Abstract
The pckA gene, encoding phosphoenolpyruvate carboxykinase, catalyzes the reversible decarboxylation and phosphorylation of oxaloacetate to form phosphoenolpyruvate. Located on the circular chromosome of Agrobacterium, this locus is adjacent to the loci chvG and chvI, encoding a two-component regulatory system that has been shown to be important in virulence. Using a reporter gene fusion, studies showed that the pckA gene is induced by acidic pH but not by acetosyringone. This acid induction is regulated by the chvG-chvI regulatory system, which controls acid-inducible genes. A pckA mutant had no demonstrable PckA enzyme activity and grew on AB minimal medium with glucose but did not grow on the same medium with succinate as the sole carbon source and was more inhibited in its growth than the wild-type strain by an acidic environment. A pckA mutant was highly attenuated in tumor-inducing ability on tobacco leaf disks and was severely attenuated in vir gene expression. Although vir gene induction was completely restored when a constitutive virG gene was introduced into the mutant strain, virulence was only partially restored. These results suggest that avirulence may be due to a combination of the inhibition of this mutant in the acidic plant wound environment and the poor induction of the vir genes.
The transfer of DNA and proteins into host cells by Agrobacterium requires that both plasmid- and tumor-inducing (Ti) plasmid-encoded genes participate (for a review, see reference 11). The virulence (vir) genes located on the Ti plasmid, defined as those under the control of the two-component regulatory system VirAG, appear to be dedicated to the processing and transfer of the transfer DNA (15). The genes that map to the chromosome (chv genes) have dual functions. They play a role in the physiology of the organism growing in the absence of its hosts, as well as in the interaction of Agrobacterium with its plant hosts (11). Perhaps the best-understood example of a chv gene product is chvE (6). This periplasmic protein binds to monosaccharides, which are components of the plant cell wall, and thereby plays a role both in the transport of these sugars into the cell and in the chemotaxis of bacteria toward these sugars. In addition, the ChvE protein, when bound to a sugar synthesized by plant cells, increases the level of induction of vir genes by binding to the periplasmic domain of the VirA sensor protein (10). In this way, Agrobacterium senses the proximity of a susceptible plant. In addition to the monosaccharides bound to the ChvE protein, Agrobacterium recognizes several other signals associated with the plant wound environment. These include acetosyringone (AS) and an acidic environment (pH 5.5) (1, 37). Upstream of each of the vir genes is a 12-base-pair consensus sequence, the vir box, which is recognized and bound by the response regulator, VirG (9). Although it is likely that all of the genes required for processing and transfer of the transfer DNA have been identified on the Ti plasmid, the identities and functions of many of the chv genes involved in plant cell transformation await elucidation.
Chromosomal virulence genes that have been identified include those that play a role in osmotic stress and attachment (5), vir gene induction (12, 23, 27), and avoidance of plant defenses (40). One of the most interesting and significant of these chromosomal virulence genes is a two-component regulatory system, chvGI, in which, by homology to other two-component systems, chvG codes for the sensor protein and chvI codes for a response regulator (7, 25). This system is of special interest because it is a global regulatory system involved in the regulation of certain acid-inducible genes (21). These include the chromosomal gene katA, which encodes a catalase that is involved in the detoxification of H2O2, presumably released during the early interaction of Agrobacterium with its host plant (40), and aopB, which encodes an outer membrane protein also required for virulence (18). Its function in virulence is not known. In addition, mutations in chvI significantly reduce the expression of virG (25), which in turn inhibits the expression of the virB and virE operons. All of these genes under the control of chvGI have at least one feature in common. They are all acid inducible, and their levels presumably increase in the environment of a wounded plant.
Mutations in the chvGI loci, apparently pleiotrophic, confer a number of distinctive properties on the cells. chvG chvI mutants cannot grow in media containing the antibiotics tetracycline, novobiocin, and carbenicillin, as well as several detergents (7). This suggests that the mutants may have defects in their cell envelopes. Furthermore, mutants cannot grow on a complex medium, a trait that is shared with another chv mutant, chvD (23).
Because of the importance of the chvGI system in the interaction of several α proteobacteria with their eukaryotic hosts, it is important to identify additional target genes of this regulatory system. It is well known that genes under the control of a two-component system often map in the vicinity of the regulatory genes (7). This potential candidate gene downstream of chvGI encodes phosphoenolpyruvate carboxykinase (PckA), the first enzyme of gluconeogenesis.
Mutations in the pckA locus have profound biological effects in a variety of other bacteria. For example, a pckA-deficient Mycobacterium bovis BCG mutant was attenuated in infection of both macrophages and mice (22). The pckA mutation in Rhizobium sp. strain NGR234 resulted in different nodulation phenotypes depending on the host plant (31). In Sinorhizobium meliloti, a pckA mutant fixed nitrogen at 70% of the level of the wild type, whereas the pckA mutant of Rhizobium leguminosarum fixed nitrogen at the same efficiency as wild-type cells.
