Abstract
The E1p enzyme is an essential part of the pyruvate dehydrogenase complex (PDHC) and catalyzes the oxidative decarboxylation of pyruvate with concomitant acetylation of the E2p enzyme within the complex. We analyzed the Corynebacterium glutamicum aceE gene, encoding the E1p enzyme, and constructed and characterized an E1p-deficient mutant. Sequence analysis of the C. glutamicum aceE gene and adjacent regions revealed that aceE is not flanked by genes encoding other enzymes of the PDHC. Transcriptional analysis revealed that aceE from C. glutamicum is monocistronic and that its transcription is initiated 121 nucleotides upstream of the translational start site. Inactivation of the chromosomal aceE gene led to the inability to grow on glucose and to the absence of PDHC and E1p activities, indicating that only a single E1p enzyme is present in C. glutamicum and that the PDHC is essential for the growth of this organism on carbohydrate substrates. Surprisingly, the E1p enzyme of C. glutamicum showed up to 51% identity to homodimeric E1p proteins from gram-negative bacteria but no similarity to E1 α- or β-subunits of heterotetrameric E1p enzymes which are generally assumed to be typical for gram-positives. To investigate the distribution of E1p enzymes in bacteria, we compiled and analyzed the phylogeny of 46 homodimeric E1p proteins and of 58 α-subunits of heterotetrameric E1p proteins deposited in public databases. The results revealed that the distribution of homodimeric and heterotetrameric E1p subunits in bacteria is not in accordance with the rRNA-based phylogeny of bacteria and is more heterogeneous than previously assumed.
The pyruvate dehydrogenase complex (PDHC) represents a member of a multienzyme complex family that also comprises the 2-oxoglutarate dehydrogenase complex (OGDHC) and the branched-chain 2-oxoacid dehydrogenase complex (BCOADHC). These enzymes catalyze the oxidative decarboxylation of pyruvate, 2-oxoglutarate, and the 2-oxo acids of the branched-chain amino acids l-leucine, l-valine, and l-isoleucine, respectively. In general, the multienzyme complexes are composed of multiple copies of three different enzymes, a thiamine pyrophosphate (TPP) containing 2-oxoacid decarboxylase (E1), a lipoic acid-containing dihydrolipoamide acyltransferase (E2), and the flavin-containing lipoamide dehydrogenase (LPD). The E1 enzyme catalyzes the irreversible, TPP-dependent oxidative decarboxylation of the 2-oxoacid, followed by the acylation of the lipoyl prosthetic group covalently attached to the E2 chain. The E2 component catalyzes the transfer of the acyl group from the lipoyl group to coenzyme A (CoA). The resulting dihydrolipoyl group is reoxidized by LPD, generating NADH and H+ from NAD+ (for a recent review, see reference 11). The E1 and E2 enzymes are specific for each of the three multienzyme complexes and therefore specified as E1p and E2p in the PDHC, E1o and E2o in the OGDHC, and E1b, and E2b in the BCOADHC. In contrast, the LPD component is common in the three multienzyme complexes in most organisms (11, 42). Depending on the organism and the type of 2-oxoacid dehydrogenase complex, the E1 enzyme exists either as a homodimer (α2) or as a heterotetramer (α2β2), the subunits of both types showing only very weak similarity to each other and not being related (11). In all known OGDHCs and in the PDHCs of all gram-negative bacteria investigated, the E1 enzyme represents a homodimeric enzyme, whereas in all known BCOADHCs and in the PDHCs of gram-positive bacteria and eukaryotes, E1 represents a heterotetramer (17, 28, 37, 42). The known exceptions from this rule are Zymomonas mobilis and Thiobacillus ferrooxidans, both of which are gram-negatives and have been shown to possess heterotetrameric E1p enzymes (37).
The genes encoding the E1p enzymes from a number of different bacteria have been cloned and (functionally) characterized (reviewed in reference 37). They are designated either aceE or pdhA in the case of the homodimeric E1p enzyme or as pdhA(α) and pdhA(β) in the case of the heterotetrameric E1p α- and β-subunits, respectively. In most organisms, the gene(s) encoding the E1p components is clustered with the gene encoding the E2 subunit of the PDHC (i.e., depending on the organism, it is designated the aceF, pdhB, or pdhC gene [37]). The LPD gene(s) in most cases is located either together with the E1p or E2p gene or in the cluster of the odh genes, which encode the enzyme components of the OGDHC (24, 37, 53, 61, 72, 73).
Corynebacterium glutamicum is an aerobic, gram-positive organism that grows on a variety of sugars and organic acids and is widely used in the industrial production of amino acids, particularly l-glutamate and l-lysine (35). Due to its importance for the carbon flux distribution within metabolism and for the precursor supply for amino acid synthesis, the phosphoenolpyruvate-pyruvate node of C. glutamicum has been intensively studied and much attention has been focused on some of the enzymes involved (reviewed in reference 49). However, despite its crucial role, the PDHC of this organism has only scarcely been investigated at the molecular and structural levels. The activity of the PDHC has been detected in various strains of C. glutamicum (8, 9, 56, 59). The activity depends on the substrate pyruvate and the cofactors TPP and Mg2+ and is higher in the presence of either cysteine or dithiothreitol (59). According to enzyme measurements with cell extracts, the C. glutamicum PDHC is not subject to any significant regulation modulating its activity (8, 9, 64). This seems surprising in view of the complex regulation of PDHCs in other bacteria as well as in eukaryotes (5, 18, 20, 23, 58). Whereas the C. glutamicum E1p and E2p proteins and their genes have not been investigated so far, Schwinde et al. (56) cloned and analyzed a functional lpd gene from C. glutamicum and purified and biochemically characterized the corresponding LDP protein. The lpd gene is monocistronic and not clustered with the genes for other enzymes of the PDHC or of the OGDHC. However, as lpd-deficient mutants of C. glutamicum have not yet been generated and analyzed, it remains unclear whether the characterized LPD protein functions as the third subunit of the PDHC and/or of another 2-oxoacid dehydrogenase complex in C. glutamicum.
The genome sequence of C. glutamicum has recently been determined and annotated (GenBank accession numbers NC_003450 and BX927147) (26, 29, 62), and an open reading frame (cg2466) coding for a protein with significant similarity to the Escherichia coli E1p enzyme has been detected and accordingly designated the aceE gene (29). In the present study we describe the genetic and functional characterization of the C. glutamicum E1 enzyme of the PDHC. It represents the first example for a homodimeric E1p protein in a gram-positive bacterium. Furthermore, we performed sequence analyses of all E1 amino acid sequences available in public databases and investigated the distribution as well as the phylogeny of homodimeric and heterotetrameric E1p proteins in prokaryotes.
MATERIALS AND METHODS
Bacteria, plasmids, oligonucleotides, and culture conditions.
All bacterial strains and plasmids and their relevant characteristics and sources are given in Table 1. The oligonucleotides used and their sequences are also listed in Table 1. The minimal medium used for C. glutamicum has been described previously (14) and contained 1% (wt/vol) acetate, lactate, pyruvate, or 2% (wt/vol) glucose as the carbon and energy source. Tryptone-yeast extract (TY) medium (48) was used as the complex medium for C. glutamicum and E. coli. When appropriate, kanamycin (50 μg ml−1) was added to the medium. If not stated otherwise, C. glutamicum was grown aerobically at 30°C and E. coli was grown aerobically at 37°C as 60-ml cultures in 500-ml baffled Erlenmeyer flasks on a rotary shaker at 120 rpm.
TABLE 1.
