Abstract
The methanogenic archaeon Methanosarcina mazei strain Gö1 uses versatile carbon sources and is able to fix molecular nitrogen with methanol as carbon and energy sources. Here, we demonstrate that when growing on trimethylamine (TMA), nitrogen fixation does not occur, indicating that ammonium released during TMA degradation is sufficient to serve as a nitrogen source and represses nif gene induction. We further report on the transcriptional regulation of soluble methyltransferases, which catalyze the initial step of methylamine consumption by methanogenesis, in response to different carbon and nitrogen sources. Unexpectedly, we obtained conclusive evidence that transcription of the mtmB2C2 operon, encoding a monomethylamine (MMA) methyltransferase and its corresponding corrinoid protein, is highly increased under nitrogen limitation when methanol serves as a carbon source. In contrast, transcription of the homologous mtmB1C1 operon is not affected by the nitrogen source but appears to be increased when TMA is the sole carbon and energy source. In general, transcription of operons encoding dimethylamine (DMA) and TMA methyltransferases and methylcobalamine:coenzyme M methyltransferases is not regulated in response to the nitrogen source. However, in all cases transcription of one of the homologous operons or genes is increased by TMA or its degradation products DMA and MMA.
The archaeon Methanosarcina mazei strain Gö1 belongs to the methylotrophic methanogens of the order Methanosarcinales. Those species have the most versatile substrate spectrum within the methanogenic archaea and are able to grow on H2 plus CO2, acetate, or methylotrophic substrates such as methanol or methylamines as sole carbon and energy sources (10, 21). We recently showed that M. mazei is able to use molecular nitrogen as sole nitrogen source when growing on methanol and characterized a single nitrogen fixation (nif) gene cluster encoding a molybdenum-containing nitrogenase (6). For M. barkeri it has been demonstrated that degradation of each trimethylamine (TMA), dimethylamine (DMA), and monomethylamine (MMA) leads to the release of ammonium into the medium (13). However, whether M. mazei fixes nitrogen when growing on methylamines or directly uses the ammonium generated by the disproportion of methylamines has never been studied.
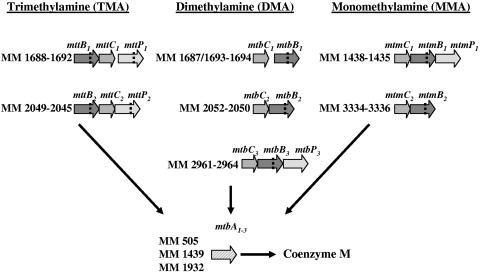
The first step of the disproportion of methylamines is catalyzed by soluble methyltransferases, which transfer the methyl group to a cognate corrinoid protein whereby each different methylamine requires its specific methyltransferase and corresponding corrinoid protein (2, 7, 8). The demethylation of the cognate corrinoid proteins is finally catalyzed by a methylcobalamine:coenzyme M (CoM) methyltransferase (9) resulting in methyl-CoM, the precursor of methane. The genes encoding these enzymes were named according to Krzycki and coworkers (15), where the final letter designates the polypeptide function. Within this nomenclature, B describes the substrate specific methyltransferase, C the corrinoid binding polypeptide, and A the CoM-methylating protein. For M. barkeri multiple and nearly identical copies of the operons encoding DMA and MMA methyltransferases and their respective corrinoid proteins have been identified (15, 19). Analyzing the genome sequence of M. mazei, seven operons coding for methylamine methyltransferases and their corresponding corrinoid proteins and three genes coding for methylcobalamine:CoM methyltransferases have been identified (4), an overview of which is depicted in Fig. 1. Although present in all Methanosarcina species, the function of those multiple homologous genes encoding methylamine methyltransferases and their corresponding corrinoid proteins remains unclear. A carbon source-dependent expression of the three homologous methanol:corrinoid methyltransferase (MtaB) proteins has been demonstrated for M. thermophila by a proteomic approach (5). Thus, a likely reason for the existence of homologous operons encoding methylamine methyltransferases may be the requirement of different methylamine methyltransferases in response to different growth conditions, which may be achieved by differential expression of the homologous operons.
FIG. 1.
Operons encoding methylamine methyltransferases in M. mazei (modified with permission from reference 14). The indicated open reading frame numbers were obtained from GenBank, of the National Center for Biotechnology (4). Genes are designated according to Paul et al. (15). The dotted lines indicate the in-frame amber codon of methylamine methyltransferases.
In order to investigate the requirement of homologous operons in M. mazei we analyzed the transcriptional regulation of genes encoding soluble methyltransferases involved in methylamine degradation in response to different nitrogen and carbon sources. The results obtained clearly demonstrate a unique nitrogen-dependent regulation of one of the two operons encoding MMA methyltransferases (mtmB2C2), whereas, if regulated, transcription of the other methyltransferases is regulated in response to the carbon source.
MATERIALS AND METHODS
Growth.
