobacter hormaechei (25), and Rhizobium leguminosarum (36). The presence of shuS homologues is relatively widespread, since uncharacterized homologues can be identified in searches of sequenced bacterial genomes, including Erwinia carotovora (2), Bartonella henselae (1), and Photorhabdus luminescens (4).
The role of ShuS in heme utilization is unknown. Stojiljkovic and Hantke (28) found that the Y. enterocolitica shuS homologue, hemS, was required for heme utilization. hemS also prevented heme toxicity when the heme receptor gene hemR was expressed from a high-copy-number vector in E. coli but not when the receptor was expressed from a low-copy-number vector. Based on these data, they proposed that the role of HemS is to protect cells from heme toxicity and speculated that it may function as a heme-degrading enzyme. Biochemical characterization of ShuS, however, showed that while it binds heme with a Kd of approximately 13 μM, it does not catalyze degradation of the heme molecule (37). Further, ShuS was purified as a large spherical complex, suggestive of the structure of ferritin.
In this communication, we report the construction and characterization of a chromosomal shuS mutant in S. dysenteriae. Although this mutant could use heme as a source of iron, it grew at a significantly reduced rate when heme was the sole iron source. We show that at low heme concentrations ShuS improves the efficiency of heme utilization and at high heme concentrations it protects cells from growth inhibition by heme.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The strains and plasmids used in this study are listed in Table 1. The iron chelator ethylenediamine di(ortho-hydroxyphenylacetic acid) (EDDA) was deferrated by the method of Rogers (24). The appropriate antibiotics were added to plasmid-containing strains at the following concentrations: 85 μg carbenicillin per ml, 50 μg kanamycin per ml, and 50 μg chloramphenicol per ml (E. coli) or 25 μg chloramphenicol per ml (S. dysenteriae).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference |
|---|---|---|
| Bacterial strains | ||
| S. dysenteriae | ||
| O4576S1 | S. dysenteriae serotype 1 | 17 |
| O4576S1A | Streptomycin-resistant derivative of O4576S1 | This study |
| AKS1 | O4576S1A entF::kan | This study |
| GLS1 | AKS1 shuS::cam | This study |
| E. coli | ||
| DH5α | Host for subcloning | 6 |
| DH5α(λpir) | Host for pCVD442 | 15 |
| Plasmids | ||
| pBluescript SK− | Cloning vector (high copy number) | Stratagene |
| pGEM-Teasy | Cloning vector (high copy number) | Promega |
| pWKS30 | Cloning vector (low copy number) | 34 |
| pCVD442 | Suicide vector pGP704 containing sacB | 3 |
| pUC4K | Kanamycin resistance cassette | Pharmacia |
| pHTL106 | BglII-EcoRI fragment containing entire heme transport locus in BamHI/EcoRI sites of pWKS30 | 38 |
| pHTL110 | pHTL106 with cam cassette inserted in SnaBI site in shuS | 38 |
| pHTL116 | EcoRV fragment containing shuA in pWKS30 | 38 |
| pSHU9 | EcoRI/PstI fragment carrying shuAS in pAT153 | 17 |
| pKAT5 | HpaI/XbaI deletion of pHTL106; remaining fragment encodes shuAS | This study |
| pDD100 | KpnI fragment from pSHU9 encoding shuS in pBluescript SK− | This study |
Growth assays.
For liquid growth assays, overnight cultures were diluted 200-fold into L broth containing the indicated supplements. All cultures were grown with aeration at 37°C, and the OD650 was measured over time.
Colony size assays were performed by diluting overnight cultures 10-fold into L broth and growing them to late log phase. Cultures were then diluted and plated at low density on L plates containing 75 μg EDDA per ml and the indicated concentration of hemin. The colony diameter was measured following 24-h incubation at 37°C. For colony size assays performed under microaerobic and anaerobic conditions, cells were spread on L plates containing 20 mM nitrate supplemented either with 20 μM FeSO4 or with 75 μg EDDA per ml with or without 15 μM hemin. Identical plates were incubated aerobically or in a sealed container made either microaerobic using a CampyGen pack (Oxoid, Basingstoke, Hampshire, England) or anaerobic using an AnaeroGen pack (Oxoid). Plates were incubated for 24 h at 37°C, except anaerobic plates, which were incubated for 48 h.
