Abstract
The cold-active α-amylase from the Antarctic bacterium Pseudoalteromonas haloplanktis (AHA) is the largest known multidomain enzyme that displays reversible thermal unfolding (around 30°C) according to a two-state mechanism. Transverse urea gradient gel electrophoresis (TUG-GE) from 0 to 6.64 M was performed under various conditions of temperature (3°C to 70°C) and pH (7.5 to 10.4) in the absence or presence of Ca2+ and/or Tris (competitive inhibitor) to identify possible low-stability domains. Contrary to previous observations by strict thermal unfolding, two transitions were found at low temperature (12°C). Within the duration of the TUG-GE, the structures undergoing the first transition showed slow interconversions between different conformations. By comparing the properties of the native enzyme and the N12R mutant, the active site was shown to be part of the least stable structure in the enzyme. The stability data supported a model of cooperative unfolding of structures forming the active site and independent unfolding of the other more stable protein domains. In light of these findings for AHA, it will be valuable to determine if active-site instability is a general feature of heat-labile enzymes from psychrophiles. Interestingly, the enzyme was also found to refold and rapidly regain activity after being heated at 70°C for 1 h in 6.5 M urea. The study has identified fundamental new properties of AHA and extended our understanding of structure/stability relationships of cold-adapted enzymes.
Cold-adapted enzymes have been isolated and characterized from a diverse range of organisms living in permanently cold environments (11) and are characterized by high activity, low activation enthalpy, and low stability at low and moderate temperatures (2, 5, 10). These properties have provided significant opportunities for exploitation of cold-active enzymes in a range of industries (3, 11).
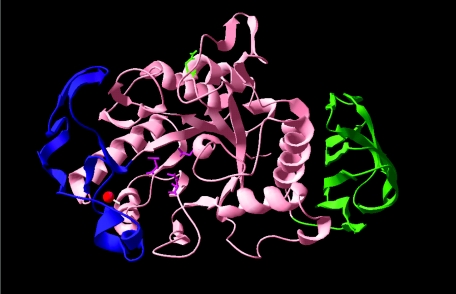
The cold-adapted α-amylase from Pseudoalteromonas haloplanktis (AHA) was the first cold-adapted enzyme to have an X-ray structure solved (1). This revealed that the enzyme consists of three domains (Fig. 1). Domain A is the N-terminal domain which is composed of a (β/α)8-barrel forming one side of the catalytic cleft (residues 1 to 86 and 130 to 356) and is able to bind a Tris molecule in the active site. Domain B (residues 87 to 129) protrudes from domain A and forms the other side of the active cleft. Domain C is the C-terminal domain consisting of residues 357 to 448. A Ca2+ ion is bound between domains A and B. The enzyme has evolved a low conformational stability and high flexibility through a reduction in the number and type of inter- and intramolecular interactions (12, 13), and the enhanced flexibility has resulted in a decreased enthalpy of activation with a concomitant improvement in kcat (6, 8).
FIG. 1.
Structure of the Pseudoalteromonas haloplanktis α-amylase. Domain A (pink, middle), domain B (blue, left), domain C (green, right), three active-site residues in domain A (magenta), residue N12 in domain A (green), and the calcium ion between domains A and B (red) are shown.
The lower stability of cold-adapted enzymes has been linked to either a general reduction in “weak interactions” causing global flexibility or weakened interactions of specific regions generating local flexibility. Site-directed mutagenesis studies of AHA have revealed a global weakening of intramolecular interactions leading to an overall decrease in thermostability (6, 14). In contrast, citrate synthase and chitobiase from Arthrobacter species, phosphoglycerate kinase from Pseudomonas species, and lactate dehydrogenases from Antarctic fish have been suggested to possibly contain distinct heat-labile and heat-stable domains (14, 19).
AHA is the largest multidomain protein (∼50 kDa) that has been shown to unfold/refold reversibly by a cooperative two-state mechanism (7, 8, 12). Biophysical measurements of unfolding/refolding have been performed by differential scanning calorimetry, circular dichroism, and fluorescence spectrophotometry (6, 12), and the spectrophotometric methods have clearly revealed cooperative unfolding through a single transition (12).
In the case of mesophilic and thermophilic α-amylases, the optimal growth temperature for activity closely corresponds to the transition temperature, illustrating that the loss in activity is due to structural unfolding (8). In contrast, AHA loses ∼50% of its enzyme activity before reaching its melting temperature (Tm) (44°C), suggesting that a local rather than global mechanism of unfolding may be involved (8). To further identify the structures that may first unfold and to examine local versus global stability, we developed transverse urea gradient (TUG) gel electrophoresis (TUG-GE) (15, 16) for the cold-active α-amylase. TUG-GE can be used to distinguish conformational states of a protein caused by different structures unfolding/folding independently at different urea concentrations, thereby identifying multistep, sequential unfolding/folding transitions. If conformations interconvert slowly within the duration of electrophoresis, unfolding/folding is characterized by a diffuse band or smear of protein that appears between folded and unfolded states. If the interconversion is very rapid compared to the duration of electrophoresis (the conformations interconvert many times during the time of electrophoresis), then protein mobility corresponds to the equilibrium distribution of conformations. This results in a sharp, continuous band between the two conformations from which the thermodynamic parameter ΔG (stability of native conformation relative to unfolded state) and urea concentration at 0 ΔG ([urea]1/2) can be calculated (15, 16).
