Abstract
We have analyzed the distribution of RNA nucleotidyltransferases from the family that includes poly(A) polymerases (PAP) and tRNA nucleotidyltransferases (TNT) in 43 bacterial species. Genes of several bacterial species encode only one member of the nucleotidyltransferase superfamily (NTSF), and if that protein functions as a TNT, those organisms may not contain a poly(A) polymerase I like that of Escherichia coli. The genomes of several of the species examined encode more than one member of the nucleotidyltransferase superfamily. The function of some of those proteins is known, but in most cases no biochemical activity has been assigned to the NTSF. The NTSF protein sequences were used to construct an unrooted phylogenetic tree. To learn more about the function of the NTSFs in species whose genomes encode more than one, we have examined Bacillus halodurans. We have demonstrated that B. halodurans adds poly(A) tails to the 3′ ends of RNAs in vivo. We have shown that the genes for both of the NTSFs encoded by the B. halodurans genome are transcribed in vivo. We have cloned, overexpressed, and purified the two NTSFs and have shown that neither functions as poly(A) polymerase in vitro. Rather, the two proteins function as tRNA nucleotidyltransferases, and our data suggest that, like some of the deep branching bacterial species previously studied by others, B. halodurans possesses separate CC- and A-adding tRNA nucleotidyltransferases. These observations raise the interesting question of the identity of the enzyme responsible for RNA polyadenylation in Bacillus.
RNA nucleotidyltransferases play an important role in the maturation, processing, and degradation of RNAs. tRNA nucleotidyltransferases (TNT) and poly(A) polymerases (PAP) are members of a superfamily of RNA nucleotidyltransferases (13, 39). tRNA nucleotidyltransferases add CCA residues to the 3′ ends of immature tRNAs and repair the 3′ ends of damaged tRNAs (36). tRNA nucleotidyltransferases (TNTs) are found in bacteria, archaeae, and eukaryotes, suggesting conservation of structure and function of this class of enzymes over the course of evolution (39). Polyadenylation of RNA 3′ ends, once thought to occur exclusively in eukaryotic cells, has now been shown definitively to occur in bacterial systems as well (7, 25, 30, 31). Escherichia coli cells contain at least two enzymes capable of adding poly(A) tails to the 3′ ends of RNA molecules. The major polyadenylating enzyme, poly(A) polymerase I (PAP I), was originally identified by virtue of its role in regulating plasmid copy number and is a product of the pcnB gene (6, 12). PAP I can utilize any of the three nucleoside triphosphates as substrates in vitro, but only A residues are added by the enzyme to RNA 3′ ends in vivo (38). Messenger RNAs and 16S and 23S ribosomal RNAs are all polyadenylated in E. coli, and this modification is an important element of the RNA degradation pathway in bacterial systems (7, 25, 30, 31). RNA 3′ ends are generated in E. coli via endonucleolytic cleavage by RNase E and RNase III, these ends are polyadenylated by PAP I, and the resulting RNAs are degraded exonucleolytically (27). Polyadenylation facilitates the exonucleolytic degradation of RNAs.
There is at least one other enzyme system in bacteria that is utilized for the synthesis of RNA 3′ tails. Mutants of E. coli lacking PAP I still retain the ability to polyadenylate RNAs. Mohanty and Kushner presented evidence indicating that the second PAP in E. coli is polynucleotide phosphorylase (PNPase) (23). PNPase normally functions as a 3′-5′-exoribonuclease in bacteria and plays a key role in the degradation of bacterial RNAs (8, 24, 25). Mohanty and Kushner argue that under appropriate conditions in vivo, PNPase can serve to degrade RNAs or synthesize poly(A) tails and that this enzyme is responsible for the C and U residues that are found at low frequency in the poly(A) tails of RNAs from wild-type E. coli (23). Recently, the function of PNPase has been studied in spinach chloroplasts and in a Synechocystis species. Both of those systems appear to lack a dedicated poly(A) polymerase like PAP I, and in both the data indicate that PNPase is the sole enzyme responsible for the synthesis of RNA 3′ tails (28, 37). Moreover, Schuster and coworkers presented evidence that PNPase is essential in Synechocystis (28). The studies of Sohlberg et al. and those from the author's laboratory have also implicated PNPase as the RNA 3′ polyribonucleotide polymerase (PAP analog) in Streptomyces (3, 33). Thus, the 3′ tails associated with RNAs in Streptomyces coelicolor and Streptomyces antibioticus are heteropolymeric, containing G, C, and U as well as A residues (3). Heteropolymeric tails are observed in mutants of E. coli that lack PAP I, and as indicated above, PNPase is thought to be responsible for the synthesis of such tails (23).
The results just cited raise the interesting question of the nature of the enzyme responsible for the synthesis of RNA 3′ tails in other bacterial systems. Moreover, several scenarios have been suggested for the evolution of CCA-adding enzymes and PAPs (22, 34). Additional comparative analyses of biochemical, structural, and phylogenetic data are required to determine whether PAPs are ancestors, descendants, or distant cousins of CCA-adding enzymes. To examine these questions further we have compiled the protein sequences for 68 members of the nucleotidyltransferase superfamily (NTSF) that includes tRNA nucleotidyltransferases and poly(A) polymerases. These sequences have been used to construct a phylogenetic tree for the bacterial NTSFs. That tree suggests functional relationships between various NTSFs that can be examined experimentally. We have examined the 3′ tails associated with RNAs from Bacillus halodurans and have overexpressed two NTSFs from that species. We show that B. halodurans lacks PAP I and that its two NTSFs are tRNA nucleotidyltransferases. Our characterization of the B. halodurans NTSFs as CC- and A-adding enzymes is the first example of organisms other than Aquifex, Deinococcus, and Synechocystis species that express these functions in separate enzymes. We suggest that PNPase or an as yet unidentified enzyme functions as the PAP in B. halodurans and perhaps in other Bacillus species as well.
MATERIALS AND METHODS
Growth of strains and RNA isolation.
Bacillus halodurans C-125 was obtained from the American Type Culture Collection and was grown on peptone-starch-carbonate medium as described by the American Type Culture Collection (www.atcc.org/SearchCatalogs). E. coli DH5α was grown in Luria-Bertani broth. Cells were grown to mid-exponential phase at 37°C and received 5 min of 10-mg ml−1 lysozyme treatment at room temperature. Total RNA was prepared using the hot phenol method described by Köhrer and Domdey (16), except that the buffer for phenol extraction was 50 mM Na-acetate, pH 5.1, 10 mM EDTA, 1% sodium dodecyl sulfate (SDS). DNA was removed by 20 min of digestion with DNase I (Promega) in New England Biolabs buffer 4. RNA integrity was checked by electrophoresis on a 1% agarose gel.
Sequences, alignment, and phylogenetic analysis.