Sensitivity to hydrogen peroxide was determined by growing AKS1/pBluescript SK−, GLS1/pBluescript SK−, and GLS1/pDD100 overnight in L broth. Then 50 μl of the culture was mixed with 20 ml L agar containing 75 μg EDDA per ml and the indicated concentration of hemin and poured into petri plates. After solidification of the agar, 10 μl of 1 M hydrogen peroxide was applied to a sterile disk placed upon the agar. The diameter of the zone of inhibition was measured following incubation for 20 h at 37°C.
Heme storage assays were performed by diluting fully grown cultures 100-fold into L broth containing 75 μg EDDA per ml and 5, 10, or 15 μM hemin and growing them overnight. Then the optical density at 650 nm (OD650) was determined and cultures were diluted into L broth containing 75 μg EDDA per ml to an approximate OD650 of 0.07. Cultures were grown with aeration at 37°C, and the OD650 was determined over time.
Western blot assays.
Overnight cultures were diluted 100-fold into L broth containing the indicated concentration of EDDA or hemin. When the OD650 reached approximately 0.5, 1 ml of culture was centrifuged for 1 min in a Microfuge and the pellet was resuspended in 0.2 ml of sodium dodecyl sulfate (SDS)-gel sample buffer. The samples were boiled for 10 min and loaded onto an SDS-12.5% polyacrylamide gel. Following electrophoresis and transfer to nitrocellulose, ShuS was visualized with a rabbit antiserum raised against purified ShuS (gift of Angela Wilks).
Strain construction.
O4576S1A is a spontaneous streptomycin-resistant derivative of S. dysenteriae strain O4576S1 (17). The growth rate of O4576S1A in L broth was indistinguishable from the parental strain. The entF mutant AKS1 was constructed in S. dysenteriae by PCR amplifying a portion of the entF gene from an O4576-derived strain using primers RIentFfor (5′ GGGATTCTACCGATTTAGCCCTTGC) and RIentFrev (5′ GGAATTCGCAGACTTCATCCAGG). The product was cloned into pGEM-Teasy (Promega) to give plasmid pENT100. pENT101 was constructed by insertion of the kanamycin resistance gene from pUC4K as a HincII fragment into the HpaI site in the entF gene. The NotI fragment containing the disrupted entF gene was made blunt with Klenow and subcloned into the SmaI site of pCVD442. This plasmid, pENT102, was introduced into S. dysenteriae strain O4576S1A by mating, and marker exchange mutations were obtained as previously described (14). The presence of an entF mutation was confirmed by PCR and phenotypic analysis.
To construct GLS1, the KpnI fragment of pHTL110 containing shuS::cam was made blunt with Klenow and subcloned into the SmaI site of pCVD442 to give pCVD442shuS::cam. This plasmid was introduced into AKS1, and marker exchange mutations were obtained as previously described (14).
RESULTS
Regulation of ShuS expression.
In gram-negative bacteria, the expression of iron acquisition genes is usually repressed in the presence of iron, an effect dependent upon the iron regulatory protein Fur. Because the shuS promoter contains a potential Fur binding site (38), we looked by Western blot analysis at the effect of iron and hemin on ShuS synthesis (Fig. 2A). An internal control was provided by a protein unrelated to ShuS that runs slightly more slowly on the gel. When cultures were grown in unsupplemented or iron-supplemented L broth, very little ShuS was visualized in the blot. However, when the iron chelator EDDA was added to the growth medium, ShuS was well expressed. At all three iron levels, the amount of ShuS synthesized was either unaffected or reduced by supplementation of the cultures with hemin, indicating that hemin does not induce ShuS synthesis. The reduced level of ShuS in the culture supplemented with EDDA and hemin could reflect hemin utilization, which leads to a lower degree of iron starvation than in cells supplemented with EDDA alone. These data are similar to those observed for shuA, which is negatively regulated by Fur in the presence of iron, and no induction occurs in the presence of hemin (16, 17).
FIG. 2.
Western blot analysis of ShuS synthesis. Total cellular proteins were separated in an SDS-12.5% polyacrylamide gel and visualized with an antiserum raised against ShuS. (A) AKS1 (ShuS+) was grown in L broth containing the indicated supplements. Concentrations of the supplements when added were 40 μM FeSO4, 10 μg EDDA per ml, and 5 μM hemin. (B) The following strains were grown in L broth plus 10 μg EDDA per ml: AKS1/pWKS30 (ShuS+), GLS1/pWKS30 (shuS::cam), GLS1/pDD100 (shuS::cam/pshuS), and GLS1/pKAT5 (shuS::cam/pshuAS).
ShuS is required for optimal hemin utilization.
