Abstract
Lactobacillus plantarum is a frequently encountered inhabitant of the human intestinal tract, and some strains are marketed as probiotics. Their ability to adhere to mannose residues is a potentially interesting characteristic with regard to proposed probiotic features such as colonization of the intestinal surface and competitive exclusion of pathogens. In this study, the variable capacity of 14 L. plantarum strains to agglutinate Saccharomyces cerevisiae in a mannose-specific manner was determined and subsequently correlated with an L. plantarum WCFS1-based genome-wide genotype database. This led to the identification of four candidate mannose adhesin-encoding genes. Two genes primarily predicted to code for sortase-dependent cell surface proteins displayed a complete gene-trait match. Their involvement in mannose adhesion was corroborated by the finding that a sortase (srtA) mutant of L. plantarum WCFS1 lost the capacity to agglutinate S. cerevisiae. The postulated role of these two candidate genes was investigated by gene-specific deletion and overexpression in L. plantarum WCFS1. Subsequent evaluation of the mannose adhesion capacity of the resulting mutant strains showed that inactivation of one candidate gene (lp_0373) did not affect mannose adhesion properties. In contrast, deletion of the other gene (lp_1229) resulted in a complete loss of yeast agglutination ability, while its overexpression quantitatively enhanced this phenotype. Therefore, this gene was designated to encode the mannose-specific adhesin (Msa; gene name, msa) of L. plantarum. Domain homology analysis of the predicted 1,000-residue Msa protein identified known carbohydrate-binding domains, further supporting its role as a mannose adhesin that is likely to be involved in the interaction of L. plantarum with its host in the intestinal tract.
The human intestinal microbiota consists of a wide variety of bacterial species, including Lactobacillus plantarum as one of the most predominant Lactobacillus species (2, 35). This versatile species has the capacity to adapt to a variety of environmental conditions. In addition, L. plantarum strains have been shown to effectively survive gastrointestinal passage after oral administration and persist in the intestine of healthy volunteers for up to 11 days after cessation (17, 24, 56). These findings have led to the selection of L. plantarum strains that are currently marketed as probiotics, claimed to be functional food components that provide beneficial effects to the consumer's health (25, 34, 42). Recently, the whole genome of L. plantarum WCFS1 has been sequenced and annotated, generating a major advantage for molecular investigation of this bacterium's behavior in the gastrointestinal tract and its potential probiotic features (14, 27). For example, a recent study revealed genes that are induced in L. plantarum WCFS1 during gastrointestinal passage (9).
Among the most prominent probiotic functions proposed for lactic acid bacteria is the inhibition of intestinal infections by enterotoxigenic Escherichia coli (ETEC), which causes travelers' diarrhea (19, 21, 43). A possible mechanism leading to inhibition could be the competitive exclusion of ETEC by recognition of the same adherence sites on the intestinal epithelial surface (39, 43). Pathogens such as ETEC express type 1 fimbriae that are involved in mannose-specific adhesion to epithelial cells (28, 58). In addition, host cell surface mannose-containing glycoconjugates also play a role as targets for the binding of many other pathogens such as Salmonella enterica serovar Enteritidis, Vibrio cholerae, and Pseudomonas aeruginosa (3, 5, 22). In analogy to its proposed probiotic effect, certain L. plantarum strains have also been shown to adhere specifically to mannose-containing sugar moieties and to human intestinal cell lines (1). This characteristic may potentially be involved in the ability of this bacterium to colonize the intestine and could possibly be relevant for competitive exclusion of pathogens at mannose-containing receptors on the epithelial surface. Recently, probiotic lactobacilli have been found to bind to mannose on the surface of the human immunodeficiency virus type 1 and were therefore proposed to possibly interfere with human immunodeficiency virus infection (L. Tao, S. I. Pavlova, S. J. Carlson, J. J. Anzinger, A. Jacobs, M. S. Caffrey, and G. T. Spear, Abstr. 104th Gen. Meet. Am. Soc. Microbiol., abstr. T-031, 2004). This concept could also be applicable for other pathogens displaying mannose residues bound to cell surface proteins such as Candida albicans (10). Therefore, the mannose-binding capacity of probiotic lactobacilli may be interesting in various perspectives. However, so-called “probiotic genes” involved in proposed probiotic features have not been the subject of detailed molecular investigations so far (29, 54).
The aim of the present study was to identify and characterize the genes involved in mannose-specific adhesion of L. plantarum. The phenotypic trait of mannose adhesion was assessed using an agglutination assay that is based on the presence of mannose-containing polysaccharides in the cell wall of Saccharomyces cerevisiae (1). The addition of mannose-adhering bacteria to these yeast cells leads to the formation of yeast cell agglutinates that are microscopically visible. This assay also has been used to determine mannose-binding properties mediated by type 1 fimbriae of E. coli and other pathogens (3, 33, 49). Therefore, its results are relevant for the assessment of the capacity of probiotic and pathogenic bacterial strains to bind to mannose receptors. Mannose adhesion was chosen as one possible example of the various bacterium-host interactions in the intestine and to discover the genetic background of this feature.
In this study, the capacity to agglutinate yeast cells in a mannose-specific manner was found to be variable among strains of L. plantarum. This phenotypic variability among the L. plantarum strains tested was correlated with a genotype diversity database for this species that was constructed on the basis of genome-wide genotyping using L. plantarum WCFS1-based DNA microarrays (33a). In silico matching of genotypic and phenotypic characteristics led to the identification of two candidate L. plantarum genes that could possibly function as mannose-specific adhesins. Gene-specific deletion and overexpression of the candidate genes and subsequent evaluation of the mutant strains in the agglutination assay indicate that the typical multidomain cell surface protein encoded by lp_1229 represents the mannose-specific adhesin (Msa) of L. plantarum.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The Lactobacillus plantarum strains used in this study are listed in Table 1. They were grown at 37°C on Man-Rogosa-Sharp (MRS) medium (Merck, Amsterdam, The Netherlands) without aeration. E. coli MC1061 was used as an intermediate cloning host for plasmid constructions and was grown aerobically at 37°C in TY medium (Difco, Surrey, United Kingdom). Saccharomyces cerevisiae was grown overnight in malt extract medium (Oxoid, Haarlem, The Netherlands) under aerobic conditions at 37°C. Yeast cells were harvested by centrifugation, washed with phosphate-buffered saline (PBS) (pH 7.4), weighed, and used in the agglutination assay (see below). When appropriate in cloning procedures, the antibiotics chloramphenicol (Cm) and erythromycin (Em) were added to the culture media (Cm at 7 and 10 μg/ml for L. plantarum gene replacement and overexpression transformants, respectively, as well as for gene replacement and overexpression of E. coli plasmid transformants; Em at 25 μg/ml during the direct L. plantarum double-crossover mutant selection procedure).
TABLE 1.