Using this approach, we were able to identify intermediate folded states and determine which domain unfolds first. Two unfolding transitions were identified, demonstrating that in contrast to thermal unfolding, urea-induced unfolding is a three-state process. By comparing the properties of the wild-type enzyme with an N12R mutant, the active site was shown to be part of the least stable structure in the enzyme. The stability data supported a model of cooperative unfolding of structures forming the active site and independent unfolding of the other more stable protein domains. AHA was also found to have a remarkable ability to refold and rapidly regain activity, being able to do so after being heated at 70°C for 1 h in 6.5 M urea. Interestingly, the findings of our study also offered support for the use of a charge/hydrophobicity ratio (25) to predict if a protein will assume partially folded conformations.
MATERIALS AND METHODS
Purification of wild-type and mutant AHA.
The recombinant wild-type α-amylase from P. haloplanktis and its N12R site-directed mutant were expressed in Escherichia coli and purified as described previously (6). The recombinant Csp from Methanogenium frigidum (22) was expressed in E. coli and purified as will be described elsewhere (L. Giaquinto et al., unpublished data). Protein concentration was determined using Bradford reagent with bovine serum albumin as a standard. Taka amylase from Aspergillus oryzae was obtained from Megazyme International (Ireland).
TUG-GE.
TUG-GE was performed by a modification of previously described methods (15, 16). A single gel assembly consisted of a ceramic-notched plate, a glass plate (10 by 8 cm), and a pair of 0.75-mm spacers held together by rubber bands. Four gel assemblies were carefully aligned relative to each other with their spacer sides at the top and bottom of the gel caster. TUG gels were prepared with a urea gradient from 0 to 6.64 M urea and an inverse acrylamide gradient of 11 to 8% using a Hoefer multiple gel caster (4 gels; 10.5 by 10.5 cm). A bottom solution (V1, 14 ml), a gradient solution plus urea (V2, 32.5 ml), a gradient solution without urea (V3, 32.5 ml), and top solution (V4, 25 ml) were prepared. Buffers (as specified) used at various concentrations and pHs are as follows: 43 mM imidazole-HEPES, pH 7.2 to 7.5; 370 mM Tris-HCl, pH 8.8; 50 mM boric acid-7.5 mM borax, pH 8.8; and 20 mM CAPS [3-(cyclohexylamino)propanesulfonic acid]-NH3, pH 10.4. V1 contained 8% acrylamide solution (40% acrylamide, 3.3% bis-acrylamide), 6.64 M urea, 23 μl of 10% ammonium persulfate, 4.5 μl of TEMED (N,N,N′,N′-tetramethlyethylenediamine), and the appropriate amount of buffer stock, and the volume was made up to 14 ml with water. V2 was made as described above for V1, except that it contained 45 μl of 10% ammonium persulfate, 9 μl of TEMED, and 50 μl of 0.4% bromophenol dye in a final volume of 32.5 ml. V3 was the same as V1, except for 11% acrylamide solution, 55 μl of 10% ammonium persulfate, and 9 μl of TEMED in a volume of 32.5 ml. V4 was the same as V1, except for 11% acrylamide solution, 70 μl of 10% ammonium persulfate, and 12 μl of TEMED in a final volume of 25 ml. V3 and V4 were degassed, and ammonium persulfate and TEMED were added immediately prior to pouring. V1 was carefully poured along the side walls of the gel assembly to cover the bottom spacer. V2 and V3 were poured into outlet and reservoir chambers, respectively, of a Hoefer Gradient Mixer (2 by 100 ml). Mixing was achieved with a magnetic stirrer. The interchamber valve was opened, and a peristaltic pump was used to pour a urea gradient from 6.64 to 0 M (from bottom to top). The remaining volume of the gel assembly was filled with V4 by peristalsis. The gel was polymerized overnight. The glass plate was lifted, leaving the gel on the ceramic plate. A single well was cut near the notched end across the urea gradient, and the glass plate was refitted to the gel assembly. Two gels were assembled in a Hoefer SE-250 electrophoresis unit connected to a temperature-controlled water bath (MultiTemp III; Pharmacia Biotech) and prerun at 50 V for 45 min using the specified buffer and temperature conditions. Electrophoresis of samples was performed at 50 to 120 V to ensure that the bromophenol dye reached the end of the gel in 3 h. All buffers and reagents, including CaCl2 (1 mM), EDTA (1 to 5 mM), and Tris base were included in the gel, tank, and sample. The sample buffer (70 μl) used for enzyme unfolding (N⇆U) consisted of 50 mM buffer, 30% glycerol, 25 to 28 μg α-amylase, and 1 μl of 0.4% bromophenol blue. The sample for enzyme folding (U⇆N) was the same, with the addition of 6.64 M urea and omission of glycerol, and the enzyme was preincubated for 3 h in the sample buffer. At the completion of electrophoresis, gels were stained with Coomassie R250, and images were collected using Imagegauge on an LAS3000 (Fujifilm, Berthold Australia, Melbourne, Australia). For amylase activity staining, gels were laid on top of an 0.8% agarose gel containing 0.1 M MOPS (morpholinepropanesulfonic acid)-NaOH, pH 7.2, 50 mM NaCl, and 1% potato starch. Zymograms containing urea were prepared by preincubating the agarose gel in urea to give a final urea concentration of 6.7 M. After 1 to 10 min of incubation, the agarose gel was negatively stained in an iodine solution (30 mg iodine, 3 mg KI, 5 mM HCl), and clear zones of starch hydrolysis were identified against a violet background of intact starch. TUG gels were analyzed as described previously (15, 16). At the urea concentration corresponding to the point of discontinuity of transition, the rate constant of unfolding (ku) was calculated as 1/t, where t is the time of electrophoresis (180 min), and half-life (t1/2) was calculated as ln 2/ku. Conformational stabilities (ΔG and [urea]1/2) were calculated based on knowing the beginning (0 M urea) and end (6.64 M urea) of the gradient from the relative position of the electrophoresis well, similar to the approach used in previous studies (4, 9, 15-17, 21, 27).