We characterized the function of the ShuS protein by constructing a defined shuS mutation in S. dysenteriae serotype 1. An entF mutation was included in this strain to reduce background growth due to siderophore production and utilization. Western blot analysis confirmed that ShuS was synthesized in the parental strain but not in the shuS mutant (Fig. 2B). When the shuS gene was supplied on plasmid pDD100, the synthesis of ShuS was restored at a level higher than that observed in the parental ShuS+ strain. ShuS synthesis was also observed at a lower level when shuA and shuS were supplied together on a plasmid.
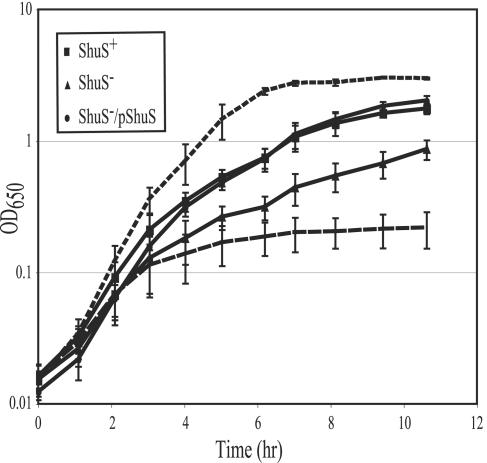
The shuS mutant was tested for the ability to use hemin as a source of iron by monitoring the growth of the shuS mutant in medium depleted of free iron but containing hemin (Fig. 3). Overnight cultures were diluted into L broth supplemented either with iron or with EDDA and hemin. As expected, all strains grew rapidly in the presence of added FeSO4 and their growth was severely limited when only the iron chelator EDDA was added to the medium. When hemin was the sole source of iron, the parental strain grew well though somewhat more slowly than in medium supplemented with FeSO4. However, the isogenic strain carrying the shuS mutation grew more slowly than its Shu+ parent and reached a lower final OD when using hemin as the source of iron. Efficient growth on hemin was restored when shuS was supplied on a plasmid (Fig. 3).
FIG. 3.
Growth of ShuS+ and ShuS− strains on hemin. Overnight cultures were diluted into L broth supplemented with 75 μg EDDA per ml and 5 μM hemin (solid lines). The cultures were grown at 37°C with aeration, and the OD650 was measured over time. The strains used were ShuS+ (AKS1/pWKS30) (▪), ShuS− (GLS1/pWKS30) (▴), and ShuS−/pShuS (GLS1/pDD100) (•). For comparison to growth with hemin, the growth of the parental strain (AKS1) in 20 μM FeSO4 (upper broken line) and in 75 μg EDDA per ml (lower broken line) is shown. Growth of the shuS mutant and the complemented strain was nearly identical to that of the parental strain under these conditions (data not shown). The data presented are the average of four independent experiments.
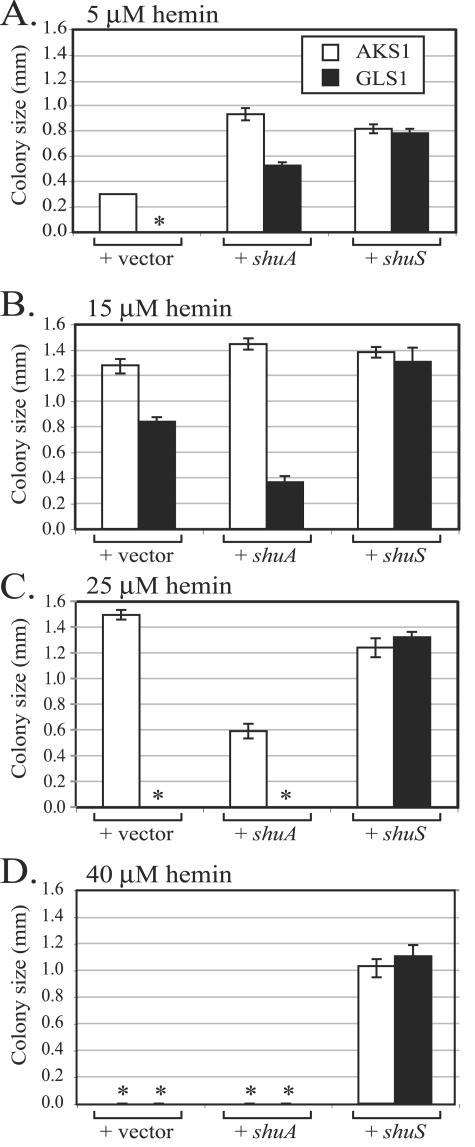
The growth defect in the shuS mutant could be due to failure to efficiently use hemin, to toxicity of the transported hemin, or both. To address these possibilities, a colony size assay was performed over a range of hemin concentrations. In this assay, cultures were diluted to a low density and spread on plates containing EDDA and hemin and the diameter of the colonies was measured at 24 h (Fig. 4). At the lowest concentration shown, the shuS mutant failed to grow while its parent strain, AKS1, formed small colonies (Fig. 4A). When shuS was provided on high-copy-number plasmid pDD100, the growth of both the shuS mutant and its shuS+ parent was stimulated. This suggests that ShuS may be limiting for utilization of hemin as an iron source at low hemin concentrations. The shuS mutant supplemented with additional shuA on a low-copy-number plasmid also formed colonies at this low hemin concentration. Supplying additional copies of shuA also increased the colony size of parental strain AKS1. We propose that at low hemin concentrations, modest overproduction of ShuA caused more hemin to be transported into the cell, thus providing more iron for growth. At the same hemin concentration (5 μM) in liquid, the shuS mutant was able to grow, albeit at a reduced rate compared with the parental strain (Fig. 3). We have found that a lower concentration of hemin is needed for growth in liquid cultures compared with growth on plates. We do not know the exact reason for this observation, but it may be related to the rate of diffusion of hemin in a plate or to differences in the efficiency of iron chelation by EDDA. Alternatively, this could be related to differences in growth rate or aeration.