Sequences were retrieved from the microbial genomes database at www.ncbi.nlm.nih.gov. The sequences of most of the proteins were used without modification for the analyses described. However, the Aquifex aeolicus, Deinococcus radiodurans, Synechocystis spp., and Nostoc spp. A-adding enzymes and the Thermotoga maritima NTSF have an N-terminal extension that has diverged significantly from the regions of the proteins that are similar to other NTSFs (34). Similar N-terminal extensions were observed for Geobacter sulfurreducens NTSFIII and Desulfovibrio vulgaris NTSFII. Those extensions were omitted in the phylogenetic analysis described here with the result that the sequences used ranged in length from 380 to 540 amino acids. Sequences were aligned using ClustalX with the default protein alignment parameters. The alignment was then fine tuned using the program TuneClustal developed by Barry G. Hall, with his permission. TuneClustal averages the quality scores produced by ClustalX. The alignment parameters were adjusted until the TuneClustal average was maximized.
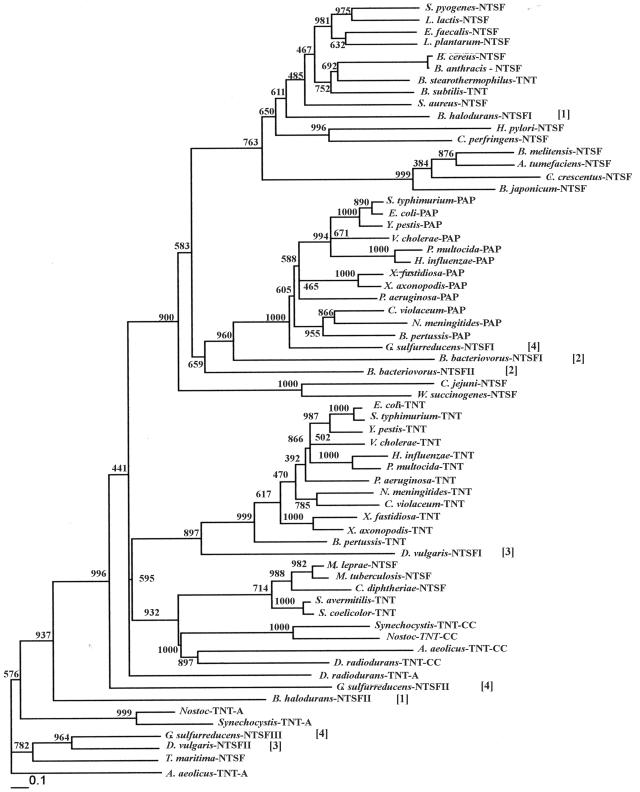
The phylogenetic tree was constructed using PHYLIP version 3.6b (11). The sequences were bootstrapped 1,000 times and jumbled once. The bootstrapped sequences were used to generate 1,000 distance matrices which were then entered into the program NEIGHBOR. The resulting trees were then entered into CONSENSE to produce an unrooted extended majority rule consensus tree. The resulting tree was then entered into FITCH to obtain the tree shown in Fig. 1, whose branch lengths reflect evolutionary distances. The bootstrap values are shown at each node in the figure, and the scale bar represents the number of amino acid substitutions per site.
FIG. 1.
Neighbor-joining phylogenetic tree relating the proteins listed in Table 2. The tree was constructed using PHYLIP (11), as indicated in Materials and Methods. Bootstrap scores are shown at each node. The bar at the bottom of the figure represents the number of amino acid substitutions per site. The numbers in brackets identify the NTSFs from B. halodurans [1], Bdellovibrio bacteriovorus [2], D. vulgaris [3], and G. sulfurreducens [4].
Determination of RNA poly(A) tail lengths.
RNA was prepared from mid-log Bacillus and E. coli cultures using the hot phenol method as described above. Poly(A) tail lengths were determined as described previously by end labeling of 20 μg of RNA with [32P]pCp and RNA ligase (4, 5). Labeled RNAs were then digested with a combination of RNase A and RNase T1, which cleaves all phosphodiester bonds in the RNAs except those between adjacent A residues. The poly(A) tails that remained following RNase digestion were displayed on 12% polyacrylamide gels and visualized by autoradiography.
RT-PCR analysis.
Reverse transcription-PCR (RT-PCR) was carried out essentially as described by Bralley and Jones (3), except that dimethyl sulfoxide was omitted from the PCR buffer. For studies of NTSFI and NTSFII expression, 5 μg of total RNA was reverse transcribed with random hexamer primers. The first round of nested PCR, carried out using primers pairs I-F1/I-R1 and II-F1/II-R1, was followed by a second round using primers I-F2/I-R2 and II-F2/II-R2 (Table 1). PCR primers were designed to be complementary to nonhomologous regions of the two proteins to avoid detecting the transcript of one gene with both sets of primers.
TABLE 1.
Primers used in this study
| Primer | Gene or protein | Sequence (5′-3′)a |
|---|---|---|
| I-F1 | NTSFI | CCATGGGTTTGAGGCCTATATC |
| I-R1 | NTSFI | CAACGGTGGCAACAGACAAGGACGC |
| II-F1 | NTSFII | GAGTGAAGAACATCAAGAATC |
| II-R1 | NTSFII | GCGAAGCGGCTCCTCAGGCACTTG |
| I-F2 | NTSFI | TATCACATCGGCCAGAAAACC |
| I-R2 | NTSFI | GCTAGCGGGAGCTTCATGCTGC |
| II-F2 | NTSFII | ATGCTAAGAGGGGTACCTGGTG |
| II-R2 | NTSFII | GATGGCGAAAGGAACGAGTGAAGCC |
| I-F3 | NTSFI | GCAGCATGAAGCTCCGCTAGC |
| I-F4 | NTSFI | GCGTCCTTGTCTGTTGCCACC |
| II-F3 | NTSFII | GGCTTCACTCGTTCCTTTCGC |
| II-F4 | NTSFII | CAAGTGCCTGAGGAGCCGC |
| BS16S | rrnB | CAGAGTGACAGGTGGTGCATG |
| ADoT | GGATCCGAATTCTCTAGAT17 | |
| AD20 | GGATCCGAATTCTCTAGA | |
| ADoCT | GGATCCGAATTCTCTAGAC10T6 | |
| I-F5 | NTSFI | AGAGCAGGTGGACATATGGATGAC |
| I-R5 | NTSFI | TTTTTAACAACGGATCCTTCACGACTCCAC |
| II-F5 | NTSFII | TAAAGGAGTCCGCATATGAGTGAA |
| II-R5 | NTSFII | CCGAGGTTTTCTGGATCCGTATCATAAGGG′ |
| 3896F1 | TNT | GTACCCTGTTTTCATATGCCGAACGCC |
| 3896R1 | TNT | CGGAAGGTCGACGGATCCACCGGCTTCTAC |
NdeI and BamHI sites are underlined.