Lactobacillus plantarum strains used in this study
|
L. plantarum
|
Origin and/or source (reference)a | |
|---|---|---|
| Original designation | NIZO culture collection | |
| WCFS1 | B1836 | Sequenced wild-type strain; single colony isolate of NCIMB 8826 from human saliva (27) |
| ATCC 8014 | B1843 | Maize ensilage, ATCC (32) |
| ATCC 14917 | B1317 | Pickled cabbage, ATCC (32) |
| 299 | B1837 | Human colon (35) |
| 299v | B2260 | Human intestine (24) |
| NCIMB 12120 | B1840 | Ogi, Nigeria; NCIMB (7) |
| CIP 104440 | B1838 | Human stool (16) |
| CIP 104441 | B2256 | Human stool (16) |
| CIP 104450 | B2257 | Human stool (16) |
| CIP 104451 | B2258 | Human urine (16) |
| CIP 104452 | B2259 | Human tooth abscess (7) |
| NC8 | B2261 | Silage (13) |
| LM3 | B2262 | Silage (37) |
| LP 80 | B2263 | Silage (7) |
| NZ7104 | Cmr; WCFS1 derivative; srt::cat (this work) | |
| NZ7510 | Cmr; WCFS1 derivative; lp_0373::cat (this work) | |
| NZ7511 | Cmr; WCFS1 derivative; msa::cat (this work) | |
| WCFS1 + pNZ7515 | Cmr; WCFS1 overexpression of lp_0373 (this work) | |
| WCFS1 + pNZ7516 | Cmr; WCFS1 overexpression of msa (this work) | |
ATCC, American Type Culture Collection, Manassas, Va.; Aberdeen, Scotland; NCIMB, National Collections of Industrial, Food and Marine Bacteria; Cmr, chloramphenicol resistant.
Agglutination assay.
The capacity of 14 L. plantarum strains to adhere mannose specifically was determined as described previously (1), with slight modifications. In brief, bacterial strains were grown overnight, washed, and suspended in 0.1 culture volume of PBS (pH 7.4). The number of CFU per ml was determined by plating of serial dilutions of the bacterial suspensions on MRS agar plates. Fifty microliters of a fourfold dilution of the initial bacterial suspensions in PBS was transferred to microtiter plates (96-well U-shaped; Greiner bio-one, Alphen a/d Rijn, The Netherlands). To each well, 50 μl PBS or PBS with methyl-α-d-mannopyranoside (final concentration, 25 mM; Sigma-Aldrich Chemie BV, Zwijndrecht, The Netherlands) was added as well as 100 μl of 1% (wt/vol) Saccharomyces cerevisiae suspended in PBS. The microtiter plates were shaken for 10 min at room temperature, and samples of 50 μl were taken from each well to examine agglutination by bright-light microscopy (200-fold magnification; Nikon Eclipse TS100 inverted microscope). The ability of each strain to induce visible yeast cell agglutination was determined in three independent experiments.
For the exemplary strain L. plantarum WCFS1, besides methyl-α-d-mannopyranoside the following inhibitory substances were tested: d(+)-mannose, methyl-α-d-glucopyranoside, d(+)-glucose, methyl-α-d-galactopyranoside, d(+)-galactose, l(+)-fucose, N-acetylglucosamine, and N-acetylgalactosamine (final concentration, 25 mM; Sigma-Aldrich Chemie BV, Zwijndrecht, The Netherlands).
When testing the mutant strains of L. plantarum WCFS1 (see below), the assay was changed in the way that a range of twofold dilutions of the bacterial suspensions of each mutant derivative was tested and for each of the dilutions agglutination capacity was examined. The agglutination titer was determined as the (log2) reciprocal of the highest dilution that still resulted in visible agglutination. Mean agglutination titers and corresponding standard deviations were calculated from three independent assays. When appropriate, the significance of the difference between the agglutination titer of the mutant strains in comparison with that of the wild-type strain was calculated using Student's t test (two sided; considered statistically significant when P is <0.05).
Identification of candidate genes involved in mannose adherence by gene-trait matching.
Candidate genes potentially involved in mannose-specific adhesion of L. plantarum were identified by in silico gene-trait matching in which results from the agglutination assay were correlated with genotypic information of the same L. plantarum strains, as previously assessed by DNA microarray-based strain-specific genotyping (33a). In these genotyping experiments, for each L. plantarum WCFS1 gene the level of confidence that this gene was present in the genome of the other L. plantarum strain was calculated (P value score per gene). In contrast to the procedure described by Molenaar et al., in the present study these P value scores were calculated using two weighting methods regarding the overlap of a gene with the clones represented on the array. In the linear method, the weight of the P value of a clone in the P value score was equal to the ratio of the overlap of clone and gene relative to the size of the clone, whereas it was equal to the square of this fraction in the quadratic method. The latter method puts an additional penalty on clones that do not completely overlap with a gene, as these may also pick up signals from other genes. Using either of these methods, gene occurrence can be predicted using a suitable P value score threshold, below which it is assumed that the gene is absent in the strain. Nevertheless, due to the complexity of the P value score calculation, it is not obvious how this P value threshold should be determined. Therefore, selection of candidate mannose adhesin-encoding genes was evaluated at different threshold values (P value score range).
The resulting data about the absence or presence of L. plantarum WCFS1 genes in other L. plantarum strains were directly correlated to the mannose adherence phenotype. The significance of the observed correlation of gene occurrence and phenotypic trait was assessed by assuming a hypergeometric distribution for the probability of the co-occurrence of genes and traits under the null hypothesis that the observed co-occurrence is caused by random processes alone, as was previously described (23).
All L. plantarum WCFS1 genes (>3,000 genes) were tested for the significance of positive correlation of gene and trait. Several hundreds of false positives may be expected when testing the null hypotheses for each gene at the same rejection level of, for example, P < 0.05. To reduce this number, the Bonferroni correction was applied to the individual hypothesis rejection level by Jim et al. (23). However, this correction is very conservative, and the probability of rejecting true positives becomes large. A higher rate of false discovery is acceptable when the decision to start experimental verification of the result is not only based on a possibly weak gene-trait correlation but also on other properties of a gene. Therefore, predicted annotation of candidate genes might be used to apply additional rational criteria to pinpoint the primary selection toward the most likely candidate genes.
DNA manipulations.
Plasmid constructions were performed with E. coli as an intermediate cloning host. Plasmids constructed in this host were eventually introduced into L. plantarum WCFS1 by electroporation (26). Molecular biology techniques such as DNA manipulations and transformation of E. coli were essentially performed according to standard procedures (48). Plasmid DNA was isolated from E. coli by using Jetstar columns, following the manufacturer's instructions (Genomed GmbH, Bad Oeynhausen, Germany). L. plantarum DNA isolation and manipulation were basically performed as described previously (4, 15). At relevant time points, the position and sequence of the genes cloned were confirmed by sequence analysis (BaseClear, Leiden, The Netherlands). Primers used in this study are listed in Table 2 and were purchased from Proligo France SAS (Paris, France). Restriction endonucleases, Taq and Pfx polymerase, and T4 DNA ligase were used as recommended by the manufacturer (Gibco BRL Life Technologies, Gaithersburg, MD; Invitrogen, Breda, The Netherlands; and New England BioLabs, Beverly, MA). The plasmids used in this study are listed in Table 3.
TABLE 2.
Primers used in this study
| Primer | Sequencea |
|---|---|
| lp_0372F | 5′-AAGGAACTTCTGATTGGGCC-3′ |
| lp_0372R | 5′-CCATTACGATGATGTTTCCC-3′ |
| lp_0374F | 5′-AATTACGGCGTCTCACTGTAGCGATTACGATG-3′ |
| lp_0374R | 5′-CGTGTTTCTTGGAAGACTTC-3′ |
| lp_0513F | 5′-GGGGTACCCCCAATGCTTCTGTCAGG-3′ |
| lp_0513R | 5′-CGGGATCCTTGCTTGGACTTCATTAATCC-3′ |
| lp_0515F | 5′-CGGGATCCCATTTTAATAACAAATATTAAC-3′ |
| lp_0515R | 5′-ATGCTCTAGAACGTGTGCCGCTTGTTGC-3′ |
| lp_1227F | 5′-TATGCAGAACTCCATGACTG-3′ |
| lp_1227R | 5′-TAGTGTGTTTTATGCTCTCC-3′ |
| lp_1230F | 5′-ATTTATGGCGTCGTCTCCTTGGTATGGCTAAG-3′ |
| lp_1230R | 5′-CATCACTCGACATGTCTTGC-3′ |
| lp_0373F | 5′-AAGGACAGAAGCGGCCGCGTTAGATAAAAGGGGTTAAAC-3′ |
| lp_0373R | 5′-ACGGGCCTCGAGATATTAGTCACGCCCTCGT-3′ |
| lp_1229F | 5′-AAGGACAGAAGCGGCCGCCGTGCGAAAGGATAGATT-3′ |
| lp_1229R | 5′-ACGGGCCTCGAGGCCCTACTCTTTGTGCT-3′ |
| 1144_BamHISalIF | 5′-GGATCCGTCGACCGCGATTTTTGTATGAGATG-3′ |
| 1144_BamHIR | 5′-GGATCCGCTGTTCGCCACCCTTTCTA-3′ |
Underlined sequences indicate restriction sites subsequently used in cloning procedures.