FIG. 4.
Growth of ShuS+ and ShuS− strains on increasing quantities of hemin. Bacteria were spread on L plates containing 75 μg EDDA per ml and the indicated concentration of hemin. After 24-h incubation at 37°C, the diameter of the colonies was measured, and the graph shows the mean diameter of 10 colonies. Open bars represent the colony size for ShuS+ strain AKS1, and filled bars represent shuS mutant strain GLS1. Each strain carries a plasmid that has the gene indicated at the bottom of the graph. The plasmids used were pWKS30 (vector only), pHTL116 (shuA), and pDD100 (shuS). An asterisk indicates that no colonies grew on the plate.
At an intermediate hemin concentration (15 μM), the shuS mutant was able to grow but showed a pronounced growth defect compared to the parental strain (Fig. 4B). When shuS was supplied on a plasmid, both the shuS mutant and the parent grew well at this hemin concentration. Additional copies of shuA on a plasmid caused the shuS mutant to form smaller colonies than those observed with the vector alone, in contrast to the result obtained with 5 μM hemin, where extra copies of shuA stimulated the growth of the shuS mutant. Unlike the shuS mutant, the parental strain formed larger colonies when additional shuA was present. These data are consistent with previously reported experiments with Y. enterocolitica which showed that hemin was toxic when the heme receptor was overproduced relative to the ShuS homologue (28).
When the hemin concentration was increased to 25 μM, ShuS was required to form a colony (Fig. 4C). Finally, at a very high concentration of hemin (40 μM), only cells that overproduced ShuS relative to other heme utilization proteins were able to grow (Fig. 4D). This suggests that ShuS can function to prevent heme toxicity.
The shuS mutant does not display increased sensitivity to hydrogen peroxide.
Since ShuS is a heme binding protein, it may prevent free heme from accumulating in the cytoplasm. Heme accumulation in the cytoplasm could result in the shuS mutant being more sensitive to conditions that promote oxidative stress. The sensitivity of the shuS mutant to hydrogen peroxide was determined by placing a disk spotted with hydrogen peroxide on hemin-EDDA agar inoculated with the bacterial strain and measuring the growth inhibition zone around the disk. The shuS mutant and its parental strain were tested in EDDA agar that contained 15, 25, or 35 μM hemin, and the presence of a shuS mutation had no effect on hydrogen peroxide sensitivity at any of these hemin concentrations (data not shown). The inoculum was sufficiently high to allow visible growth of the bacteria at all hemin concentrations in the absence of hydrogen peroxide.
ShuS is needed for optimal heme utilization under microaerobic and anaerobic conditions.
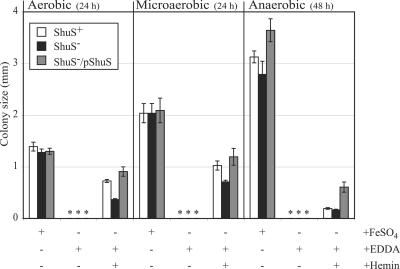
If the failure of the shuS mutant to grow optimally with hemin as the sole iron source is due to unbound heme leading to the accumulation of reactive products, it is possible that this effect would be suppressed by restricting the amount of oxygen in the environment. To test this, the parent, the shuS mutant, and the complemented shuS mutant were spread at a low density on L plates containing sodium nitrate. Following incubation under aerobic, microaerobic, or anaerobic conditions, the colony size was determined (Fig. 5).