RESULTS
TUG-GE of AHA.
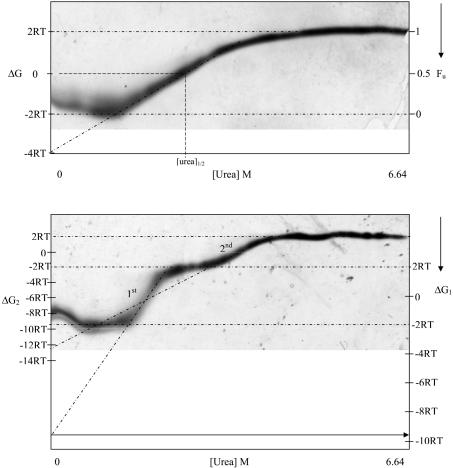
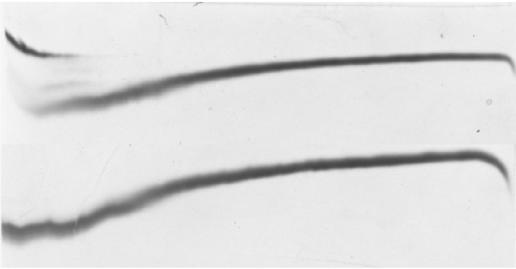
Plotting of the mean hydrophobicity of a protein versus mean net charge can be used to predict whether an enzyme unfolds with or without intermediate partially folded states (25). AHA has a mean hydrophobicity of 0.458 and a mean charge of 0.046 at pH 7 and lies in the phase space where proteins are predicted to unfold with the accumulation of partially folded conformations. This suggests that the enzyme may not always display a single unfolding transition in spite of the fact that AHA has overall low core hydrophobicity (6). TUG-GE of AHA gave a clear banding pattern that readily distinguished two-state reversible unfolding (Fig. 2a) from reversible, sequential unfolding with two transitions (Fig. 2b), enabling ΔG and [urea]1/2 to be calculated.
FIG. 2.
TUG-GE of AHA. (A) Two-state reversible unfolding at 30°C in the presence of EDTA (pH 8.8). (B) Reversible sequential unfolding at 12°C in the presence of 1 mM Ca2+, pH 8.8. Fu, fraction unfolded; ΔG, free-energy difference between folded and unfolded protein; [urea]1/2, urea concentration at 0 ΔG; R, universal gas constant (8.314 kJ mol−1); T, absolute temperature; arrow, direction of electrophoresis down the gel. An inverse acrylamide gradient from 11 to 8% across the gel is shown.
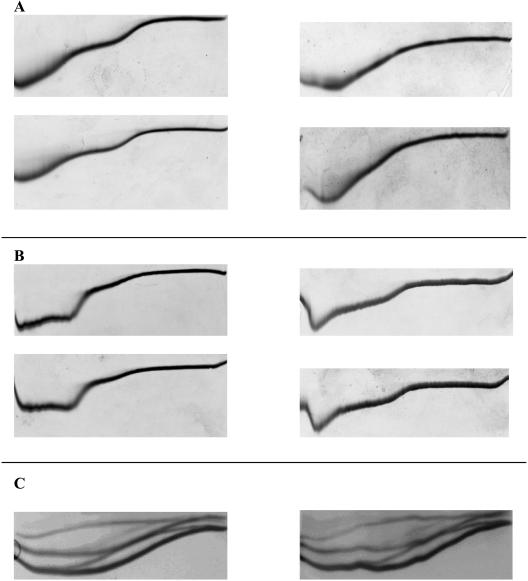
The reproducibility of TUG-GE profiles was good. This is demonstrated by the consistency of profiles obtained when AHA was loaded in its folded (N⇆U) or unfolded (U⇆N) form and subjected to electrophoresis at 12°C or 30°C (Fig. 3a). At 12°C, AHA showed sequential unfolding with [urea]1/2 corresponding to 1.3 M and 3.6 M (Fig. 3a, left), and at 30°C, it unfolded with a single transition corresponding to [urea]1/2 of 2.5 M (Fig. 3a, right). Consistent profiles were also obtained from gels run on different days under the same conditions of electrophoresis (Fig. 3b). Sequential unfolding in pH 7.5 buffer (Fig. 3b, left panels) with [urea]1/2 corresponding to 2.1 M and 3.5 M and pH 10.4 buffer (Fig. 3b, right panels) with [urea]1/2 corresponding to 0.9 M and 3.2 M were obtained when TUG-GE was performed on different days (Fig. 3b, top and bottom panels). We also demonstrated that TUG-GE gave fully reversible unfolding (Fig. 3c) for a single domain, small (7.8-kDa) cold shock protein (Csp) from the cold-adapted archaeon Methanogenium frigidum (22). The TUG-GE of the Csp revealed a number of isomers with reproducible two-state unfolding most clearly revealed by the more intense lower band (Fig. 3c). The Csp was used as a control in this study and will be described fully elsewhere (Giaquinto et al., unpublished). All TUG gels generated a characteristic edge effect around 0 M and 6.64 M urea that is caused during electrophoresis by current leakage down the sides of the gel (21). This does not affect the interpretation of unfolding transitions or the calculation of conformational stabilities (4, 9, 17, 21, 27). Moreover, while the absolute values of thermodynamic properties have been accurately determined from the TUG gels, our study focuses on the relative differences between gels with the intent of demonstrating multiple unfolding transitions and identifying which domain unfolds first.