Methanosarcina mazei strain Gö1 (DSM 3647) was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ). Cells were grown without shaking at 37°C in 5- or 50-ml closed growth tubes on 150 mM methanol and 40 mM acetate, which is defined as growth on methanol in the following or 25 mM trimethylamine (TMA) in a mineral medium under a nitrogen atmosphere containing 20% CO2 as described previously (3, 6). Nitrogen sufficiency (+NH4+) is defined as conditions under which 9 mM ammonium is added to an otherwise nitrogen-free medium, whereas nitrogen limitation (−NH4+) is defined in the following as conditions under which ammonium was omitted from the medium and molecular nitrogen in the gas atmosphere was the sole nitrogen source available. Using the Ammonium Assay kit (Sigma-Aldrich), the amount of free ammonium in nitrogen-limited medium was determined to be below 10 μM, irrespectively of whether the medium was additionally supplemented with TMA as carbon source or not. Thus, TMA and TMA-supplemented nitrogen-limited medium were not contaminated with free ammonium. For ammonium up-shift experiments cells were grown with methanol under nitrogen limitation in 1-liter cultures until they reached a turbidity of 0.3 to 0.4 at 600 nm (exponential growth phase). Thirty-milliliter aliquots were supplemented with 15 mM ammonium (ammonium up-shift), whereas the nitrogen-limited culture served as control. Cultures were further incubated for 1 h before RNA was isolated as described below.
RNA isolation.
For quantitative reverse transcriptase (RT)-PCR experiments, M. mazei cultures were grown anaerobically until cells reached a turbidity of 0.26 to 0.29 (methanol, −NH4+) or 0.45 to 0.6 at 600 nm (methanol, +NH4+; and TMA, ±NH4+), which corresponds to mid-exponential growth phase under the respective conditions. In order to prevent RNA degradation during harvest, 70-ml cell cultures were rapidly chilled for 10 min on ice before centrifugation of the anaerobic culture tubes at 4°C for 20 min at 3,000 rpm. When TMA served as carbon source, cells were additionally washed under anaerobic conditions with 50 ml ice-cold medium without a carbon source to minimize TMA contaminations, which otherwise negatively affected quantitative RT-PCRs. Cell pellets were resuspended in 125 μl cold 0.01 M sodium acetate (pH 5.2) supplemented with 0.3 M sucrose. After the addition of 125 μl 0.01 M sodium acetate (pH 5.2) containing 2% sodium dodecyl sulfate followed by an incubation at 65°C for 1.5 min, RNA was isolated by phenol extraction (18). Isolated RNA was incubated with 40 U DNaseI (Roche) for 30 min at room temperature to avoid contamination with genomic DNA. After removal of DNaseI by chloroform-isoamylalcohol (24:1, vol/vol) extractions followed by ethanol precipitation, the RNA was finally resuspended in 40 μl diethyl pyrocarbonate-water. Concentration and purity of extracted RNA were determined by measuring the absorption at 260 and 280 nm. Control PCRs using RNA as template in the absence of reverse transcriptase confirmed that the isolated RNA was free of contaminating genomic DNA. For the analysis of potential growth-phase-dependent transcription of methylamine methyltransferases, RNA was isolated from cells grown with 12.5 mM TMA as carbon source. Cells for RNA isolation were harvested in lag phase (turbidity of 0.2 at 600 nm), exponential phase (turbidity of 0.45 at 600 nm), and stationary phase (turbidity of 0.65 at 600 nm).
Quantitative reverse transcriptase (RT)-PCR analysis.
Quantitative RT-PCR assays were performed using the QuantiTect Probe RT-PCR kit (QIAGEN) and the iCycler system from Bio-Rad. For each condition, at least three independent experiments using RNA isolated from independently grown cultures were performed for the respective genes of interest. Reactions were prepared according to the manufacturer's protocol using 400 ng of total M. mazei RNA per reaction in a total volume of 25 μl. In general, 55°C was used for primer annealing. Using specific primer sets to amplify the genes of interest and control genes (summarized in Table 1) yielded amplification products, the size of which varied from 100 to 150 bp in length. For negative controls, water was used instead of the RNA template. At the end of each run a melting curve was plotted to verify the specificity of the amplification product. The cycle threshold (Ct), which defines the cycle in which the first significant increase in fluorescence is detected, was calculated by the iCycler software (Bio-Rad) and was used to determine the fold change in gene expression. A high Ct value of a certain gene corresponds to a low mRNA amount of the respective gene, whereas a low Ct value indicates a high abundance of the specific mRNA. Ct values were normalized in respect to the corresponding Ct values obtained from the same RNA for three genes (MM 1621, MM 2181, MM 1215; see Table 1), which were shown to be transcribed to the same amount irrespective of the nitrogen or carbon availability in microarray experiments (K. Veit and R. A. Schmitz, unpublished data). The fold change was calculated using the formula fold change = 2−ΔΔCt, as described by Talaat et al. (20).
TABLE 1.
