Abstract
The cold-adapted α-amylase from Pseudoalteromonas haloplanktis unfolds reversibly and cooperatively according to a two-state mechanism at 30°C and unfolds reversibly and sequentially with two transitions at temperatures below 12°C. To examine the role of the four disulfide bridges in activity and conformational stability of the enzyme, the eight cysteine residues were reduced with β-mercaptoethanol or chemically modified using iodoacetamide or iodoacetic acid. Matrix-assisted laser desorption-time of flight mass spectrometry analysis confirmed that all of the cysteines were modified. The iodoacetamide-modified enzyme reversibly folded/unfolded and retained approximately one-third of its activity. Removal of all disulfide bonds resulted in stabilization of the least stable region of the enzyme (including the active site), with a concomitant decrease in activity (increase in activation enthalpy). Disulfide bond removal had a greater impact on enzyme activity than on stability (particularly the active-site region). The functional role of the disulfide bridges appears to be to prevent the active site from developing ionic interactions. Overall, the study demonstrated that none of the four disulfide bonds are important in stabilizing the native structure of enzyme, and instead, they appear to promote a localized destabilization to preserve activity.
Pseudoalteromonas haloplanktis is a psychrophilic bacterium isolated from Antarctica (9, 18). Numerous cold-active enzymes (e.g., α-amylase, DNA ligase, and xylanase) from P. haloplanktis have been isolated and characterized (17, 21). These enzymes and other cold-active enzymes isolated from a broad range of cold-adapted organisms tend to have low activation enthalpy, low stability at low and moderate temperatures, and high activity at low temperatures (6). The unique ability of these enzymes to perform catalysis effectively at low temperature has provided a range of opportunities for applications in an expanding biotechnology market (6, 7, 22).
The P. haloplanktis α-amylase (AHA) has been the subject of extensive biochemical and biophysical characterization (9-12, 19), and its X-ray structure has been solved (1, 2). The enzyme has three domains (19) and is the largest multidomain enzyme (∼50 kDa) that unfolds reversibly (27). Recently, transverse urea gradient gel electrophoresis (TUG-GE) analysis revealed that the active site was part of the least stable structure in the enzyme (34a). Cooperative unfolding of structures forming the active site was shown to precede independent unfolding of other more stable protein domains, and it was proposed that active-site instability may be typical of cold-active enzymes.
The X-ray structure of AHA (1, 2) is similar to that of α-amylases from animals. It is most similar to mammalian (e.g., pig) α-amylases (13, 31) and may have arisen in diverse organisms through horizontal gene transfer (8). As is typical for mesophilic counterparts of cold-adapted enzymes, the pig enzyme is inherently more stable than AHA (11, 19). Comparative analysis of the structures of the two enzymes identified a disulfide bridge as potentially contributing to differences in protein stability. Eight cysteine residues are present in AHA, compared to 10 in the pig α-amylase (1, 2, 10, 31). In AHA, the eight cysteine residues are involved in four disulfide bridges which are conserved in all chloride-dependent α-amylases from animals and some bacteria. Two are intradomain A, one is intradomain B, and one is intradomain C (1, 2, 13). The extra disulfide bridge in the pig enzyme may link domains A and B near the entrance of the catalytic cleft and led to the hypothesis that it provided thermal stability (10, 13). The introduction of a fifth disulfide bridge in a site-directed mutant of AHA resulted in increased conformational stability with a concomitant decrease in activity (10).
While the stability characteristics of wild-type AHA, mutant AHA, and the pig enzyme are consistent with the fifth disulfide bridge stabilizing the active site, the role of the other four disulfide bridges in AHA (which are also present in the pig enzyme) is not clear. It has been argued that they must provide some selective advantage to the enzyme or they would have been eliminated through an absence of evolutionary selection pressure (16, 17). In general, disulfide bridges may enhance the conformational stability of a protein by decreasing the flexibility and entropy of the unfolded state (29). Alternatively, disulfide bridges may destabilize folded states entropically by decreasing the number of water molecules ordered in the unfolded state (14). While it has been argued that disulfide bonds contribute to higher stability by reducing the unfolding entropy of proteins and increasing the rigidity of the molecule (3, 30), this may rarely occur because favorable orientation of cysteine pairs can be restricted by spatial constraints (3, 38). Cytosolic enzymes tend to maintain more reduced cysteine residues than secreted enzymes (4). As a result, it has been suggested that secreted enzymes such as α-amylases may be stabilized by maintaining disulfide bridges (4). Removal of disulfide bridges has been shown to cause loss of enzyme activity (36) and stability (35, 37), and the formation and reduction of disulfide bridges may function as an on-off switch to regulate enzyme activity in response to stress (4).
To further our understanding of the roles of disulfide bridges in activity and stability of AHA, we used β-mercaptoethanol (BME) or chemical modification (carboxymethylation) with iodoacetamide (IA) or iodoacetic acid (IAA) to disrupt disulfide bonds and examined conformational stability, unfolding/folding transitions, and enzyme kinetics. The extent of modification was assessed by matrix-assisted laser desorption-time of flight mass spectrometry (MALDI-TOF-MS), which provided an accurate assessment of the degree of modification of all eight cysteine residues. The studies demonstrated that the disulfide bridges are not involved in stabilizing the enzyme and instead support their role in promoting a localized destabilization of the domains containing the active site.
MATERIALS AND METHODS
Chemical modification.