Strains, plasmids and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Relevant characteristic(s) or sequence | Source/reference or purpose |
|---|---|---|
| Strains | ||
| E. coli DH5α | supE44 hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 21 |
| C. glutamicum | ||
| WT | WT strain ATCC 13032 | American Type Culture Collection |
| WT ΔaceE mutant | WT strain with deletion of aceE, encoding the E1 enzyme of the PDHC | This work |
| Plasmids | ||
| pK19mobsacB | Kmr, mobilizable (oriT), oriV | 50 |
| pK19mutaceE | pK19mobsacB carrying a 1,155-bp insert with a truncated aceE gene, shortened by 2,077 bp | This work |
| pET2 | Promoter probe vector carrying the promoterless cat gene, Kmr | 66 |
| pET-PaceE | pET2 containing a 409-bp insert of the aceE promoter region | This work |
| Oligonucleotides | ||
| delaceE1 | 5′-ACGCGTCGACCACCAAAAGGACATCAGACC-3′ | Primer for deletion in aceE, SalI site (underlined) |
| delaceE3 | 5′-GGATAGGTGATTGGAAGTTGGGCAAACGAAGCATGAGGTAACG-3′ | Primer for deletion in aceE, crossover overlap (italicized) |
| delaceE4.1 | 5′-CCAACTTCCAATCACCTATCCGGGCATCTACCTCTACTC-3′ | Primer for deletion in aceE, crossover overlap (italicized) |
| delaceE5.3 | 5′-CGCGGATCCGCGGGATTTATCTGTCCC-3′ | Primer for deletion in aceE, BamHI site (underlined) |
| aceEprom1 | 5′-ACGCGTCGACCACCAAAAGGACATCAGACC-3′ | Primer for amplification of the aceE promotor, SalI site (underlined) |
| aceEprom2 | 5′-CGCGGATCCACACCTCCTGTTGGAATG-3′ | Primer for amplification of the aceE promotor, BamHI site (underlined) |
| CM4 | 5′-GAAAATCTCGTCGAAGCTCG-3′ | 67 |
| CM5 | 5′-AAGCTCGGCGGATTTGTC-3′ | This work |
DNA preparation and transformation.
The isolation of chromosomal DNA and plasmids from C. glutamicum was performed as described previously (15). Plasmid isolation from E. coli was carried out according to the method of Birnboim (4). DNA transfer into C. glutamicum was performed by electroporation, and the recombinant strains were selected on LBBHIS agar plates containing kanamycin (15 μg ml−1) (65). Electroporation of E. coli was performed with competent cells according to the method of Dower et al. (12).
PCR techniques.
PCR experiments were performed in a Biometra personal cycler (Biotron, Göttingen, Germany). Amplification of DNA was carried out with Vent polymerase (New England Biolabs). Buffers and deoxynucleoside triphosphates were taken from MBI-Fermentas (St. Leon-Rot, Germany). Oligonucleotides (primers) were obtained from MWG-Biotech (Ebersberg, Germany). Cycling times and temperatures were chosen according to fragment length and primer constitution. PCR products were purified from agarose gels using the Nucleospin extract II kit (Macherey & Nagel, Düren, Germany).
DNA manipulation and Southern hybridization.
Restrictions enzymes, T4 DNA ligase, calf intestinal phosphatase, RNase A, proteinase K, and Taq polymerase were purchased from MBI-Fermentas and used according to the instructions of the manufacturer. After restriction digests, DNA was separated on agarose gels and purified with the Nucleospin extract II kit. DNA hybridization experiments were performed as previously described (15). An aceE-specific 580-bp DNA fragment was amplified from chromosomal DNA of wild-type (WT) C. glutamicum by PCR with the primers delaceE1 and delaceE3 and used as a probe. Labeling, hybridization, washing, and detection were conducted using the nonradioactive DNA labeling and detection kit and the instructions from Roche Diagnostics (Penzberg, Germany).
Construction of a C. glutamicum aceE deletion mutant.
Inactivation of the chromosomal aceE gene in C. glutamicum was performed as described previously (38), using crossover PCR and the suicide vector pK19mobsacB. aceE-specific DNA fragments were generated using the primer pairs delaceE1-delaceE3 and delaceE4.1-delaceE5.3. Fragment 1 covers 393 bp upstream of aceE and 187 bp of the 5′ end of aceE, and fragment 2 covers 547 bp of the 3′ end of aceE and 28 bp downstream of the aceE stop codon (see Fig. 2). The two fragments were purified, mixed in equal amounts, and subjected to crossover PCR using primers delaceE1 and delaceE5.3. The resulting fusion product (containing the aceE gene with a deletion of 2,077 bp) was digested with BamHI/SalI, ligated into the BamHI/SalI-restricted plasmid pK19mobsacB, and transformed into E. coli. The recombinant plasmid was isolated from E. coli and electroporated into WT C. glutamicum. By application of the method described by Schäfer et al. (50), the intact chromosomal aceE gene in WT C. glutamicum was replaced by the truncated aceE gene via homologous recombination (double crossover). The screening of the aceE mutants was done on LB agar plates containing 0.5% (wt/vol) glucose and 10% (wt/vol) sucrose. The replacement at the chromosomal locus was verified by PCR using primers delaceE1/delaceE5.3 and by Southern blot analysis. For the latter, a labeled aceE probe was hybridized to SalI-restricted and size-fractionated chromosomal DNA from WT C. glutamicum and the C. glutamicum ΔaceE mutant, resulting in one signal of about 8.3 kb with the DNA from the WT strain and one signal of about 6.2 kb with the DNA from the ΔaceE mutant.
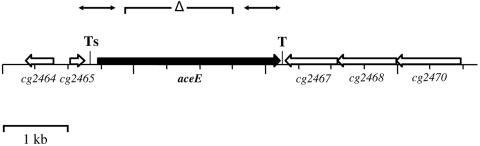
FIG. 2.
Genomic locus of the aceE gene in C. glutamicum. The arrows represent the coding regions of aceE and adjacent genes. The transcriptional start site and the terminator structure are indicated as Ts and T, respectively. The double-headed arrows indicate the fragments used for Southern and Northern blot hybridizations and for the construction of the aceE deletion mutant.
Cloning of the aceE promoter.
The aceE promoter fragment was amplified from chromosomal DNA of WT C. glutamicum by PCR with the primers aceEprom1 and aceEprom2. The PCR product was digested with SalI and BamHI, ligated into SalI/BamHI-restricted plasmid pET2, and transformed into E. coli. The recombinant plasmid pET-PaceE was then isolated from E. coli and introduced into C. glutamicum by electroporation.
RNA techniques.
For RNA isolation, C. glutamicum cells grown to the exponential phase (optical density at 600 nm of about 3) were treated with 1 volume of ice-cold killing buffer (20 mM Tris-HCl, pH 8.0, 20 mM NaN3, MgCl2), harvested, and resuspended in 100 μl ice-cold RNase-free water. The cell suspension was transferred to 2-ml screw-cap vials containing 500 mg glass beads (150 to 212 μm; Sigma), 0.5 ml RNeasy lysis buffer (QIAGEN, Hilden, Germany), and 0.5 ml acidic phenol (pH 5.5) and then mechanically disrupted by incubation three times for 45 s at 4°C in a RiboLyser (Hybaid, Heidelberg, Germany) at setting 6.5. After disruption, glass beads and cellular debris were removed by centrifugation (10,000 × g, 4°C). The supernatant was extracted with hot phenol as described by Schwinde et al. (55), and then the RNA was ethanol precipitated and dissolved in 400 μl RNase-free water. After treatment with 30 units RNase-free DNase (MBI Fermentas) for 15 min at 37°C, the sample was applied to an RNeasy Midi spin column (QIAGEN) and purified by following the instructions of the manufacturer. Aliquots of RNA were stored at −70°C until use.
For Northern (RNA) hybridization, a digoxigenin-dUTP-labeled aceE-specific 580-bp DNA probe was generated as described above. For hybridization, 10 μg of total RNA from WT C. glutamicum was separated on an agarose gel containing 17% (vol/vol) formaldehyde and transferred onto a nylon membrane (15). Hybridization (at 50°C, in the presence of 50% formamide, vol/vol), washing, and detection were carried out using a nucleic acid detection kit according to the instructions from Roche Diagnostics (Penzberg, Germany). The size marker was the 0.24- to 9.5-kb RNA ladder from GibcoBRL.