Primer sets used for quantitative RT-PCR analysis
| Descriptiona | Gene and/or protein | Sequence (5′ → 3′) |
|---|---|---|
| MM1621 for.rt | Putative CobW protein | TAGGAGGTTTTCTCGGAAGCG |
| MM1621 rev.rt | Putative CobW protein | AAGCGTATCTCCATCAAGCCC |
| MM2181 for.rt | Fructose-1,6-bisphosphatase | GCCTCCATGAGAAGAATGCTC |
| MM2181 rev.rt | Fructose-1,6-bisphosphatase | CTTCAAGGTCTCCAACTCCTG |
| MM1215 for.rt | Hexulose-6-phosphate synthase | TCAAGAGCGAGGGCATGAATG |
| MM1215 rev.rt | Hexulose-6-phosphate synthase | GCACTACCGAGAACAATAGCC |
| Mm nifH for.rt | nifH; nitrogenase iron protein | CCACGCAGAATCTTACTGCAG |
| Mm nifH rev.rt | nifH; nitrogenase iron protein | AGCACGGTTTTCTGGTTCAGG |
| MM1438 for.rt | mtmC1; MMA corrinoid protein 1 | GGGCTTGTGTCCAGCATAAAC |
| MM1438 rev.rt | mtmC1; MMA corrinoid protein 1 | CCTCCGTTTCTGTCGAAAACG |
| MM3334 for.rt | mtmC2; MMA corrinoid protein 2 | GAAAGAAGTGATTATGGGATGAC |
| MM3334 rev.rt | mtmC2; MMA corrinoid protein 2 | AGAGGGGAAATTCCTGCATC |
| MM2052 for.rt | mtbC2; DMA corrinoid protein 2 | TGGGGGTTGAGAATGACAAGC |
| MM2052 rev.rt | mtbC2; DMA corrinoid protein 2 | AGGCTCCATAACCTGCTTTGC |
| MM1688 for.rt | mttB1; TMA methyltransferase 1 | CTTATTCGGAGGTTACAACCCTGG |
| MM1688 rev.rt | mttB1; TMA methyltransferase 1 | GAATTGCTTTGAGTTCATCAGTGG |
| MM2049 for.rt | mttB2; TMA methyltransferase 2 | CCTGAAGCAAGGCAGATCTTC |
| MM2049 rev.rt | mttB2; TMA methyltransferase 2 | CAGAGAACGAACCTTGAAGGC |
| MM505 for.rt | mtbA1; methylcobalamine:CoM methyltransferase 1 | GGAACCGTAGAGCTTATGGAC |
| MM505 rev.rt | mtbA1; methylcobalamine:CoM methyltransferase 1 | AGTGGACAGCTTCAAAGCCTG |
| MM1439 for.rt | mtbA2; methylcobalamine:CoM methyltransferase 2 | TATCGGGTCCATGATAGGTCC |
| MM1439 rev.rt | mtbA2; methylcobalamine:CoM methyltransferase 2 | GCTATGTTGAAGTCTGCGCAG |
| MM1932 for.rt | mtbA3; methylcobalamine:CoM methyltransferase 3 | CTGTTCTGTTACCCAGACTGG |
| MM1932 rev.rt | mtbA3; methylcobalamine:CoM methyltransferase 3 | CCCACATTCTCAAATCCTGC |
Mm, M. mazei; for.rt, forward primer; rev.rt, reverse primer.
Construction of plasmids.
Operons encoding each of the methylamine methyltransferases and their respective corrinoid proteins were amplified from genomic M. mazei DNA with specific homologous primers. The PCR products were poly(A) tailed using 0.2 mM dATP and Taq polymerase and were cloned into pDrive using the PCR Cloning kit according to the manufacturer's instructions (QIAGEN). The resulting plasmids were verified by sequencing and designated pRS249 and pRS250 for the MMA methyltransferase operons, pRS254, pRS255, and pRS256 for the DMA methyltransferase operons, and pRS251 and pRS252 for the TMA methyltransferase operons (see Table 2 for details).
TABLE 2.
Plasmids constructed to analyze the specificity of primer sets used in quantitative RT-PCR analysis
| Plasmid | Descriptiona |
|---|---|
| pDrive | General cloning vector (QIAGEN) |
| pRS249 | orfs MM1436 to MM1438 cloned in pDrive |
| pRS250 | orfs MM3334 to MM3336 cloned in pDrive |
| pRS251 | orfs MM1687 to MM1690 cloned in pDrive |
| pRS252 | orfs MM2047 to MM2049 cloned in pDrive |
| pRS254 | orfs MM1693 to MM1694 cloned in pDrive |
| pRS255 | orfs MM2050 to MM2052 cloned in pDrive |
| pRS256 | orfs MM2961 to MM2963 cloned in pDrive |
ORFs, open reading frames.
Specificity of primers used for quantitative RT-PCR.
The operons encoding the homologous methylamine methyltransferases are highly similar to each other. In order to obtain specific PCR products by quantitative RT-PCR at least one of the primers had to be designed directly upstream of the translational start codon of the corresponding gene or in regions that were unique to one of the homologous genes. To verify the specificity of the primer sets, plasmids pRS249 to pRS256 (Table 2) were used as templates in PCR amplifications. In all cases a single PCR product was obtained when using the specific primers (Table 1) (data not shown). The specificity of the primer sets was further confirmed by cloning and sequencing the amplification products from the corresponding quantitative RT-PCR assays for all different methylamine methyltransferases. These analyses clearly demonstrated that for all primer sets exclusively the requested product was amplified.
M. mazei mtbC1, mttB1, mttC1, mttP, and mtbB1 (MM 1687 to MM 1694) have been shown to form a transcriptional unit (K. Veit and R. A. Schmitz, unpublished), which is in accordance to the genomic organization in M. barkeri (15). Thus, in the following experiments the mttB1 gene was exemplarily investigated for the regulation of the complete operon encoding DMA and TMA methyltransferase 1, their corresponding corrinoid proteins, and a potential TMA permease. We were able to design specific primer sets for each of the methylamine methyltransferases except for the mtbB3C3 operon, though multiple primer sets were generated and analyzed for specificity.
RESULTS
Soluble methyltransferases are involved in the degradation of different methylamines, TMA, DMA, or MMA, by catalyzing the initial methyl group transfer onto the respective corrinoid proteins. This degradation of methylamines finally leads to the release of ammonium. In order to investigate the functional role of the different homologous methylamine methyltransferases we (i) analyzed growth of M. mazei on TMA as carbon plus energy source under different nitrogen availabilities and (ii) focused on the influence of different carbon and nitrogen sources on the transcriptional regulation of soluble methyltransferases.