In this paper, we demonstrate that pckA is indeed under the control of chvGI and that, like other genes under this control system, the expression of pckA is induced by acid and is important for maximum virulence. Other properties of the gene and its protein product are described.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are listed in Table 1. Agrobacterium tumefaciens strains were grown in Mg/L or AB minimal medium at 28°C with shaking (4). Escherichia coli strain DH5α was grown in LB medium at 37°C with shaking (34). The following antibiotics were used at the indicated concentrations (in μg/ml): for A. tumefaciens, carbenicillin (100), kanamycin (100), and gentamicin (50); for E. coli, carbenicillin (100), kanamycin (100), and gentamicin (10). vir gene induction was analyzed in cells grown in induction broth as previously described (28).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Agrobacterium | ||
| C58 | Wild-type strain | 14 |
| At11063 | C58 chvG292::aac1 | 7 |
| A136 | C58; Ti plasmid cured | 35 |
| C58ΔpckA | C58 (ΔpckA) | This study |
| E. coli strain DH5α | endA1 hsdR17 supE44 thi-1 recA1 gyrA96 relA1 Δ(argF-lacZYA)U169 φ80dlacZΔM15 | Invitrogen |
| Plasmids | ||
| pSM243cd | virB::lacZ fusion of pTiA6 in pVK102 | 36 |
| pSM358cd | virE::lacZ fusion of pTiA6 in pVK102 | 36 |
| pWT160 | virG::lacZ fusion | 33 |
| pEX18Gm | GmroriT+sacB+; gene replacement vector from pUC18 | 16 |
| pBBR1MCS4 | Broad-host-range vector | 20 |
| pAB2002 | Vector for lacZ fusion | 3 |
| pSY204 | virG constitutive construction | 19 |
| pLP200 | pBBR1MCS-4 with 1.5-kb pckA fragment | This study |
| pLP201 | pWT200 with lacZ-Gmr fragment | This study |
| pEX18Gm-pckA | Construct for deleting pckA | This study |
The detergents sodium dodecyl sulfate (SDS), sodium deoxycholate (DOC), and Sarkosyl were used at concentrations (in mg/ml) of 0.2, 2, and 2, respectively (7).
Construction of pckA-lacZ fusion.
To determine whether pckA is acid inducible, the pckA gene was fused with the lacZ reporter gene. To make this construct, two primers, pckA1 (CGTTCGATCTCGAGTTGCGTTTCCAAAAGCTG) and pckA2 (GCTAGTTAGAATTCTCATTCAGCCGCGAGCAG), were used to amplify the pckA gene from A. tumefaciens genomic DNA, using Taq-plus polymerase. XhoI and EcoRI restriction sites were then introduced into primers pckA1 and pckA2, respectively. The 1.5-kb pckA PCR product was digested with XhoI and EcoRI and ligated to a 4.95-kb XhoI-EcoRI fragment of pBBR1MCS-4 to create pLP200. The 3.5-kb EcoRI fragment of pAB2002 containing the lacZ and gentamicin resistance genes was cloned into pLP200, which was then digested with EcoRI, creating pLP201.
Construction of the pckA in-frame deletion mutant.
The unmarked A. tumefaciens pckA deletion mutant was generated as described previously (16). In brief, 1.5-kb regions were amplified from the upstream and downstream regions flanking the region targeted for replacement, using primers that included specific restriction enzyme sites. After restriction enzyme digestion, the upstream and downstream fragments were ligated into the vector pEX18Gm using a directional three-way ligation. The plasmid was introduced into strain C58 by electroporation, and after incubation for 3 h to allow homologous recombination, the cells were plated on LB medium with 5% sucrose for the first selection. Colonies growing on the sucrose plates were streaked onto plates of Mg/L medium and Mg/L medium plus 25 μg/ml gentamicin for the second selection. The deletion mutant cannot grow on Mg/L medium containing 25 μg/ml gentamicin. Putative mutations were verified by sequencing the junction fragment generated by the deletion using PCR fragments that spanned the open reading frame selected for deletion. All experiments that involved a mutation of the pckA locus used an in-frame deletion mutation of the entire gene unless otherwise indicated.
Gene expression measurements.
Expression of the pckA gene was measured as β-galactosidase activity using the pckA::lacZ fusion. The expression of the virG, virB, and virE genes was assayed by using the virG::lacZ, virB::lacZ, and virE::lacZ fusions, respectively (Table 1). For the assay of β-galactosidase, A. tumefaciens cells containing the appropriate fusions were grown in AB minimal liquid medium with shaking at 28°C overnight and then transferred to induction broth (4) at pH 5.5 or fresh AB minimal medium (pH 7.0) and incubated for 24 h. The β-galactosidase activity reported was assayed as described previously (28). All β-galactosidase activities represent an average of three independent determinations.
Virulence assay.
Virulence assays were performed on Nicotiana tabacum leaf disks according to the method of Banta et al. (2). Briefly, A. tumefaciens cells were grown in liquid Mg/L medium to mid-log phase and harvested by centrifugation, and the cell concentration was adjusted to an optical density at 600 nm (OD600) of 0.4 to 0.5 with MS medium (29). The cells were cocultivated with 40 leaf squares of N. tabacum in 20 ml of hormone-free MS liquid medium (29) in a petri dish supplemented with 300 μM AS. After 2 days, 40 leaf squares were transferred to hormone-free MS medium containing vancomycin (200 μg/ml) and timentin (200 μg/ml) and cultured at 25°C in the dark. The tumors on each leaf disk were observed after 14 days.
Complementation.
The plasmid pSY204 (24), containing the constitutive virG gene, which induces the vir regulon in the absence of AS and sugar and at a neutral pH, was electroporated into C58 containing the virB-lacZ or virE-lacZ construct.
Assay of PckA enzyme activity.