Primer extension reactions were carried out as described previously (30) with IRD800-labeled primers CM4 and CM5 and RNA from C. glutamicum (pET-PaceE). The IRD800-labeled primers were obtained from MWG Biotech. Primers CM4 and CM5 are complementary to regions from nucleotides (nt) 45 to 64 and 34 to 51, respectively, downstream of the BamHI site of the multiple cloning site in plasmid pET2. Primer extension products were analyzed with an automatic sequencer (LI-COR 4000L; Licor, Inc.) using a 6% (wt/vol) polyacrylamide gel at 1,500 V and 50°C. For the exact localization of the transcriptional start site, sequencing reactions using plasmid pET-PaceE and the same oligonucleotide used for the respective primer extension reaction were coelectrophoresed.
Enzyme assays.
To determine PDHC activity in cell extracts, C. glutamicum cells were harvested in the exponential growth phase (optical density at 600 nm of about 8), washed twice in 100 mM Tris-HCl, pH 7.2, 3 mM l-cysteine, 10 mM MgCl2, and resuspended in 0.5 ml of the same buffer. The cell suspension was transferred to 2-ml screw-cap vials together with 250 mg glass beads (150 to 212 μm; Sigma) and subjected five times for 30 s to mechanical disruption with a RiboLyser at 4°C, with intermittent cooling on ice for 2 min. After disruption, the glass beads and the cellular debris were removed by centrifugation (10,000 × g, 4°C, 15 min) and the supernatant was used for the assay. The protein concentration was determined by a bicinchoninic acid protein assay reagent kit (Pierce, Bonn, Germany) with bovine serum albumin as the standard. PDHC enzyme activities were determined photometrically according to the method described by Guest and Creaghan (19). One unit of activity is defined as 1 μmol NADH formed per min at 30°C.
For the determination of chloramphenicol acetyltransferase (CAT) activity, crude extracts were prepared as described above, except that cells were washed twice in 50 mM Tris-HCl, pH 7, and resuspended in 0.5 ml of the same buffer containing 10 mM MgCl2, 1 mM EDTA, and 30% (vol/vol) glycerol. CAT activity was assayed photometrically at 412 nm as described by Shaw (57) in 1 ml 100 mM Tris-HCl, pH 7.8, 1 mM 5,5′-dithiobis-2-nitrobenzoic acid, 0.1 mM acetyl-CoA, and 0.25 mM chloramphenicol. One unit of CAT activity is defined as 1 μmol chloramphenicol acetylated per min at 37°C.
For the determination of E1p activity, crude extracts were prepared as described above, except that cells were washed twice in 50 mM Tris-HCl, pH 7.5, 10% (vol/vol) glycerol, and resuspended in 0.5 ml of the same buffer. E1p activity was assayed photometrically according to the method described by Schwartz and Reed (54). One unit of activity corresponds to 2 μmol ferricyanide reduced per min at 30°C.
Computational analysis.
MFold (75) was the software used for the calculation of the ΔGo′ value (free energy under standard conditions) of the aceE terminator structure. Databank searches and alignments were carried out by using BLAST and CLUSTALW (2, 63). Phylogenetic trees were generated using the neighbor-joining method (46), including 100 bootstrap replicates. The accession numbers for all homodimeric E1p and heterotetrameric E1pα and E1bα sequences used for the phylogenetic analysis were as follows. The homodimeric E1p sequences were Acinetobacter sp. strain ADP1 (YP_047975), Actinobacillus pleuropneumoniae (ZP_00134360), Anopheles gambiae (EAL42079), Azotobacter vinelandii (ZP_00342533), Blochmannia floridanus (NP_878459), Bordetella parapertussis (NP_883760), Buchnera aphidicola (NP_660552), Burkholderia cepacia (ZP_00212749), Chromobacterium violaceum (AAQ58203), Corynebacterium glutamicum (CAF20589), Coxiella burnetii (NP_819497), Dechloromonas aromatica (ZP_00150166), Deinococcus radiodurans (NP_293980), Erwinia carotovora (YP_051878), Escherichia coli (NP_414656), Haemophilus influenzae (ZP_00154971), Legionella pneumophila (YP_126868), Leifsonia xyli (YP_062215), Mannheimia succiniciproducens (YP_088528), Methylobacillus flagellatus (ZP_00172315), Methylococcus capsulatus (YP_115388), Microbulbifer degradans (ZP_00315287), Mycobacterium tuberculosis (NP_336771), Neisseria meningitidis (NP_274360), Nitrosomonas europaea (NP_840448), Nocardia farcinica (YP_117823), Pasteurella multocida (NP_245832), Photobacterium profundum (YP_131304), Photorhabdus luminescens (CAE15996), Polaromonas sp. strain JS666 (ZP_00362465), Propionibacterium acnes (YP_055700), Pseudomonas aeruginosa (NP_253702), Psychrobacter sp. strain 273-4 (ZP_00146054), Ralstonia metallidurans (ZP_00271472), Rhodopirellula baltica (NP_865502), Rubrivivax gelatinosus (ZP_00245303), Salmonella enterica serovar Typhimurium (NP_459157), Shewanella oneidensis (NP_716061), Shigella flexneri (NP_706068), Streptomyces coelicolor (NP_626618), Thermus thermophilus (YP_005770), Tropheryma whipplei (NP_789444), Vibrio cholerae (AAF95557), Wautersia eutropha (AAA21598), Xanthomonas axonopodis (NP_640929), Xylella fastidiosa (NP_297959), and Yersinia pestis (NP_406881). The heterotetrameric E1pα sequences were Acholeplasma laidlawii (P35485), Agrobacterium tumefaciens (NP_532119), Arabidopsis thaliana (AAB86803), Azorhizobium caulinodans (AAG38097), Bacillus subtilis (DEBSPA), Bartonella henselae (YP_033409), Bradyrhizobium japonicum (NP_771423), Brucella melitensis (NP_539771), Candida albicans (EAK96452), Caulobacter crescentus (NP_420534), Coxiella burnetii (NP_819670), Cyanidium caldarium (AAF12897), Cytophaga hutchinsonii (ZP_00308483), Danio rerio (NP_001002399), Enterococcus faecalis (AAO81144), Exiguobacterium sp. (ZP_00182969), Geobacter sulfurreducens (NP_953699), Gloeobacter violaceus (NP_924475), Gracilaria tenuistipitata (YP_063628), Haloferax volcanii (AAD34202), Lactobacillus plantarum (NP_785659), Lactococcus lactis (NP_266218), Leuconostoc mesenteroides (ZP_00062839), Listeria monocytogenes (NP_464577), Magnetospirillum magnetotacticum (ZP_00208699), Mesorhizobium sp. (ZP_00196269), Methylobacterium extorquens (AAN03811), Mus musculus (NP_032837), Mycoplasma capricolum (AAC44342), Nostoc sp. (NP_486748), Novosphingobium aromaticivorans (ZP_00303573), Oceanobacillus iheyensis (NP_692333), Oenococcus oeni (ZP_00320046), Pediococcus pentosaceus (ZP_00323580), Porphyra purpurea (P51267), Prochlorococcus marinus (NP_875753), Rattus norvegicus (NP_446446), Rhodobacter sphaeroides (ZP_00007453), Rhodopseudomonas palustris (NP_948208), Rhodospirillum rubrum (ZP_00268857), Rickettsia rickettsii (ZP_00153395), Saccharomyces cerevisiae (NP_011105), Silicibacter sp. (ZP_00339083), Sinorhizobium meliloti (NP_385551), Staphylococcus aureus (YP_040480), Synechococcus elongatus (YP_172860), Synechocystis sp. (BAA18592), Thermosynechococcus elongatus (BAC08721), Thermus thermophilus (YP_144205), Trichodesmium erythraeum (ZP_00327615), and Zymomonas mobilis (AAC70361). The BCOADHC E1α sequences were B. subtilis (C69593), L. monocytogenes (NP_464897), M. musculus (NP_031559), O. iheyensis (NP_692787), Pseudomonas putida (P09060), Streptomyces avermitilis (AAA66072), and T. thermophilus (1UMD_C).