TMA is used as carbon and nitrogen source in M. mazei.
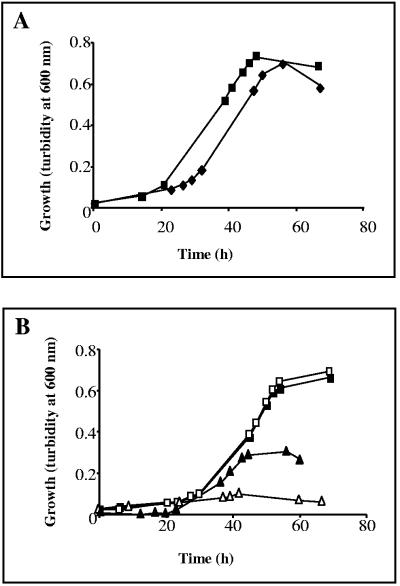
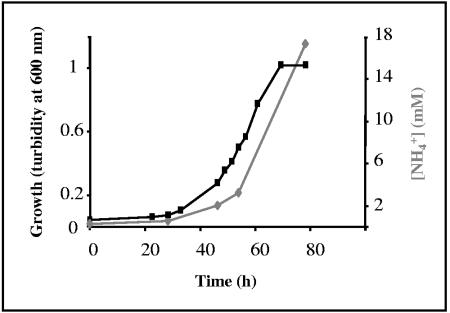
To determine whether M. mazei uses the ammonium released during degradation of methylamines, growth on TMA was analyzed under different nitrogen availabilities in comparison to growth on methanol as the sole carbon and energy source. Comparing growth with the two different carbon sources under nitrogen-sufficient conditions (+NH4+) demonstrated that the doubling time (td) and cell yield obtained under either condition was in the same range (tdmethanol, +NH4+ = 11 ± 1 h; tdTMA, +NH4+ = 11.5 ± 1.4 h) (Fig. 2A). However, under nitrogen-limiting conditions (with molecular nitrogen as sole nitrogen source), cells provided with TMA reached a significantly higher cell yield than cells grown on methanol, although the respective doubling times were nearly identical (tdTMA, −NH4+ = 12.2 ± 0.8 h; tdmethanol, −NH4+ = 12.6 ± 0.8 h) (Fig. 2B, filled symbols). When in addition molecular nitrogen in the gas atmosphere was changed to argon, methanol-supplemented cells were not able to grow at all (Fig. 2B, empty triangle) (6), confirming that the medium was free of nitrogen contaminants. However, cells provided with TMA were growing with nearly the same doubling time (Fig. 2B, empty squares) as observed for growth with ammonium, even though no additional nitrogen source was present under those conditions. These findings indicate that TMA not only served as carbon and but also as nitrogen source. In order to further investigate whether genes required for nitrogen fixation (nif genes) are repressed under those conditions, transcription of the structural nifH gene encoding the iron protein of nitrogenase was analyzed. RNA purified from methanol-grown cells with molecular nitrogen as sole nitrogen source served as control for nif gene induction. Quantitative RT-PCR analysis of RNA isolated from cells grown under nitrogen limitation demonstrated that transcription of nifH is significantly decreased when TMA is the carbon source (CtTMA, −NH4+ = 25 ± 1.3; CtTMA, +NH4+ = 26 ± 1.9; Ctmethanol, +NH4+ = 27 ± 0.9 h; Ctmethanol, −NH4+ = 14 ± 0.7). As TMA-complemented nitrogen-limited medium did not contain significant amounts of free ammonium (see Materials and Methods), this finding indicates that the release of ammonium during methylamine degradation represses nif gene induction. Further confirmation was obtained by demonstrating that up to 16.5 mM of free ammonium was accumulated in the medium during growth on TMA (Fig. 3).
FIG. 2.
Growth of M. mazei with different carbon and nitrogen sources. Growth was analyzed under nitrogen sufficiency (A) and nitrogen limitation (B) with methanol (triangles) or TMA (squares) as carbon source. Under nitrogen limitation (B) different gas atmospheres were used, nitrogen gas atmosphere containing 20% CO2 (filled symbols) or an argon gas atmosphere containing 20% CO2 (empty symbols).
FIG. 3.
Determination of ammonium in nitrogen-free medium during growth of M. mazei on TMA. The amount of free ammonium was determined at different time points during growth of M. mazei in nitrogen-free medium supplemented with 25 mM TMA as carbon source using the Ammonium Assay kit (Sigma-Aldrich). Squares, growth curve; rhombuses, concentration of free ammonium (in millimolars).
Regulation of soluble methyltransferases in response to different nitrogen and carbon sources.
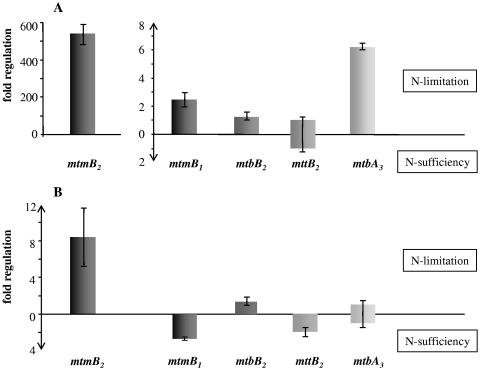
A nitrogen-dependent differential transcription of the two operons encoding a soluble MMA methyltransferase and the corresponding corrinoid protein (mtmB1C1 and mtmB2C2) was recently observed in genome-wide microarray experiments. Both operons were shown to be significantly up-regulated (approximately 25-fold) when molecular nitrogen was used as the sole nitrogen source and methanol as the carbon source (K. Veit and R. A. Schmitz, unpublished). To verify this finding and examine the potential regulation of other methyltransferases in response to different nitrogen and carbon sources, quantitative RT-PCR analysis for cultures grown under various growth conditions was carried out using specific primers for the respective genes (see Materials and Methods and Table 1). The data obtained from at least three independent RNA preparations were normalized in respect to three genes, which were transcribed to the same amount irrespective of the nitrogen or carbon availability (see Materials and Methods).