FIG. 5.
Growth of the shuS mutant under microaerobic and anaerobic conditions. Bacteria were spread on L plates containing sodium nitrate, 75 μg EDDA per ml, and 15 μM hemin. Control L plates contained sodium nitrate and 20 μM FeSO4. Plates were incubated either aerobically or in sealed chambers made either microaerobic or anaerobic. The diameter of the colonies was measured after 24-h (aerobic and microaerobic) or 48-h (anaerobic) incubation. The strains used were AKS1/pWKS30 (ShuS+), GLS1/pWKS30 (ShuS−), and GLS1/pDD100 (ShuS−/pShuS). An asterisk indicates that no colonies grew on the plate.
Under aerobic conditions, the shuS mutant formed a smaller colony on hemin than the parental strain or the mutant strain carrying shuS on a plasmid, as shown in Fig. 4. The S. dysenteriae strain grew well under microaerobic conditions, but ShuS was still required for optimal growth on hemin. Under anaerobic conditions, the strains grew very well in the presence of iron. However, growth with hemin as the sole iron source was very poor. After 48 h of incubation, both the parental strain and the shuS mutant produced colonies near the lower limit of visibility. The difference in colony size between these two strains was not statistically significant. Interestingly the synthesis of higher levels of ShuS from the plasmid significantly stimulated growth under anaerobic conditions. This suggests a role for ShuS in hemin utilization even under conditions where the amount of available molecular oxygen is severely restricted.
ShuS does not store heme for subsequent use as an iron source.
A possible function of a heme binding protein is that it could store heme in a nontoxic form and then release the bound heme for use as an iron source under iron-restricted conditions. To test this possibility, cells were grown overnight with 5, 10, or 15 μM hemin as the sole source of iron. They were then diluted into medium in which the available iron was restricted by the iron chelator EDDA and the OD was measured over time. The shuS mutant, its isogenic parent, and the complemented mutant all grew at the same rate and reached a plateau after approximately a fourfold increase in cell density, indicating that the cells contained enough iron to divide two times (data not shown). The fourfold increase in cell density was observed in all experiments regardless of the hemin concentration in the overnight culture. Thus, the presence of ShuS did not apparently increase the amount of stored iron available for cellular growth, suggesting that ShuS does not function as a heme storage protein.
DISCUSSION
Proteins involved in the transport of heme into the cell have been identified in many bacteria, but the function of the cytoplasmic heme binding protein ShuS and its homologues in the transport and utilization of heme has remained elusive. In this work, we have created a shuS mutant in S. dysenteriae serotype 1. At a low heme concentration, the shuS mutant grew more slowly than the parent strain in liquid medium when hemin was the sole iron source (Fig. 3). On solid medium, the shuS mutant failed to grow when a low concentration of hemin was the sole iron source. Under these conditions, growth of the shuS mutant was promoted either by increasing the heme concentration in the medium or by supplying additional copies of shuA on a plasmid, actions expected to increase the amount of heme transported into the cell (Fig. 4A and B). Thus, the shuS mutant has a reduced ability to use heme for cellular growth.
Growth of the shuS mutant was inhibited at high heme concentrations. At 25 μM hemin, the shuS mutant failed to grow though the parental strain grew well (Fig. 4C). At a very high hemin concentration of 40 μM, only cells overexpressing shuS from a plasmid were able to grow (Fig. 4D). This observation suggests that ShuS can reduce the toxicity of the transported heme. It was shown previously that Y. enterocolitica HemS prevented heme toxicity when the heme receptor was carried on a high-copy-number plasmid in E. coli (28), but to our knowledge this is the first study showing heme sensitivity in a shuS mutant. It is difficult to compare the role of ShuS in S. dysenteriae with observations of Yersinia spp. In Y. enterocolitica, a hemS mutation is lethal (28); however, a Y. pestis hmuS mutant was constructed and found to have no defect in growth or heme utilization, although use of some heme proteins was affected when hmuTUV were also deleted (30). Both HemS and HmuS have greater than 65% amino acid identity with ShuS (38), and HemS and HmuS are nearly 90% identical to each other (30). While it is possible that the different phenotypes are the result of differences in the activities of the ShuS homologues, the high sequence similarity makes it more likely that these phenotypic differences are due to differences in the genetic backgrounds of these three species.