RESULTS
PDHC activity in C. glutamicum.
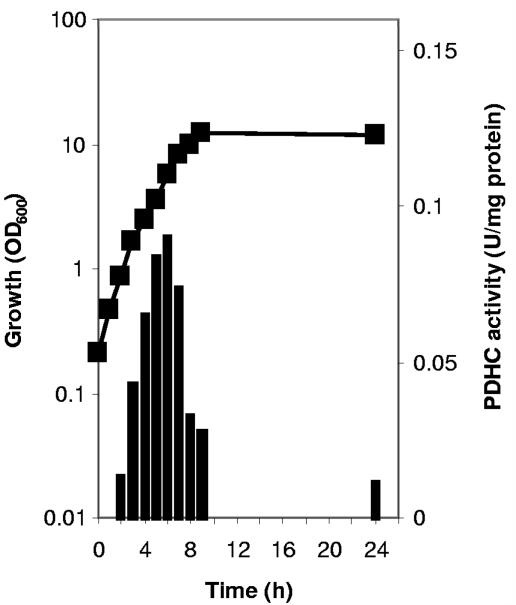
The specific activity of the PDHC was determined in cell extracts of WT C. glutamicum grown in TY complex medium and in minimal medium containing glucose or acetate as the carbon source and harvested at the mid-exponential growth phase. The highest specific activity (0.087 U mg protein−1) was found in cells grown in TY medium (Table 2). The activity was about two- to threefold lower when the cells were grown in media containing glucose or acetate. The dependency of the PDHC activity on the growth phase was elucidated by measuring the specific activity in cell extracts from WT C. glutamicum grown to the stationary growth phase in TY medium and harvested in 2-hour intervals (Fig. 1). The highest specific activity was found when the cells were harvested in the mid- to late exponential growth phase; much lower activities were found in cells of the stationary phase. These results suggest that the PDHC in C. glutamicum is weakly regulated by the growth medium and is significantly regulated dependent on the growth phase. In view of the central position of the PDHC in the metabolism, this regulation might be significant for the carbon flux in C. glutamicum.
TABLE 2.
Specific activities of the PDHC in cell extracts of WT C. glutamicum and the C. glutamicum ΔaceE mutant grown on different mediaa
| Medium | Sp act of PDHC (U/mg protein)b
|
|
|---|---|---|
| C. glutamicum | C. glutamicum ΔaceE mutant | |
| TY | 0.087 | ND |
| TY + glucose | 0.029 | ND |
| TY + acetate | 0.032 | <0.001 |
| MM + glucose | 0.036 | NG |
| MM + acetate | 0.034 | <0.001 |
| MM + glucose + acetate | 0.034 | <0.001 |
TY medium without and with 1% glucose or 0.5% acetate; minimal medium (MM) containing 2% glucose, 1% acetate, or 1% glucose plus 1% acetate as the carbon source. The cells were harvested in the mid-exponential growth phase.
The values are means obtained from at least two independent cultivations and two determinations per experiment. The standard deviations were in all cases below 10%. ND, not determined; NG, no growth.
FIG. 1.
Growth (black squares) and pyruvate dehydrogenase complex (PDHC) activity (black bars) of WT C. glutamicum grown on TY complex medium. The standard deviations of the enzyme determinations were below 10%. OD600, optical density at 600 nm.
The analysis of the PDHC activity in extracts of WT C. glutamicum revealed an apparent Km value of about 1.7 mM for pyruvate, which is about twofold higher than that (0.8 mM) reported by Shiio et al. (59). In accordance with these authors, the WT C. glutamicum enzyme required Mg2+ and TPP for maximal activity.
Nucleotide sequence of the C. glutamcium aceE gene and analysis of the deduced E1p amino acid sequence.
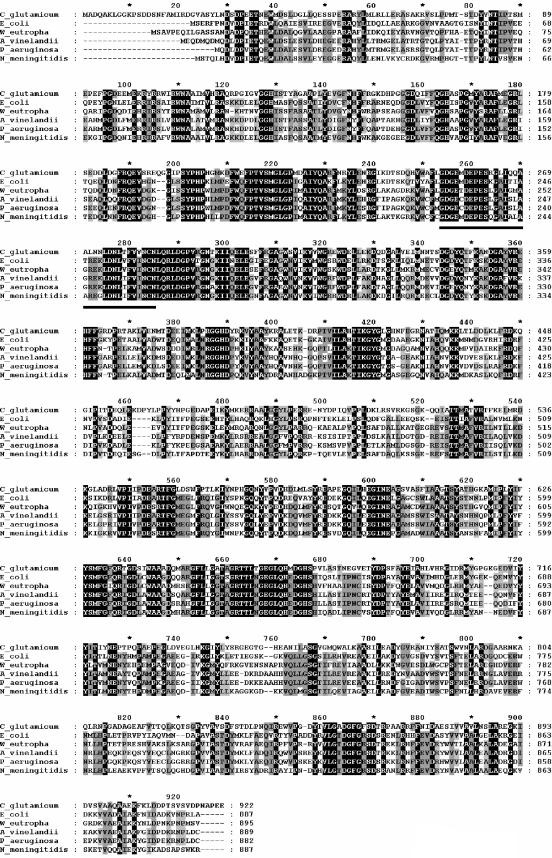
Due to sequence similarity to the E. coli aceE gene, the cg2466 gene of the C. glutamicum genome has recently been annotated as an aceE gene, putatively coding for the E1 enzyme of the C. glutamicum PDHC (29). The aceE gene of C. glutamcium consists of 2,766 bp and is preceded by a typical ribosomal binding site (AGGAGG). Centered 38 bp downstream of the aceE stop codon, a region of dyad symmetry followed by several T residues similar to rho-independent transcription terminators was found. The mRNA hairpin loop predicted from this sequence has a ΔGo′ value of −24.0 kcal/mol at 25°C. This result indicates transcriptional termination downstream of the aceE gene. According to the C. glutamicum genome sequence (GenBank accession numbers NC_003450 and BX927147 [26, 29]), three divergently orientated open reading frames, cg2467, cg2468, and cg2470, are located downstream of the aceE gene (Fig. 2). The deduced amino acid sequences of these open reading frames show significant similarities to components of ABC transporters (ATP binding protein, permease, and substrate binding protein, respectively). Two open reading frames (cg2464 and cg2465) were identified in the region up to 1,000 bp upstream of the aceE gene, both annotated as coding for hypothetical (unknown) proteins. According to the nucleotide sequence, the C. glutamicum aceE gene encodes a polypeptide of 922 amino acids with a predicted molecular mass of 102.8 kDa. Alignment studies revealed the 49% to 51% identity of the C. glutamicum protein to the functionally well-characterized homodimeric E1 enzymes of PDHCs from the gram-negatives E. coli, Wautersia (Ralstonia) eutropha, Azotobacter vinelandii, Pseudomonas aeruginosa, and Neisseria meningitidis (Fig. 3). The alignment of the C. glutamicum enzyme with the homodimeric E1p proteins revealed highly conserved residues and sequence motifs, especially in those regions suggested to be essential for cofactor binding and catalytic activity (3). These motifs include the TPP binding motif GDG…X26…NCN (residues 253 to 284 in the C. glutamicum sequence) located in the N-terminal part of all homodimeric E1p enzymes, binding sites for divalent cations (Asp254, Asn284, and Gln286), and several residues proposed to be involved in cofactor binding and catalysis (His128, His164, His668, Glu550, Tyr201, and Asp549) (Fig. 3). Surprisingly, the databank searches of the C. glutamicum genome revealed no putative proteins showing similarity to the E1α or E1β subunits of heterotetrameric E1p enzymes, generally assumed to be typical for gram-positive bacteria (11, 17, 37). In summary, sequence analysis of the cg2466 gene suggests the presence of a homodimeric E1p enzyme in the PDHC of gram-positive C. glutamicum.