FIG. 3.
Reproducibility of TUG-GE. (A) TUG-GE of cold-adapted α-amylase at 12°C (left panels) and 30°C (right panels) in Tris-glycine (pH 8.8)-EDTA (2 mM) buffer showing complete reversibility when the protein was loaded in its folded (N⇆U [upper panels]) or unfolded (U⇆N [lower panels]) state. The folded and unfolded forms were electrophoresed at the same time in a dual-electrophoresis unit. (B) TUG-GE of AHA at 12°C in imidazole-HEPES (pH 7.5) and Ca2+ (1 mM) (left panels) and in CAPS-NH3 (pH 10.4)-EDTA (2 mM) (right panels) performed on two different days (upper versus lower panels). (C) TUG-GE of a single-domain, small (7.8 kDa) Csp from the cold-adapted archaeon Methanogenium frigidum (22) performed at 11°C in Tris-glycine (pH 8.8) buffer carried out on different days (left versus right panel).
Effect of temperature on unfolding.
In the presence of urea at 30°C in pH 7.5 (Fig. 4a), pH 8.8 (Fig. 4b), or pH 10.4 (Fig. 4c) buffer and at 42 and 53°C in pH 7.5 (Fig. 5) buffer, the enzyme folded/unfolded reversibly with a single transition, although at 53°C, the unfolding curve became too flat to distinguish a sharp and distinct transition. Previous spectrophotometric studies showed that at temperatures ≥40°C or at 20°C in the presence of guanidine hydrochloride, the enzyme unfolds according to a two-state mechanism with a single transition (8, 12).
FIG. 4.
Urea unfolding curves at different temperatures and in different buffers. Shown are results with imidazole-HEPES, pH 7.5 (A), Tris-glycine, pH 8.8 (B), and CAPS-NH3, pH 10.4 (C). Concentrations are as follows: EDTA, 2 mM; CaCl2, 1 mM. nd, not determined. Urea gradient was 0 to 6.64 M (left to right), and the direction of electrophoresis is from top to bottom.
FIG. 5.
Urea unfolding curves and activity staining profiles at different temperatures in imidazole-HEPES (pH 7.5). P. haloplanktis cold-active α-amylase and A. oryzae Taka amylase in the presence of 1 mM CaCl2 are shown. Gels were stained with Coomassie R250 for total protein or with iodine for starch hydrolysis activity. Urea gradient was 0 to 6.64 M (left to right), and the direction of electrophoresis is from top to bottom. N⇆U, TUG-GE for rows 1, 2, 3, and 5; U⇆N, TUG-GE for row 4.
In contrast, AHA unfolded reversibly and sequentially with two transitions when subjected to electrophoresis at 12°C in buffers at pH 7.5, pH 8.8, and pH 10.4 (Fig. 4). The presence of EDTA or Ca2+ (Fig. 4) or Ca2+ plus Tris base at pH 7.2 (Fig. 6) did not alter the pattern. Moreover, the patterns were superimposable when the folded enzyme (N⇆U) and the unfolded enzyme preincubated in 6.64 M urea (U⇆N) were run under parallel conditions (Fig. 3a). These data imply that the transitions were fully reversible at 12°C and 30°C and further illustrate that at 12°C, the enzyme unfolds sequentially, whereas at 30°C, it unfolds cooperatively as a single unit.
FIG. 6.
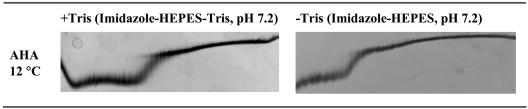
Effect of Tris on urea unfolding curves. Shown is TUG-GE in imidazole-HEPES (pH 7.2)-1 mM CaCl2 in the absence or presence of Tris (10 mM). Urea gradient was 0 to 6.64 M (left to right), and the direction of electrophoresis is shown from top to bottom.
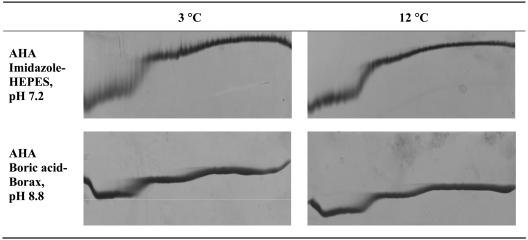
Decreasing the temperature to 3°C resulted in the first transition showing a smear, reflecting that there was a relatively slow interconversion of the conformations within the duration of TUG-GE (Fig. 7). The duration of electrophoresis is 3 h, whereas the t1/2 of interconversion is ∼125 min, indicating that the protein underwent an N⇆U transition only once during the duration of TUG-GE (15). This is apparent from the TUG gel by comparing the smear at a low urea concentration at 3°C to the band at 12°C at both pH 7.2 and pH 8.8 (Fig. 7). This indicates that the domains involved in the first transition are cold denatured at 3°C. Previous studies examining the bell-shaped stability curves of thermally adapted α-amylases found that AHA was a cold-labile enzyme, with a low Tm of approximately −10°C (8). The TUG gel results (at 3°C) demonstrate that in the presence of urea, the cold denaturation Tm of a single domain is shifted to a higher temperature while that of the rest of the protein remains low; a single domain is destabilized independently from the rest, thus yielding a two-transition unfolding at low temperature in the presence of urea (Fig. 4 to 7). This contrasts to enzyme unfolding with a single transition at temperatures between 30°C and 53°C (Fig. 4 and 5).