Cell extracts were prepared from cells grown in 50 ml AB medium grown to the stationary phase. Cells were harvested by centrifugation in a Sorvall RC-58 centrifuge (10 min; 4°C; 10,000 × g) and then washed twice with cold 20 mM Tris-HCl buffer containing 1 mM MgCl2 (pH 7.4) and resuspended in the same buffer. The cells were sonicated on ice six times for 15 seconds each time. The crude extract was clarified by centrifugation (15 min; 13,000 × g), and the supernatant was assayed for PckA activity by a procedure described previously (17).
RESULTS
Identification and characterization of the pckA gene.
The pckA gene encodes the enzyme phosphoenolpyruvate (PEP) carboxykinase, which catalyzes the decarboxylation and phosphorylation of oxaloacetic acid to produce the glycolytic intermediate PEP. This enzyme is the first enzyme in gluconeogenesis. The gene, coding for a predicted protein of 537 amino acids, was initially identified in Agrobacterium from the similarity of its nucleotide sequence to those of previously identified genes. The predicted protein encoded by the gene has 52.4% identity to the E. coli (26) and 78.3% identity to the S. meliloti PckA proteins (30). The PckA enzyme has the specific domain (IGGTSYAGE-KKS; 190 to 202), which is required for its activity (30), and a phosphate-binding site (G--G-GKT; 236 to 243). A divalent or transition metal ion binding site (G--EG) could also be identified in residues 226 to 229. A BLAST search of the C58 genome did not reveal any other copies of this gene (39). Nutritional and biochemical data support the contention that this protein, identified as PckA by a bioinformatics analysis, is the only protein in Agrobacterium strain C58 with PckA enzyme activity. This activity is not activated by calcium or cell growth to stationary phase (Table 2), which differs from what has been observed in E. coli (26). Furthermore, crude extracts from cells with a deletion of the gene lack PckA enzyme activity (Table 2). Cells with a mutation in the gene cannot grow on succinate as a sole carbon and energy source, presumably because they lack this key enzyme in gluconeogenesis and therefore cannot synthesize the sugars required for macromolecule synthesis. This suggests that an alternative pathway from oxaloacetate to PEP does not exist in A. tumefaciens strain C58. The genome sequence indicates that two putative malic enzymes that convert malate to pyruvate are encoded (atu1652 and atu3356), but no evidence for phosphoenolpyruvate synthase, the enzyme that synthesizes PEP from pyruvate, could be seen from a BLAST search of the genome.
TABLE 2.
PEP carboxykinase activities under different growth conditionsa
| Conditions | Sp act (nmol of ADP/mg of protein/min)
|
|
|---|---|---|
| C58 | pckA mutant | |
| AB medium | 7.0 ± 0.3 | 0.3 ± 0.2 |
| + 6 mM MgCl2 | 7.6 ± 0.4 | |
| + 6 mM MgCl2 and 6 mM CaCl2 | 8.5 ± 0.4 | |
| + 1 mM MnCl2 | 7.2 ± 0.3 | |
| + 1 mM MnCl2 and 6 mM CaCl2 | 6.8 ± 0.3 | |
| Log phase | 6.7 ± 0.2 | |
A. tumefaciens strains were cultured to stationary phase in AB minimal medium (pH 7.0) plus different compounds. The values in the table are means ± standard deviations from at least three independent experiments.
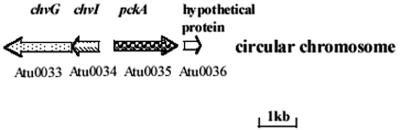
The pckA gene maps to the circular chromosome and is downstream of genes encoding a two-component system, chvG and chvI (7, 25, 39), but is transcribed in a direction opposite to that of the regulatory genes (Fig. 1). Downstream of the pckA gene is a locus that by BLAST analysis codes for a conserved hypothetical protein and is transcribed in the same direction as the pckA gene.
FIG. 1.
Location of pckA on the circular chromosome of A. tumefaciens C58.
Regulation of pckA expression.
Since two-component regulatory systems often regulate genes that map nearby, we determined whether chvGI regulates the expression of pckA. Since a previous study (21) showed that this two-component system regulates many acid-inducible genes in Agrobacterium, we first determined whether pckA is acid inducible. To test this possibility, pckA was fused with the reporter gene lacZ; the pckA-lacZ fusion was then introduced into strain C58, and the β-galactosidase activity was assayed. As shown in Table 3, pckA-lacZ expression increased 4.2-fold when the cells were grown in minimal medium at pH 5.5 compared with growth in the same medium at pH 7.0. Moreover, AS in the induction medium did not affect the expression of pckA (data not shown). This observation was expected, since no vir box, which is found in the promoter regions of AS-induced genes, could be identified in the upstream region of the gene. These results indicate that pckA is an acid-inducible gene.
TABLE 3.
Effects of chvG on the expression of the acid-inducible gene pckA, determined with the pckA-lacZ fusiona
| Strain | β-Galactosidase activity
|
Change (fold) | |
|---|---|---|---|
| pH 5.5 | pH 7.0 | ||
| C58 (wild type) | 2,468 ± 145 | 583 ± 45 | 4.2 |
| chvG mutant | 27 ± 8 | 31 ± 9 | 0 |
All strains were grown for 20 to 24 h in AB minimal medium at the pH indicated. The cultures were then assayed for β-galactosidase activity as described in Materials and Methods. The data represent the average of three independent experiments. Data are expressed in Miller units (28).