TABLE 3.
Plasmids used in this study
| Plasmids | Relevant characteristicsa | Source or reference |
|---|---|---|
| pCR-Blunt | Kmr, cloning vector | Invitrogen |
| pUC18 | Apr, cloning vector | 59 |
| pNZ7101 | Cmr Emr, vector for construction of L. plantarum gene replacement mutants | 8 |
| pNZ7105 | Kmr, pCR-Blunt derivative containing 5′-flanking region of srtA | This work |
| pNZ7106 | Apr, pUC18 derivative containing 3′-flanking region of srtA | This work |
| pNZ7104 | Cmr Emr, pNZ7101 derivative containing pNZ7105- and pNZ7106-derived flanking regions of srtA | This work |
| pNZ7510 | Cmr Emr, pNZ7101 derivative containing flanking regions of L. plantarum WCFS1 lp_0373 | This work |
| pNZ7511 | Cmr Emr, pNZ7101 derivative containing flanking regions of L. plantarum WCFS1 lp_1229 (msa) | This work |
| pNZ7512 | Kmr, pCR-Blunt derivative containing L. plantarum WCFS1 lp_0373 | This work |
| pNZ7513 | Kmr, pCR-Blunt derivative containing L. plantarum WCFS1 lp_1229 (msa) | This work |
| pNZ273 | Cmr, vector containing gusA reporter gene | 40 |
| pNZ7514 | Cmr, pNZ273 derivative containing constitutive promoter upstream of gusA reporter gene | This work |
| pNZ7515 | Cmr, pNZ7514 derivative containing L. plantarum WCFS1 lp_0373 | This work |
| pNZ7516 | Cmr, pNZ7514 derivative containing L. plantarum WCFS1 lp_1229 (msa) | This work |
Apr, ampicillin resistant; Cmr, chloramphenicol resistant; Emr, erythromycin resistant; Kmr, kanamycin resistant.
cat replacement of srtA and adhesin candidate genes.
For the construction of gene replacement mutants of the sortase-encoding gene srtA as well as the adhesin candidate genes lp_0373 and lp_1229, the mutagenesis vector pNZ7101 was used, which had been constructed previously in our laboratory for single-step selection of cat replacement mutants in L. plantarum (8). This vector contains a pACYC184-derived origin of replication, which is a functional replicon in E. coli but does not support replication in L. plantarum (47). In addition, this vector harbors two resistance marker genes: a Cm resistance cassette consisting of cat-194 (20) under control of the strong lactococcal P32 promoter (52) and an Em resistance gene, ery, under control of its own promoter (53). Both of these markers can be selected after a single-copy insertion in the L. plantarum chromosome (8).
As a first step, the chromosomal 5′- and 3′-flanking regions of the target genes were amplified by PCR using Pfx polymerase in order to clone them in the mutagenesis vector. In this PCR, L. plantarum WCFS1 chromosomal DNA was used as a template with the locus-specific primer combinations lp_0513F/lp_0513R and lp_0515F/lp_0515R for srtA::cat replacement, lp_0372F/lp_0372R and lp_0374F/lp_0374R for lp_0373::cat replacement, and lp_1227F/lp_1227R and lp_1230F/lp_1230R for lp_1229::cat replacement, respectively. For srtA replacement, the 0.7-kb 5′-PCR product was subsequently ligated into a pCR-Blunt vector (Invitrogen, Breda, The Netherlands); the 0.7-kb 3′-PCR product was digested with XbaI and BamHI (sites introduced into primers) and ligated into similarly digested pUC18 (59), yielding the plasmids pNZ7105 and pNZ7106, respectively. The 3′-flanking region was then retrieved as a PvuII fragment from pNZ7106 and the 5′-flanking region as an Ecl136II-EcoRV fragment from pNZ7105. The 3′-flanking region was then cloned into SmaI-digested pNZ7101 before the 5′-flanking region was ligated into the PvuII site of the resulting plasmid. For replacement of the adhesin candidate genes, first the ∼0.8-kb PCR products representing the 5′-flanking regions were cloned into SmaI-digested pNZ7101. Subsequently, the ∼0.8-kb PCR products representing the 3′-flanking regions were digested with BsaHI (sites introduced in forward primers). The pNZ7101 derivatives already containing the 5′-flanking regions of the same target locus were digested with PvuII and BsaHI and ligated with the corresponding 3′-flanking regions. Proper ligation of all fragments in the desired orientation—the 5′-flanking regions upstream of P32-cat and the 3′-flanking regions between cat and ery—was confirmed by PCR, restriction, and sequence analyses (data not shown). The resulting srtA, lp_0373, and lp_1229 knockout plasmids were designated pNZ7104, pNZ7510, and pNZ7511, respectively, harboring the chloramphenicol resistance cassette flanked by the 5′- and 3′-flanking regions of the L. plantarum WCFS1 target genes.
These vectors were used for stable double-crossover cat replacement of the target genes in the chromosome of L. plantarum WCFS1. For this purpose, they were introduced into competent cells of L. plantarum WCFS1 by electroporation and primary integrants were selected on MRS agar containing 7 μg/ml Cm. Candidate double-crossover colonies were selected by replica plating to MRS agar containing 25 μg/ml Em, respectively, selecting for those colonies that displayed Cm resistance and Em sensitivity. The anticipated chromosomal organization in the mutant strains was confirmed by PCR and Southern blot analysis following standard procedures (data not shown). Finally, mutants with the correct double-crossover gene replacement genotype were selected and designated L. plantarum strains NZ7104 (srtA::cat), NZ7510 (lp_0373::cat), and NZ7511 (lp_1229::cat), respectively.
Overexpression of adhesin candidate genes.
Overexpression of the candidate adhesin proteins was achieved by cloning the corresponding gene into a high-copy vector downstream of a constitutive promoter region. First, the vector pNZ7514 containing the promoter region was constructed. A 100-bp promoter fragment upstream of lp_1144 of L. plantarum WCFS1, annotated as the DNA helicase pcrA, was amplified by PCR containing BamHI sites at both ends using L. plantarum WCFS1 genomic DNA as a template and the primers 1144_BamHISalHIF and 1144_BamHIR (sites introduced in primers). This fragment was cloned in pCR-Blunt and recovered from the resulting vector as a BamHI fragment, which was subcloned in the BglII-digested gusA (β-glucuronidase) reporter vector pNZ273 (40), yielding pNZ7514. The constitutive characteristics of the promoter present in the amplified lp_1144 fragment could be confirmed using gusA as a reporter (12, 40).