FIG. 7.
Effect of low temperature on urea unfolding curves. Shown is TUG-GE at 3°C and 12°C in imidazole-HEPES (pH 7.2)-1 mM CaCl2 or boric acid-borax (pH 8.8)-1 mM CaCl2. Row 1 (N⇆U), TUG-GE for AHA showing unfolding; row 2 (U⇆N), TUG-GE for unfolded enzyme showing folding. Urea gradient was 0 to 6.64 M (left to right), and the direction of electrophoresis is shown from top to bottom.
Effect of pH on stability.
At 30°C in the presence of 1 mM Ca2+, enzyme stability decreased with increasing pH from 7.5 to 10.4 (Fig. 4 and Table 1). Moreover, there was an overall decrease in stability between pH 7.5 and pH 10.4 of 0.2 M ([urea]1/2) and 3.1 kJ mol−1 (ΔG). In the pH 8.8 buffer, Tris and chloride ions would be bound by the enzyme (see below).
TABLE 1.
Thermodynamic parameters for AHA unfoldinga
| Temperature (°C) and additive | Buffer
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Imidazole-HEPES, pH 7.2
|
Imidazole-HEPES, pH 7.5
|
Tris-glycine, pH 8.8
|
Boric acid-borax, pH 8.8
|
CAPS-NH3, pH 10.4
|
||||||||||||||||
| Transition | ΔG | [urea]1/2 | Fig. | Transition | ΔG | [urea]1/2 | Fig. | Transition | ΔG | [urea]1/2 | Fig. | Transition | ΔG | [urea]1/2 | Fig. | Transition | ΔG | [urea]1/2 | Fig. | |
| 3 | ||||||||||||||||||||
| Ca2+ | 1st (irrev) | ND | ND | 7 | ND | ND | 1st (irrev) | ND | ND | 7 | ND | |||||||||
| 2nd | 20.4 | 3.7 | 7 | 2nd | 33 | 3.7 | 7 | |||||||||||||
| 12 | ||||||||||||||||||||
| Ca2+ | 1st | 32.0 | 1.8 | 6 | 1st | 33 | 2.1 | 4 | 1st | 23 | 1.8 | 4 | 1st | 36 | 1.9 | 7 | 1st | 19 | 1.3 | 4 |
| 2nd | 18.7 | 3.5 | 6 | 2nd | 26 | 3.5 | 4 | 2nd | 30 | 3.5 | 4 | 2nd | 46.5 | 3.6 | 7 | 2nd | 40 | 3.4 | 4 | |
| Ca2+ + Tris | 1st | 44.6 | 2.7 | 6 | ND | ND | ND | ND | ||||||||||||
| 2nd | 24.0 | 4.3 | 6 | |||||||||||||||||
| EDTA | ND | 1st | 9.5 | 1.4 | 4 | 1st | 8.4 | 1.3 | 4 | ND | ND | 1st | 12 | 0.9 | 4 | |||||
| 2nd | 24 | 3.5 | 4 | 2nd | 26 | 3.6 | 4 | 2nd | 35 | 3.3 | 4 | |||||||||
| 30 | ||||||||||||||||||||
| Ca2+ | ND | Single | 8.7 | 1.6 | 4 | Single | 7.5 | 1.7 | 4 | ND | Single | 5.6 | 1.4 | 4 | ||||||
| EDTA | ND | ND | Single | 9.6 | 2.5 | 4 | ND | ND | ||||||||||||
ND, not determined; irrev, irreversible within the duration of TUG-GE (3 h); Fig., number of the figure used for calculating values.
In the presence of Ca2+ at 12°C, the stability of the domain undergoing transition 1 decreased with increasing pH from 7.5 to 10.4, whereas the stability of the domain undergoing transition 2 increased with increasing pH (Fig. 4 and Table 1). This indicates that the domains undergoing the second transition are not only more thermally stable, they are more alkali stable than the domain undergoing the first transition.
Tris contribution to stability.
Tris is a competitive inhibitor of AHA and may stabilize the active site (1). Protection from unfolding would be afforded by the cationic form of Tris, which would predominate below its pKa of 8.2 at 25°C (1). The stabilizing effect of Tris (10 mM) was examined by TUG-GE at 12°C in imidazole-HEPES, pH 7.2, in the presence and absence of the competitive inhibitor (Fig. 6 and Table 1). For the domain undergoing the first transition in the presence and absence of Tris, the [urea]1/2 was 2.7 and 1.8 M, and ΔG was 45 and 32 kJ mol−1, respectively (Fig. 6 and Table 1). At this pH, the domain undergoing the second transition was also stabilized (Fig. 6 and Table 1); however, the difference in Δ[urea]1/2 and ΔΔG for the domain undergoing the first transition was much greater than that for the domain undergoing the second transition. This indicates that the first transition arises from the unfolding of the region in the active site that binds Tris.
Effect of calcium.
In the AHA structure (1, 13), a calcium ion is located between domains A and B approximately 12 Å away from the active site (Fig. 1). The calcium is bound with a 2,000-fold lower affinity than for equivalent metal ions bound by a mesophilic porcine α-amylase (1). At pH 7.5 (Fig. 4a) and pH 8.8 (Fig. 4b) at 12°C, the structure undergoing the first transition moves to a higher urea concentration, reflecting that the initial conformation becomes significantly more stable in the presence of Ca2+ than in its absence (presence of EDTA). In contrast, little effect is observed on the domains undergoing the second transition. Similar results were obtained at 12°C in pH 10.4 buffer (Fig. 4c and Table 1). The data indicate that the first transition corresponds to the unfolding of structures involved in Ca2+ binding.