To determine whether ChvGI plays a role in the regulation of pckA, the pckA-lacZ fusion was introduced into a chvG Tn-phoA insertion mutant (7). We found that the expression of pckA in this chvG mutant was reduced about 90-fold compared to its expression in wild-type cells grown at pH 5.5 (2,468 versus 27) (Table 3). Moreover, the expression of pckA in the chvG mutant is the same in cells growing at pH 5.5 and pH 7.0 but is 10-fold lower than the expression in the wild-type strain growing at pH 7.0. It appears that chvG is very important for the expression of pckA.
In Sinorhizobium meliloti, the expression of pckA is modulated by the carbon source, and arabinose is a stronger inducer of pckA expression than glucose (30). To determine if the expression of pckA in Agrobacterium is also controlled by the carbon source, cells containing the pckA-lacZ fusion were cultured in AB minimal medium at pH 7.0 with either glucose, arabinose, glycerol, or sucrose as the sole carbon source. No difference in the expression of pckA was seen in cells grown on these different carbon sources (data not shown).
The pckA mutant is inhibited by an acidic environment.
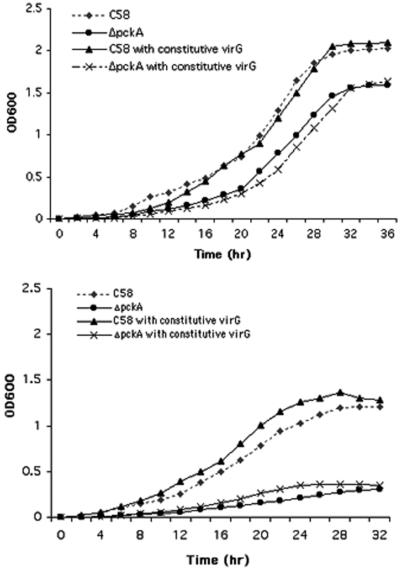
Since pckA is an acid-inducible gene, we next determined if a pckA deletion mutant was acid sensitive by monitoring the growth behavior of both the pckA mutant and its parent strain, C58 (Fig. 2). The pckA mutant grew more slowly than its parent at pH 7.0, and this growth defect was even more pronounced at pH 5.5. Furthermore, we grew the pckA mutant and wild-type C58 in the acidic medium and adjusted to the same OD and then plated serial dilutions of the bacteria on AB solid medium at pH 7.0; the number of pckA mutants growing on AB solid medium was threefold less than that of the wild-type C58. These data suggest that the pckA mutant is indeed inhibited by acid. The fact that gene expression is acid inducible suggests that the gene plays a role in overcoming growth inhibition under acidic conditions. Therefore, it is not surprising that a strain lacking the gene would be acid sensitive.
FIG. 2.
Growth curve of ΔpckA in AB minimal medium. Log-phase cultures of C58, C58 with constitutive virG, ΔpckA, and ΔpckA with constitutive virG were diluted in AB minimal medium (top, pH 7.0; bottom, pH 5.5) with a starting OD600 calculated to be 0.001. The cells were grown at 28°C with shaking. The OD600 was measured at 2-h intervals over a 36-h period.
Virulence of pckA mutant.
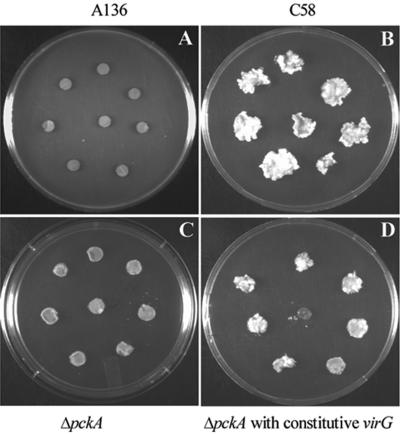
To determine if the pckA locus is important in tumor formation, we inoculated tobacco leaf disks with the pckA deletion mutant, as well as the C58 strain and the C58 strain cured of its Ti plasmid (A136), as described in Materials and Methods. The results are shown in Fig. 3 (compare B and C). The pckA deletion mutant was significantly less virulent than the wild-type C58 strain.
FIG. 3.
Virulence assay of pckA mutant on tobacco leaf disks. A. tumefaciens cells were grown and inoculated onto tobacco leaf disks as described in Materials and Methods. Tumor formation on leaf disks (n = 40) is shown.
vir gene induction of mutant.
The attenuated virulence of the pckA mutant may be due to the poor growth of the mutant in the acidic plant wound environment, which likely alters the overall physiology of the cell, and/or some other requirement for tumor formation. A readily testable possibility is that the mutation reduces the level of vir gene induction in some way. This seemed like a real possibility, since so many other mutations (ivr211, miaA, and chvD), and another gene in carbohydrate metabolism (38), also reduced vir gene induction. The plasmid pSM243cd containing the virB::lacZ translational fusion was introduced into the pckA deletion mutant and the wild-type C58 strain. As shown in Table 4, expression of the virB-lacZ fusion was reduced 84% in the pckA deletion mutant compared to the wild-type strain. We also introduced a virE-lacZ fusion on a plasmid (pSM358cd) into the same strain. Under optimal inducing conditions, the expression of virE-lacZ was reduced approximately the same amount, 80%. These results show that the expression of both virB and virE decreased in the pckA mutant to an extent that could likely explain its attenuated phenotype. To further explore this possibility, a virG-lacZ translational fusion, pWT160, was introduced into the pckA deletion mutant. In the presence of AS, the expression level of virG-lacZ decreased 88% compared with the wild type under inducing conditions (Table 4). This greatly reduced induction of the response regulator virG could certainly account for the reduced level of expression of virB and virE and thereby play a role in the attenuated virulence of the pckA mutant.