For the oligo(dT)-based method used in 3′ tail analysis, reverse transcription was carried out using primer ADoT (Table 1). This primer contains an oligo(dT)17 sequence that can anneal to poly(A) tails. cDNA amplification used gene-specific forward primers and reverse primer AD20 (Table 1). The AD20 primer sequence is identical to the 5′ sequence of ADoT (Table 1). cDNAs obtained using rRNA templates were amplified by one round of PCR using a single gene-specific forward primer (BS16S) complementary to the B. halodurans 16S rrnB gene. Amplification of cDNAs derived from mRNAs required seminested PCR utilizing two gene-specific forward primers (I-F3, I-F4 for NTSFI and II-F3, II-F4 for NTSFII; Table 1) and reverse primer AD20. Amplification products were cloned using the TOPO-TA Cloning kit (Invitrogen) and then sequenced by the Emory University DNA Core Facility.
The polyguanylation method used in 3′ tail analysis was adapted from Kusov et al. (17). Fifteen micrograms of total RNA was dissolved in 14 μl water, heated for 5 min at 65°C, and then snap chilled on ice. The reaction mix (25 μl) contained 0.6 mM GTP, 2,175 U of yeast poly(A) polymerase, and 1× reaction buffer (Amersham Bioscience). After a 2-h incubation at 37°C, 750 U of enzyme (1 μl) was added and incubation was continued for 2 h. The reaction was stopped by phenol-chloroform and chloroform extractions and ethanol precipitation. The RNA pellet was resuspended in 20 μl of water and reverse transcribed with Superscript II RNase H− (Invitrogen) and primer ADoCT following the manufacturer's protocol. Seminested PCR and cloning were as described above.
Cloning and overexpression of the B. halodurans NTSFs.
As indicated below, B. halodurans genes encode two members of the nucleotidyltransferase superfamily that includes TNTs and PAPs. We designated these two proteins NTSFI (the open reading frame [ORF] is annotated in the B. halodurans genome sequence as BH1684) and NTSFII (BH2881). The NTSFI ORF is 1,128 bp in length, and that of NTSFII is 1,284 bp. PCR primers corresponding to the 5′ and 3′ ends of the two genes were designed and used in the PCR as described previously (15). Primers I-F5 and R5 were used to amplify NTSFI, and II-F5 and R5 were used for NTSFII (see Table 1). In each case, the 5′ primer introduced an NdeI site, overlapping the translation start codon, into the PCR product, and the 3′ primer introduced a BamHI site just downstream of the stop codon for each gene (Table 1). The PCR products were cloned in the pCR2.1-TOPO cloning vector (Invitrogen) to generate pJSE4100 and pJSE4101 for NTSFI and NTSFII, respectively. The NTSFI and NSTFII genes were excised by digestion with NdeI and BamHI. The resulting fragments were then cloned in pET19B (Novagen) to generate pJSE4102 (NTSFI) and pJSE4103 (NTSFII). Cloning in this vector attaches a decahistidine tag to the N terminus of the cloned protein. pJSE4102 was used to transform E. coli BL21(DE3)pLysS, and the protein was overexpressed by induction with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at an A600 of ca. 0.7. Cells were harvested from two 500-ml cultures 4 h after IPTG addition, washed with Buffer I composed of 50 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 5% glycerol, 1 mM dithiothreitol, and stored frozen. The cells were disrupted following the protocol that accompanies the Talon column procedure (BD Biosciences Clontech), and the protein was purified by Talon column chromatography following the manufacturer's instructions. As a control for our analyses, we also cloned the Streptomyces coelicolor tRNA nucleotidyltransferase (SCO3896) in pET19B (yielding pJSE3560), using primers 3896F1 and R1 (Table 1) for PCR, and purified that protein by Talon column chromatography.
The N-terminally his-tagged version of NTSFII did not adhere to the Talon column. Therefore, the NdeI-BamHI fragment bearing the open reading frame was cloned into pET11A to generate pJSE4113 and the protein was overexpressed by IPTG induction as described above. NTSFII was purified by a combination of ion exchange and hydrophobic interaction chromatography. NTSFI and NTSFII were concentrated following chromatography by dialysis against Buffer I containing 25% glycerol. SCO3896 was concentrated by dialysis against ammonium sulfate without stirring. Details of these purifications will be presented in a subsequent publication.
Enzyme assays.
TNT assays (60 μl) contained 50 mM Tris-HCl or 50 mM glycine-NaOH; 10 mM MgCl2; 0.2 mM nonradioactive ATP or CTP; 20 μCi ml−1 of [α-32P]ATP or CTP (3,000 Ci/mmole; Amersham Pharmacia); 100 μg ml−1 bovine serum albumin; and 2 mg ml−1 E. coli tRNA (Sigma). In the experiments of Fig. 6, the pH of the buffers was varied as indicated using Tris-HCl from pH 6.5 to 8.5 and glycine-NaOH at pH 9 and 9.5. Reaction mixtures contained 6.7 μM NTSFI, 18.4 μM NTSFII, or 15.7 μM SCO3896. Duplicate reaction mixtures were incubated for 15 min at 37°C, and then 50 μl was removed and the reaction products were collected by precipitation with trichloroacetic acid. Precipitates on glass fiber filters were analyzed by liquid scintillation counting. E. coli poly(A) polymerase I was obtained from Ambion, and assays were performed according to the supplier's instructions using tRNA as the acceptor. The same tRNA preparation was used in the TNT and PAP assays. To examine the products of the TNT and PAP reactions, large-scale mixtures (60 to 240 μl) were incubated as described above and extracted with phenol-chloroform. Labeled RNAs were precipitated with ethanol with 10 μg of E. coli tRNA added as a carrier. RNAs were examined by electrophoresis on 10% denaturing polyacrylamide gels along with Ambion Century-Plus size markers.
FIG. 6.
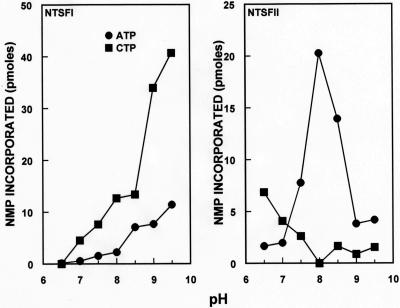
pH dependency of the tRNA nucleotidyltransferase assays of B. halodurans NTSFI and NTSFII. Left panel, NTSFI assayed with ATP and CTP; right panel, NTSFII assayed with ATP and CTP. Assays were performed as described in Materials and Methods, and results were corrected by subtracting a zero enzyme control value from each experimental value. Results are expressed as picomoles of AMP or CMP incorporated in a 60-μl reaction mixture.
Miscellaneous methods.
Protein was determined by the method of Bradford (2) using the Bio-Rad assay reagent with bovine serum albumin as a standard. SDS-polyacrylamide gel electrophoresis (PAGE) was performed essentially as described by Laemmli (18), and gels were stained with Coomassie brilliant blue.
RESULTS AND DISCUSSION
Distribution and sequence analyses of enzymes modifying bacterial RNA 3′ ends.