In contrast to activity at 12°C, at 30°C and pH 8.8, the Ca2+-complexed enzyme showed less stability ([urea]1/2, 1.7 M; ΔG, 7.5 kJ mol−1) than the apo enzyme ([urea]1/2, 2.5 M; ΔG, 7.5 kJ mol−1). Previous studies also recorded that the Tm of apo enzyme was slightly higher than that for the Ca2+-loaded enzyme (12). The present data indicate that at 30°C, temperature-dependent unfolding of domains A and B may displace Ca2+ from its binding site. This is consistent with a previous suggestion that in the absence of Ca2+, the Ca2+-binding ligands may form more stable bonds with neighboring residues (12).
Unfolding of AHA initiates from the catalytic domain.
Analysis of the preceding TUG-GE data indicates that at 12°C or 3°C, AHA unfolds with two sequential transitions. The effects of calcium indicate that the domain undergoing the first transition participates in Ca2+ binding, and the Ca2+-binding site is known to be located near the active site (1). The Tris-induced thermodynamic stabilization of the domain undergoing the first transition also indicates that the active site is the structure which initially unfolds. In order to prove that the first transition corresponds to unfolding of structures from domain A, which bears the catalytic residues, we applied TUG-GE to an N12R mutant of the α-amylase (6). The N12R mutation is in domain A (Fig. 1), which results in N12R forming an extra salt bridge with Asp15, thereby decreasing the unfolding cooperativity of the enzyme (6, 7). Differential scanning calorimetry has shown that the N12R mutant exhibits a predominantly irreversible pattern of unfolding (6, 7). In fact, this mutant has been used in lieu of wild-type AHA to study kinetic stability due to its irreversible unfolding (8).
The mutant AHA run at 12°C showed an unfolding pattern similar to that of the wild-type enzyme (unpublished results); therefore, drastic TUG-GE conditions were chosen to enable the structural differences between the wild-type and mutant proteins to become evident. The TUG gel was run at 12°C for 45 min for the separation of the domains undergoing sequential transitions and then shifted to 47°C for 90 min to modify the unfolding mechanism and facilitate the split in the curve due to irreversible denaturation (Fig. 8, top). The break, which occurred at 1 M urea, was caused by low mobility resulting from the increased hydrodynamic volume of the irreversibly unfolded form of the enzyme (Fig. 8, top). This demonstrates that an extra salt bridge in domain A produces an irreversible unfolding within the duration of TUG-GE that corresponds to the first transition. It can therefore be concluded that cold-adapted α-amylase has two stability domains. The cold-labile domain contains the active site (effect of Tris), the Ca2+-binding site (that is part of the active site), and the N-terminal part of domain A, which contains the catalytic acid residues (effect of both Tris and N12R), whereas the more stable domains (part of domain A and domain C) correspond to the structural elements undergoing the second transition.
FIG. 8.
Effect of N12R mutation on urea unfolding curves. Shown is TUG-GE in imidazole-HEPES (pH 7.2)-1 mM CaCl2 at 12°C for 45 min followed by a rapid shift to 47°C and electrophoresis for 90 min. N12R mutant, upper broken band; wild-type enzyme, lower continuous band. The upper-band breakpoint for the N12R mutant corresponds to 1 M urea. The unfolding curves for both the wild type and the N12R mutant are superimposable and have been placed one above the other in the figure for comparison purposes.
A high level of unfolding reversibility at high temperatures.
Multidomain enzymes typically unfold irreversibly to produce two distinct bands on TUG gels that correspond to fully folded (high Rf) and fully unfolded (low Rf) enzymes (24). As a result, even if the enzyme is subsequently incubated under nondenaturing conditions, only the bands corresponding to the fully folded form of the enzyme are expected to demonstrate activity in an activity stain (23). This was established for the Taka amylase from Aspergillus oryzae, which showed activity following TUG-GE at 53°C, pH 7.2, at concentrations of urea up to 2.6 M (Fig. 5, bottom).
When AHA was stained after TUG-GE at 12°C, pH 7.5, and incubated for 1 to 2 min on starch-containing agarose at 25°C, activity was detected along the full length of the urea curve (Fig. 5). Inclusion of 6.7 M urea in the starch agarose prevented activity, demonstrating that the unfolded enzyme needed to refold in order to be active. Increasing the TUG-GE temperature to 53°C led to a flattening of the urea curve; however, the zymogram illustrated that the enzyme could refold.
To rigorously test that the enzyme could refold from a completely unfolded state, it was heated at 70°C for 1 h in 6.5 M urea-1 mM Ca2+, pH 7.5, prior to being run on TUG gels at 12°C and activity staining (Fig. 5). At low urea concentrations, two distinct bands were present. The upper band demonstrated reversible unfolding. In contrast, the lower band was discontinuous, implying that it unfolded irreversibly in the time scale of electrophoresis. In addition, the structure undergoing the second transition (corresponding to high urea concentration) was smeary, indicating a slow rate of folding. The pattern of bands in the second transition was indicative of a heterogeneous population with one form of the enzyme refolding rapidly and the other form refolding slowly (16).
Our data have demonstrated AHA to be a multidomain enzyme that reversibly unfolds even under very harsh denaturing conditions. It is striking that an enzyme from a psychrophile has the capacity to refold and regain activity after such treatment.