The ability of ShuS homologues to protect cells from heme toxicity has led to the model in which their primary role is to prevent the toxic effects of transported heme (29). In this model, the lack of heme binding by ShuS leads to the accumulation of free heme in the cell, which presumably would promote oxidative damage, such as peroxidation of lipids. Since the toxic effects of heme, however, were only observed in very high concentrations of hemin or when the heme receptor gene was overexpressed from a plasmid, it is unclear whether these assays reflect the role of ShuS under more physiologically relevant conditions. Although the experiments in this work do not rigorously disprove the model in which the reduced growth of the shuS mutant at low and moderate hemin concentrations is caused by heme toxicity, they do not support it. The shuS mutation had reduced growth in concentrations of hemin that were low enough to be limiting for cell growth (Fig. 4A). This result suggests that ShuS functions in some way to improve the utilization of heme. Further, the observations that the shuS mutant does not have increased sensitivity to hydrogen peroxide and that ShuS promoted growth in a moderate concentration of hemin when the availability of molecular oxygen is reduced suggest that this mutant is not subject to oxidative stress. For these reasons, we believe that ShuS most likely directly promotes the utilization of heme. It is possible that ShuS has two separate functions, one that promotes heme utilization when the heme concentration is limiting and another that protects the cell from toxic effects at high heme concentrations. The more parsimonious model, however, is that ShuS has a single activity that simultaneously increases the utilization of heme and also prevents accumulation of free heme to a toxic level.
The mechanism by which ShuS promotes heme utilization is not known. Several models are consistent with our data. One possibility is that ShuS has a role in the trafficking of transported heme from the membrane to heme-containing or heme-degrading proteins. The fact that free heme is toxic and minimally soluble in water suggests that such a heme carrier protein may be needed to maintain transported heme in a soluble, nontoxic form and to transport the heme to the appropriate cellular location. This model is consistent with work with purified proteins which showed that heme bound to ShuS is rapidly transferred to a heme oxygenase (A. Wilks, personal communication). Another model consistent with our data is that ShuS may work in cooperation with another protein to form heme-degrading activity or that it may allow cells to use heme in the absence of heme oxygenase activity.
It is unknown whether enteric pathogens require a heme-degrading enzyme for heme utilization. No heme oxygenase activity has been detected in these species, and no potential heme oxygenase gene has been identified in the sequenced genomes. However, heme oxygenases have been identified in several other pathogenic bacteria, and in those species the heme oxygenase is required for efficient heme utilization (27, 40, 41). All known heme oxygenases require molecular oxygen for enzymatic activity, and there is little information on the function of heme-degrading enzymes under anaerobic conditions. Both ShuS+ and ShuS− S. dysenteriae strains used hemin very poorly under anaerobic conditions, while overproduction of ShuS strongly enhanced the use of heme under these conditions (Fig. 5). This is consistent with models in which heme oxygenase activity is rate limiting under anaerobiosis, and ShuS either enhances heme oxygenase activity or allows heme utilization to occur in the absence of heme oxygenase activity.
A potential role of a cytoplasmic heme binding protein is to store heme for subsequent use during conditions of iron starvation. ShuS is a large, ring-shaped complex (37), suggesting that by analogy with ferritin, it could be a storage site for excess heme. However, ShuS did not increase growth under iron-depleted conditions following prolonged growth with excess hemin. This may indicate that ShuS has a function other than iron storage and sequestration.
A heme transport locus in Vibrio cholerae also encodes a cytoplasmic heme binding protein designated HutZ (39). Although it has no sequence identity with ShuS, HutZ is required for efficient utilization of heme (9, 18, 39), and the absorbance spectra produced when heme is bound to ShuS or HutZ are nearly identical (37, 39). Interestingly, many of the heme transport loci contain either a hutZ or a shuS homologue, and we are not aware of any loci that contain both. These data suggest that ShuS and HutZ may have some common functions; however, unlike ShuS, HutZ appears to function as a heme storage protein (37, 39). In addition, shuS did not complement a hutZ mutation in a liquid assay and hutZ only partially complemented the shuS mutant in a colony size assay (data not shown). This suggests they have incompletely overlapping functions or have interactions with proteins that are not conserved between S. dysenteriae and V. cholerae.
The fate of heme after it has been transported into the cytoplasm is one of the unsolved problems in understanding heme transport. Characterizing the role of ShuS in facilitating iron usage and the prevention of heme toxicity will further elucidate the strategies used by pathogens in the scavenging of iron for growth in the human host.