Under nitrogen limitation, a 543-fold up-regulation of the mtmB2C2 operon encoding MMA methyltransferase 2 was obtained when methanol was used as carbon source, whereas transcription of the homologous mtmB1C1 operon occurred at a constant level independently of the nitrogen source (Fig. 4A). This finding is in contrast to the results obtained by the DNA microarray experiments, indicating that the DNA microarray technique we used does not allow differentiating between the homologous methylamine methyltransferase operons. The use of specific primer sets in quantitative RT-PCR analysis, however, overcomes this problem. In order to further confirm the nitrogen-dependent regulation of the mtmB2C2 operon, an ammonium up-shift experiment was carried out in which M. mazei was grown under nitrogen limitation using methanol as carbon source. In exponential growth phase the culture was divided in two parts, one of which was exposed to 15 mM ammonium. Both cultures were further incubated for an hour before RNA was isolated and analyzed. Comparing the data from three independent experiments demonstrated that the mean Ct value obtained for RNA isolated from the ammonium-shifted cultures was significantly higher (Ct = 24 ± 0.4) than the one for the nitrogen-limited culture (Ct = 17 ± 2.1). This significant decrease of mtmB2C2 mRNA amount upon the ammonium up-shift within an hour again strongly indicated the nitrogen-dependent regulation of the mtmB2C2 operon. When cells were grown on TMA, the mtmB2C2 operon was only slightly up-regulated under nitrogen limitation (approximately eightfold; Fig. 4B), which is apparently based on the release of free ammonium during TMA degradation.
FIG. 4.
Transcriptional regulation of genes encoding methylamine methyltransferases in response to different carbon and nitrogen sources. RNA was isolated from cultures grown with methanol (A) or TMA (B) as carbon source in exponential growth phase and subjected to quantitative RT-PCR. The regulation (fold) under the different nitrogen availabilities was calculated for the genes indicated as described in Materials and Methods.
In general, genes encoding DMA and TMA methyltransferases and their respective corrinoid proteins were not differentially transcribed in response to the different nitrogen availabilities with methanol as carbon source as exemplarily indicated for the mtbB2 and mttB2 genes in Fig. 4A. Further, transcription of only one of three homologous genes encoding methylcobalamine:CoM methyltransferase (mtbA2) was slightly increased under nitrogen limitation (approximately sixfold; Fig. 4A), whereas the other two homologous genes were transcribed independently of the nitrogen source (data not shown). Also, when using TMA as carbon source, transcription of the mtmB1C1 operon as well as transcription of the operons encoding DMA and TMA methyltransferases and genes encoding methylcobalamine:CoM methyltransferase were not affected by the different nitrogen availabilities (Fig. 4B, which exemplarily shows one of the respective homologous operons or genes).
We further investigated whether a regulation of soluble methylamine methyltransferases occurs in response to the carbon source. RNA was isolated from cultures grown either on methanol or TMA under nitrogen sufficiency. Quantitative RT-PCR analysis demonstrated that in general, only one of the homologous genes or operons encoding a specific methyltransferase was carbon-dependent regulated: mtmB1C1, mtbB1, mttB1C1, and mtbA2 (Table 3). Transcription of the respective operons or genes was apparently increased in the presence of TMA or its degradation products DMA and MMA. In order to exclude a growth-phase-dependent expression of the different homologous genes, RNA was further isolated from TMA-grown cells at different growth phases. Exemplarily, transcription of the two mtmBC operons during lag, exponential, or stationary phase was analyzed and compared to transcription of a control gene (MM 1621). This analysis showed that the observed changes in expression of the mtmBC operons during lag- or stationary-phase growth compared to exponential- phase growth were subjected to the same influences as determined for the control gene (data not shown). This finding indicated that the observed differential transcription of the mtmB1C1 operon on TMA versus methanol is not based on growth-phase-dependent expression.
TABLE 3.
Transcriptional regulation of genes encoding methylamine methyltransferases (MT) in cells growing on TMA or methanol in the presence of ammonium
| Genea | Regulation (fold) (TMA versus methanol) |
|---|---|
| MMA-MT | |
| mtmB1C1 | 1,091 ± 367 |
| mtmB2C2 | Approximately 1 |
| DMA-MT | |
| mtbB1 | 179.2 ± 84.5 |
| mtbB2C2 | Approximately 1 |
| TMA-MT | |
| mttB1C1 | 179.2 ± 84.5 |
| mttB2C2 | Approximately 1 |
| Methylcobalamine:CoM-MT | |
| mtbA1 | Approximately 1 |
| mtbA2 | 61.9 ± 3 |
| mtbA3 | Approximately 1 |
mtbB1 and mttB1C1 form a transcriptional unit (MM1687 to MM1694). Only mttB1C1 was analyzed to determine the transcriptional regulation of the complete operon.