The availability of genome sequences for a large number of bacterial species makes it possible to compare the sequences of the RNA 3′ end-modifying enzymes described above. For this analysis, the genome sequences of 43 bacterial species were examined. Representatives of the α-, β-, γ-, δ-, and ɛ-proteobacteria, the low-GC gram-positive (LGC+) bacteria, the high-GC gram-positive (HGC+) bacteria, and cyanobacteria appear in Table 2, as do several species postulated to be near the base of the bacterial clade based on rRNA phylogenies (Aquifex aeolicus, Deinococcus radiodurans, Thermotoga maritima) (1, 21). Relevant observations derived from the analyses of data for these species are as follows.
TABLE 2.
Distribution of NTSFs in bacteria
| Speciesb | NTSF annotation(s)a | Biochemical function |
|---|---|---|
| Deep branching | ||
| Aquifex aeolicus (2)c | Two PAPs | TNT (CC and A addition) |
| Deinococcus radiodurans (2) | TNT, TNT/PAP | TNT (CC and A addition) |
| Thermotoga maritima (1) | TNT | TNT (CCA addition) |
| Cyanobacteria | ||
| Synechocystis sp. (2)c | Two PAPs | TNT (CC and A addition) |
| Nostoc sp. strain PCC 7120 (2) | Two PAPs | Unknown |
| HGC+ | ||
| Corynebacterium diphtheriae (1) | TNT | Unknown |
| Mycobacterium leprae (1) | PAP | Unknown |
| Mycobacterium tuberculosis (1) | PAP | Unknown |
| Streptomyces avermitilis (1) | PAP | Probable TNTd |
| Streptomyces coelicolor (1) | NTSF | TNT (CCA addition) |
| LGC+ | ||
| Bacillus anthracis (1) | PAP | Unknown |
| Bacillus cereus (1) | PAP | Unknown |
| Bacillus halodurans (2) | PAP, unknown NTSF | TNT (CC and A addition) |
| Bacillus subtilis (1) | PAP | TNT (CCA addition) |
| Bacillus stearothermophilus (1) | PAP | TNT (CCA addition) |
| Clostridium perfringens (1) | PAP | Unknown |
| Enterococcus faecalis (1) | PAP | Unknown |
| Lactobacillus plantarum (1) | PAP | Probable TNTe |
| Lactococcus lactis (1) | PAP | Unknown |
| Staphylococcus aureus (1) | PAP | Unknown |
| Streptococcus pyogenes (1) | PAP | Unknown |
| α-Proteobacteria | ||
| Agrobacterium tumefaciens (1) | PAP | Unknown |
| Bradyrhizobium japonicum (1) | PAP | Unknown |
| Brucella melitensis (1) | PAP/TNT | Unknown |
| Caulobacter crescentus (1) | PAP | Unknown |
| β-, γ-Proteobacteria | ||
| Bordetella pertussis (2) | TNT and PAP | Unknown |
| Chromobacterium violaceum (2) | TNT and PAP | Unknown |
| Neisseria meningitides (2) | TNT and PAP | Unknown |
| Escherichia coli (2) | TNT and PAP | TNT and PAP |
| Haemophilus influenzae (2) | TNT and PAP | Unknown |
| Pasteurella multocida (2) | TNT and PAP | Unknown |
| Pseudomonas aeruginosa (2) | TNT and PAP | Unknown |
| Salmonella enterica serovar Typhimurium (2) | TNT and PAP | Unknownf |
| Vibrio cholerae (2) | TNT and PAP | Unknown |
| Xanthomonas axonopodis (2) | TNT and PAP | Unknown |
| Xylella fastidiosa (2) | TNT and PAP | Unknown |
| Yersinia pestis (2) | TNT and PAP | Unknown |
| δ-Proteobacteria | ||
| Bdellovibrio bacteriovorus (2) | PAP, TNT/PAP | Unknown |
| Desulfovibrio vulgaris (2) | TNT, PAP family | Unknown |
| Geobacter sulfurreducens (3) | One PAP, two PAP family | Unknown |
| ɛ-Proteobacteria | ||
| Campylobacter jejuni (1) | NTSF | Unknown |
| Helicobacter pylori (1) | PAP | Unknown |
| Wolinella succinogenes (1) | TNT | Unknown |
The annotation of the NTSFs in the genome database. In some cases the ORF is annotated as a TNT and in others as a PAP. Some annotations describe the NTSF as a member of the TNT/PAP or PAP family, and others list it as an unknown NTSF.
The numbers in parentheses after the species names indicate the number of NTSFs in the genome of that species.
The functions of the NTSFs from A. aeolicus, D. radiodurans, T. maritima, and Synechocystis spp. have been determined.
Based on the sequence similarity between the S. avermitilis and S. coelicolor NTSFs, the former NTSFI is thought to be a TNT.
The NTSF of Lactobacillus acidophilus is a TNT (19).
E. coli pcnB hybridizes to a DNA fragment from S. enterica serovar Typhimurium (10).
(i) All eubacterial genomes examined encode at least one member of the nucleotidyltransferase superfamily that includes TNTs and PAPs.
As shown in Table 2, a search of the genome sequences of the species listed identified at least one protein with significant similarity to a TNT or a PAP. The genomes of several species encode two such proteins. Thus, A. aeolicus, D. radiodurans, Synechocystis sp., and Nostoc sp. each contain two NTSFs. In the first three of these four species, it is known that both NTSFs function as TNTs. These organisms have evolved an interesting biochemical system for maturing and repairing tRNA 3′ ends, a system that utilizes one enzyme to add the two C's to such ends and a second enzyme to add the terminal A residue (34, 35). The biochemical function of the Nostoc enzymes has not been studied, but the phylogenetic analysis presented below strongly suggests that it too contains separate CC- and A-adding enzymes. The β- and γ-proteobacteria also contain two NTSFs. In E. coli, it has been shown that one of the NTSFs functions as a TNT and the other as a PAP (6, 9). The genome annotations for all the β- and γ-proteobacterial species identify the NTSFs in this fashion. Thus, all such species are indicated in Table 2 as containing both a TNT and a PAP. It should be noted, however, that to our knowledge, among the β- and γ-proteobacteria, PAP activity has only been demonstrated in E. coli. We discuss other species that encode more than one NTSF in greater detail below.
In the case of Bacillus subtilis and Bacillus stearothermophilus, it has been shown that the one NTSF encoded by the genomes of these organisms is a TNT (20, 26). Streptomyces coelicolor gene SCO3896 encodes a TNT as well (33). The genomes of the α- and ɛ-proteobacterial species examined encode only one NTSF. The biochemical function of these proteins is unknown. With the exceptions just noted and a few other species in which TNT activity has been demonstrated, the function of the NTSFs listed in Table 2 is unknown.
(ii) The NTSF sequences can be used to construct an NTSF phylogeny.