Acknowledgments
We thank Ashima Sharma and Erin Murphy for strain AKS1, Donald Duncan for pDD100, and Bonnie Reus for O4576S1A. We are indebted to Angela Wilks for ShuS antiserum and for communication of data prior to publication.
The work was supported by National Institutes of Health grant AI16935.
REFERENCES
- 1.Alsmark, C. M., A. C. Frank, E. O. Karlberg, B. A. Legault, D. H. Ardell, B. Canback, A. S. Eriksson, A. K. Naslund, S. A. Handley, M. Huvet, B. La Scola, M. Holmberg, and S. G. Andersson. 2004. The louse-borne human pathogen Bartonella quintana is a genomic derivative of the zoonotic agent Bartonella henselae. Proc. Natl. Acad. Sci. USA 101:9716-9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell, K. S., M. Sebaihia, L. Pritchard, M. T. Holden, L. J. Hyman, M. C. Holeva, N. R. Thomson, S. D. Bentley, L. J. Churcher, K. Mungall, R. Atkin, N. Bason, K. Brooks, T. Chillingworth, K. Clark, J. Doggett, A. Fraser, Z. Hance, H. Hauser, K. Jagels, S. Moule, H. Norbertczak, D. Ormond, C. Price, M. A. Quail, M. Sanders, D. Walker, S. Whitehead, G. P. Salmond, P. R. Birch, J. Parkhill, and I. K. Toth. 2004. Genome sequence of the enterobacterial phytopathogen Erwinia carotovora subsp. atroseptica and characterization of virulence factors. Proc. Natl. Acad. Sci. USA 101:11105-11110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duchaud, E., C. Rusniok, L. Frangeul, C. Buchrieser, A. Givaudan, S. Taourit, S. Bocs, C. Boursaux-Eude, M. Chandler, J. F. Charles, E. Dassa, R. Derose, S. Derzelle, G. Freyssinet, S. Gaudriault, C. Medigue, A. Lanois, K. Powell, P. Siguier, R. Vincent, V. Wingate, M. Zouine, P. Glaser, N. Boemare, A. Danchin, and F. Kunst. 2003. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat. Biotechnol. 21:1307-1313. [DOI] [PubMed] [Google Scholar]
- 5.Genco, C., and D. W. Dixon. 2001. Emerging strategies in microbial haem capture. Mol. Microbiol. 39:1-11. [DOI] [PubMed] [Google Scholar]
- 6.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 7.Hantke, K., G. Nicholson, W. Rabsch, and G. Winkelmann. 2003. Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc. Natl. Acad. Sci. USA 100:3677-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 9.Henderson, D. P., E. E. Wyckoff, C. E. Rashidi, H. Verlei, and A. L. Oldham. 2001. Characterization of the Plesiomonas shigelloides genes encoding the utilization of iron from heme. J. Bacteriol. 183:2715-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kammler, M., C. Schon, and K. Hantke. 1993. Characterization of the ferrous iron uptake system of Escherichia coli. J. Bacteriol. 175:6212-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kehres, D. G., A. Janakiraman, J. M. Slauch, and M. E. Maguire. 2002. SitABCD is the alkaline Mn2+ transporter of Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:3159-3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawlor, K. M., P. A. Daskaleros, R. E. Robinson, and S. M. Payne. 1987. Virulence of iron transport mutants of Shigella flexneri and utilization of host iron compounds. Infect. Immun. 55:594-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawlor, K. M., and S. M. Payne. 1984. Aerobactin genes in Shigella spp. J. Bacteriol. 160:266-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mey, A. R., and S. M. Payne. 2001. Haem utilization in Vibrio cholerae involves multiple TonB-dependent haem receptors. Mol. Microbiol. 42:835-849. [DOI] [PubMed] [Google Scholar]
- 15.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae required toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mills, M., and S. M. Payne. 1997. Analysis of shuA, the gene encoding the heme receptor of Shigella dysenteriae, and analysis of invasion and intracellular multiplication of a shuA mutant. Infect. Immun. 65:5358-5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mills, M., and S. M. Payne. 1995. Genetics and regulation of heme iron transport in Shigella dysenteriae and detection of an analogous system in Escherichia coli O157:H7. J. Bacteriol. 177:3004-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mourino, S., C. R. Osorio, and M. L. Lemos. 2004. Characterization of heme uptake cluster genes in the fish pathogen Vibrio anguillarum. J. Bacteriol. 186:6159-6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy, E. R., R. E. Sacco, A. Dickenson, D. J. Metzger, Y. Hu, P. E. Orndorff, and T. D. Connell. 2002. BhuR, a virulence-associated outer membrane protein of Bordetella avium, is required for the acquisition of iron from heme and hemoproteins. Infect. Immun. 70:5390-5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ochsner, U. A., Z. Johnson, and M. L. Vasil. 2000. Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiology 146:185-198. [DOI] [PubMed] [Google Scholar]
- 21.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 22.Postle, K., and R. A. Larsen. 2004. The TonB, ExbB and ExbD proteins, p. 96-112. In J. H. Crosa, A. R. Mey, and S. M. Payne (ed.), Iron transport in bacteria. ASM Press, Washington, D.C.