Recombinant wild-type AHA was prepared as described previously (9). IAA (Merck, Sydney, Australia) was recrystallized to maximize purity and stored dry at −20°C. IA (Sigma Chemical Co., Sydney, Australia) and recrystallized IAA were used to convert thiol groups in cysteine residues to S-CH2CONH2 and S-CH2COO−, respectively, using a previously described method (25). AHA (0.34 mg) in 0.1 M Tris-HCl, pH 8.5, containing 1 mM dithiothreitol and 6 M urea was incubated at 4°C for 18 h. IA or IAA (0.1 M) was made fresh in 0.1 M Tris-HCl, pH 8.5, and 250 μl of this IA solution (20 mM final concentration) was added to 1 ml of reduced AHA. The reaction was carried out in the dark at 25°C for 15 min. The carboxymethylated enzyme was dialyzed against 50 mM MOPS (morpholinepropanesulfonic acid)-NaOH (pH 7.2)-50 mM NaCl at 5°C for 2 h with four changes of buffer. Prior to TUG-GE, the modified amylase was further dialyzed with water and freeze-dried in 28-μg aliquots. For MALDI-TOF-MS analysis, modified enzymes were desalted using PD10 Sephadex G25M columns (Pharmacia, Sydney, Australia), 0.5-ml fractions were collected, and the A280 was recorded. The protein-containing fractions were pooled and freeze-dried. Endoproteinase Lys-C (EndoLysC) proteolysis (0.25 μg) of alkylated AHA (10 μg) was performed in 25 mM Tris (pH 8.5)-1 M urea at 37°C for 24 h. The resulting hydrolysate was used for MALDI-TOF-MS analysis.
MALDI-TOF-MS of AHA EndoLysC peptides.
MALDI analyses of AHA EndoLysC peptides were performed with a Voyager DE STR mass spectrometer with a nitrogen laser (337 nm, 2-ns pulse) (PE Biosystems, Framingham, MA). Data acquisition was performed in the positive-ion mode, and the instrument was calibrated immediately prior to each analysis. Three-point external calibration was performed using Calmix 1 from the Sequazyme kit (Applied Biosystems, Melbourne, Australia). Calmix 1 was used with dihydroxybenzoic acid (DHB) matrix (Sigma Chemical Co., Sydney, Australia), and monoisotopic masses of the singly protonated molecular ions of des-Arg-Bradykinin ([M + H]+ = 904.47), angiotensin 1 ([M + H]+ = 1,296.69), and Glu-fibrinopeptide B ([M + H]+ = 1,570.68) were used during calibration. All analyses were performed in reflectron-delayed extraction mode. Peptides were extracted from the proteolysis mixture using C4 zip tips (Millipore, Sydney, Australia), washed with acetonitrile-trifluoroacetic acid-water (2:0.1:98, vol/vol/vol), and eluted directly onto a MALDI target with 1.5 μl of acetonitrile-water-trifluoroacetic acid (80:20:0.1, vol/vol/vol) containing DHB matrix (10 mg/ml). Once applied to the target, all samples were air dried at ambient temperature prior to analysis. PAWS software (5) was utilized to identify peptide masses observed in the spectra. Using the Voyager Data Explorer software (PE Biosystems), spectra were adjusted by noise removal (2 standard deviations) and Gaussian smoothing (SM25) in order to improve peak centroid assignment. Water used for all mass spectrometry work was purified by a Millipore water purification system and had a resistivity of >18 MΩ/cm.
Enzyme kinetics and thermodynamics.
To determine the effect of substrate concentration, α-amylase assays were carried out at 3°C and 25°C in 0.1 M MOPS-NaOH (pH 7.2) buffer containing 50 mM NaCl and various concentrations (0.13 mg ml−1 to 0.73 mg ml−1) of potato starch (Fluka, Sydney, Australia). Kinetic and thermodynamic properties of reduced unmodified AHA were determined by including BME in the assay mixture at a concentration of 1% (vol/vol). Two batches of substrate solution (1 ml each) were equilibrated at 3°C and 25°C, and the reaction was initiated by the addition of unmodified or modified enzyme (2.6 × 10−5 to 1.1 × 10−3 mg). After 10 min, the reaction was terminated by the addition of 1 ml of dinitrosalicylic acid reagent (28). The solution was boiled for 5 min and cooled to room temperature, and the A540 was measured against a reagent blank. The A540 units were converted to units per milligram of protein, using ɛmM of 1.25, determined from a maltose standard curve using a flat-bottom microtiter plate. The kinetic data (starch versus units per milligram) of unmodified and modified α-amylases at 3°C and 25°C was slightly sigmoidal and was fitted to a Hill's plot rather than a Michaelis-Menten plot using Enzyme Kinetics Module 1.1 linked to Sigma Plot 8.02. To determine the effect of temperature, α-amylase assays were performed as described above using 1% starch at temperatures ranging from 3°C to 36°C. Substrate solutions were equilibrated at the respective temperatures prior to commencement of the assay. Thermodynamic values were calculated from kinetic data by plotting activity as a function of temperature (ln units milligram−1 versus 1/T) according to the Arrhenius equation using Origin Software. Activation energy (Ea) of starch hydrolysis was determined from the slopes of the Arrhenius plots, and thermodynamic parameters of activation (#) were determined using the transition state theory (15, 33, 34) using the following equations: ΔH# (enthalpy of activation) = Ea − RT, ΔG# (free energy of activation) = −RT ln [(kcat · h)/(KB · T)], and ΔS# (entropy of activation) = (ΔH# − ΔG#)/T, where T is the absolute temperature, h is the Planck constant (6.63 × 10−34 Js), KB is the Boltzman constant (1.38 × 10−23 kJ−1), and R is the universal gas constant (8.314 kJ−1 mol−1).