The two candidate genes lp_0373 and lp_1229 were amplified by PCR using L. plantarum WCFS1 DNA and the primer combinations lp_0373F/lp_0373R or lp_1229F/lp_1229R (NotI sites integrated in forward primers, XhoI sites in reverse primers). The PCR products of approximately 3.7 kb (lp_0373) and 3.1 kb (lp_1229) were cloned into a pCR-Blunt vector, resulting in the plasmids pNZ7512 and pNZ7513, respectively. These plasmids were subsequently digested with NotI and XhoI, yielding fragments consisting of lp_0373 and lp_1229, respectively. These fragments were ligated into pNZ7514 after gusA had been removed from this plasmid by digestion with NotI and XhoI. Replacement of gusA with lp_0373 or lp_1229 downstream of the constitutive promoter region resulted in the vectors pNZ7515 and pNZ7516, respectively. These high-copy plasmids were introduced into L. plantarum WCFS1; the presence of the anticipated vector in selected Cm-resistant colonies was validated by PCR (data not shown).
RESULTS
Mannose adhesion is a variable phenotype in L. plantarum.
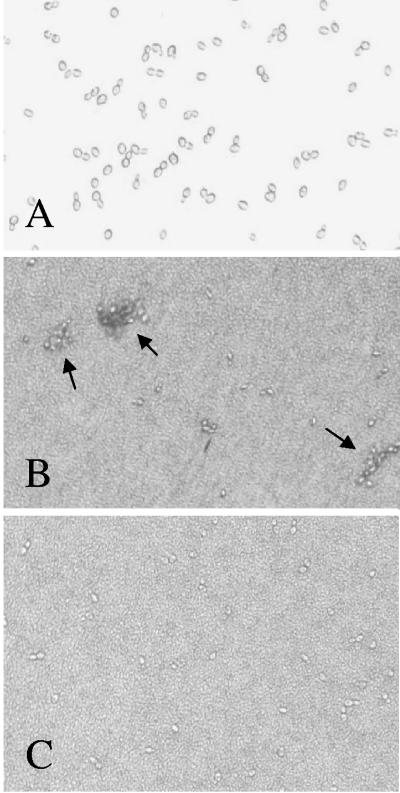
Initial experiments were aimed at the determination of the mannose-specific adherence capacity of different L. plantarum strains. For this purpose, an in vitro agglutination assay was used (1). Overnight cultures of L. plantarum strains were diluted to approximately 2.5 × 109 CFU/ml, as confirmed by serial plating on MRS agar (data not shown), and their ability to induce yeast cell agglutination was determined (Table 4). The L. plantarum strains tested varied remarkably in their mannose adhesion ability. For all but one strain that scored positive in this test, agglutination could be inhibited by the addition of methyl-α-d-mannopyranoside to the assay buffer, confirming a mannose-specific mechanism (data not shown). NCIMB 12120, the strain displaying agglutination that could not be inhibited mannose specifically, was excluded from further analysis. Importantly, the sequenced strain WCFS1 was among the strains capable of inducing agglutination of S. cerevisiae (Fig. 1). Mannose specificity of the agglutination observed was corroborated by the finding that agglutination could only be prevented by methyl-α-d-mannopyranoside or d(+)-mannose, as assessed with the exemplary strain WCFS1 but not by any of the other substances tested, which included methyl-α-d-glucopyranoside, d(+)-glucose, methyl-α-d-galactopyranoside, d(+)-galactose, l(+)-fucose, N-acetylglucosamine, and N-acetylgalactosamine (data not shown).
TABLE 4.
Agglutination of Saccharomyces cerevisiae mediated by various L. plantarum strainsa
| L. plantarum strain | Agglutination |
|---|---|
| WCFS1 | + |
| ATCC 8014 | − |
| ATCC 14917 | + |
| 299 | + |
| 299v | + |
| NCIMB 12120 | + |
| CIP 104440 | − |
| CIP 104441 | − |
| CIP 104450 | − |
| CIP 104451 | − |
| CIP 104452 | + |
| NC8 | + |
| LM3 | + |
| LP80 | + |
Results are from cultures 4× diluted. Results from three independent experiments are shown. Only agglutination induced by strain NCIMB 12120 could not be inhibited by α-methylmannoside.
FIG. 1.
Agglutination of Saccharomyces cerevisiae mediated by mannose-specific adhering lactobacilli (4× diluted), as visualized by bright-light microscopy (magnification, ×200). (A) S. cerevisiae with no bacterial cells added did not agglutinate. (B) The wild-type strain L. plantarum WCFS1 induced agglutination of S. cerevisiae (see arrows). (C) L. plantarum ATCC 8014 did not cause agglutination of S. cerevisiae.
Identification of candidate genes involved in mannose adherence by gene-trait matching.
The measured yeast agglutination capacity of the L. plantarum strains was used to identify candidate mannose adhesin-encoding genes by correlation of these phenotypic data to whole-genome genotyping information of the same strains, which was determined by Molenaar et al. (33a). Considering that the sequenced L. plantarum strain WCFS1 and several of the other strains tested displayed the yeast agglutination capacity while other strains lacked this ability, the correlation applied was the logical selection of those WCFS1 genes whose presence and absence in the different strains were significantly correlated with the presence and absence, respectively, of the agglutination phenotype observed for the same strains. A linear weighting method and a quadratic weighting method were used to calculate the P value scores indicating the presence of WCFS1 genes in the other L. plantarum strains tested.
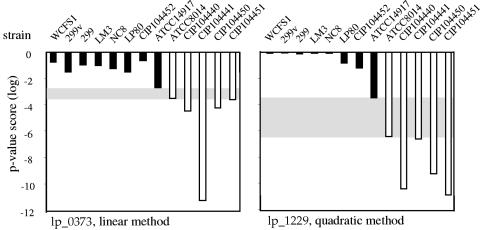
Using these methods, hypothetical mannose adhesin-encoding genes whose presence correlated with the agglutination phenotype were identified. With a P value score threshold of 1.0 × 10−3, the linear weighting method yielded three genes that displayed a 100% gene-trait match (P = 0.00078). These genes were lp_0373, annotated as a putative cell surface protein containing an LPxTG motif; lp_0402 with unknown function; and lp_0403, encoding PlnR, which is a protein involved in plantaricin biosynthesis. For lp_0373, the gene-trait correlation was valid over a narrow range of relatively high P value scores (P value score range, 1.8 × 10−3 to 3.0 × 10−4; see Fig. 2), indicating that for some strains it is less clear whether an lp_0373 homologue is present or absent. The quadratic weighting method yielded with a P value score threshold of 1.0 × 10−4 gene lp_1229 as the best candidate gene (P = 0.00078). In fact, within a wide range of P value scores (P value score range, 3.2 × 10−4 to 3.7 × 10−7, see Fig. 2) lp_1229 was the only gene with 100% gene-trait match out of all genes in L. plantarum. The lp_1229 gene is predicted to encode a putative cell surface protein precursor with LPxTG anchor. Except for these four primary candidate genes, no other 100% gene-trait matches were identified.
FIG. 2.
Data upon which the putative candidate genes were selected. For each gene in each strain, the P value scores averaged over overlapping clones were calculated, indicating confidence in the presence of that gene in the strain. Two calculation methods were used (see text). Indicated are the P value scores for lp_0373 using the linear method (left panel) and for lp_1229 (msa) using the quadratic method (right panel). Strains able to induce agglutination of S. cerevisiae are depicted by filled bars; strains not having this ability are represented by open bars. Gray areas indicate the P value score ranges within which perfect matches between the presence of the genes and the presence of the mannose adhesion phenotype are found.