- 23.Reeves, S. A. 2001. Iron acquisition in the intracellular environment of the host: multiple iron transport systems in Shigella dysenteriae. Ph.D. dissertation. The University of Texas, Austin.
- 24.Rogers, H. J. 1973. Iron-binding catechols and virulence in Escherichia coli. Infect. Immun. 7:445-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roggenkamp, A., H. Hoffmann, and M. W. Hornef. 2004. Growth control of small-colony variants by genetic regulation of the hemin uptake system. Infect. Immun. 72:2254-2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Runyen-Janecky, L. J., S. A. Reeves, E. G. Gonzales, and S. M. Payne. 2002. Contribution of the Shigella flexneri Sit, Iuc, and Feo iron acquisition systems to growth in the intracellular environment of the host. Infect. Immun. 71:1919-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmitt, M. P. 1997. Utilization of host iron sources of Corynebacterium diphtheriae: identification of a gene whose product is homologous to eukaryotic heme oxygenases and is required for acquisition of iron from heme and hemoglobin. J. Bacteriol. 179:838-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stojiljkovic, I., and K. Hantke. 1994. Transport of haemin across the cytoplasmic membrane through a haemin-specific periplasmic binding-protein-dependent transport system in Yersinia enterocolitica. Mol. Microbiol. 13:719-732. [DOI] [PubMed] [Google Scholar]
- 29.Stojiljkovic, I., and D. Perkins-Balding. 2002. Processing of heme and heme-containing proteins by bacteria. DNA Cell Biol. 21:281-295. [DOI] [PubMed] [Google Scholar]
- 30.Thompson, J. M., H. A. Jones, and R. D. Perry. 1999. Molecular characterization of the hemin uptake locus (hmu) from Yersinia pestis and analysis of hmu mutants for hemin and hemoprotein utilization. Infect. Immun. 67:3879-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Torres, A. G., and S. M. Payne. 1997. Haem iron transport system in enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 23:825-833. [DOI] [PubMed] [Google Scholar]
- 32.Vanderpool, C. K., and S. K. Armstrong. 2001. The Bordetella bhu locus is required for heme iron utilization. J. Bacteriol. 183:4278-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wandersman, C., and I. Stojiljkovic. 2000. Bacterial heme sources: the role of heme, hemoprotein receptors and hemophores. Curr. Opin. Microbiol. 3:215-220. [DOI] [PubMed] [Google Scholar]
- 34.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 35.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wexler, M., K. H. Yeoman, J. B. Stevens, N. G. de Luca, G. Sawers, and A. W. Johnston. 2001. The Rhizobium leguminosarum tonB gene is required for the uptake of siderophore and haem as sources of iron. Mol. Microbiol. 41:801-816. [DOI] [PubMed] [Google Scholar]
- 37.Wilks, A. 2001. The ShuS protein of Shigella dysenteriae is a heme-sequestering protein that also binds DNA. Arch. Biochem. Biophys. 387:137-142. [DOI] [PubMed] [Google Scholar]
- 38.Wyckoff, E. E., D. Duncan, A. G. Torres, M. Mills, K. Maase, and S. M. Payne. 1998. Structure of the Shigella dysenteriae haem transport locus and its phylogenetic distribution in enteric bacteria. Mol. Microbiol. 28:1139-1152. [DOI] [PubMed] [Google Scholar]
- 39.Wyckoff, E. E., M. Schmitt, A. Wilks, and S. M. Payne. 2004. HutZ is required for efficient heme utilization in Vibrio cholerae. J. Bacteriol. 186:4142-4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu, W., D. J. Hunt, A. Richardson, and I. Stojiljkovic. 2000. Use of heme compounds as iron sources by pathogenic neisseriae requires the product of the hemO gene. J. Bacteriol. 182:439-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu, W., A. Wilks, and I. Stojiljkovic. 2000. Degradation of heme in gram-negative bacteria: the product of the hemO gene of neisseriae is a heme oxygenase. J. Bacteriol. 182:6783-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]