Figure 1 presents an unrooted Neighbor-joining phylogenetic tree, constructed using PHYLIP (11), relating the NTSFs from the species listed in Table 2. As the bootstrap scores at several of the ancestral nodes of Fig. 1 are low, we use the results of Fig. 1 to suggest possible functional relationships between the NTSFs. Relevant features of the phylogenetic analysis are as follows. First, the A-adding enzymes of those species known to possess two TNTs are related and cluster together in the phylogenetic tree. The T. maritima TNT is closely related to the A-adding TNTs from A. aeolicus and the cyanobacteria. Second, the CC-adding enzymes are clearly closely related and are components of a clade that also includes the proteins from the high-GC gram-positive species. Third, one of the Nostoc NTSFs is closely related to the Synechocystis A-adding enzyme and the other is related to the Synechocystis CC-adding enzyme. Thus, it seems highly likely that Nostoc also contains separate CC- and A-adding TNTs. Fourth, the NTSF from Helicobacter pylori is more closely related to the NTSFs from the low-GC gram-positive bacteria, specifically to that from Clostridium perfringens, than to those of the β-, γ-, or ɛ-proteobacteria. This observation strongly suggests that the NTSF of H. pylori may have arisen by horizontal gene transfer from one of the low-GC gram-positive species. Fifth, the TNTs from the β- and γ-proteobacteria are related to each other, as are the PAPs from those same species. PAPs in the β- and γ-proteobacteria probably resulted from gene duplication of an ancestral TNT gene or horizontal gene transfer of such an ancestral gene.
Finally, several species other than the species known to possess two TNTs and the β- and γ-proteobacteria contain more than one NTSF. These species are indicated by numbers in brackets in Fig. 1. B. halodurans [1], Bdellovibrio bacteriovorus [2], and Desulfovibrio vulgaris [3] each encode two ORFs annotated as NTSFs, and Geobacter sulfurreducens [4] contains three such ORFs. B. halodurans NTSFI appears to be related to the NTSFs from other bacilli, while NTSFII is most closely related to the tRNA nucleotidyltransferases from the species with separate CC- and A-adding enzymes. D. vulgaris NTSFI appears to have arisen from an ancestor that also gave rise to the TNTs of the β- and γ-proteobacteria, while NTSFII from this species is related to the A. aeolicus A-adding enzyme, the T. maritima NTSF, and G. sulfurreducens NTSFIII. There is the formal possibility, therefore, that D. vulgaris contains separate CC- and A-adding enzymes, as is the case for Aquifex spp., Deinococcus spp., and the cyanobacteria. Both B. bacteriovorus NTSFs appear to have descended from an ancestor of the PAPs. G. sulfurreducens NTSFI, like the B. bacteriovorus NTSFs, appears most closely related to the PAPs. Martin and Keller have recently proposed a consensus NTSF sequence, [LIV]-[LIV]-G-[RK]-F-x-[LIV]-h-[HLQ]-[LIV] (alternative amino acids are in brackets), that they argue is diagnostic of PAPs (22). G. sulfurreducens NTSFI possesses such a sequence, LVGRRFRLAHI, that is nearly identical to the signature sequence proposed for E. coli PAP I, LVGRRFRLAHV. B. bacteriovorus NTSFI also possesses such a sequence, VIGRRFKLVLV. However, neither G. sulfurreducens NTSFII or NTSFIII, B. bacteriovorus NTSFII, nor either of the D. vulgaris NTSFs possess such a sequence. G. sulfurreducens NTSFII and NTSFIII are related to the NTSFs from the species with separate CC- and A-adding enzymes. Thus, it is possible that δ-proteobacterial NTSFs represent additional variations on the general themes suggested above for the species with separate CC- and A-adding enzymes (two TNTs, no PAP I-like enzyme), the LGC+ and HGC+ bacteria and the α- and ɛ-proteobacteria (one TNT, no PAP I-like enzyme), and the β- and γ-proteobacteria (one TNT, one PAP). We note that Martin and Keller have also presented a phylogenetic analysis of the bacterial NTSFs (22). However, their analysis involved fitting the NTSF sequences into a tree based on rRNA sequences and did not show the relationships between NTSFs demonstrated in the tree shown in Fig. 1.
The genes for B. halodurans NTSFI and NTSFII are transcribed in vivo.
We have begun the characterization of the NTSFs from other species' genomes that encode more than one member of the superfamily by examining B. halodurans. In addition to the observation that B. halodurans contains two NTSFs, there is evidence that another Bacillus species, B. subtilis, contains a poly(A) polymerase (29). Thus, it was of interest to examine B. halodurans to determine whether its genome encodes both a TNT and a PAP. Figure 2 shows an alignment of the two NTSFs from B. halodurans C-125 along with several other Bacillus NTSFs. The sequences were aligned using ClustalX and maximized using TuneClustal as described in Materials and Methods. It is apparent that the B. halodurans NTSFs bear significant sequence similarity to those from the other species shown. NTSFI is 32% identical, 52% similar to the NTSF from B. subtilis (over a stretch of 394 amino acids), and NTSFII is 27% identical, 46% similar to the B. subtilis NTSF (over a stretch of 219 amino acids). NTSFI and NTSFII are 25% identical, 42% similar to each other (over 399 amino acids). As indicated above, the B. subtilis and B. stearothermophilus NTSFs are known to be TNTs (20, 26).
FIG. 2.
Sequence alignment of Bacillus NTSFs. The alignment was performed using ClustalX as described in Materials and Methods. Identical residues are indicated by an asterisk. Similar residues are indicated by one or two dots, depending on the degree of similarity of the relevant amino acids. BAN, Bacillus anthracis; BTH, Bacillus thuringiensis; BCE, Bacillus cereus; BST, B. stearothermophilus; BSU, B. subtilis; BHA, B. halodurans.
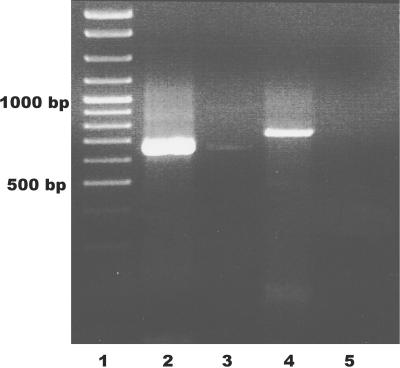
As the next step in characterizing the NTSFs of B. halodurans, we asked whether both NTSFI and NTSFII were expressed in vivo. This was accomplished by reverse transcribing total RNA and performing nested PCR with gene-specific primers. The predicted size of the amplification products was 659 bp and 730 bp for NTSFI and NTSFII, respectively. Products with these predicted sizes were found (Fig. 3, lanes 2 and 4), suggesting that both genes are expressed. Performing PCR without reverse transcription showed little DNA contamination of RNA preparations (lanes 3 and 5). The expression of both genes was later confirmed through the cloning and sequencing of cDNAs (see below).
FIG. 3.
RT-PCR of B. halodurans total RNA. Fifteen microliters of nested PCR products, prepared as described in Materials and Methods, were run on a 1% agarose gel. Lane 1, size standards; lanes 2 and 4, NTSFI and NTSFII RT-PCR products; lanes 3 and 4, parallel RT-PCRs run without reverse transcriptase.