Remarkably, there were two prominent candidate genes with a 100% gene-trait match among the four primary hypothetical mannose adhesin-encoding genes resulting from the two weighting methods: i.e., lp_0373 and lp_1229. Both of these genes are annotated as putative cell surface proteins containing an LPxTG anchor. The linear weighting method yielded lp_3114 as a third gene coding for a protein of this class; however, this gene did not match 100% with either of the P value score thresholds tested. Regarding the trait mannose adhesion, exposure of the predicted adhesin on the cell surface is likely, which adds a supporting rational criterion to the primary selection of candidate genes. The LPxTG motif is present slightly modified in 25 L. plantarum WCFS1 proteins (27). The surface anchoring of this class of surface proteins of many species has been shown to depend on the transpeptidase sortase, which cleaves polypeptides with a conserved LPxTG consensus sequence and anchors them to the cell wall (38, 50). To validate the role of sortase target proteins for mannose adhesion at first, a sortase-lacking mutant strain of L. plantarum WCFS1 was constructed by stable gene replacement. Importantly, evaluation of this mutant strain (NZ7104) in the agglutination assay showed that this strain, in contrast to the wild type, was not able to agglutinate yeast cells (data not shown). This result indicates the involvement of 1 of the 25 sortase target proteins of L. plantarum WCFS1 in mannose adhesion and thereby consolidated the proposed role of the candidate genes lp_0373 and lp_1229. Consequently, the function of these two genes was assessed by gene-specific DNA manipulation in L. plantarum WCFS1, followed by determination of the mutant strains' mannose adherence capacity.
Verification of lp_1229 as the mannose-specific adhesin of L. plantarum.
To evaluate the postulated role of lp_0373 and lp_1229 in the mannose-specific adhesion phenotype of L. plantarum, derivatives of L. plantarum WCFS1 with gene-specific deletions of these genes were constructed, yielding strains NZ7510 (lp_0373::cat) and NZ7511 (lp_1229::cat), respectively. The mannose-specific agglutination titers of these mutant strains were recorded in the yeast agglutination assay as the (log2) reciprocal of the highest dilution of bacterial suspensions that still resulted in visible agglutination. A mean agglutination titer of 5.0 ± 0.0 (log2 ± standard deviation) was determined in three independent experiments for the wild-type strain, WCFS1. Replacement of lp_0373 (NZ7510) did not affect the yeast agglutination capacity (agglutination titer, 5.0 ± 0.0), as compared to that of the wild-type strain, whereas deletion of lp_1229 (NZ7511) resulted in a complete loss of the capacity to agglutinate S. cerevisiae (agglutination titer, 0.0 ± 0.0). These results indicate a direct involvement of lp_1229 in mannose-specific adhesion of L. plantarum. To further validate the role of lp_1229 in this phenotype, overexpression of the two candidate genes was achieved by introduction of a multicopy expression vector, containing the lp_0373 (pNZ7515) or lp_1229 (pNZ7516) gene under control of an active L. plantarum promoter, into the wild-type strain L. plantarum WCFS1. In analogy with the results obtained with the deletion derivatives, overexpression of lp_0373 did not affect the yeast agglutination capacity of L. plantarum (agglutination titer, 5.0 ± 0.0), confirming that this gene does not play a role in mannose adhesion. Overexpression of lp_1229 resulted in a slight, but significant enhancement of agglutination capacity compared to that of the wild-type strain (agglutination titer, 6.3 ± 0.6; P = 0.016). These results confirm the observations made with the deletion derivatives and support a direct role of lp_1229 in the phenotype investigated. Overall, we concluded that lp_1229 encodes the mannose-specific adhesin of L. plantarum and renamed the gene accordingly: msa. The trait encoded by the msa gene may be of interest regarding adhesion of L. plantarum in the gastrointestinal tract.
In silico analysis of msa and its protein product.
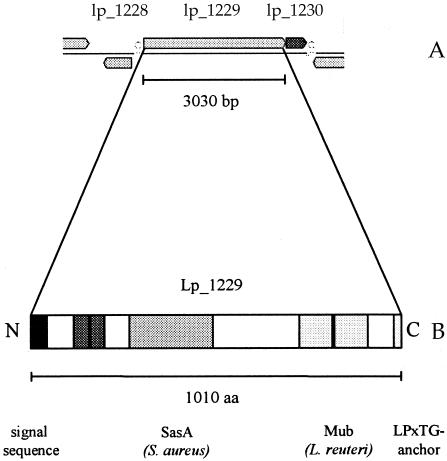
Following identification of the msa gene of L. plantarum, more detailed analyses of this gene and the characteristics of its encoded protein were performed using web-based bioinformatic tools, aiming for structure-function insight (Fig. 3). Regarding the genetic surroundings, the neighboring gene (lp_1228) is located 261 bp upstream of msa and is divergently transcribed. This finding, combined with the identification of a 6-bp stem-loop structure upstream of msa (ΔG free energy, −24.1 kcal), indicates that msa does not form an operon structure with upstream genes. Nevertheless, no obvious candidate promoter sequence could be identified upstream of msa. Almost directly downstream of msa (separated by 11 bp), a predicted transcriptional regulator (lp_1230) has been annotated that is transcribed in the same direction as msa. The lp_1230 gene is directly followed by a potential termination sequence (ΔG free energy, −30.6 kcal). Moreover, the subsequent downstream gene (lp_1231) is convergently oriented relative to msa and lp_1230, indicating that no additional downstream genes belong to this operon. This genetic organization of msa and lp_1230 suggests that the two genes are transcribed as a bicistronic mRNA. It is tempting to speculate that the lp_1230 gene product is involved in transcriptional regulation of the expression of the msa operon. Notably, lp_1230 was also among the candidate genes resulting from the gene-trait correlation based on both the linear and the quadratic weighting methods, although not with a 100% gene-trait match. This supports the prediction that msa and lp_1230 may be organized in an operon and form a functional unit in which the protein encoded by lp_1230 possibly fulfills an autoregulatory role.
FIG. 3.
(A) Genetic organization of the msa locus in the genome of L. plantarum WCFS1. (B) Molecular structure of the encoded protein Msa, implying a plausible domain organization for a bacterial adhesin. Domains with sequence similarity to proteins possibly being informative for functional domain annotations are indicated.
The 1,010-amino-acid protein encoded by msa, Msa, displays the characteristic multidomain structure observed for many cell surface proteins of L. plantarum (Fig. 3). As stated above, it contains the typical elements associated with sortase-dependent surface proteins, including an N-terminal signal peptide and a hydrophobic C-terminal stop transfer sequence that is preceded by the typical L. plantarum sortase target-anchoring sequence LPQTxE (27). Moreover, straightforward sequence alignment of Msa with known proteins, using web-based tools like BLAST or FASTA, revealed sequence similarities to other cell surface proteins of different gram-positive bacteria. Msa contains perfect repeat sequences, which partly coincide with the domains that display the highest similarity to other known proteins. Close examination showed that the region close to the C terminus of Msa (residues 720 to 916) encompasses two 97-residue copies of a mucus-binding domain previously identified in a cell surface protein (Mub) of L. reuteri (46) (GenBank accession no. AF120104). A domain with sequence similarity to SasA, a ConA-like lectin, from Staphylococcus aureus (44, 55) was identified, for which, due to analysis with hidden Markov models from the Superfamily database, no exact borders could be determined. This domain consists of approximately 255 amino acids (in the region of residues 262 to 517) and contains an insertion in the L. plantarum sequence of about 42 residues. Since both the mucus-binding protein Mub and ConA-like lectins are known to bind to carbohydrates such as mannose (46, 57), these domains of Msa are likely to be relevant for mannose recognition and adhesion. Close to the N terminus, for two repeats of 35 residues no homologies to known proteins could be identified. In conclusion, the domain structure of Msa supports a role of this protein in carbohydrate recognition and binding and therefore provides additional evidence supporting its designation as the mannose adhesin of L. plantarum.