Protein electrophoresis.
TUG-GE was performed as described previously (23, 24, 34a). Native gel electrophoresis and 7 M urea gel electrophoresis were performed based on previously described methods (23). The gels (7.5% acrylamide) were run in a discontinuous Tris-glycine buffer system at pH 6.8 and 8.8. For urea gels, 7 M urea was used in stacking and resolving gels and sample buffer. For electrophoresis of the BME-treated enzyme, BME was added to the sample buffer at a concentration of 2.5% (vol/vol) but not to the transverse urea gradient or native gels, as it inhibited polymerization. Proteins were detected using Coomassie R250 (32) and visualized using a Fujifilm LAS-3000 (Berthold Australia, Melbourne, Australia).
RESULTS AND DISCUSSION
Modified AHA lacking disulfide bonds.
By employing 5,5′-dithiobis-(2-nitrobenzoic acid) titration, it was previously shown that AHA contains no free thiol groups, and all eight cysteine residues are linked in four disulfide bonds (10, 20). In order to break disulfide bonds, BME was used to reduce cysteines to thiol groups. Alternatively, disulfide bonds were irreversibly modified by reducing cysteines with dithiothreitol and blocking with IA or IAA. IA modification produced a neutral modification, whereas coupling to IAA gave a negative charge at pH ≥7 due to the low pKa (∼5) of the IAA carboxyl group.
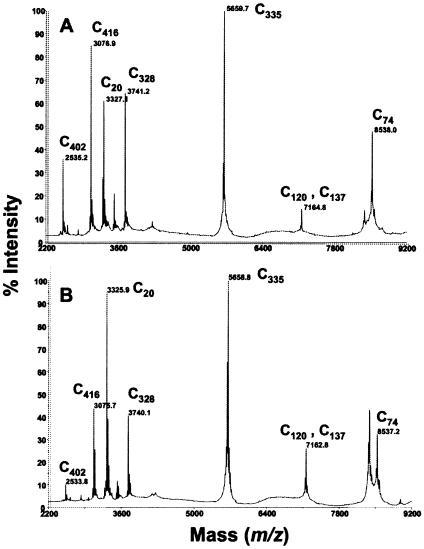
MALDI-TOF-MS analysis revealed that all eight cysteine residues were modified following both the IAA and IA alkylations (Table 1 and Fig. 1). The single-dalton difference in theoretical mass between IAA- and IA-treated peptides was reflected in the observed mass values (Table 1). The eight cysteine residues in AHA are predicted to occur in individual peptides following EndoLysC proteolysis, with the exception of C120 and C137, which are both present in peptides 107 to 169. Complete coverage of all cysteine-containing peptides was achieved using EndoLysC proteolysis. Unmodified cysteine-containing peptides were not present in the spectra (Fig. 1). Proteolysis using endoproteinase Asp-N was also used for generating multiple peptides in the 1-to 3-kDa-mass range. This approach provided additional data on five cysteine-containing peptides, confirming the findings of the EndoLysC proteolysis (data not shown).
TABLE 1.
MALDI-TOF peptide mass mapping of alkylated AHA using endolysC proteolysis
| Protein sequence | Position of cysteine residues | Theoretical average mass ([M + H]+) (Da)
|
Observed average mass ([M + H]+) (Da)
|
||
|---|---|---|---|---|---|
| IAA | IA | IAA | IA | ||
| 1-27 | C20 | 3,326.7 | 3,325.7 | 3,327.4 | 3,326.3 |
| 28-106 | C74 | 8,537.4 | 8,536.4 | 8,538.6 | 8,538.7 |
| 107-169 | C120, C137 | 7,163.7 | 7,161.8 | 7,164.6 | 7,161.8 |
| 301-334 | C328 | 3,740.0 | 3,739.0 | 3,741.5 | 3,740.4 |
| 335-383 | C335 | 5,659.1 | 5,658.1 | 5,659.7 | 5,659.0 |
| 384-406 | C402 | 2,534.8 | 2,533.8 | 2,534.9 | 2,533.8 |
| 415-443 | C416 | 3,076.4 | 3,075.4 | 3,077.3 | 3,075.2 |
FIG. 1.
MALDI-TOF mass spectra of EndoLysC peptides of modified AHA. Enzyme alkylated with IAA (A) or IA (B) are shown. Analyses were performed using reflectron-delayed extraction and in positive-ion mode using DHB matrix. Masses represent average protonated molecular ions [M + H]+ of EndoLysC peptides. A mass error of <400 ppm is typical with the instrument acquisition parameters used.
Mobility of modified AHA on native and urea gels.