TABLE 4.
vir gene induction by AS and acidic pH in a pckA deletion mutant
| Plasmid | β-Galactosidase activitya
|
|||||
|---|---|---|---|---|---|---|
| C58
|
ΔpckA
|
ΔpckA with constitutive virG
|
||||
| pH 7.0 | pH 5.5 + AS | pH 7.0 | pH 5.5 + AS | pH 7.0 | pH 5.5 + AS | |
| virB::lacZ | 3 ± 2 | 579 ± 15 | 5 ± 4 | 95 ± 11 (83.5%) | 523 ± 56 | 592 ± 76 |
| virE::lacZ | 11 ± 2 | 629 ± 23 | 5 ± 3 | 126 ± 10 (80.0%) | 579 ± 67 | 606 ± 45 |
| virG::lacZ | 7 ± 5 | 1,043 ± 25 | 21 ± 5 | 125 ± 15 (88%) | 907 ± 134 | 1,278 ± 63 |
A. tumefaciens strains were cultured in AB minimal medium (pH 7.0) or induction medium at pH 5.5 and assayed for β-galactosidase activity between 20 and 24 hours. The data are means ± standard errors of the mean calculated from three independent experiments. The percentages in parentheses represent the reduction in expression in the ΔpckA mutant relative to the C58 wild-type strain grown at the same pH. Data are expressed in Miller units (28).
Effect of constitutive virG on vir gene induction, virulence, and growth.
To gain some insight into whether the reduced vir gene expression was responsible for the attenuated tumor phenotype and whether this reduction was upstream or downstream of the VirA-VirG signaling cascade, we introduced a constitutive virG locus (N54D) on the high-copy-number vector pSY204 into the pckA mutant. The mutant virG does not require AS, monosaccharides, or acidic conditions to induce the vir genes (19). These observations were confirmed (Table 4). However, although vir gene induction was raised to the wild-type level (Table 4), the pckA mutant still was not as virulent as the parent. Thus, pSY204 can only partially restore virulence (Fig. 3). Moreover, the constitutive virG mutation did not change the growth rate of the pckA mutant significantly in acidic or neutral glucose salts medium (Fig. 2). These results strongly suggest that reduced vir gene expression is only partially responsible for the attenuated avirulence of the pckA mutant. Presumably, the acid sensitivity of the mutant may also play a role in its reduced virulence.
The phenotype of the pckA mutant.
A mutation in chvGI confers a number of distinct properties on the cell. Since chvGI is a positive regulator of pckA, some of these properties could be due to reduced expression of this gene. chvGI mutants are sensitive to the detergents SDS, DOC, and Sarkosyl. We investigated whether the pckA mutant displayed these properties by plating the pckA insertion mutant on solid AB minimal media with one of the three detergents (0.1 g/liter SDS, 1 g/liter DOC, or 2 g/liter Sarkosyl). No difference was observed in growth on media with and without these detergents (data not shown). Furthermore, the chvGI mutants cannot grow on a complex medium, although they grow well at pH 7.0 on minimal media. The pckA mutant grows well on a complex medium, and therefore, this mutation cannot account for the phenotype (data not shown).
DISCUSSION
This study was initiated in order to identify additional genes that are under the control of the two-component regulatory system, ChvGI, that had previously been shown to be important in the virulence of Agrobacterium (7, 25). This regulatory system is especially important to understand because it controls many acid-inducible genes, both chromosomal and on the Ti plasmid, all of which (at least those so far identified) play some role in the virulence of Agrobacterium. Furthermore, it appears to function upstream of the critically important two-component system VirAG, which responds to environmental signals and activates all vir genes on the Ti plasmid. Since acidic conditions represent one of the key environmental signals for a plant wound site, the identification and characterization of genes that respond to acid conditions is important to an overall understanding of the physiology of Agrobacterium, both as a ubiquitous inhabitant of soils, which often are acidic, and as a plant pathogen, which requires an acidic environment to activate the entire pathogenesis program. The importance of the chvGI regulatory system is further underscored by its importance in the interaction of other α proteobacteria with their hosts. In S. meliloti, the synthesis of succinoglycan, which is required for the nodulation of alfalfa, is under the control of ExoS/ChvI, the homologs of ChvGI (8). In Brucella, BvrS/BvrR, the homolog of ChvGI, controls the synthesis of two outer membrane proteins, at least one of which is required for virulence (13).
The pckA locus falls into the class of acid-inducible genes under the control of ChvGI, which are important in virulence. Although only acid-inducible genes have been found thus far to be under the control of chvGI, not all such genes are regulated by chvGI. For example, the chromosomally encoded acvB locus, which is induced about threefold at pH 5.5 compared to pH 7.0 and which plays a role in virulence, is expressed to the same extent in a wild-type cell and a chvI mutant (P. Liu, unpublished observation). A purine biosynthesis gene, purB, is also acid inducible but is not under the control of chvGI (Y. Liang and P. Liu, unpublished observation).