FIG.3.
Amino acid sequence alignment of the E1p enzymes of C. glutamicum (accession number CAF20589), E. coli (NP_414656), Wautersia eutropha (AAA21598), Azotobacter vinelandii (ZP_00342533), Pseudomonas aeruginosa (NP_253702), and Neisseria meningitidis (NP_274360). Amino acids identical in all sequences are shaded in black, and amino acids identical in at least five sequences are shaded in gray. The TPP signature motif is underlined.
Inactivation of the chromosomal aceE gene.
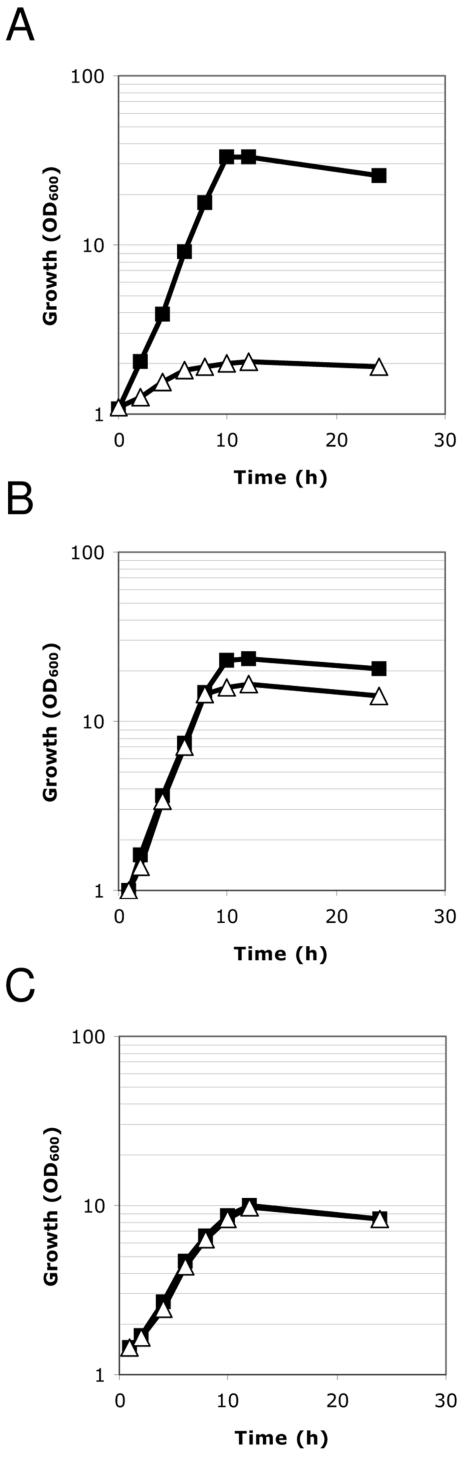
To study whether C. glutamicum requires the aceE gene for growth and for PDHC activity, the chromosomal aceE locus was partially deleted and the resulting strain, the C. glutamicum ΔaceE mutant, was tested for growth in different media and for PDHC activity. The growth of the C. glutamicum ΔaceE mutant was negligible in TY medium, but the strain grew as well as WT C. glutamicum on TY medium supplemented with acetate. Furthermore, the C. glutamicum ΔaceE mutant was unable to grow in minimal medium with glucose, pyruvate, or lactate as the sole carbon source but grew almost identically to WT C. glutamicum in minimal medium containing glucose plus acetate as carbon sources (Fig. 4). In accordance with these findings, the growth of the C. glutamicum ΔaceE mutant was indistinguishable from that of the WT strain in minimal medium containing acetate as the sole carbon source (Fig. 4). These results indicate that the aceE gene is essential for the growth of C. glutamicum in minimal medium containing glucose or other substrates entering the central metabolism as intermediates of glycolysis but not for growth in minimal medium with acetate as the carbon and energy source.
FIG. 4.
Growth of WT C. glutamicum (filled squares) and the C. glutamicum ΔaceE mutant (open triangles) on minimal medium containing 2% glucose (A), 1% glucose plus 1% acetate (B), and 1% acetate (C). OD600, optical density at 600 nm.
To investigate whether the aceE gene in fact encodes the E1 enzyme of the PDHC in C. glutamicum, both the specific PDHC and E1p activities were determined in cell extracts of WT C. glutamicum and the C. glutamicum ΔaceE mutant grown in complex and minimal media containing acetate. As shown in Table 2, the C. glutamicum ΔaceE mutant was devoid of any detectable PDHC activity (<0.01 U mg protein−1), whereas the WT strain showed 0.032 U mg protein−1 and 0.034 U mg protein−1, respectively. Furthermore, no E1p activitiy (<0.01 U mg protein−1) was detected in extracts of C. glutamicum ΔaceE mutant cells grown in TY medium supplemented with acetate (0.5%, wt/vol), whereas an activity of 0.017 U mg protein−1 was measured in extracts of WT C. glutamicum. These findings indicate (i) that the aceE gene in fact encodes the E1 enzyme of the PDHC in C. glutamicum and (ii) that there are no isoenzymes for the E1p protein in C. glutamicum. Furthermore, our findings represent the first example of a homodimeric E1 enzyme of a PDHC in a gram-positive bacterium.
Transcriptional analysis of the aceE gene.
Northern (RNA) hybridization experiments were performed in order to analyze the size of the aceE transcript. For this purpose, total RNA from WT C. glutamicum was isolated, size-fractionated, transferred onto a nylon membrane, and hybridized to an aceE-specific digoxigenin-UTP-labeled DNA probe. The hybridization revealed a signal at about 2.9 kb (data not shown), which corresponds well to the size of the aceE gene (2.8 kb). This result indicates that the C. glutamicum aceE transcript is monocistronic.
To confirm the presence of a promoter and to investigate transcriptional regulation of the aceE gene, a transcriptional fusion between the putative aceE promoter region and the promoterless chloramphenicol acetyltransferase (CAT) gene was constructed in the promoter probe vector pET2. The resulting plasmid, pET2-PaceE, was transformed into WT C. glutamicum and CAT activity was determined in the plasmid-carrying strain during growth in TY medium and in minimal medium containing glucose or acetate as the carbon source. Whereas C. glutamicum carrying the host plasmid pET2 showed no CAT activity (<0.01 U mg protein−1) on either medium, the strain carrying pET2-PaceE showed 0.92 U mg protein−1 during growth in TY medium and approximately 1.5-fold-lower CAT activity during growth in minimal medium containing glucose or acetate (0.61 and 0.58 U mg protein−1, respectively). These results confirm the presence of a promoter immediately upstream of the aceE gene and indicate weak transcriptional control of the aceE gene by the carbon source in the growth medium.