Analysis of the 3′ tails associated with RNAs from B. halodurans.
We analyzed RNAs from B. halodurans C-125 as described previously (3, 5) and in Materials and Methods to determine whether those RNAs possessed poly(A) tails. Total RNA was end labeled with [32P]pCp and digested with a combination of RNases A and T1. This digestion procedure cleaves all phosphodiester bonds except those between adjacent A residues, leaving only the poly(A) tails at the 3′ ends of the labeled RNA intact. Products of this digestion were displayed on a 12% denaturing polyacrylamide gel as shown in Fig. 4. Lane 1 of the figure shows a size standard, oligo(dT)18. Lane 2 shows the poly(A) tails obtained from RNA isolated from E. coli DH5α, and lane 3 shows the results of labeling and digestion of RNAs from B. halodurans C-125. It is clear that, like E. coli, RNAs from B. halodurans have poly(A) tails at their 3′ ends. Tails at least 30 A residues in length are observed in Fig. 4, and longer exposures of this and other autoradiograms showed tails of >40 residues associated with RNAs from B. halodurans.
FIG. 4.
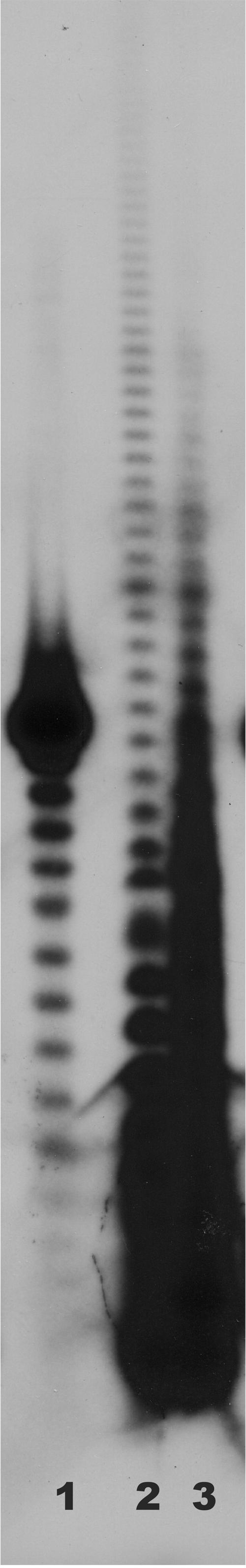
Measurement of the lengths of oligo(A) and poly(A) stretches found in the 3′ tails of RNAs from E. coli DH5α (lane 2) and B. halodurans C-125 (lane 3). Lane 1 contains an oligo(dT)18 size standard. Labeled 3′ tails were fractionated on a 12% denaturing polyacrylamide gel.
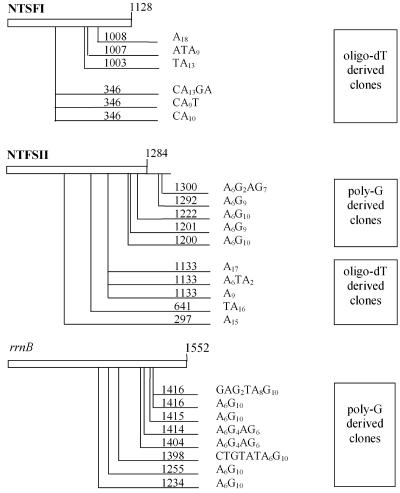
To confirm the presence of poly(A) tails and to analyze those tails for sequence heterogeneity, we cloned cDNAs using two RT-PCR protocols. In the first method an oligo(dT)17 primer designed to hybridize to poly(A) tail sequences was used for reverse transcription of total RNA from B. halodurans. This primer (ADoT; Table 1) also contained an adaptor sequence identical to the reverse primer AD20 that was subsequently used along with gene-specific primers in PCR amplification as described in Materials and Methods. We targeted the mRNAs of the NTSFI and NTSFII genes as well as the 16S rRNA of rrnB. Amplified cDNAs were then cloned and sequenced. Since we had previously used this method to determine that streptomycete poly(A) tails are heteropolymeric (3, 4), we anticipated finding stretches of poly(A) sequence 17 bases or longer. However, many tail sequences were shorter than expected, suggesting that the reverse primer was annealing to poly(A) tail sequences as short as A9 (Fig. 5). Thus, we used a second method designed to identify the 3′ end of poly(A) tails by enzymatically polyguanylating the 3′ ends of RNAs prior to RT-PCR amplification. Polyguanylation would presumably create a poly(A)-poly(G) junction which could be targeted during reverse transcription with primer ADoCT (Table 1). The PCR amplification was performed as before. An A6G10 sequence should then identify the 3′ end of any poly(A) tails.
FIG. 5.
Location and composition of 3′ tails of cDNA clones derived from B. halodurans NTSFI, NTSFII, and 16S rRNA. The position of each tail is reckoned from the translation start codon of mRNAs or the mature 5′ end of the 16S rRNA transcript and is indicated by the number on the line preceding the cDNA tail sequence. The number of bases to the translation stop codon or the mature 3′ end of rRNA is indicated by the number at the top right hand end of each gene box. The method used to create the cDNAs [either oligo(dT) or polyguanylation-poly(G)] is indicated to the right. Genes and tail locations are not drawn to scale.
Twenty-four independent cDNA clones, with roughly half derived from either the oligo(dT) or poly(G) method, revealed short poly(A) tails with low levels of heterogeneity (Fig. 5). These tails occur predominantly on probable decay intermediates as attachment sites map to internal ORF sequences. However, two sites are located downstream of the translation stop codon of NTSFII. While these tail lengths are rather short compared to lengths observed in the tail-length assay (compare Fig. 4 and 5), we suspect this result reflects the fact that tails of shorter length predominate in the total pool of poly(A) tails associated with the bacterial RNAs (Fig. 4). While these data reflect a limited number of genes, they do show that 3′ tails are associated with translated and nontranslated RNAs in B. halodurans, and the occurrence of heterogeneous tail sequences is consistent with results obtained for other organisms' genomes that do not encode a dedicated poly(A) polymerase (3, 4, 28, 37).
Overexpression and assay of B. halodurans NTSFI and NTSFII.
As described in Materials and Methods, PCR products corresponding to the open reading frames for NTSFI and NTSFII were prepared and cloned into pET19B. NTSFI was overexpressed and purified as an N-terminal his-tagged derivative by immobilized metal ion affinity chromatography. We prepared a his-tagged derivative of NTSFII as well, but this derivative did not bind to the metal ion affinity column. Thus, the NTSFII open reading frame was cloned into pET11A and the protein was overexpressed and purified by a combination of ion exchange and hydrophobic interaction chromatography. We also cloned, overexpressed, and purified an N-terminally his-tagged derivative of the tRNA nucleotidyltransferase from S. coelicolor, the product of the SCO3896 gene. An SDS-PAGE gel of the purified proteins showed that nearly homogenous proteins were obtained in each case. The N-terminal his-tagged forms of SCO3896 and NTSFI have expected molecular weights of 57,166 and 45,011, respectively, and the untagged NTSFII has an expected Mr of 48,500. The migration positions observed for the purified proteins were consistent with these expected molecular weights (data not shown).