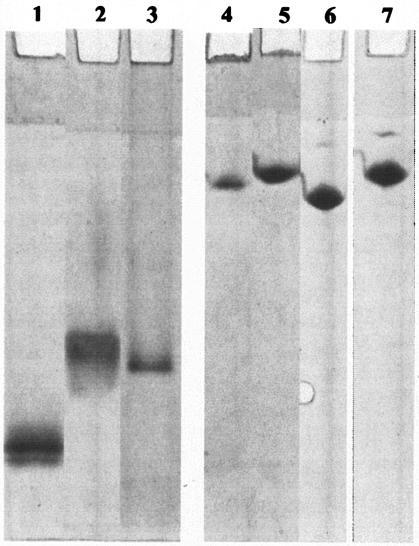
The mobility of modified and nonmodified AHA in native gel electrophoresis was examined to gauge the impact of the modifications on protein structure. Net protein movement results from a competition between hydrodynamic volume, which retards migration, and negative charge, which propels the protein towards the anode. In the native gel, the unmodified enzyme (pI 4.8) migrated with the highest Rf (Fig. 2, lane 1), while the enzymes in the presence of BME (Fig. 2, lane 2) or modified by IAA (Fig. 2, lane 3) migrated with a reduced Rf. This is likely to reflect unfolding of the modified proteins causing an increase in hydrodynamic volume. This implies that removing the disulfide bridges leads to a protein with a less compact structure. When the samples were run in 7 M urea gels, a further reduction in mobility was observed, consistent with an even greater increase in hydrodynamic volume caused by urea unfolding (Fig. 2, lanes 4 to 7). The IAA- and IA-modified enzymes showed a single band on 7 M urea gels (Fig. 2, lanes 6 and 7). The presence of a single band is indicative of complete modification of IAA- and IA-modified AHA and is consistent with our findings from MALDI-TOF-MS analysis (above).
FIG. 2.
Native gel electrophoresis and urea gel electrophoresis of AHA. Native gel electrophoresis (lanes 1 to 3) and 7 M urea gel electrophoresis (lanes 4 to 7) were performed in Tris-glycine buffer, pH 8.8. Unmodified AHA, lanes 1 and 4; AHA treated with 2.5% BME, lanes 2 and 5; AHA modified with IAA, lanes 3 and 6; AHA modified with IA, lane 7.
BME-modified AHA: effect on conformational stability.
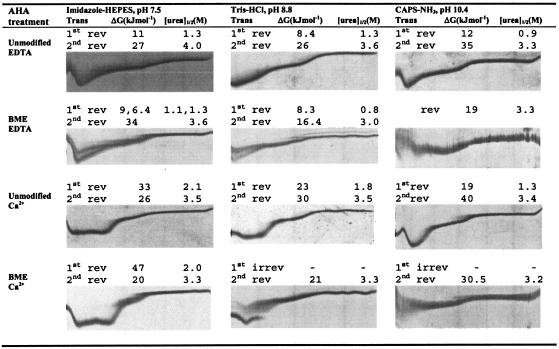
The conformational stability of BME-treated AHA was assessed using TUG-GE, at pH 7.5, 8.8, and 10.4, in the presence or absence of Ca2+ (presence of EDTA) at 12°C (Fig. 3). We have demonstrated that TUG-GE provided a reproducible means of evaluating unfolding transitions and conformational stabilities of AHA (34a). At pH 7.5 in the absence of Ca2+, the unmodified enzyme showed a single curve undergoing two transitions, with the first transition occurring at 1.3 M urea and the second transition occurring at 4 M urea (Fig. 3). This is consistent with our findings that the first transition was found to result from the unfolding of the most labile protein structures within domains A and B, which includes the active site of AHA (34a). Under equivalent TUG-GE conditions, the reduced enzyme (presence of BME) resolved into several bands, implying that the protein population was heterogeneous with individual species unfolding at different urea concentrations. This heterogeneity may be due to a rapid thiol-disulfide interchange, resulting in disulfide mismatches (4) resulting from BME being added to the sample buffer and not to the gel. At least one band demonstrated slow interconversion between conformations within the duration of the TUG-GE, and two bands demonstrated reversible unfolding.
FIG. 3.
Urea unfolding curves and thermodynamic parameters of unmodified and BME-modified AHA. TUG-GE was performed at pH 7.5, 8.8, or 10.4 in the absence of Ca2+ at 12°C. The urea gradient was 0 to 6.64 M (left to right); direction of electrophoresis was from top to bottom; Trans, first or second folding/unfolding transition; rev, reversible transition; ΔG, free energy of unfolding between two states; [urea]1/2, urea concentration at 0 ΔG; CAPS, 3-(cyclohexylamino)propanesulfonic acid.
It has been shown that at low temperature and in the presence of Ca2+, the structures undergoing the first transition showed slow interconversion of conformations within the duration of the TUG-GE (34a). In the presence of Ca2+ at pH 7.5, the structure undergoing the first transition of the BME-treated AHA was more stable (Δ[urea]1/2, −0.1 M; ΔΔG, +14 kJ mol−1), and the initial conformation undergoing the second transition was slightly destabilized (Δ[urea]1/2, −0.2 M; ΔΔG, −6 kJ mol−1) compared with the unmodified enzyme in the presence of Ca2+ (Fig. 3). At pH 8.8 and 10.4, the initial conformation undergoing the first transition became irreversibly unfolded and the initial conformation undergoing the second transition was destabilized within the duration of the TUG-GE (Δ[urea]1/2, −0.20 M; ΔΔG, −9 kJ mol−1). As the pKa of SH groups is ∼8.3, at pH 7.5, the repulsion from opposing S− groups should be minimal, whereas at pH 8.8 and 10.4, the enzyme is likely to be destabilized by mutual charge repulsion of ionized SH and COOH groups.