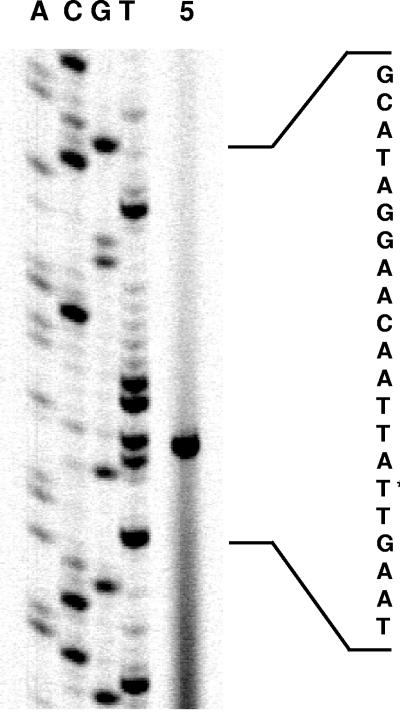
To identify the transcriptional initiation site of the C. glutamcium aceE gene and to localize the aceE promoter, primer extension experiments were performed. Using oligonucleotide CM4 and 50 μg total RNA isolated from C. glutamicum pET2-PaceE, a major signal corresponding to an A residue 121 nucleotides upstream of the aceE translational start was obtained (Fig. 5). This result was confirmed in an independent experiment with primer CM5 (data not shown). At an appropriate distance (8 bp) upstream of the transcriptional initiation site of the aceE gene, a sequence, TATCCT, with significant similarity to the consensus −10 region (TAT/CAAT) of C. glutamicum promoters (40, 41) is present. No apparent −35 region (TTGCCA) can be recognized, which is a common feature of C. glutamicum promoters (40, 41). In addition to the main signal at position −121, with both primer CM4 and primer CM5 we obtained a less prominent primer extension signal which corresponded to an A residue 54 nucleotides upstream of the aceE translational start site. However, analysis of the DNA sequence upstream of this site revealed no motifs which are similar to the consensus −10 and −35 regions of C. glutamicum vegetative promoters.
FIG. 5.
Primer extension analysis of the transcriptional start site in front of the aceE gene. The primer extension product is shown in lane 5. Lanes A, C, G, and T represent the products of sequencing reactions with the same primer used for the primer extension reaction. The relevant DNA sequence (coding strand) is shown on the right, and the transcriptional start site is indicated by an asterisk.
Sequence comparisons and phylogenetic analyses.
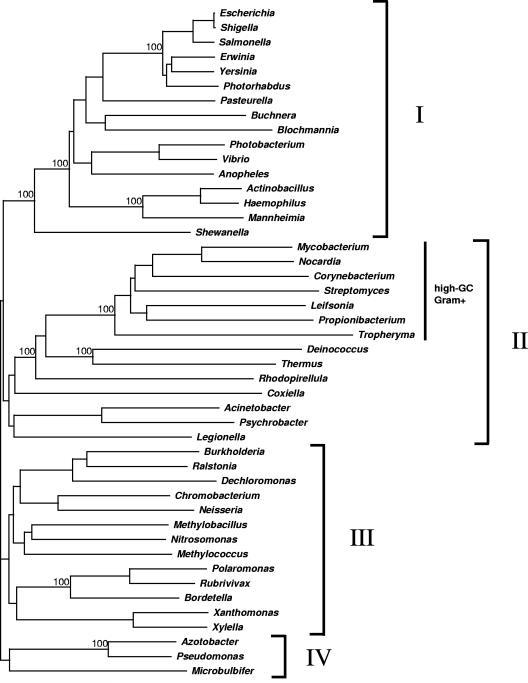
Databank searches with the amino acid sequence of homodimeric E1p of C. glutamicum revealed, aside from similarities to the functionally proven E1p proteins of different gram-negatives (Fig. 3), significant levels of identity (≥40%) to putative or hypothetical E1p proteins from other bacteria, e.g., from “high-GC-content gram-positives,” deinococci, planctomycetes, several proteobacteria, and, surprisingly, the eukaryotic Anopheles gambiae (Table 3). To elucidate the phylogeny of the homodimeric E1p protein family, a phylogenetic tree based on comparison of the deduced amino acid sequences of 46 E1p genes currently available in public databases was constructed. As shown in Fig. 6, the phylogenetic tree of homodimeric E1p proteins contains four major clusters. Clusters I and IV predominantly entail sequences of the γ-proteobacteria, whereas cluster II comprises all sequences of the high-GC-content gram-positives, deinococci, and planctomycetes and several sequences of γ-proteobacteria and cluster III contains mainly proteins of the β-proteobacteria.
TABLE 3.
Protein sequence identities among representative homodimeric E1p enzymes
| Organisma | % Identity
|
|
|---|---|---|
| Escherichia coli | Corynebacterium glutamicum | |
| Corynebacterium glutamicum | 49 | |
| Streptomyces coelicolor | 47 | 61 |
| Mycobacterium tuberculosis | 49 | 66 |
| Propionibacterium acnes | 50 | 63 |
| Deinococcus radiodurans | 48 | 47 |
| Thermus thermophilus | 52 | 52 |
| Rhodopirellula baltica | 55 | 51 |
| Escherichia coli | 49 | |
| Vibrio cholerae | 72 | 51 |
| Burkholderia cepacia | 59 | 50 |
| Coxiella burnetii | 51 | 43 |
| Chromobacterium violaceum | 62 | 52 |
| Anopheles gambiae | 75 | 52 |
For accession numbers, see Materials and Methods.
FIG. 6.
Phylogenetic tree of the homodimeric E1 protein family. Numbers at the nodes give the bootstrap values. Organisms are listed in Materials and Methods.
To elucidate also the distribution of heterotetrameric E1p proteins in bacteria, databank searches were carried out using the functionally proven E1p α-subunits of Z. mobilis (37) and Bacillus subtilis (24) (Table 4). The E1p α-subunits of Z. mobilis showed highest similarity (≥40% identity) to putative proteins of many α-proteobacteria, sphingobacteria, and several eukaryotes, e.g., Mus musculus, Rattus norvegicus, and Saccharomyces cerevisiae. In contrast, the B. subtilis protein showed only relatively weak similarity to the Z. mobilis enzyme, but high levels of identity (≥40%) to the described E1p α-subunits from Acholeplasma laidlawii (68) and to putative E1p α-subunits from several low-GC-content gram-positives, γ-proteobacteria, deinococci, and the archaeon Haloferax volcanii. Surprisingly, the B. subtilis E1p α-subunit showed only weak similarity to the functionally proven protein of the low-GC-content gram-positive bacterium Mycoplasma capricolum (74).
TABLE 4.
Protein sequences identities among representative heterotetrameric E1p α-subunits
| Organisma | % Identity
|
|
|---|---|---|
| Zymomonas mobilis | Bacillus subtilis | |
| Zymomonas mobilis | 33 | |
| Rickettsia rickettsii | 53 | 29 |
| Caulobacter crescentus | 59 | 30 |
| Bradyrhizobium japonicum | 59 | 30 |
| Agrobacterium tumefaciens | 58 | 32 |
| Cytophaga hutchinsonii | 47 | 28 |
| Mus musculus | 49 | 26 |
| Rattus norvegicus | 51 | 27 |
| Saccharomyces cerevisiae | 49 | 28 |
| Acholeplasma laidlawii | 33 | 42 |
| Listeria monocytogenes | 28 | 74 |
| Oceanobacillus iheyensis | 30 | 72 |
| Lactobacillus plantarum | 29 | 53 |
| Bacillus subtilis | 33 | |
| Mycoplasma capricolum | 29 | 32 |
| Geobacter sulfurreducens | 28 | 44 |
| Coxiella burnetii | 30 | 40 |
| Thermus thermophilus | 33 | 42 |
| Haloferax volcanii | 29 | 41 |
| Synechocystis sp. | 39 | 27 |
| Prochlorococcus marinus | 43 | 30 |
| Synechococcus elongatus | 42 | 28 |
| Arabidopsis thaliana | 43 | 29 |
For accession numbers, see Materials and Methods.