The relationship between acid inducibility and growth inhibition of mutations at pH 5.5 is variable. A gene that is induced at pH 5.5 is likely to be especially important in a physiological process that occurs at that pH. This process might be related to the growth or survival of the bacteria in acidic soils or in the interaction with wounded plants in an acidic environment. In either case, a mutation in the gene would be magnified by a greater inhibitory effect on growth at the lower pH and perhaps a loss of virulence. This is the situation that has been observed. Thus, mutants of pckA and another chv gene, acvB, are inhibited in their growth at pH 5.5 much more than are their wild-type parents (M. Brodhagen, unpublished observation). It might be expected that a chv gene that plays a role in the physiology of Agrobacterium in the absence of a wounded plant would have several functions, only one of which is related to virulence. It would not be surprising for a mutation in a gene whose function is important for bacterial physiology under acidic conditions to result in a greatly reduced rate of growth. On the other hand, mutants of virG or virB grow as well as the wild-type strain at pH 5.5 and 7.0. A mutation in a vir gene dedicated solely to virulence would not be expected to exhibit general physiological changes manifested by a lower growth rate at pH 5.5.
At least two factors apparently play roles in the attenuation of virulence in pckA mutants. One may be related to the general physiological consequences of the mutation and the other to effects on a specific interaction with a host plant. The first relates to the inhibition of growth of the pckA mutant at pH 5.5, which approximates the acidic conditions at the wound site on a plant. Although wild-type Agrobacterium does not grow as well at pH 5.5 as it does at pH 7.0, the mutation in pckA amplifies this growth inhibition. This increased generation time certainly must alter the overall physiology of the cell significantly, which in turn might lead to reduced DNA transfer. However, a reduced growth rate does not always reduce virulence. Liu et al. (23) reported that a mutation in chvD resulted in an avirulent strain that grew slowly in a complex rich medium but could be complemented with a plasmid that restored virulence without affecting the generation time of the cells. In the present study, the constitutive virG locus significantly enhanced the virulence of the pckA mutant without significantly affecting its growth rate (Fig. 2 and 3).
The second, and probably more interesting, factor that plays a role in reduced virulence in the pckA mutant is the major reduction in the expression of virG under acidic conditions with AS. This reduction in virG expression, in turn, results in the reduced expression of all vir genes. Thus, the induction of virB and virE is reduced over 80%. The basis for this reduction is not at all clear, but it apparently relates to an early step in the signaling process. Since a constitutive virG gene can overcome the reduction in the expression of all three vir genes, the pckA gene in some way must be involved in the signaling cascade upstream of virG gene expression. Furthermore, since the restoration of vir gene expression is not accompanied by a complete restoration of virulence, an additional factor(s) must be involved in reducing virulence. One possibility, as already discussed, is the sensitivity of the pckA mutant to growth under acid conditions.
The reason(s) why a mutation in pckA reduces signal transduction so significantly is not at all clear. This enzyme is the first step in gluconeogenesis, and therefore, the synthesis of sugars would be reduced. Although sugars are critical for vir gene induction, especially in strain C58, glucose was supplied, which should have provided the sugar needed for induction. Thus, the pckA mutant can be added to the list of chromosomal-gene mutants in Agrobacterium that are significantly reduced in vir gene induction. These include the chvD (23), ivr211 (27), and miaA mutants (12). In none of these cases is the basis for this reduction understood. We have observed recently that a mutation in citrate synthase also results in reduced vir gene induction (38). The question of whether the reductions in vir gene induction in the two mutations in carbohydrate metabolism share a common basis awaits further study.
One of the interesting features that these studies have revealed is the difference in regulation of pckA shown by Agrobacterium compared to all of the other prokaryotes studied thus far. There is no evidence for catabolite repression by glucose, sucrose, or glycerol in Agrobacterium, all of which are found in Rhizobium. Furthermore, in contrast to Rhizobium, there is no evidence from the lacZ fusion expression data in pckA that the enzyme is induced in the stationary phase of growth (data not shown). This induction in Rhizobium requires cyclic AMP (32), but there is no evidence for a cyclic AMP binding site in the promoter region of the pckA gene of Agrobacterium.
The biological significance of the divalent cation binding site is also uncertain. In E. coli, Ca2+ binds to this site and activates the enzyme allosterically (26). We could not demonstrate that Ca2+ activates the pckA enzyme in Agrobacterium. We conclude that the control of pckA differs in Agrobacterium and in E. coli. This may reflect the possibility that this enzyme plays somewhat different roles in Agrobacterium, Rhizobium, and E. coli. In all cases, PckA is the first enzyme in gluconeogenesis, and all three organisms can synthesize sugars from succinate. However, in Agrobacterium, it likely plays an additional as-yet-undefined role, which may involve the interaction of Agrobacterium with its hosts.
Although we can add the pckA gene to the list of genes that are under the control of the ChvGI regulatory system, it is clear that additional genes remain to be identified. This conclusion is based on the phenotypic characterization of mutants of chvGI and the mutants known to be under its control. chvGI mutants grow poorly under acidic conditions, do not grow on a complex medium, and are inhibited in their growth by detergents; the last phenotype suggests a defect in the cell envelope. Some of the mutants under the control of ChvGI have these properties. The pckA mutant grows poorly under acidic conditions, and the aopB mutation involves a protein on the surface of the cell. However, whether it confers sensitivity to detergents has not been reported. Since the ChvGI regulatory system plays a crucial role in the acidic signaling process between plants and Agrobacterium, it is of considerable interest to identify all of the genes under its control. Probably the most direct approach is to identify genes that are up or down regulated in a chvI mutant, using microarray technology. Such experiments are under way.