It is noteworthy that at pH 7.5, disulfide bond breakage conferred stability to the structure undergoing the first transition, whereas the initial conformation undergoing the first transition became irreversibly denatured within the duration of the TUG-GE at pH 8.8 and 10.4. The reversible unfolding/folding of the initial conformation undergoing the first transition has been proposed to result from a minimal number of ionic interactions that are required for the refolding of the enzyme (9, 34a). This has been supported by studies of site-directed mutants that produced extra ionic interactions (9) and resulted in irreversible unfolding within the duration of the TUG-GE. As the initial conformation undergoing the first transition was irreversible within the duration of the TUG-GE at pH 8.8 and 10.4 (Fig. 3), this may imply that the ionized SH groups mediated additional ionic interactions with positively charged side chains or Ca2+. Reduced disulfide bonds have previously been reported to coordinate metal ions (26). A single Ca2+ ion is capable of binding at the interface between domains A and B and stabilizing the enzyme (1, 2). Consistent with this, both initial conformations described by these transitions of the BME-treated AHA were stabilized in the presence of Ca2+ (Fig. 3).
IA-modified AHA: effect on conformational stability.
A possible complication with the use of BME for interpreting the effects of disulfide bond breakage is that rapid thiol-disulfide interconversion can lead to the transient formation of nonnative bridges (4). This potential problem was prevented by trapping the ionized S groups with a blocking reagent as soon as the cysteine residues were reduced. Modifying with IA also ensured that no extra charges were introduced.
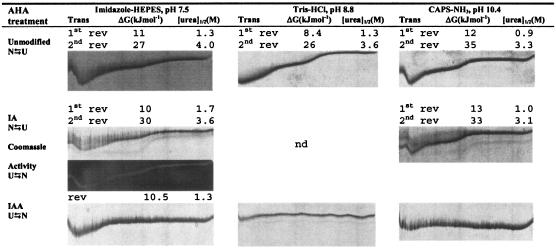
Compared to the unmodified enzyme, in the absence of Ca2+ at pH 7.5, an increase in Δ[urea]1/2 of 0.4 M for the structures undergoing the first transition indicated that the IA modification produced a slight stabilization (Fig. 4). Under the same conditions, the structures of the IA-modified AHA undergoing the second transition unfolded at a 0.4 M lower urea concentration than with the unmodified enzyme (Fig. 4). At pH 8.8 (data not shown) and pH 10.4 (Fig. 4), ΔG and [urea]1/2 corresponding to both transitions were similar in the IA-modified and unmodified enzymes. The initial conformation of the IA-modified enzyme undergoing the first transition was reversible irrespective of pH; however, unfolding occurred at a lower urea concentration at pH 10.4 (1.0 M) compared to pH 7.5 (1.7 M). Moreover, activity staining demonstrated that the IA-modified enzyme retained activity throughout the complete unfolding/folding curve, highlighting that unfolding was fully reversible (Fig. 4).
FIG. 4.
Urea unfolding curves and thermodynamic parameters of unmodified, IA-modified, and IAA-modified AHA. TUG-GE was performed at pH 7.5, 8.8, or 10.4 in the absence of Ca2+ at 12°C. U⇆N, TUG-GE for unfolded enzyme showing folding; N⇆U, TUG-GE for folded enzyme showing unfolding. Urea gradient was 0 to 6.64 M (left to right); direction of electrophoresis was from top to bottom. nd, not determined; Trans, first or second folding/unfolding transition; rev, reversible transition; ΔG, free energy of unfolding between two states; [urea]1/2, urea concentration at 0 ΔG; CAPS, 3-(cyclohexylamino)propanesulfonic acid.
The structure undergoing the first transition of the IA-modified enzyme (Fig. 4) was more stable ([urea]1/2 and ΔG) than the BME-modified AHA (Fig. 3) in the absence of Ca2+ at pH 7.5. At pH 10.4, the structures undergoing the second transition were also more stable in the IA-modified than in the BME-modified enzyme. This indicates that free thiols present in the structures undergoing first and second transitions may have a destabilizing effect that is overcome when they are completely blocked.
An interesting activity property of the IA-modified enzyme was that its Topt (25°C) was 7°C lower than that of the unmodified enzyme, despite the conformational stability being comparable to or greater than that of the unmodified enzyme (Fig. 3 and 4). This parallels similar findings of a site-directed mutant (C95S) of a β-amylase which exhibited a 20°C reduced Topt (35). In this case, the wild-type and mutant enzymes had similar circular dichroism spectra, implying that removal of the disulfide bridge did not change the conformation of the enzyme.
IAA-modified AHA: effect on conformational stability.
The pKa of IAA is 3.1, resulting in the IAA-modified AHA having a high proportion of negatively charged alkylated cysteines at neutral pH. At pH 7.5, TUG-GE analysis showed that the IAA-modified AHA folds by a single, two-state mechanism and that the enzyme denatures at 1.3 M urea, 0.4 M urea lower than the IA-modified form (Fig. 4). At higher pH values (8.8 and 10.4), the flat unfolding/folding curve demonstrated that the IAA-modified enzyme was totally denatured at all urea concentrations (Fig. 4). The data indicate that removing the ability to form disulfide bridges and incorporating modifications that produce increasing negative charge as pH is increased greatly destabilizes the protein. In this context, it is, however, noteworthy that at pH 7.5, the stability of the protein structures (including the active site) undergoing the first transition are as stable as in the unmodified enzyme ([urea]1/2, 1.3 M) (Fig. 4).