DISCUSSION
In the present study, in silico phenotype-genotype matching has been used to identify a bacterial gene responsible for a specific phenotypic trait. For this strategy, a simple and rapid assay was employed to screen a number of bacterial strains for the trait of interest. In addition, whole-genome genotyping data of the same bacterial strains were available for correlation of the presence of genes with the presence of the phenotypic characteristic. This study provides evidence that such a correlation strategy can be applied to identify the genetic factors involved in phenotypic traits, in order to search for “genes for a function” rather than for “functions for genes” (6). The example described here revealed the mannose-specific adhesin of L. plantarum.
Other attempts have been made to correlate genomic diversity to phenotypic features. Genomic comparisons of Brucella species, Bacillus fumarioli strains, and citrus-associated bacteria, respectively, have been performed recently to identify genes or operons relevant for bacterial adaptations to their environment (11, 36, 41). In these studies, genomic diversity among bacterial strains has been investigated using genomic microarrays, comparable to the L. plantarum microarray employed in this study, and other techniques to characterize genome differences. However, the resulting data have not been exploited to identify a gene responsible for one specific, chosen function by correlation with phenotypic data, as in the present study. Moreover, the biodiversity-based approach chosen here included a gene-trait-matching method similar to that described by Levesque et al. (30) and Jim et al. (23), who—in contrast to the present study—used only fully sequenced bacterial genomes. Remarkably, in the approach described here, a rather simple and straightforward correlation strategy was applied successfully. This method was effective despite the use of a small sample of strains, relatively crude genotyping procedures, and basic gene-trait analysis software. Additional information on the annotation of the resulting candidate genes was valuable to interpret the primary data and to proceed with the most promising candidates. Still, there are some general limitations of the phenotype-genotype-matching method described here. One restriction is that genes unique to other bacterial strains than the exemplary strain cannot be detected in this manner. In addition, this approach cannot be successful if the variation of the phenotypic trait of interest is based on differences in gene expression. Furthermore, a small mutation of the gene in question for instance might cause a frameshift leading to an entire loss of the gene's function, even when the gene is present. Variations in gene sequence and protein processing among different strains might have to be considered in the interpretation of results.
Various lines of evidence support the role of the lp_1229 gene of L. plantarum as the mannose-specific adhesin (msa) gene. The genetic organization of the region surrounding msa indicates a genetic linkage with lp_1230 in a bicistronic operon, in which the latter gene might encode its transcriptional autoregulator. Since the presence of lp_1230 is also, although more weakly, correlated with the trait of yeast agglutination, the transcription of the proposed msa operon may be autonomous, supporting the belief that this gene is the only determinant required for mannose adhesion capacity.
In silico sequence analysis of the mannose-specific adhesin (Msa) revealed that this protein has a characteristic domain organization observed for many adhesins of gram-positive bacteria (46). Some domains possibly create spacer regions through the peptidoglycan structures to the cell surface. Homology searches indicated a similarity of domains from Msa to a mucus-binding protein (Mub) from Lactobacillus reuteri (46) and to SasA, an LPxTG-containing cell surface protein from Staphylococcus aureus (44, 45), referred to as a ConA-like lectin (55). One or both of these domains might be directly involved in mannose recognition by Msa: Mub has been identified to adhere to carbohydrate structures in mucus (46), and ConA-like lectin domains such as in SasA are also known to bind to mannose and are often involved in the recognition and adhesion processes of, for instance, bacterial toxins (57). Moreover, the ConA-like domain of Msa might harbor hydrolase activity, since those domains can also be found in bacterial glucanases (18). Hydrolase activity could possibly be involved in the release of sugar moieties from more complex carbohydrates, thereby releasing fermentable carbohydrates that would be available for growth of L. plantarum. No correlation was found for the presence of msa and the ability to metabolize α-methylmannoside or any other mono- or disaccharide, as indicated by API 50CH metabolic profiling of the strains tested in this study (data not shown). Nevertheless, a role of Msa in utilization of more complex carbohydrates (oligo- or polysaccharides) can certainly not be excluded. BLAST analysis of the Mub domain of Msa revealed homologies to potential adhesins from other bacterial species: for example, a hypothetical protein from Lactococcus lactis Il1403 (L39650) and Mlp, a surface protein from Lactobacillus fermentum BR11 with an LPxTG-motif (51). Furthermore, Mub-like domains can also be identified in other L. plantarum WCFS1 genes: e.g., lp_1643, lp_3059, and lp_3114, all annotated as cell surface proteins with an LPxTG anchor. These findings suggest that these binding domains might be commonly used in proteins among different bacterial species and might be directly involved in recognition of (complex) carbohydrates.
Finally, the results of this study provide insight into the genetic background of bacterial adhesion, a potential probiotic feature of L. plantarum. In contrast to the isogenic, gene-specific mutant strains constructed in this study, formerly spontaneous mutants were studied that bear undefined genetic alterations, such as an uncharacterized mutant of L. plantarum 299v that lacks the ability to agglutinate S. cerevisiae (31). It would be very interesting to analyze the msa locus of strain 299v and its agglutination-negative derivative. The importance of Msa for intestinal colonization and its potential probiotic effects will have to be validated in future studies. The mutant strains of L. plantarum WCFS1 constructed in this study will be advantageous to evaluate the potential role of Msa in this process. So far, there are only speculations about the structure of the mannose-containing receptors that is recognized by ETEC and L. plantarum (1). Future experiments will have to determine whether the mannose moieties on yeast cells are appropriate representatives of intestinal receptors in vivo.
A primary indication of the importance of Msa in terms of intestinal colonization could be deduced from recent mouse passage experiments performed with the srtA mutant of L. plantarum WCFS1 (NZ7104) in comparison to the wild-type strain (8). Since the proper surface exposure of the Msa protein depends on sortase activity, a similar experiment using the msa-lacking mutant strain NZ7511 would expectedly generate the same results. In the mouse passage experiments mentioned, the srtA mutant and wild-type strains were equally and simultaneously administered to the same BALB/c mice and differentially enumerated in time course fecal samples by counting CFU and using quantitative PCR detection of wild-type and mutant chromosomal genetic loci. These experiments revealed that srtA deletion does not have a significant effect on the survival and persistence of NZ7104 in the intestinal tract of mice; wild-type and sortase mutant strains displayed the same characteristics in this model. These results suggest that the sortase-dependent surface proteins, including msa, do not significantly add to these features of L. plantarum. Nevertheless, this mouse model is not the preferred system to study potential functional properties of msa. For instance, the presence of mannose receptors on the intestinal epithelium differs in various organisms. Mannose adhesion is possibly not a relevant mechanism in the mouse intestine, which would be in agreement with the observation that there is no suitable mouse model for E. coli diarrhea. However, the measurements of survival and persistence are unlikely to reflect adherence properties adequately. These data do not exclude a role of the Msa protein as an adhesin that might be of interest regarding competitive exclusion of pathogens in the human intestine.
In conclusion, the identification of the mannose adhesin-encoding gene in L. plantarum WCFS1 contributes to an improved understanding of bacterial adhesion processes. Furthermore, the mutant strains constructed here allow evaluation of the importance of the mannose adherence capacity of this species in terms of the proposed competitive exclusion of E. coli. Future investigations will aim at further characterization of this adhesin's potential probiotic function and more detailed insights into its molecular structure.
Acknowledgments
This work was supported by the Centre for Human Nutrigenomics, The Netherlands.
The authors thank Alexander Lindenbergh for construction of NZ7104 and Jos Boekhorst and Roland J. Siezen for their assistance with in silico analysis of the msa gene and its product.