Transcription of pyl genes does not appear to be affected by nitrogen limitation and the presence of TMA.
All methylamine methyltransferase transcripts possess an internal amber codon (UAG) (15), which encodes the recently discovered amino acid pyrrolysine (12, 19). Thus, synthesis of functional methylamine methyltransferases is dependent on the expression of the pyl genes, whose gene products are responsible for the synthesis of pyrrolysyl-tRNAPyl. In order to investigate whether the high up-regulation of the mtmB2C2 operon under nitrogen limitation with methanol as carbon source also gives rise to an elevated transcription level of the pyl genes, quantitative RT-PCR analysis for the pylT and pylS genes was performed. The results obtained demonstrated that transcription of the pyl genes is not regulated in accordance to the mtmB2C2 operon, as they were transcribed to the same amount independently of the nitrogen source (CtpylT,+NH4+ = 23 ± 0.6; CtpylT, −NH4+ = 23 ± 0.4). Moreover, preliminary data from a genome-wide transcriptional analysis indicated that even with TMA as sole carbon source, conditions under which methylamine methyltransferases are highly required, transcription of the pyl genes was not increased compared to methanol as carbon source (K. Veit and R. A. Schmitz, unpublished). This finding was confirmed by quantitative RT-PCR for the pylS and pylT genes, which did not show differential transcription in response to the carbon source (e.g., CtpylT, TMA = 23 ± 0.6; CtpylT, methanol = 22 ± 0.6).
DISCUSSION
Multiple homologous genes encoding methylamine methyltransferases have been identified in the genomes of Methanosarcina species (4, 11, 15) (M. thermophila strain TM-1, DuPont, unpublished data). However, the biological significance and functional role of those homologs remains unclear. As the degradation of methylamines results in the release of ammonium, it appeared to be likely that different methylamine methyltransferases are expressed in response to the nitrogen availability, which was the focus of this analysis.
Potential roles of the multiple homologs encoding methylamine methyltransferases.
We obtained conclusive evidence that MMA methyltransferase 2 is highly up-regulated under nitrogen limitation when methanol is the carbon source. However, when TMA served as carbon source, the nitrogen effect was nearly abolished, which is due to the fact that sufficient ammonium is available through TMA degradation. The finding that, under nitrogen limitation, MMA methyltransferase 2 expression is up-regulated in the absence of methylamines, the substrate of methylamine methyltransferases, indicates that adaptability to different growth substrates is a likely reason for the existence of homologous genes. A similar finding was observed by Ferry and coworkers (5) in M. thermophila by a proteomic approach. One of the three methanol methyltransferases (MtaB3C3) involved in methanol degradation is expressed to the same amount independent of whether methanol or acetate is the carbon source (5). The authors hypothesize that expression of the methanol-degrading methyltransferase MtaB3C3 on acetate is an adaptation strategy to improve the ability to be prepared for a potential carbon source switch to methanol, which is the favorable energy source (22). Thus, it is tempting to speculate that in M. mazei a similar need for adaptability is the reason for the up-regulation of the mtmB2C2 operon under nitrogen limitation as the ammonium generated by degradation of methylamines is energetically more favorable than reduction of molecular nitrogen. This is further supported by taking into account that in the natural environment methylamines are likely to become available. We further observed that transcription of only one of the homologous operons encoding the MMA, DMA, TMA, or methylcobalamine:CoM methyltransferases was increased in the presence of TMA or its degradation products DMA and MMA (Table 4). This elevated transcription occurred in direct response to the carbon source (TMA), as we excluded growth-phase-dependent expression of the homologous methylamine methyltransferases. The finding that the mtmB2C2 operon is nitrogen regulated, whereas the homologous mtmB1C1 operon is regulated by the carbon source (Table 4), again indicates that the two homologous operons apparently have distinct functions.
TABLE 4.
Transcriptional regulation of genes encoding soluble methylamine methyltransferases and genes involved in pyrrolysine synthesisa
| Gene | Nitrogen source
|
Carbon source
|
Regulation of transcription | ||
|---|---|---|---|---|---|
| TMA | N2 | TMA | Methanol | ||
| MMA-MT | |||||
| mtmB1C1 | − | − | ++ | − | Increased in the presence of TMA |
| mtmB2C2 | +/− | ++ | − | − | Increased by N-limitation |
| DMA-MT | |||||
| mtbB1 | − | − | ++ | − | Increased in the presence of TMA |
| mtbB2C2 | − | − | − | − | Not differentially transcribed |
| TMA-MT | |||||
| mttB1C1 | − | − | ++ | − | Increased in the presence of TMA |
| mttB2C2 | − | − | − | − | Not differentially transcribed |
| Methylcobalamine:CoM-MT | |||||
| mtbA1 | − | − | − | − | Not differentially transcribed |
| mtbA2 | − | +/− | ++ | − | Increased in the presence of TMA |
| mtbA3 | − | − | − | − | Not differentially transcribed |
| Genes involved in pyrrolysine synthesis | |||||
| pylS, pylT | − | − | − | − | Not differentially transcribed |
| lysRS1, lyrRS2 | − | − | − | − | Not differentially transcribed |
+/−, Slight increase; ++, strong increase of transcription under the respective conditions; −, not differentially transcribed.