It was formally possible that NTSFI, NTSFII, or both might function as poly(A) polymerases in B. halodurans. We first assayed NTSFI and NTSFII under conditions that would detect tRNA nucleotidyltransferase activity, since those conditions might also be expected to support poly(A) polymerase activity. We observed that both NTSFI and NTSFII were active under the conditions used for the tRNA nucleotidyltransferase assays, albeit with different substrate specificities. Those differences were readily apparent in experiments performed to determine the pH optimum for incorporation of radioactive substrate by each enzyme. As shown in Fig. 6, NTSFI was highly active with CTP as a substrate at pH 9.5 but only 25% as active at this pH with ATP as a substrate. On the other hand, NTSFII showed an optimum for ATP incorporation around pH 8. At this pH there was very little incorporation of CTP by NTSFII.
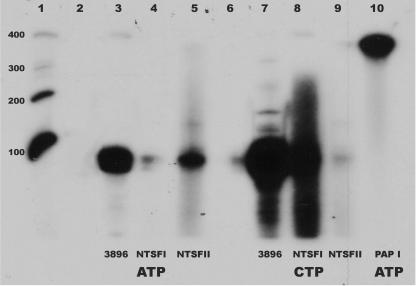
It is known that E. coli PAP I will utilize tRNA as an acceptor for polyadenylation (6). Thus, it was necessary to confirm that the data of Fig. 6 reflected the addition of CCA residues to the tRNA 3′ ends rather than the synthesis of longer polymers. To this end, products of assays like those performed to generate the data in Fig. 6 were collected by phenol extraction and ethanol precipitation, and the resulting labeled RNAs were analyzed by polyacrylamide gel electrophoresis on 10% denaturing gels. In these experiments, we also analyzed the products formed using a commercial preparation of E. coli poly(A) polymerase. The results of these analyses are shown in Fig. 7. As shown in lanes 2 and 6, omission of enzyme from the reaction mixtures resulted in no synthesis of labeled RNA. As expected, the S. coelicolor tRNA nucleotidyltransferase utilized both ATP and CTP as substrates for the addition of C and A residues to the 3′ end of the tRNA acceptor (Fig. 7, lanes 3 and 7). Figure 7, lanes 5 and 8, shows that B. halodurans NTSFI and NTSFII are tRNA nucleotidyltransferases. The products of reactions utilizing these two enzymes are of approximately the same size as those obtained using the S. coelicolor TNT, significantly smaller than the higher molecular weight species obtained from reactions containing PAP I (lane 10). Moreover, the data strongly suggest that NTSFI is primarily involved in C addition, whereas NTSFII appears to be an A-adding enzyme. Consistent with the results shown in Fig. 6, NTSFI did not use ATP efficiently as a substrate (Fig. 7, lane 4) and NTSFII did not utilize CTP efficiently (lane 9). Varying the ATP concentration in the NTSFI assays or the CTP concentrations in the NTSFII assays between 50 and 500 μM did not increase the activity of the enzymes with these substrates. We speculate that the low levels of activity observed for NTSFI with ATP as a substrate (lane 4) and for NTSFII with CTP as a substrate (lane 9) reflect relaxed specificities of the two enzymes in vitro. Thus, our results are consistent with the conclusion that, like A. aeolicus, D. radiodurans, and at least some cyanobacteria (32, 34, 35), B. halodurans contains separate enzymes for the addition of A and C residues to tRNA in vivo. It is possible that the smear above the major band of lane 8 may reflect the synthesis of poly(C), as has been observed for other TNTs (14, 32).
FIG. 7.
Autoradiogram of a 10% denaturing polyacrylamide gel of the reaction products from assays of SCO3896, B. halodurans NTSFI and NTSFII, and E. coli PAP I. Lane 1 contains end-labeled RNA size standards of 400, 300, 200, and 100 bases. Lanes 2 and 6, products of reactions with no added enzyme; lanes 3 and 7, products of assays of SCO3896 with ATP and CTP, respectively; lanes 4 and 8, products of assays of NTSFI with ATP and CTP, respectively; lanes 5 and 9, products of assays of NTSFII with ATP and CTP, respectively; lane 10, products of the PAP I assay. Other conditions were as described in Materials and Methods.
Since B. halodurans NTSFI and NTSFII are tRNA nucleotidyltransferases, the question remains as to what enzyme functions in poly(A) tail synthesis in bacilli. That enzyme might be PNPase, but we will present evidence in a subsequent publication that B. subtilis contains a poly(A) polymerase that is distinct from PAP I and PNPase. We are examining the activities of the NTSFs from the other species whose genomes encode more than one such protein.
Acknowledgments
This work was supported by grant number MCB-0133520 from the National Science Foundation.
We thank Shozo Yokoyama for expert advice on the performance and interpretation of the phylogenetic analyses.