DISCUSSION
Our study revealed several new findings about the unfolding characteristics of AHA. First, reducing the temperature from 30°C to 3°C changed the unfolding mechanism of AHA from two-state unfolding, to sequential unfolding, to a slow interconversion between conformations at low urea concentration. Second, the initial structures to unfold were found to be part of the active site. Third, AHA has a striking ability to refold and regain activity following severe denaturation.
AHA shows a reversible two-state unfolding/folding pattern between 30°C and 43°C. The sequential unfolding with two transitions only becomes apparent when the temperature is decreased to 12°C, even though AHA is expected to show the absence of unfolding intermediates due to overall low core hydrophobicity (6). Decreasing the temperature to 3°C does not affect sequential unfolding; however, the domain undergoing the first transition (corresponding to low urea concentrations) showed a slow folding/unfolding interconversion within the duration of TUG-GE. This implies that the domain undergoing the first transition is cold sensitive. The effects of Tris, Ca2+, and the response of the N12R mutant indicate that the structures undergoing the first transition are part of the active site. This is consistent with previous findings that the activity of AHA was lost before the protein unfolds during heat-induced denaturation (8, 11). Previously, it was also argued that the α-amylase exhibited a global reduction in ionic interactions (12, 14, 19).
The present study has shown that the active site is located within the least stable structure of the protein. This demonstrates a region of localized lability within the enzyme and provides a structural basis for the cold denaturation. The results therefore provide experimental support for the hypothesis that cold adaptation of AHA (25), and possibly other enzymes from psychrophiles, may proceed via localized increases in conformational flexibility and that active-site flexibility is a key parameter of biocatalysis at low temperature (11).
A wide variety of TUG-GE conditions (pH, temperature, metal ion, and buffer composition) were used to examine unfolding of this three-domain protein (1). Despite the ability to characterize two unfolding transitions, a third transition was not detected. Our results show that the catalytic region is the most cold labile. In the structure of the protein (1), the catalytic pocket is formed at the interface of domains A and B (Fig. 1). From the structural data, we surmise that the N-terminal part of domains A and B (active site) unfold cooperatively (first transition), with the more stable regions (C-terminal part of domain A and domain C) unfolding independently at a higher urea concentration (second transition).
Proteins that have equilibrium intermediates are more hydrophobic and in general have a smaller net charge than those not able to form distinct intermediate states (25, 26). This may indicate that such proteins are strengthened by hydrophobic interactions at moderately low and moderately high temperatures and are less perturbed by electrostatic repulsion (2, 18). The reversible unfolding of AHA results from the presence of a minimal number of these types of interactions which are simultaneously disrupted (6, 8, 14).
It was noteworthy that the protein structures undergoing the first transition become irreversibly denatured within the duration of TUG-GE at very low temperatures (3°C [N⇆U]), as well as when preincubation was performed in 6.5 M urea at very high temperatures (70°C [U⇆N]) and when the gel was run at 12°C. Although the mechanisms of cold unfolding are still poorly understood, our data are consistent with irreversibility at 3°C being caused by the hydration of ionic and polar groups (solvation penalty), and to a lesser extent aromatic interactions, rendering the energy cost of the formation of salt bridges and some hydrophobic interactions exceedingly high for refolding (20).
A consequence of having evolved structural characteristics to enable cold activity was the striking ability to rapidly refold and regain activity. Typically, an unfolded protein would aggregate through the interaction of hydrophobic surfaces when placed under conditions that may allow it to refold (18). The fact that AHA is able to successfully refold may reflect an overall reduction in hydrophobic interactions. This study has identified surprising new features of this otherwise-well-characterized cold-adapted enzyme, highlighting the need to examine the folding/unfolding properties of other cold-adapted enzymes using a similar approach.
Acknowledgments
The work was supported by the Australian Research Council.
Thanks to Lily Ting for careful reading of the manuscript and Paul Curmi and Anne Poljak for helpful discussions. S.D. is a postdoctoral researcher from the Fonds National de la Recherche Scientifique (FNRS, Belgium). We express our gratitude to the reviewers for their particular insight and constructive comments.
REFERENCES
- 1.Aghajari, N., G. Feller, C. Gerday, and R. Haser. 1998. Crystal structure of the psychrophilic α-amylase from Alteromonas haloplanctis in its native form and complexed with an inhibitor. Protein Sci. 7:564-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cavicchioli, R., and K. S. Siddiqui. 2004. Cold-adapted enzymes, p. 615-638. In A. Pandey, C. Webb, C. R. Soccol, and C. Larroche (ed.), Enzyme technology. AsiaTech Publishers, New Delhi, India.