As the mtmB2C2 operon is responsible for the first step within the transfer of the methyl group to CoM, an up-regulation of at least one of the three genes encoding methylcobalamine:CoM methyltransferases (mtbA) was expected under nitrogen limitation. However, significant but only slight increase of mtbA2 transcription was obtained under nitrogen limitation (Table 4). This finding indicates that the basal transcription of one of the mtbA genes may allow sufficiently high methylcobalamine:CoM methyltransferase activity to provide the cell with sufficient ammonium from MMA.
It still has to be confirmed on the protein level which of the methylamine methyltransferases are synthesized and active on methanol under different nitrogen availabilities. Therefore, a proteomic approach and the determination of enzyme activities have to follow. However, the results obtained by quantitative RT-PCR strongly indicate that synthesis of methylamine methyltransferases is induced on methanol under nitrogen limitation, again supporting the hypothesis of Ding et al. (5) that multiple homologs are differentially expressed to facilitate the switch between growth substrates.
Synthesis of pyrrolysine does not appear to be regulated in accordance to methylamine methyltransferases.
The recently discovered amino acid pyrrolysine, which is encoded by the internal amber codon UAG, is so far only known to be present in methylamine methyltransferases (12, 19). The gene encoding the respective amber supressor tRNA (tRNAPyl encoded by pylT) was identified in M. barkeri, where it is cotranscribed with pylS, pylB, and pylC (19). One would expect an induction of these genes during growth on TMA compared to growth on methanol, as methylamine methyltransferases are partially up-regulated on TMA (Table 4). However, we did not obtain evidence for a transcriptional up-regulation of the pylT and pylS genes in dependence of the carbon source or in dependence of the extremely elevated expression of the mtmB2C2 operon under nitrogen limitation (Table 4). Considering the level of pyl gene transcription, it appears that these basal transcriptional levels ensure pyrrolysine formation under all circumstances. However, we cannot rule out that differences in transcription in dependence of the carbon source might occur either during early lag or early exponential phase, as the RNA analyzed was exclusively isolated in mid-exponential phase. Recently, two groups showed independently that PylS is an aminoacyl-tRNA synthetase that charges pyrrolysine to tRNAPyl in vitro (1, 16). In addition, Krzycki and coworkers demonstrated PylS charging activity in vivo (1). Evidence for two potential pathways of aminoacylation of the suppressor tRNAPyl were obtained in vitro (16), leaving the question open as to which are used in vivo. Currently, it is speculated that the existence of two routes prevents pyrrolysine limitation to ensure proper methylamine methyltransferase formation (16). As one pathway involves the formation of lysyl-tRNAPyl by LysRS1 and LysRS2 (17) prior to the suggested modification to pyrrolysyl-tRNAPyl by additional uncharacterized pyl genes (19), we investigated the regulation of the corresponding genes in M. mazei as well. Similar to the pylS and pylT genes, they were not regulated under any of the conditions examined but appear to be expressed in all cases (Table 4). This observation indicates that neither one nor the other pathway is preferentially used under the conditions analyzed.
Acknowledgments
We thank Gerhard Gottschalk for continuous support and helpful discussions. We further thank Armin Ehrenreich and Uwe Deppenmeier for collaboration in the genome-wide transcriptional analysis using microarrays mentioned as unpublished results in this work.
This work was supported by the Deutsche Forschungsgemeinschaft (SCHM1052/6-1 and 6-2) and by a Ph.D. fellowship to C.E. from the Fonds der Chemischen Industrie.
Footnotes
Dedicated to Gerhard Gottschalk on the occasion of his 70th birthday.
REFERENCES
- 1.Blight, S. K., R. C. Larue, A. Mahapatra, D. G. Longstaff, E. Chang, G. Zhao, P. T. Kang, K. B. Green-Church, M. K. Chan, and J. A. Krzycki. 2004. Direct charging of tRNA(CUA) with pyrrolysine in vitro and in vivo. Nature 431:333-335. [DOI] [PubMed] [Google Scholar]
- 2.Burke, S. A., and J. A. Krzycki. 1997. Reconstitution of Monomethylamine:Coenzyme M methyl transfer with a corrinoid protein and two methyltransferases purified from Methanosarcina barkeri. J. Biol. Chem. 272:16570-16577. [DOI] [PubMed] [Google Scholar]
- 3.Deppenmeier, U., M. Blaut, A. Mahlmann, and G. Gottschalk. 1990. Reduced coenzyme F420: heterodisulfide oxidoreductase, a proton-translocating redox system in methanogenic bacteria. Proc. Natl. Acad. Sci. USA 87:9449-9453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deppenmeier, U., A. Johann, T. Hartsch, R. Merkl, R. A. Schmitz, R. Martinez-Arias, A. Henne, A. Wiezer, S. Baumer, C. Jacobi, H. Bruggemann, T. Lienard, A. Christmann, M. Bomeke, S. Steckel, A. Bhattacharyya, A. Lykidis, R. Overbeek, H. P. Klenk, R. P. Gunsalus, H. J. Fritz, and G. Gottschalk. 2002. The genome of Methanosarcina mazei: evidence for lateral gene transfer between Bacteria and Archaea. J. Mol. Microbiol. Biotechnol. 4:453-461. [PubMed] [Google Scholar]