REFERENCES
- 1.Adlerberth, I., S. Ahrné, M. L. Johansson, G. Molin, L. Å. Hanson, and A. E. Wold. 1996. A mannose-specific adherence mechanism in Lactobacillus plantarum conferring binding to the human colonic cell line HT-29. Appl. Environ. Microbiol. 62:2244-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahrné, S., S. Nobaek, B. Jeppsson, I. Adlerberth, A. E. Wold, and G. Molin. 1998. The normal Lactobacillus flora of healthy human rectal and oral mucosa. J. Appl. Microbiol. 85:88-94. [DOI] [PubMed] [Google Scholar]
- 3.Aslanzadeh, J., and L. J. Paulissen. 1992. Role of type 1 and type 3 fimbriae on the adherence and pathogenesis of Salmonella enteritidis in mice. Microbiol. Immunol. 36:351-359. [DOI] [PubMed] [Google Scholar]
- 4.Bernard, N., T. Ferain, D. Garmyn, P. Hols, and J. Delcour. 1991. Cloning of the D-lactate dehydrogenase gene from Lactobacillus delbrueckii subsp. bulgaricus by complementation in Escherichia coli. FEBS Lett. 290:61-64. [DOI] [PubMed] [Google Scholar]
- 5.Bhattacharjee, J. W., and B. S. Srivastava. 1978. Mannose-sensitive haemagglutinins in adherence of Vibrio cholerae eltor to intestine. J. Gen. Microbiol. 107:407-410. [DOI] [PubMed] [Google Scholar]
- 6.Bork, P., T. Dandekar, Y. Diaz-Lazcoz, F. Eisenhaber, M. Huynen, and Y. Yuan. 1998. Predicting function: from genes to genomes and back. J. Mol. Biol. 283:707-725. [DOI] [PubMed] [Google Scholar]
- 7.Bringel, F., P. Quénée, and P. Tailliez. 2001. Polyphasic investigation of the diversity within Lactobacillus plantarum related strains revealed two L. plantarum subgroups. Syst. Appl. Microbiol. 24:561-571. [DOI] [PubMed] [Google Scholar]
- 8.Bron, P. A. 2004. The molecular response of Lactobacillus plantarum to intestinal passage and conditions. Ph.D. thesis. Wageningen University, Wageningen, The Netherlands.
- 9.Bron, P. A., C. Grangette, A. Mercenier, W. M. de Vos, and M. Kleerebezem. 2004. Identification of Lactobacillus plantarum genes that are induced in the gastrointestinal tract of mice. J. Bacteriol. 186:5721-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dalle, F., T. Jouault, P. A. Trinel, J. Esnault, J. M. Mallet, P. d'Athis, D. Poulain, and A. Bonnin. 2003. β-1,2- and α-1,2-linked oligomannosides mediate adherence of Candida albicans blastospores to human enterocytes in vitro. Infect. Immun. 71:7061-7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Clerck, E., D. Gevers, K. Sergeant, M. Rodríguez-Díaz, L. Herman, N. A. Logan, J. Van Beeumen, and P. De Vos. 2004. Genomic and phenotypic comparison of Bacillus fumarioli isolates from geothermal Antarctic soil and gelatine. Res. Microbiol. 155:483-490. [DOI] [PubMed] [Google Scholar]
- 12.De Ruyter, P. G. G. A., O. P. Kuipers, M. M. Beerthuyzen, I. van Alen-Boerrigter, and W. M. de Vos. 1996. Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J. Bacteriol. 178:3434-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derzelle, S., B. Hallet, K. P. Francis, T. Ferain, J. Delcour, and P. Hols. 2000. Changes in cspL, cspP, and cspC mRNA abundance as a function of cold shock and growth phase in Lactobacillus plantarum. J. Bacteriol. 182:5105-5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Vos, W. M., P. A. Bron, and M. Kleerebezem. 2004. Post-genomics of lactic acid bacteria and other food-grade bacteria to discover gut functionality. Curr. Opin. Biotechnol. 15:86-93. [DOI] [PubMed] [Google Scholar]
- 15.Ferain, T., D. Garmyn, N. Bernard, P. Hols, and J. Delcour. 1994. Lactobacillus plantarum ldhL gene: overexpression and deletion. J. Bacteriol. 176:596-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gasser, F., and M. Sebald. 1966. Nucleic acid composition of bacteria of the Lactobacillus genus. Ann. Inst. Pasteur 110:261-275. [PubMed] [Google Scholar]
- 17.Goossens, D., D. Jonkers, M. Russel, E. Stobberingh, A. Van Den Bogaard, and R. Stockbrügger. 2003. The effect of Lactobacillus plantarum 299v on the bacterial composition and metabolic activity in faeces of healthy volunteers: a placebo-controlled study on the onset and duration of effects. Aliment. Pharmacol. Ther. 18:495-505. [DOI] [PubMed] [Google Scholar]
- 18.Hahn, M., J. Pons, A. Planas, E. Querol, and U. Heinemann. 1995. Crystal structure of Bacillus licheniformis 1,3-1,4-β-D-glucan 4-glucanohydrolase at 1.8 Å resolution. FEBS Lett. 374:221-224. [DOI] [PubMed] [Google Scholar]
- 19.Hart, C. A., R. M. Batt, and J. R. Saunders. 1993. Diarrhoea caused by Escherichia coli. Ann. Trop. Paediatr. 13:121-131. [DOI] [PubMed] [Google Scholar]
- 20.Horinouchi, S., and B. Weisblum. 1982. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J. Bacteriol. 150:815-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hove, H., H. Nørgaard, and P. B. Mortensen. 1999. Lactic acid bacteria and the human gastrointestinal tract. Eur. J. Clin. Nutr. 53:339-350. [DOI] [PubMed] [Google Scholar]
- 22.Imberty, A., M. Wimmerová, E. P. Mitchell, and N. Gilboa-Garber. 2004. Structures of the lectins from Pseudomonas aeruginosa: insight into the molecular basis for host glycan recognition. Microbes Infect. 6:221-228. [DOI] [PubMed] [Google Scholar]
- 23.Jim, K., K. Parmar, M. Singh, and S. Tavazoie. 2004. A cross-genomic approach for systematic mapping of phenotypic traits to genes. Genome Res. 14:109-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johansson, M.-L., G. Molin, B. Jeppsson, S. Nobaek, S. Ahrné, and S. Bengmark. 1993. Administration of different Lactobacillus strains in fermented oatmeal soup: in vivo colonization of human intestinal mucosa and effect on the indigenous flora. Appl. Environ. Microbiol. 59:15-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johansson, M. L., S. Nobaek, A. Berggren, M. Nyman, I. Björck, S. Ahrné, B. Jeppsson, and G. Molin. 1998. Survival of Lactobacillus plantarum DSM 9843 (299v), and effect on the short-chain fatty acid content of faeces after ingestion of a rose-hip drink with fermented oats. Int. J. Food Microbiol. 42:29-38. [DOI] [PubMed] [Google Scholar]
- 26.Josson, K., T. Scheirlinck, F. Michiels, C. Platteeuw, P. Stanssens, H. Joos, P. Dhaese, M. Zabeau, and J. Mahillon. 1989. Characterization of a gram-positive broad-host-range plasmid isolated from Lactobacillus hilgardii. Plasmid 21:9-20. [DOI] [PubMed] [Google Scholar]
- 27.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. Fiers, W. Stiekema, R. M. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krogfelt, K. A., H. Bergmans, and P. Klemm. 1990. Direct evidence that the FimH protein is the mannose-specific adhesin of Escherichia coli type 1 fimbriae. Infect. Immun. 58:1995-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, Y. K., and K. Y. Puong. 2002. Competition for adhesion between probiotics and human gastrointestinal pathogens in the presence of carbohydrate. Br. J. Nutr. 88(Suppl. 1):S101-S108. [DOI] [PubMed] [Google Scholar]