- 3.Cavicchioli, R., K. S. Siddiqui, D. Andrews, and K. R. Sowers. 2002. Low-temperature extremophiles and their applications. Curr. Opin. Biotechnol. 13:253-261. [DOI] [PubMed] [Google Scholar]
- 4.Creighton, T. E., and R. H. Paine. 1980. Unfolding and refolding of Staphylococcus aureus penicillinase by urea-gradient electrophoresis. J. Mol. Biol. 137:431-436. [DOI] [PubMed] [Google Scholar]
- 5.D'Amico, S., P. Claverie, C. Collins, D. Georlette, E. Gratia, A. Hoyoux, M.-A. Meuwis, G. Feller, and C. Gerday. 2002. Molecular basis of cold adaptation. Philos. Trans. R. Soc. Lond. B 357:917-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D'Amico, S., C. Gerday, and G. Feller. 2001. Structural determinants of cold adaptation and stability in a large protein. J. Biol. Chem. 276:25791-25796. [DOI] [PubMed] [Google Scholar]
- 7.D'Amico, S., C. Gerday, and G. Feller. 2003. Temperature adaptation of proteins: engineering mesophilic-like activity and stability in a cold-adapted α-amylase. J. Mol. Biol. 332:981-988. [DOI] [PubMed] [Google Scholar]
- 8.D'Amico, S., J. C. Marx, C. Gerday, and G. Feller. 2003. Activity-stability relationship in extremophilic enzymes. J. Biol. Chem. 278:7891-7896. [DOI] [PubMed] [Google Scholar]
- 9.Evans. R. W., and J. Williams. 1980. The electrophoresis of transferrins in urea/polyacrylamide gels. Biochem. J. 189:541-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feller, G. 2003. Molecular adaptations to cold in psychrophilic enzymes. Cell. Mol. Life Sci. 60:648-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feller, G., and C. Gerday. 2003. Psychrophilic enzymes: hot topics in cold adaptation. Nat. Rev. Microbiol. 1:200-208. [DOI] [PubMed] [Google Scholar]
- 12.Feller, G., D. D'Amico, and C. Gerday. 1999. Thermodynamic stability of a cold-active α-amylase from Antarctic bacterium Alteromonas haloplanctis. Biochemistry 38:4613-4619. [DOI] [PubMed] [Google Scholar]
- 13.Feller, G., F. Payan, F. Theys, M. Qian, R. Hasser, and C. Gerday. 1994. Stability and structural analysis of α-amylase from the Antarctic psychrophile Alteromonas haloplanctis A23. Eur. J. Biochem. 222:441-447. [DOI] [PubMed] [Google Scholar]
- 14.Georlette, D., V. Blaise, T. Collins, S. D'Amico, E. Gratia, A. Hoyoux, J.-C. Marx, G. Sonan, G. Feller, and C. Gerday. 2004. Some like it cold: biocatalysis at low temperatures. FEMS Microbiol. Rev. 28:25-42. [DOI] [PubMed] [Google Scholar]
- 15.Goldenberg, D. P. 1989. Analysis of protein conformation by gel electrophoresis, p. 225-250. In T. E. Creighton (ed.), Protein structure: a practical approach. IRL Press, Oxford, United Kingdom.
- 16.Goldenberg, D. P., and T. E. Creighton. 1984. Gel electrophoresis in studies of protein conformation and folding. Anal. Biochem. 138:1-18. [DOI] [PubMed] [Google Scholar]
- 17.Kashiwagi, K., K. Shiba, K. Fukami-Kobayashi, T. Noda, K. Nishikawa, and H. Noguchi. 2003. Characterization of folding pathways of the type-1 and type-2 periplasmic binding proteins MglB and ArgT. J. Biochem. 133:371-376. [DOI] [PubMed] [Google Scholar]
- 18.Kumar, S., J.-G. Tsai, and R. Nussinov. 2002. Maximal stabilities of reversible two-state proteins. Biochemistry 41:5359-5374. [DOI] [PubMed] [Google Scholar]
- 19.Lonhienne, T., C. Gerday, and G. Feller. 2000. Psychrophilic enzymes: revisiting the thermodynamic parameters of activation may explain local flexibility. Biochim. Biophys. Acta 1543:1-10. [DOI] [PubMed] [Google Scholar]
- 20.Makhtadze, G. I., and P. L. Privalov. 1994. Hydration effects in protein unfolding. Biophys. Chem. 51:291-309. [DOI] [PubMed] [Google Scholar]
- 21.Mast, A. E., J. J. Enghild, S. V. Pizzo, and G. Salvesen. 1991. Analysis of the plasma elimination kinetics and conformational stabilities of native, proteinase-complexed, and reactive site cleaved serpins: comparison of α1-proteinase inhibitor, α1-antichymotrypsin, antithrombin III, α2-antiplasmin, angiotensinogen, and ovalbumin. Biochemistry 30:1723-1730. [DOI] [PubMed] [Google Scholar]
- 22.Saunders, N., T. Thomas, P. M. G. Curmi, J. S. Mattick, E. Kuczek, R. Slade, J. Davis, P. D. Franzmann, D. Boone, K. Rusterholtz, R. Feldman, C. Gates, S. Bench, K. Sowers, K. Kadner, A. Aerts, P. Dehal, C. Detter, T. Glavina, S. Lucas, P. Richardson, F. Larimer, L. Hauser, M. Land, and R. Cavicchioli. 2003. Mechanisms of thermal adaptation revealed from the genomes of the Antarctic Archaea, Methanogenium frigidum and Methanococcoides burtonii. Genome Res. 13:1580-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siddiqui, K. S. 1990. Proteolytic nicking and carboxyl group modification of Arthrobacter D-xylose isomerase. Ph.D. thesis. London University, London, United Kingdom.
- 24.Siddiqui, K. S., M. Rangarajan, B. S. Hartley, A. Kitmitto, M. Panico, I. P. Blench, and H. R. Morris. 1993. Arthrobacter D-xylose isomerase: partial proteolysis with thermolysin. Biochem. J. 289:201-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uversky, V. N. 2002. Cracking the folding code. Why do some proteins adopt partially folded conformations, whereas others don't? FEBS Lett. 514:181-183. [DOI] [PubMed] [Google Scholar]
- 26.Uversky, V. N., J. R. Gillespie, and A. J. Fink. 2000. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins 41:415-427. [DOI] [PubMed] [Google Scholar]
- 27.Yamashita, H., T. Nakatsuka, and M. Hirose. 1995. Structural and functional characteristics of partially disulfide-reduced intermediates of ovotransferrin N lobe. J. Biol. Chem. 270:29806-29812. [DOI] [PubMed] [Google Scholar]