Elimination of disulfide bridges reduced activity.
The kinetic and thermodynamic properties of activity at 3°C and 25°C were calculated for the unmodified, BME-modified, and IA-modified AHA (Table 2). The Km for the BME-treated AHA decreased at both temperatures, whereas the Km for the IA-modified form dropped marginally at 3°C and increased at 25°C (Table 2). At both assay temperatures, the BME- and IA-modified enzymes retained 46% and 32% activity (kcat), respectively, compared to the unmodified enzyme (Table 2). In contrast, the IAA-modified form was inactive (data not shown). Elimination of disulfide bonds has previously been reported to reduce enzyme activity to a similar or greater extent than what we observed (4, 26, 35, 36); for example, the removal of all (two) disulfide bridges from RNase T1 resulted in a severe reduction in melting temperature, conformational stability, and activity (29).
TABLE 2.
Kinetic and thermodynamic properties of AHA activity
| Kinetic or thermodynamic parameter | Treatment
|
|||||
|---|---|---|---|---|---|---|
| 3°C
|
25°C
|
|||||
| Unmodified | BMEa | IA | Unmodified | BME | IA | |
| kcat (s−1) | 840 | 370 | 267 | 2321 | 1062 | 713 |
| Km (mg ml−1) | 2.3 | 0.78 | 1.7 | 2.9 | 1.3 | 5.6 |
| ΔG# (kJ mol−1) | 52 | 54 | 55 | 54 | 56 | 57 |
| ΔH# (kJ mol−1) | 26 | 13b | 33 | 26 | 13b | 33 |
| ΔS# (J mol−1 K−1) | −93 | −149 | 79 | −94 | −144 | −81 |
BME, 1% in assay mixture.
Arrhenius plot was nonlinear at all temperatures.
For the IA-modified enzyme, decreasing the temperature from 25°C to 3°C led to a decrease in activity and an increase in activation enthalpy (ΔH#), a response that is characteristic of the mesophilic pig α-amylase and the disulfide AHA site-directed mutant (11). In contrast, lower activity at 3°C was accompanied by reduction in ΔH# for the BME-modified enzyme, implying that the decrease in activity was entropically driven (Table 2). The result for the BME-modified enzyme should, however, be treated with some caution, as the Arrhenius plot was nonlinear at all temperatures below 25°C. This suggested that the rate of catalysis was continuously changing during the assay and may have resulted from the conformation of the enzyme changing due to thiol-disulfide interchange.
The functional role of disulfide bridges in AHA.
The contribution of disulfide bridges to the activity-flexibility-stability relationship of thermally adapted enzymes has been extensively discussed (9, 16); however, little experimental work has been performed. It was therefore striking to find that TUG-GE analysis demonstrated that AHA can fold and remain functional at physiological pH in the absence of all four native disulfide bridges. A loss of activity and stability was achieved only when disulfide bridges were broken and blocked with negatively charged IAA (Fig. 4). Based on sequence comparisons of chloride-dependent α-amylases from animals and some extremophilic gram-negative bacteria, it was proposed that the function of these bridges is probably to provide the required conformation at and around the active site and is not related to temperature adaptation (12, 13). Our results demonstrate experimentally that none of the four disulfide bridges are important in stabilizing the native structure of AHA.
The function of the disulfide bridges may in fact be to promote a localized destabilizing effect. The structures undergoing the first transition (including the active site) were stabilized in the IA-modified enzyme (Fig. 4), and the enzyme was less active and had a higher ΔH# than the unmodified enzyme (Table 2).
This can also be considered by comparing the activity of the unmodified and IA-modified enzymes at 3°C and 25°C. At 3°C, the difference was more pronounced, indicating that blocking the cysteine residues made the enzyme less flexible at lower temperature. Two disulfide bridges are intradomain A and form part of the catalytic cleft, and one disulfide bridge is intradomain B and forms part of the other side of the catalytic cleft (1, 2). If disulfide bridges contributed a stabilizing role, their removal would be predicted to produce a more flexible structure (1, 2, 17). In contrast, we observed that the conformations for the IA-modified enzyme undergoing the first transition became less flexible than those in the unmodified enzyme.
The TUG-GE analysis has also aided in separating the contributions of disulfide bridges to activity versus stability. The effect of the BME and IA modifications was greater on enzyme activity than on stability (particularly the active-site region) (Fig. 3 and 4). These effects on AHA are consistent with effects of mutations causing loss of disulfide bonds in β-amylase from soybean and Bacillus polymyxa which resulted in a large reduction in activity with no apparent change in stability (35, 36). The effects on AHA are particularly interesting in view of the lack of primary sequence similarity or placement of disulfide bridges between α- and β-amylases.
The decrease of ΔH# is the major adaptive characteristic for cold-adapted enzymes to function effectively at low temperature and is achieved by decreasing the number of enthalpy-driven interactions that have to be broken during formation of the transition state. These interactions also contribute to the stability of the folded conformation, and as a result, the region of the enzyme containing the active site should be less stable. These are the key features that explain the activity-stability trade-off in cold-adapted enzymes (12, 16). The IA-modified enzyme is less active and displays an increased ΔH# at low temperature (Table 2). This indicates that the modified enzyme has more enthalpy-driven interactions than the unmodified enzyme. According to this reasoning, the intradomain A and intradomain B disulfide bridges may be positioned in AHA to prevent the active site from developing ionic interactions. Future site-directed mutagenesis studies examining the conversion of cysteine residues to serine residues will help to clarify this.