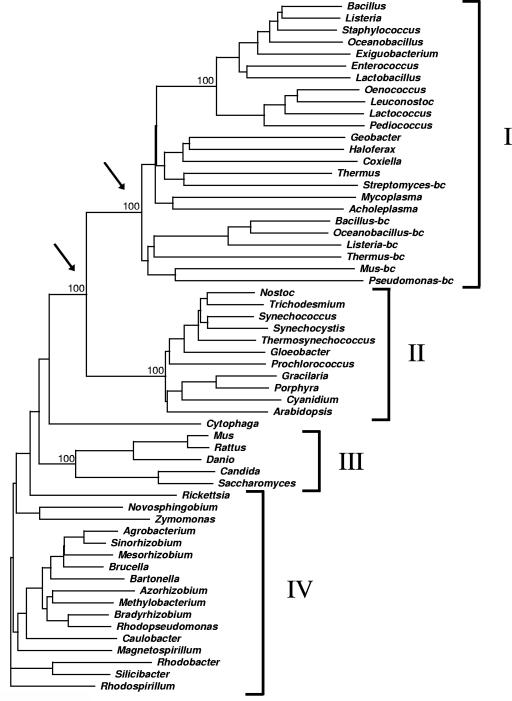
A phylogenetic tree of the heterotetrameric E1p protein family was constructed using the deduced amino acid sequences of 58 E1p α-subunits deposited in public databases. As depicted in Fig. 7, this tree contains four major clusters broadly corresponding to the low-GC-content gram-positive bacteria (cluster I), cyanobacteria, and chloroplasts (cluster II), the mitochondria (cluster III), and the α-proteobacteria (cluster IV).
FIG. 7.
Phylogenetic tree of the heterotetrameric E1α protein family. Numbers at the nodes give the bootstrap values. Organisms are listed in Materials and Methods. Arrows denote proposed gene duplications. Abbreviation: -bc, E1 α-subunit of a branched-chain 2-oxoacid dehydrogenase complex.
Surprisingly, databank searches and sequence comparisons revealed that the α-subunits of the known heterotetrameric E1p proteins of Z. mobilis, B. subtilis, M. capricolum, A. laidlawii, and Synechocystis sp. show significant similarity (28 to 42% identity) to the known heterotetrameric E1 α-subunits of the BCOADHC from B. subtilis (69), S. avermitilis (60), P. putida (6), T. thermophilus (36), and M. musculus (10). All these BCOADHC E1 α-proteins cluster with the PDHC E1 α-proteins of the low-GC-content gram-positive bacteria (Fig. 7, cluster I).
In summary, the sequence comparisons and phylogenetic studies suggest that (i) homodimeric E1p proteins are typical for the PDHCs of β- and γ-proteobacteria, the deinococci, and the high-GC-content gram-positive bacteria and (ii) heterotetrameric E1p proteins are typical for the PDHCs of the α-proteobacteria, the low-GC-content gram-positive bacteria, and the cyanobacteria.
DISCUSSION
It has generally been assumed that aerobic gram-negative bacteria possess homodimeric and that gram-positive bacteria possess heterotetrameric E1p enzymes in their PDHCs. The present study describes for the first time the genetic and functional characterization of an E1p enzyme of the homodimeric type from a gram-positive bacterium, i.e., from C. glutamicum.
DNA sequence analysis of the C. glutamicum E1p gene (aceE) and adjacent open reading frames and comparisons with the respective gene loci in other bacteria highlighted some differences between the chromosomal organization of the C. glutamicum aceE locus and that of functionally proven aceE (pdhA) genes in other microorganisms. Except in Z. mobilis, the genes for the (homodimeric and heterotetrameric) E1p and the E2p enzymes of the PDHCs are clustered in the genomes of all bacteria studied so far (37). Additionally, a gene encoding an LPD can be found downstream of the E1p and E2p genes in some of these bacteria (37). A functional lpd gene (cg0441) putatively coding for the LPD enzyme of the PDHC has been identified in the C. glutamicum genome; however, it is not located in the region of the aceE locus and it is not clustered with genes for other subunits of the PDHC or the OGDHC (13, 56). Other genes (i.e., cg2194, cg0790, and cg3339) coding for proteins with some similarity (24 to 29% identity) to LPD proteins from other bacteria are also not clustered with the aceE gene or with a gene possibly coding for an E2 protein. A single gene (cg2421 or NCgl2126) for a protein with homology to the E2 subunits of PDHCs or OGDHCs can be found in the C. glutamicum genome (29); however, so far this gene has not been studied and it remains to be proven whether it in fact represents a functional E2 gene. Thus, although a functional E2p gene (aceF or pdhB) so far has not yet been identified in C. glutamicum, it is obvious from sequence analyses up- and downstream of the aceE gene that in this organism the genes encoding the E1 and E2 enzymes of the PDHC are also not clustered. In E. coli, the structural genes for the PDHC are transcribed in a single operon, together with an upstream gene encoding the regulatory protein PdhR (44). This regulator negatively controls the expression of the whole pdh operon, probably with pyruvate serving as an inducing effector (44). The C. glutamicum aceE gene is expressed as an independent single cistron, and from genome and sequence analyses, there is no indication for a PdhR homologue. All these findings indicate that the chromosomal aceE locus in C. glutamicum does not resemble any of the aceE or pdhA loci analyzed so far from other organisms and that the transcriptional organization in this organism is different.
The enzymatic characterization of the defined C. glutamicum ΔaceE mutant unequivocally demonstrated that the aceE gene in fact codes for the E1 subunit of the PDHC. Furthermore, the analysis revealed that the PDHC is essential for the growth of C. glutamicum on minimal medium containing glucose, pyruvate, or lactate as the carbon and energy source. These findings indicate that under the conditions tested, the PDHC is the main enzyme responsible for providing acetyl-CoA during the growth of C. glutamicum on substrates or intermediates that do not enter the central metabolism via acetyl-CoA. In contrast, the PDHC is dispensable for the growth of C. glutamicum on minimal medium containing acetate as the sole or as an additional carbon and energy source. This substrate is activated in C. glutamicum to acetyl-CoA by acetate kinase and phosphotransacetylase (45, 71) and then, independently of the PDHC, channeled into the tricarboxylic acid cycle. In accordance with our results, the known PDHC-negative mutants of E. coli, Pseudomonas aeruginosa, and Salmonella enterica serovar Typhimurium also show an acetate-auxotrophic phenotype (1, 25, 27, 33). Very recently, we identified, purified, and characterized a pyruvate:quinone oxidoreductase (PQO) in C. glutamicum (52). This enzyme is a homotetrameric flavoprotein containing TPP, and it catalyzes the oxidative decarboxylation of pyruvate to acetate and CO2, probably using a menaquinone as the physiological electron acceptor. Using 2,6-dichloroindophenol as an artificial electron acceptor, the PQO activity was highest (about 0.055 U/mg of protein) in C. glutamicum cells grown on complex medium and about threefold lower when glucose was added to the complex medium or when the cells were grown in minimal medium containing different carbon sources (52). The PQO thus represents an additional pyruvate-decarboxylating enzyme in C. glutamicum, and we therefore speculated that the PQO reaction, together with the reactions of the acetate kinase and the phosphotransacetylase, might be able to bypass the PDHC reaction (52). However, the inability of the PDHC-negative C. glutamicum mutant to grow on minimal medium containing glucose indicated that the PQO is not able to compensate for a PDHC under the conditions generally used to cultivate C. glutamicum. The inability to replace the PDHC might be due to the relatively low affinities of the PQO for pyruvate (Km = 30 mM [52]) and/or of the acetate kinase for acetate (Km = 7.9 mM [45]). However, in nature or under different culture conditions there might be conditions when the intracellular pyruvate concentration in C. glutamicum rises to values well above the Km value, and then the PQO might substitute for the PDHC.