Acknowledgments
This work was supported by a grant from the National Institutes of Health to E. W. Nester (GM 32618).
We are grateful to T. Charles for Agrobacterium strain At11063.
REFERENCES
- 1.Ankenbauer, R. G., and E. W. Nester. 1993. The Agrobacterium Ti plasmid and crown gall tumorigenesis: a model for signal transduction in host-pathogen interactions, p. 67-104. In J. Kurjan and B. L. Taylor (ed.), Signal transduction. Academic Press Inc., San Diego, Calif.
- 2.Banta, L. M., R. D. Joerger, V. R. Howitz, A. M. Campbell, and A. N. Binns. 1994. Glu-255 outside the predicted ChvE binding site in virA is crucial for sugar enhancement of acetosyringone perception by Agrobacterium tumefaciens. J. Bacteriol. 176:3242-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker, A., M. Schmidt, W. Jager, and A. Puhler. 1995. New gentamicin-resistance and lacZ promoter-probe cassettes suitable for insertion mutagenesis and generation of transcriptional fusions. Gene 162:37-39. [DOI] [PubMed] [Google Scholar]
- 4.Cangelosi, G. A., E. A. Best, G. Martinetti, and E. W. Nester. 1991. Genetic analysis of Agrobacterium. Methods Enzymol. 204:384-397. [DOI] [PubMed] [Google Scholar]
- 5.Cangelosi, G. A., G. Martinetti, J. A. Leigh, C. C. Lee, C. Theines, and E. W. Nester. 1989. Role of Agrobacterium tumefaciens ChvA protein in export of B-1,2-glucan. J. Bacteriol. 171:1609-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cangelosi, G. A., R. G. Ankenbauer, and E. W. Nester. 1990. Sugars induce the Agrobacterium virulence genes through a periplasmic binding protein and a transmembrane signal protein. Proc. Natl. Acad. Sci. USA 87:6708-6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charles, T. C., and E. W. Nester. 1993. A chromosomally encoded two-component sensory transduction system is required for virulence of Agrobacterium tumefaciens. J. Bacteriol. 175:6614-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng, H. P., and G. C. Walker. 1998. Succinoglycan production by Rhizobium meliloti is regulated through the ExoS-ChvI two-component regulatory system. J. Bacteriol. 180:20-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das, A. S., S. Stachel, P. Ebert, P. Allenza, A. Montoya, and E. W. Nester. 1986. Promoters of Agrbacterium tumefaciens Ti-plasmid virulence gene. Nucleic Acids Res. 14:2869-2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doty, S. L., M. C. Yu, J. I. Lundin, J. D. Heath, and E. W. Nester. 1996. Mutational analysis of the input domain of the VirA protein of Agrobacterium tumefaciens. J. Bacteriol. 178:961-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gelvin, S. B. 2003. Agrobacterium-mediated plant transformation: the biology behind the “Gene-Jockeying” tool. Microbiol. Mol. Biol. Rev. 67:16-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray, J., J. Wang, and S. B. Gelvin. 1992. Mutation of the miaA gene of Agrobacterium tumefaciens results in reduced vir gene expression. J. Bacteriol. 174:1086-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guzman-Verri, C., L. Manterola, A. Sola-Landa, A. Parra, A. Cloeckaert, J. Garin, J. P. Gorvel, I. Moriyon, E. Moreno, and I. Lopez-Goni. 2002. The two-component system BvrR/BvrS essential for Brucella abortus virulence regulates the expression of outer membrane proteins with counterparts in members of the Rhizobiaceae. Proc. Natl. Acad. Sci. USA 99:12375-12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamilton, R. H., and M. Z. Fall. 1971. The loss of tumor-initiating ability in Agrobacterium tumefaciens by incubation at high temperature. Experientia 27:229-230. [DOI] [PubMed] [Google Scholar]
- 15.Heath, J. D., T. C. Charles, and E. W. Nester. 1995. Ti plasmid and chromosomally encoded two-component systems important in plant cell transformation by Agrobacterium species, p. 367-385. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. ASM Press, Washington, D.C.