- 30.Levesque, M., D. Shasha, W. Kim, M. G. Surette, and P. N. Benfey. 2003. Trait-to-gene: a computational method for predicting the function of uncharacterized genes. Curr. Biol. 13:129-133. [DOI] [PubMed] [Google Scholar]
- 31.Mack, D. R., S. Ahrné, L. Hyde, S. Wei, and M. A. Hollingsworth. 2003. Extracellular MUC3 mucin secretion follows adherence of Lactobacillus strains to intestinal epithelial cells in vitro. Gut 52:827-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayo, B., S. Derzelle, M. Fernández, C. Léonard, T. Ferain, P. Hols, J. E. Suárez, and J. Delcour. 1997. Cloning and characterization of cspL and cspP, two cold-inducible genes from Lactobacillus plantarum. J. Bacteriol. 179:3039-3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mirelman, D., G. Altmann, and Y. Eshdat. 1980. Screening of bacterial isolates for mannose-specific lectin activity by agglutination of yeasts. J. Clin. Microbiol. 11:328-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33a.Molenaar, D., F. Bringel, F. H. Schuren, W. M. de Vos, R. J. Siezen, and M. Kleerebezem. 2005. Exploring Lactobacillus plantarum genome diversity by using microarrays. J. Bacteriol. 187:6120-6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Molin, G. 2001. Probiotics in foods not containing milk or milk constituents, with special reference to Lactobacillus plantarum 299v. Am. J. Clin. Nutr. 73:380S-385S. [DOI] [PubMed] [Google Scholar]
- 35.Molin, G., B. Jeppsson, M. L. Johansson, S. Ahrné, S. Nobaek, M. Ståhl, and S. Bengmark. 1993. Numerical taxonomy of Lactobacillus spp. associated with healthy and diseased mucosa of the human intestines. J. Appl. Bacteriol. 74:314-323. [DOI] [PubMed] [Google Scholar]
- 36.Moreira, L. M., R. F. De Souza, N. F. Almeida, Jr., J. C. Setubal, J. C. Oliveira, L. R. Furlan, J. A. Ferro, and A. C. Da Silva. 2004. Comparative genomics analyses of citrus-associated bacteria. Annu. Rev. Phytopathol. 42:163-184. [DOI] [PubMed] [Google Scholar]
- 37.Muscariello, L., R. Marasco, M. De Felice, and M. Sacco. 2001. The functional ccpA gene is required for carbon catabolite repression in Lactobacillus plantarum. Appl. Environ. Microbiol. 67:2903-2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Navarre, W. W., and O. Schneewind. 1999. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63:174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neeser, J. R., D. Granato, M. Rouvet, A. Servin, S. Teneberg, and K. A. Karlsson. 2000. Lactobacillus johnsonii La1 shares carbohydrate-binding specificities with several enteropathogenic bacteria. Glycobiology 10:1193-1199. [DOI] [PubMed] [Google Scholar]
- 40.Platteeuw, C., G. Simons, and W. M. de Vos. 1994. Use of the Escherichia coli β-glucuronidase (gusA) gene as a reporter gene for analyzing promoters in lactic acid bacteria. Appl. Environ. Microbiol. 60:587-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rajashekara, G., J. D. Glasner, D. A. Glover, and G. A. Splitter. 2004. Comparative whole-genome hybridization reveals genomic islands in Brucella species. J. Bacteriol. 186:5040-5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reid, G. 1999. The scientific basis for probiotic strains of Lactobacillus. Appl. Environ. Microbiol. 65:3763-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reid, G., and J. Burton. 2002. Use of Lactobacillus to prevent infection by pathogenic bacteria. Microbes Infect. 4:319-324. [DOI] [PubMed] [Google Scholar]
- 44.Robinson, D. A., and M. C. Enright. 2004. Evolution of Staphylococcus aureus by large chromosomal replacements. J. Bacteriol. 186:1060-1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roche, F. M., R. Massey, S. J. Peacock, N. P. Day, L. Visai, P. Speziale, A. Lam, M. Pallen, and T. J. Foster. 2003. Characterization of novel LPXTG-containing proteins of Staphylococcus aureus identified from genome sequences. Microbiology 149:643-654. [DOI] [PubMed] [Google Scholar]
- 46.Roos, S., and H. Jonsson. 2002. A high-molecular-mass cell-surface protein from Lactobacillus reuteri 1063 adheres to mucus components. Microbiology 148:433-442. [DOI] [PubMed] [Google Scholar]
- 47.Rose, R. E. 1988. The nucleotide sequence of pACYC184. Nucleic Acids Res. 16:355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 49.Schembri, M. A., E. V. Sokurenko, and P. Klemm. 2000. Functional flexibility of the FimH adhesin: insights from a random mutant library. Infect. Immun. 68:2638-2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ton-That, H., G. Liu, S. K. Mazmanian, K. F. Faull, and O. Schneewind. 1999. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc. Natl. Acad. Sci. USA 96:12424-12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Turner, M. S., L. M. Hafner, T. Walsh, and P. M. Giffard. 2003. Peptide surface display and secretion using two LPXTG-containing surface proteins from Lactobacillus fermentum BR11. Appl. Environ. Microbiol. 69:5855-5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van der Vossen, J. M., D. van der Lelie, and G. Venema. 1987. Isolation and characterization of Streptococcus cremoris Wg2-specific promoters. Appl. Environ. Microbiol. 53:2452-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Kranenburg, R., J. D. Marugg, I. I. van Swam, N. J. Willem, and W. M. de Vos. 1997. Molecular characterization of the plasmid-encoded eps gene cluster essential for exopolysaccharide biosynthesis in Lactococcus lactis. Mol. Microbiol. 24:387-397. [DOI] [PubMed] [Google Scholar]
- 54.Vaughan, E. E., M. C. de Vries, E. G. Zoetendal, K. Ben-Amor, A. D. Akkermans, and W. M. de Vos. 2002. The intestinal LABs. Antonie Leeuwenhoek 82:341-352. [PubMed] [Google Scholar]
- 55.Velloso, L. M., K. Svensson, G. Schneider, R. F. Pettersson, and Y. Lindqvist. 2002. Crystal structure of the carbohydrate recognition domain of p58/ERGIC-53, a protein involved in glycoprotein export from the endoplasmic reticulum. J. Biol. Chem. 277:15979-15984. [DOI] [PubMed] [Google Scholar]
- 56.Vesa, T., P. Pochart, and P. Marteau. 2000. Pharmacokinetics of Lactobacillus plantarum NCIMB 8826, Lactobacillus fermentum KLD, and Lactococcus lactis MG 1363 in the human gastrointestinal tract. Aliment. Pharmacol. Ther. 14:823-828. [DOI] [PubMed] [Google Scholar]
- 57.Wedekind, J. E., C. B. Trame, M. Dorywalska, P. Koehl, T. M. Raschke, M. McKee, D. FitzGerald, R. J. Collier, and D. B. McKay. 2001. Refined crystallographic structure of Pseudomonas aeruginosa exotoxin A and its implications for the molecular mechanism of toxicity. J. Mol. Biol. 314:823-837. [DOI] [PubMed] [Google Scholar]
- 58.Wold, A. E., M. Thorssén, S. Hull, and C. S. Edén. 1988. Attachment of Escherichia coli via mannose- or Galα1→4Galβ-containing receptors to human colonic epithelial cells. Infect. Immun. 56:2531-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]