- 5.Ding, Y. H., S. P. Zhang, J. F. Tomb, and J. G. Ferry. 2002. Genomic and proteomic analyses reveal multiple homologs of genes encoding enzymes of the methanol:coenzyme M methyltransferase system that are differentially expressed in methanol- and acetate-grown Methanosarcina thermophila. FEMS Microbiol. Lett. 215:127-132. [DOI] [PubMed] [Google Scholar]
- 6.Ehlers, C., K. Veit, G. Gottschalk, and R. A. Schmitz. 2002. Functional organization of a single nif cluster in the mesophilic archaeon Methanosarcina mazei strain Gö1. Archaea 1:143-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferguson, D. J., Jr., N. Gorlatova, D. A. Grahame, and J. A. Krzycki. 2000. Reconstitution of dimethylamine:coenzyme M methyl transfer with a discrete corrinoid protein and two methyltransferases purified from Methanosarcina barkeri. J. Biol. Chem. 275:29053-29060. [DOI] [PubMed] [Google Scholar]
- 8.Ferguson, D. J., Jr., and J. A. Krzycki. 1997. Reconstitution of trimethylamine-dependent coenzyme M methylation with the trimethylamine corrinoid protein and the isozymes of methyltransferase II from Methanosarcina barkeri. J. Bacteriol. 179:846-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferguson, D. J., Jr., J. A. Krzycki, and D. A. Grahame. 1996. Specific roles of methylcobamide:coenzyme M methyltransferase isozymes in metabolism of methanol and methylamines in Methanosarcina barkeri. J. Biol. Chem. 271:5189-5194. [DOI] [PubMed] [Google Scholar]
- 10.Ferry, J. G. 1999. Enzymology of one-carbon metabolism in methanogenic pathways. FEMS Microbiol. Rev. 23:13-38. [DOI] [PubMed] [Google Scholar]
- 11.Galagan, J. E., C. Nusbaum, A. Roy, M. G. Endrizzi, P. Macdonald, W. FitzHugh, S. Calvo, R. Engels, S. Smirnov, D. Atnoor, A. Brown, N. Allen, J. Naylor, N. Stange-Thomann, K. DeArellano, R. Johnson, L. Linton, P. McEwan, K. McKernan, J. Talamas, A. Tirrell, W. Ye, A. Zimmer, R. D. Barber, I. Cann, D. E. Graham, D. A. Grahame, A. M. Guss, R. Hedderich, C. Ingram-Smith, H. C. Kuettner, J. A. Krzycki, J. A. Leigh, W. Li, J. Liu, B. Mukhopadhyay, J. N. Reeve, K. Smith, T. A. Springer, L. A. Umayam, O. White, R. H. White, E. Conway de Macario, J. G. Ferry, K. F. Jarrell, H. Jing, A. J. Macario, I. Paulsen, M. Pritchett, K. R. Sowers, R. V. Swanson, S. H. Zinder, E. Lander, W. W. Metcalf, and B. Birren. 2002. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 12:532-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hao, B., W. Gong, T. K. Ferguson, C. M. James, J. A. Krzycki, and M. K. Chan. 2002. A new UAG-encoded residue in the structure of a methanogen methyltransferase. Science 296:1462-1466. [DOI] [PubMed] [Google Scholar]
- 13.Hippe, H., D. Caspari, K. Fiebig, and G. Gottschalk. 1979. Utilization of trimethylamine and other N-methyl compounds for growth and methane formation by Methanosarcina barkeri. Proc. Natl. Acad. Sci. USA 76:494-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hovey, R., S. Lentes, A. Ehrenreich, K. Salmon, K. Saba, G. Gottschalk, R. P. Gunsalus, and U. Deppenmeier. 2005. DNA microarray analysis of Methanosarcina mazei Gö1 reveals adaptation to different methanogenic substrates. Mol. Genet. Genomics, 273:225-239. [DOI] [PubMed]
- 15.Paul, L., D. J. Ferguson, Jr., and J. A. Krzycki. 2000. The trimethylamine methyltransferase gene and multiple dimethylamine methyltransferase genes of Methanosarcina barkeri contain in-frame and read-through amber codons. J. Bacteriol. 182:2520-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polycarpo, C., A. Ambrogelly, A. Berube, S. M. Winbush, J. A. McCloskey, P. F. Crain, J. L. Wood, and D. Soll. 2004. An aminoacyl-tRNA synthetase that specifically activates pyrrolysine. Proc. Natl. Acad. Sci. USA 101:12450-12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Polycarpo, C., A. Ambrogelly, B. Ruan, D. Tumbula-Hansen, S. F. Ataide, R. Ishitani, S. Yokoyama, O. Nureki, M. Ibba, and D. Soll. 2003. Activation of the pyrrolysine suppressor tRNA requires formation of a ternary complex with class I and class II lysyl-tRNA synthetases. Mol. Cell 12:287-294. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 19.Srinivasan, G., C. M. James, and J. A. Krzycki. 2002. Pyrrolysine encoded by UAG in Archaea: charging of a UAG-decoding specialized tRNA. Science 296:1459-1462. [DOI] [PubMed] [Google Scholar]
- 20.Talaat, A. M., S. T. Howard, W. Hale, R. Lyons, H. Garner, and S. A. Johnston. 2002. Genomic DNA standards for gene expression profiling in Mycobacterium tuberculosis. Nucleic Acids Res. 30:e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thauer, R. K. 1998. Biochemistry of methanogenesis: a tribute to Marjory Stephenson. 1998 Marjory Stephenson Prize Lecture. Microbiology 144:2377-2406. [DOI] [PubMed] [Google Scholar]
- 22.Zinder, S. H., and A. F. Elias. 1985. Growth substrate effects on acetate and methanol catabolism in Methanosarcina sp. strain TM-1. J. Bacteriol. 163:317-323. [DOI] [PMC free article] [PubMed] [Google Scholar]