- 16.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 17.Inui, M., K. Nakata, J. H. Roh, K. Zahn, and H. Yukawa. 1999. Molecular and functional characterization of the Rhodopseudomonas palustris no. 7 phosphoenolpyruvate carboxykinase gene. J. Bacteriol. 181:2689-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jia, Y. H., L. P. Li, Q. M. Hou, and S. Q. Pan. 2002. An Agrobacterium gene involved in tumorigenesis encodes an outer membrane protein exposed on the bacterial cell surface. Gene 284:113-124. [DOI] [PubMed] [Google Scholar]
- 19.Jin, S., Y. Song, S. Q. Pan, and E. W. Nester. 1993. Characterization of a virG mutation that confers constitutive virulence gene expression in Agrobacterium. Mol. Microbiol. 7:555-562. [DOI] [PubMed] [Google Scholar]
- 20.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotics-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 21.Li, L., Y. Jia, Q. Hou, T. C. Charles, E. W. Nester, and S. Q. Pan. 2002. A global pH sensor: Agrobacterium sensor protein ChvG regulates acid-inducible genes on its two chromosomes and Ti plasmid. Proc. Natl. Acad. Sci. USA 99:12369-12374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, K., J. Yu, and D. G. Russell. 2003. pckA-deficient Mycobacterium bovis BCG shows attenuated virulence in mice and in macrophages. Microbiology 149:1829-1835. [DOI] [PubMed] [Google Scholar]
- 23.Liu, Z., M. Jacobs, D. A. Schaff, C. A. McCullen, and A. N. Binns. 2001. ChvD, a chromosomally encoded ATP-binding cassette transporter-homologous protein involved in regulation of virulence gene expression in Agrobacterium tumefaciens. J. Bacteriol. 183:3310-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lohrke, S. M., S. Nechaev, H. Yang, K. Severinov, and S. J. Jin. 1999. Transcriptional activation of Agrobacterium tumefaciens virulence gene promoters in Escherichia coli requires the A. tumefaciens RpoA gene, encoding the alpha subunit of RNA polymerase. J. Bacteriol. 181:4533-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mantis, N. J., and S. C. Winans. 1993. The chromosomal response regulatory gene chvI of Agrobacterium tumefaciens complements an Escherichia coli phoB mutation and is required for virulence. J. Bacteriol. 175:6626-6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medina, V., R. Pontarollo, D. Glaeske, H. Tabel, and H. Goldie. 1990. Sequence of the pckA gene of Escherichia coli K-12: relevance to genetic and allosteric regulation and homology of E. coli phosphoenolpyruvate carboxykinase with the enzymes from Trypanosoma brucei and Saccharomyces cerevisiae. J. Bacteriol. 172:7151-7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Metts, J., J. West, S. H. Doares, and A. G. Matthysse. 1991. Characterization of three Agrobacterium tumefaciens avirulent mutants with chromosomal mutations that affect induction of vir genes. J. Bacteriol. 173:1080-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Murashige, T., and F. Skong. 1962. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant 15:473-497. [Google Scholar]
- 30.Osteras, M., B. T. Driscoll, and T. M. Finan. 1995. Molecular and expression analysis of the Rhizobium meliloti phosphoenolpyruvate carboxykinase (pckA) gene. J. Bacteriol. 177:1452-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osteras, M., T. M. Finan, and J. Stanley. 1991. Site-directed mutagenesis and DNA sequence of pckA of Rhizobium NGR234, encoding phosphoenolpyruvate carboxykinase: gluconeogenesis and host-dependent symbiotic phenotype. Mol. Gen. Genet. 230:257-269. [DOI] [PubMed] [Google Scholar]
- 32.Park, E. A., A. L. Gurney, S. E. Nizielski, P. Hakimi, Z. Cao, A. Moorman, and R. W. Hanson. 1993. Relative roles of CCAAT/enhancer-binding protein beta and cAMP regulatory element-binding protein in controlling transcription of the gene for phosphoenolpyruvate carboxykinase (GTP). J. Biol. Chem. 268:613-619. [PubMed] [Google Scholar]
- 33.Peng, W. T., Y. W. Lee, and E. W. Nester. 1998. The phenolic recognition profiles of the Agrobacterium tumefaciens VirA protein broadened by the high level of the sugar binding protein ChvE. J. Bacteriol. 180:5632-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Sciaky, D., A. L. Montoya, and M. D. Chilton. 1978. Fingerprints of Agrobacterium Ti plasmids. Plasmid 1:238-253. [DOI] [PubMed] [Google Scholar]
- 36.Stachel, S. E., and P. C. Zambryski. 1986. virA and virG control the plant-induced activation of the T-DNA transfer process of A. tumefaciens. Cell 46:325-333. [DOI] [PubMed] [Google Scholar]
- 37.Stachel, S. E., E. Nessebsm, M. Van Montagu, and P. C. Zambryski. 1985. Identification of the signal molecules produced by wounded plant cells that activate T-DNA transfer in Agrobacterium tumefaciens. Nature 318:624-629. [Google Scholar]
- 38.Suksomtip, M., P. Liu, T. Anderson, S. Tungpradabkul, D. W. Wood, and E. W. Nester. 2005. Citrate synthase mutants of Agrobacterium are attenuated in virulence and display reduced vir gene induction. J. Bacteriol. 187:4844-4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wood, D. W., J. C. Setubal, R. Kaul, D. E. Monks, J. P. Kitajima, V. K. Okura, Y. Zhou, L. Chen, G. E. Wood, N. F. Almeida, L. Woo, Y. Chen, I. T. Paulsen, J. A. Eisen, P. D. Karp, D. Bovee, P. Chapman, J. Clendenning, G. Deatherage, W. Gillet, C. Grant, T. Kutyavin, R. Levy, M.-J. Li, E. McClelland, A. Palmieri, C. Raymond, G. Rouse, C. Saenphimmachak, Z. Wu, P. Romero, D. Gordon, S. Zhang, H. Yoo, Y. Tao, P. Biddle, M. Jung, W. Krespan, M. Perry, B. Gordon-Kamm, L. Liao, S. Kim, C. Hendrick, Z.-Y. Zhao, M. Dolan, F. Chumley, S. V. Tingey, J.-F. Tomb, M. P. Gordon, M. V. Olson, and E. W. Nester. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317-2323. [DOI] [PubMed] [Google Scholar]
- 40.Xu, X. Q., and S. Q. Pan. 2000. An Agrobacterium catalase is a virulence factor involved in tumorigenesis. Mol. Microbiol. 35:407-414. [DOI] [PubMed] [Google Scholar]