Acknowledgments
The work was supported by the Australian Research Council. Mass spectrometric analysis for the work was carried out at the Bioanalytical Mass Spectrometry Facility, UNSW, and was supported in part by grants from the Australian Government Systemic Infrastructure Initiative and Major National Research Facilities Program (UNSW node of the Australian Proteome Analysis Facility) and by the UNSW Capital Grants Scheme.
Thanks go to Paul Curmi for helpful discussion. S.D. is a postdoctoral researcher from the Fonds National de la Recherche Scientifique (FNRS, Belgium).
REFERENCES
- 1.Aghajari, N., G. Feller, C. Gerday, and R. Haser. 1998. Crystal structure of the psychrophilic α-amylase from Alteromonas haloplanctis in its native form and complexed with an inhibitor. Protein Sci. 7:564-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aghajari, N., G. Feller, C. Gerday, and R. Haser. 1998. Structures of the psychrophilic Alteromonas haloplanctis α-amylase give insights into cold adaptation at a molecular level. Structure 6:1503-1516. [DOI] [PubMed] [Google Scholar]
- 3.Arpigny, J. L., G. Feller, S. Davail, S. Genicot, E. Narinx, Z. Zekhnini, and C. Gerday. 1994. Molecular adaptations of enzymes from thermophilic and psychrophilic organisms. Adv. Comp. Environ. Physiol. 20:269-295. [Google Scholar]
- 4.Aslund, F., and J. Beckwith. 1999. Bridge over troubled waters: sensing stress by disulfide-bond formation. Cell 96:751-753. [DOI] [PubMed] [Google Scholar]
- 5.Beavis, R. 2000. PAWS-freeware computer program. Edition for Windows NT. ProteoMetrics (http://www.proteometrics.com).
- 6.Cavicchioli, R., and K. S. Siddiqui. 2004. Cold-adapted enzymes, p. 615-638. In A. Pandey, C. Webb, C. R. Soccol, and C. Larroche (ed.), Enzyme technology. AsiaTech Publishers, New Delhi, India.
- 7.Cavicchioli, R., K. S. Siddiqui, D. Andrews, and K. R. Sowers. 2002. Low-temperature extremophiles and their applications. Curr. Opin. Biotechnol. 13:253-261. [DOI] [PubMed] [Google Scholar]
- 8.Da Lage, J. L., G. Feller, and S. Janecek. 2004. Horizontal gene transfer from Eukarya to bacteria and domain shuffling: the α-amylase model. Cell. Mol. Life Sci. 61:97-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D'Amico, S., C. Gerday, and G. Feller. 2001. Structural determinants of cold adaptation and stability in a large protein. J. Biol. Chem. 276:25791-25796. [DOI] [PubMed] [Google Scholar]
- 10.D'Amico, S., C. Gerday, and G. Feller. 2002. Dual effects of an extra disulfide-bond on the activity and stability of a cold-adapted α-amylase. J. Biol. Chem. 277:46110-46115. [DOI] [PubMed] [Google Scholar]
- 11.D'Amico, S., C. Gerday, and G. Feller. 2003. Temperature adaptation of proteins: engineering mesophilic-like activity and stability in a cold-adapted α-amylase. J. Mol. Biol. 332:981-988. [DOI] [PubMed] [Google Scholar]
- 12.D'Amico, S., J. C. Marx, C. Gerday, and G. Feller. 2003. Activity-stability relationship in extremophilic enzymes. J. Biol. Chem. 278:7891-7896. [DOI] [PubMed] [Google Scholar]
- 13.D'Amico, S., C. Gerday, and G. Feller. 2000. Structural similarities and evolutionary relationships in chloride-dependent α-amylases. Gene 253:95-105. [DOI] [PubMed] [Google Scholar]
- 14.Doig, A. J., and D. H. Williams. 1991. Is the hydrophobic effect stabilizing or destabilizing in proteins? The contribution of disulphide bonds to protein stability. J. Mol. Biol. 217:389-398. [DOI] [PubMed] [Google Scholar]
- 15.Eyring, H., and A. E. Stearn. 1939. The application of the theory of absolute reaction rates to proteins. Chem. Rev. 24:253-270. [Google Scholar]
- 16.Feller, G. 2003. Molecular adaptations to cold in psychrophilic enzymes. Cell. Mol. Life Sci. 60:648-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feller, G., and C. Gerday. 2003. Psychrophilic enzymes: hot topics in cold adaptation. Nat. Rev. Microbiol. 1:200-208. [DOI] [PubMed] [Google Scholar]
- 18.Feller, G., E. Narinx, J. L. Arpigny, Z. Zekhnini, J. Swings, and C. Gerday. 1994. Temperature dependence of growth, enzyme secretion and activity of psychrophilic Antarctic bacteria. Appl. Microbiol. Biotechnol. 41:477-479. [Google Scholar]
- 19.Feller, G., D. d'Amico, and C. Gerday. 1999. Thermodynamic stability of a cold-active α-amylase from Antarctic bacterium Alteromonas haloplanctis. Biochemistry 38:4613-4619. [DOI] [PubMed] [Google Scholar]
- 20.Feller, G., F. Payan, F. Theys, M. Qian, R. Haser, and C. Gerday. 1994. Stability and structural analysis of an α-amylase from the Antarctic psychrophile Alteromonas haloplanctis A23. Eur. J. Biochem. 222:441-447. [DOI] [PubMed] [Google Scholar]
- 21.Georlette, D., V. Blaise, T. Collins, S. D'Amico, E. Gratia, A. Hoyoux, J.-C. Marx, G. Sonan, G. Feller, and C. Gerday. 2004. Some like it cold: biocatalysis at low temperatures. FEMS Microbiol. Rev. 28:25-42. [DOI] [PubMed] [Google Scholar]
- 22.Gerday, C., M. Aittaleb, M. Bentahir, J. P. Chessa, P. Claverie, T. Collins, S. D'Amico, J. Dumont, G. Garsoux, A. Georlette-Hoyoux, T. Lonhienne, M. A. Meuwis, and G. Feller. 2000. Cold-adapted enzymes: from fundamentals to biotechnology. TIBTECH 18:103-107. [DOI] [PubMed] [Google Scholar]
- 23.Goldenberg, D. P. 1989. Analysis of protein conformation by gel electrophoresis, p. 225-250. In T. E. Creighton (ed.), Protein structure: a practical approach. IRL Press, Oxford, United Kingdom.