REFERENCES
- 1.Bocchetta, M., S. Gribaldo, A. Sanangelantoni, and P. Cammarano. 2000. Phylogenetic depth of the bacterial genera Aquifex and Thermotoga inferred from analysis of ribosomal protein, elongation factor and RNA polymerase subunit sequences. J. Mol. Evol. 50:366-380. [DOI] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein using the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Bralley, P., and G. H. Jones. 2002. cDNA cloning confirms the polyadenylation of RNA decay intermediates in Streptomyces coelicolor. Microbiology 148:1421-1425. [DOI] [PubMed] [Google Scholar]
- 4.Bralley, P., and G. H. Jones. 2003. Overexpression of the polynucleotide phosphorylase gene (pnp) of Streptomyces antibioticus affects mRNA stability and poly(A) tail length but not ppGpp levels. Microbiology 149:2173-2182. [DOI] [PubMed] [Google Scholar]
- 5.Bralley, P., and G. H. Jones. 2001. Poly(A) polymerase activity and RNA polyadenylation in Streptomyces coelicolor A3(2). Mol. Microbiol. 40:1155-1164. [DOI] [PubMed] [Google Scholar]
- 6.Cao, G. J., and N. Sarkar. 1992. Identification of the gene for an Escherichia coli poly(A) polymerase. Proc. Natl. Acad. Sci. USA 89:10380-10384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carpousis, A. J., N. F. Vanzo, and L. C. Raynal. 1999. mRNA degradation: a tale of poly(A) and multiprotein machines. Trends Genet. 15:24-28. [DOI] [PubMed] [Google Scholar]
- 8.Coburn, G. A., and G. A. Mackie. 1999. Degradation of mRNA in Escherichia coli: an old problem with some new twists. Prog. Nucleic Acids Res. Mol. Biol. 62:55-105. [DOI] [PubMed] [Google Scholar]
- 9.Cudny, H., and M. Deutscher. 1986. High-level overexpression, rapid purification and properties of Escherichia coli tRNA nucleotidyltransferase. J. Biol. Chem. 261:6450-6453. [PubMed] [Google Scholar]
- 10.Cudny, H., J. R. Lupski, G. N. Godson, and M. Deutscher. 1986. Cloning, sequencing, and species relatedness of the Escherichia coli cca gene encoding the enzyme tRNA nucleotidyltransferase. J. Biol. Chem. 261:6444-6449. [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1989. PHYLIP - phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 12.He, L., F. Soderbom, E. G. Wagner, U. Binnie, N. Binns, and M. Masters. 1993. PcnB is required for the rapid degradation of RNAI, the antisense RNA that controls the copy number of ColE1-related plasmids. Mol. Microbiol. 9:1131-1142. [DOI] [PubMed] [Google Scholar]
- 13.Holm, L., and C. Sander. 1995. DNA polymerase beta belongs to an ancient nucleotidyltransferase superfamily. Trends Biochem. Sci. 20:345-347. [DOI] [PubMed] [Google Scholar]
- 14.Hou, Y.-M. 2000. Unusual synthesis by the Escherichia coli CCA-adding enzyme. RNA 6:1031-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones, G. H., M. F. Symmons, J. S. Hankins, and G. A. Mackie. 2003. Overexpression and purification of untagged polynucleotide phosphorylases. Protein Expr. Purif. 32:202-209. [DOI] [PubMed] [Google Scholar]
- 16.Köhrer, K., and H. Domdey. 1991. Preparation of high molecular weight RNA. Methods Enzymol. 194:398-405. [DOI] [PubMed] [Google Scholar]
- 17.Kusov, Y. Y., G. Shatirishvili, G. Dzagurov, and V. Gauss-Müller. 2001. A new G-tailing method for the determination of the poly(A) tail length applied to hepatitis A virus RNA. Nucleic Acids Res. 29:E57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laemmli, U. 1970. Cleavage of structural protein during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 19.Leineweber, M., and G. R. Phillipps. 1978. Comparison of tRNA nucleotidyltransferase from Escherichia coli and Lactobacillus acidophilus. Hoppe-Seyler's Z. Physiol. Chem. 359:473-480. [DOI] [PubMed] [Google Scholar]
- 20.Li, F., Y. Xiong, J. Wang, H. D. Cho, K. Tomita, A. M. Weiner, and T. A. Steitz. 2002. Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and its complexes with ATP or CTP. Cell 111:815-824. [DOI] [PubMed] [Google Scholar]
- 21.Ludwig, W., and K. H. Schleifer. 1994. Bacterial phylogeny based on 16S and 23S rRNA sequence analysis. FEMS Microbiol. Rev. 15:155-173. [DOI] [PubMed] [Google Scholar]
- 22.Martin, G., and W. Keller. 2004. Sequence motifs that distinguish ATP(CTP):tRNA nucleotidyl transferases from eubacterial poly(A) polymerases. RNA 10:899-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohanty, B. K., and S. R. Kushner. 2000. Polynucleotide phosphorylase functions both as a 3′-5′ exonuclease and a poly(A) polymerase in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:11966-11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohanty, B. K., and S. R. Kushner. 2000. Polynucleotide phosphorylase, RNase II and RNase E play different roles in the in vivo modulation of polyadenylation in Escherichia coli. Mol. Microbiol. 36:982-994. [DOI] [PubMed] [Google Scholar]
- 25.Rauhut, R., and G. Klug. 1999. mRNA degradation in bacteria. FEMS Microbiol. Rev. 23:353-370. [DOI] [PubMed] [Google Scholar]
- 26.Raynal, L. C., H. M. Krisch, and A. J. Carpousis. 1998. The Bacillus subtilis nucleotidyl transferase is a tRNA CCA-adding enzyme. J. Bacteriol. 180:6276-6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Régnier, P., and C. M. Arraiano. 2000. Degradation of mRNA in bacteria: emergence of ubiquitous features. Bioessays 22:235-244. [DOI] [PubMed] [Google Scholar]
- 28.Rott, R., G. Zipor, V. Portnoy, V. Liveanu, and G. Schuster. 2003. RNA polyadenylation and degradation in cyanobacteria are similar to the chloroplast but different from E. coli. J. Biol. Chem. 278:15771-15777. [DOI] [PubMed] [Google Scholar]
- 29.Sarkar, B., G. J. Cao, and N. Sarkar. 1997. Identification of two poly(A) polymerases in Bacillus subtilis. Biochem. Mol. Biol. Int. 41:1045-1050. [DOI] [PubMed] [Google Scholar]
- 30.Sarkar, N. 1996. Polyadenylation of mRNA in bacteria. Microbiology 142:3125-3133. [DOI] [PubMed] [Google Scholar]
- 31.Sarkar, N. 1997. Polyadenylation of mRNA in prokaryotes. Annu. Rev. Biochem. 66:173-197. [DOI] [PubMed] [Google Scholar]
- 32.Seth, M., D. L. Thurlow, and Y.-M. Hou. 2002. Poly(C) synthesis by class I and class II CCA-adding enzymes. Biochemistry 41:4521-4532. [DOI] [PubMed] [Google Scholar]
- 33.Sohlberg, B., J. Huang, and S. N. Cohen. 2003. The Streptomyces coelicolor polynucleotide phosphorylase homologue, and not the putative poly(A) polymerase can polyadenylate RNA. J. Bacteriol. 185:7273-7278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomita, K., and A. M. Weiner. 2002. Closely related CC- and A-adding enzymes collaborate to construct and repair the 3′-terminal CCA of tRNA in Synechocystsis sp. and Deinococcus radiodurans. J. Biol. Chem. 277:48192-48198. [DOI] [PubMed] [Google Scholar]
- 35.Tomita, K., and A. M. Weiner. 2001. Collaboration between CC- and A-adding enzymes to build and repair the 3′ terminal CCA of tRNA in Aquifex aeolicus. Science 294:1334-1336. [DOI] [PubMed] [Google Scholar]
- 36.Weiner, A. M. 2004. tRNA maturation: RNA polymerization without a nucleic acid template. Curr. Biol. 14:R883-R885. [DOI] [PubMed] [Google Scholar]
- 37.Yehudai-Resheff, S., M. Hirsh, and G. Schuster. 2001. Polynucleotide phosphorylase functions as both an exonuclease and a poly(A) polymerase in spinach chloroplasts. Mol. Cell. Biol. 21:5408-5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yehudai-Resheff, S., and G. Schuster. 2000. Characterization of the E. coli poly(A) polymerase: nucleotide specificity, RNA-binding affinities and RNA structure dependence. Nucleic Acids Res. 28:1139-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yue, D., N. Maizels, and A. M. Weiner. 1996. CCA-adding enzymes and poly(A) polymerases are all members of the same nucleotidyl transferase superfamily: characterization of the hyperthermophile Sulfolobus shibatae. RNA 2:895-908. [PMC free article] [PubMed] [Google Scholar]