- 24.Goldenberg, D. P., and T. E. Creighton. 1984. Gel electrophoresis in studies of protein conformation and folding. Anal. Biochem. 138:1-18. [DOI] [PubMed] [Google Scholar]
- 25.Hollecker, M. 1989. Counting integral number of residues by chemical modification, p. 145-153. In T. E. Creighton (ed.), Protein structure: a practical approach. IRL Press, Oxford, United Kingdom.
- 26.Jakob, U., W. Muse, M. Eser, and J. C. Bardwell. 1999. Chaperone activity with a redox switch. Cell 96:341-352. [DOI] [PubMed] [Google Scholar]
- 27.Kumar, S., J.-G. Tsai, and R. Nussinov. 2002. Maximal stabilities of reversible two-state proteins. Biochemistry 41:5359-5374. [DOI] [PubMed] [Google Scholar]
- 28.Miller, G. L. 1959. Use of dinitrosalicylic acid reagent for the determination of reducing sugar. Anal. Chem. 31:426-428. [Google Scholar]
- 29.Pace, C. N., G. R. Grimsley, J. A. Thomson, and B. J. Bernett. 1988. Conformational stability and activity of ribonuclease T1 with zero, one, and two intact disulfide-bonds. J. Biol. Chem. 263:11820-11825. [PubMed] [Google Scholar]
- 30.Privalov, P. L. 1979. Stability of proteins. Adv. Protein Chem. 33:167-241. [DOI] [PubMed] [Google Scholar]
- 31.Qian, M., R. Haser, and F. Payan. 1993. Structure and molecular model refinement of pig pancreatic α-amylase at 2.1 Å resolution. J. Mol. Biol. 231:785-799. [DOI] [PubMed] [Google Scholar]
- 32.See, Y. P., and G. Jackowski. 1989. Estimating molecular weights of polypeptides by SDS gel electrophoresis, p. 1-21. In T. E. Creighton (ed.), Protein structure: a practical approach. IRL Press, Oxford, United Kingdom.
- 33.Siddiqui, K. S., A. A. Najmus-Saquib, M. H. Rashid, and M. I. Rajoka. 2000. Carboxyl group modification significantly altered the kinetic properties of purified carboxymethylcellulase from Aspergillus niger. Enzyme Microb. Technol. 27:467-474. [DOI] [PubMed] [Google Scholar]
- 34.Siddiqui, K. S., R. Cavicchioli, and T. Thomas. 2002. Thermodynamic activation properties of elongation factor 2 (EF-2) proteins from psychrotolerant and thermophilic archaea. Extremophiles 6:143-150. [DOI] [PubMed] [Google Scholar]
- 34a.Siddiqui, K. S., G. Feller, S. D'Amico, C. Gerday, L. Giaquinto, and R. Cavicchioli. 2005. The active site is the least stable structure in the unfolding pathway of a multidomain cold-adapted α-amylase. J. Bacteriol. 187:6197-6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Totsuka, A., V. H. Nong, H. Kadokawa, C. S. Kim, Y. Itoh, and C. Fukazawa. 1994. Residues essential for catalytic activity of soybean β-amylase. Eur. J. Biochem. 221:649-654. [DOI] [PubMed] [Google Scholar]
- 36.Uozumi, N., T. Matsuda, N. Tsukagoshi, and S. Udaka. 1991. Structural and functional roles of cysteine residues of Bacillus polymyxa β-amylase. Biochemistry 30:4594-4599. [DOI] [PubMed] [Google Scholar]
- 37.Vogl, T., R. Brengelmann, H.-J. Hinz, M. Scharf, M. Lotzbeyer, and J. W. Engels. 1995. Mechanism of protein stabilization by disulfide-bridges: calorimetric unfolding studies of disulfide-deficient mutants of the α-amylase inhibitor tendamistat. J. Mol. Biol. 254:481-496. [DOI] [PubMed] [Google Scholar]
- 38.Wetzel, R. 1987. Harnessing disulfide-bonds using protein engineering. Trends Biochem. Sci. 12:478-482. [Google Scholar]