Although the PDHC has a crucial role within the metabolism of C. glutamicum, relatively little effort has been devoted to the regulation of this enzyme complex on the genetic level. The analysis of the specific PDHC activities in extracts of cells grown on different media indicated that the synthesis or the assembly of the PDHC in C. glutamicum is only very weakly regulated by the carbon source in the growth medium. In accordance with our findings, enzyme measurements with extracts of C. glutamicum cells cultivated in a chemostat at various dilution rates on minimal media containing glucose, pyruvate, or lactate revealed no significant differences in the specific activities under all conditions applied (8, 9, 64). These results suggested that the PDHC is constitutively, and with about the same specific activity, present in C. glutamicum cells grown in minimal medium containing different carbon sources. This is in contrast to the situation with E. coli, where two- to fourfold-higher activity and about-twofold-lower activity was observed in minimal medium containing pyruvate and acetate, respectively, than in minimal medium containing glucose (34, 43, 44). However, under batch culture conditions, we found the specific PDHC activity in C. glutamicum to be significantly dependent on the growth phase. The low PDHC activity at the beginning and at the end of growth might be explained by an ineffective or unbalanced assembly of the multienzyme complex or by low de novo synthesis of one or more enzymes of the PDHC in the respective growth phases. However, the transcriptional-fusion experiments showed no correlation between the specific PDHC activity and the aceE promoter activity in the course of the growth. Thus, it seems likely that the amount of functional PDHC in C. glutamicum depends on the expression of the gene(s) encoding E2p and/or the LPD and, thus, on the availability of these enzymes. However, this hypothesis and the nature and mechanism of the genetic (co)control of the E1p, E2p, and the LPD genes in C. glutamicum still have to be elucidated.
Our compilation of E1 sequences deposited in public databases suggests that the distribution of homodimeric and heterotetrameric E1p subunits in bacteria is not in accordance with the rRNA-based phylogeny of bacteria and, instead, is much more heterogeneous than previously described (11, 17, 37). The analyses indicate that either homodimeric or heterotetrameric E1p enzymes are present in a given bacterial group; e.g., homodimeric enzymes are found mainly in β- and γ-proteobacteria, in high-GC-content gram-positive bacteria, and in deinococci, whereas heterotetrameric enzymes are found predominantly in α-proteobacteria, cyanobacteria, and the low-GC-content gram-positive bacteria. A simple explanation for this heterogeneous distribution of the two E1p types is that homodimeric and heterotetrameric E1 progenitors have existed in ancestral bacteria and that later, in the process of diversification, the enzymes in different bacterial groups acquired specialized (e.g., pyruvate-specific) functions. This hypothesis would explain (i) that homodimeric and heterotetrameric E1p proteins show no obvious sequence similarity (11, 17), (ii) that both E1 types possess completely different sequence motifs important for catalysis (17), and (iii) that some bacteria (e.g., C. burnetii and T. thermophilus) (Fig. 6 and 7) possess genes for proteins with significant similarity to both homodimeric and heterotetrameric E1p enzymes.
Analysis of the phylogeny of the homodimeric E1p family (Fig. 6) reveals that the cluster comprising the enzymes of the high-GC-content gram-positive bacteria and deinococci also contains several enzymes of γ-proteobacteria, e.g., Legionella and Acinetobacter (cluster II). Thus, those proteins are probably more closely related to the proteins of the high-GC-content gram-positive bacteria and deinococci than to those of other γ-proteobacteria, e.g., E. coli or Haemophilus. Furthermore, the homodimeric E1p proteins of the γ-proteobacteria Azotobacter, Microbulbifer, and Pseudomonas are located in a completely independent cluster (cluster IV in Fig. 6). This heterogeneity suggests that the enzymes may originate from independent gene transfer events from ancient bacteria into ancestral γ-proteobacteria. Further analysis of the phylogeny of the homodimeric E1p family revealed that the eukaryotic Anopheles gambiae protein is positioned within the γ-proteobacterial cluster (cluster I in Fig. 6) and that the three γ-proteobacterial Xanthomonas, Xylella, and Methylococcus proteins are located within the main β-proteobacterial cluster of enzymes (cluster III in Fig. 6). These findings probably represent examples for more recent gene transfers from bacteria to eukaryotes and from γ-proteobacteria to β-proteobacteria.
The phylogeny of the heterotetrameric E1 family (Fig. 7) relates mitochondrial and chloroplast E1p α-subunits with E1p α-subunits of the α-proteobacteria and cyanobacteria, respectively, which is in agreement with endosymbiotic gene transfer from bacteria to eukaryotes (7, 16, 51). Furthermore, the putative heterotetrameric E1p α-subunit of the archaeon Haloferax is positioned within the cluster containing mainly proteins of the low-GC-content gram-positive bacteria (cluster I in Fig. 7), suggesting that this E1p α-subunit gene has been recruited via gene transfer from the gram-positive bacteria. The obvious absence of PDHCs in other archaea (70) is in agreement with the finding that the oxidative decarboxylation of pyruvate is generally catalyzed by pyruvate:ferredoxin oxidoreductases in archaea (31, 32, 47).
Interestingly, the phylogenetic analysis revealed that all heterotetrameric E1p α-subunits of the low-GC-content gram-positive bacteria cluster with the known heterotetrameric E1 α-subunits of the BCOADHC in many bacteria (Fig. 7, cluster I). This clustering indicates that the heterotetrameric E1p proteins of this lineage are more closely related to E1b proteins than to heterotetrameric E1p enzymes in α-proteobacteria and cyanobacteria. Thus, we conclude that it is likely that the E1p proteins of the low-GC-content gram-positive lineage are derived from E1b proteins of a BCOADHC via a relatively recent gene duplication event in an ancestral low-GC-content gram-positive bacterium. This hypothesis is corroborated by the finding that B. subtilis E1p catalyzes both the decarboxylation of pyruvate (Km = 10.2 μM, Vmax = 60 mU/mg protein) and of branched-chain 2-oxoacids (Km = 33.6 to 88.8 μM, Vmax = 11.8 to 53.0 mU/mg protein) (39). The cyanobacterial heterotetrameric E1p α-subunits also are more closely related to E1b α-subunits than to the respective E1p α-subunits of α-proteobacteria, suggesting that the cyanobacterial heterotetrameric E1p proteins also arose from E1b proteins via a less recent gene duplication event in an ancestral strain. The hypothesis that the heterotetrameric E1p proteins derived from E1b proteins, rather than the reverse direction, is in agreement with the assumption that early environments have been rich in amino acids and thus that amino acid-degrading enzymes such as BCOADHCs should be an earlier development in the evolution than the “aerobic” PDHCs. The notion that BCOADHCs have existed early in evolution is substantiated by the fact that BCOADHCs are present in archaea, which indicates that BCOADHC have developed before the divergence of bacteria and archaea (22). The finding that heterotetrameric PDHCs are abundant only in α-proteobacteria, low-GC-content gram-positive bacteria, and cyanobacteria thus suggests that heterotetrameric PDHCs trace back to developments that probably occurred late during the diversification of bacteria. In conclusion, we propose that heterotetrameric E1p subunits in bacteria arose from ancient E1b proteins via multiple independent gene duplication events.
Acknowledgments
We thank Joy Schreiner for critically reading the manuscript.
The support of the EC (grant VALPAN, QLK3-2000-00497), the Fachagentur Nachwachsende Rohstoffe of the BMVEL (grant 04NR004/22000404), and the Grant Agency of the Czech Republic (grant 525/04/0548) is gratefully acknowledged.
REFERENCES
- 1.Abdel-Hamid, A. M., M. M. Attwood, and J. R. Guest. 2001. Pyruvate oxidase contributes to the aerobic growth efficiency of Escherichia coli. Microbiology 147:1483-1498. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Arjunan, P., N. Nemeria, A. Brunskill, K. Chandrasekhar, M. Sax, Y. Yan, F. Jordan, J. R. Guest, and W. Furey. 2002. Structure of the pyruvate dehydrogenase multienzyme complex E1 component from Escherichia coli at 1.85 Å resolution. Biochemistry 41:5213-5221. [DOI] [PubMed] [Google Scholar]
- 4.Birnboim, H. C. 1983. A rapid alkaline extraction method for the isolation of plasmid DNA. Methods Enzymol. 100:243-255. [DOI] [PubMed] [Google Scholar]
- 5.Bosma, H. J., A. C. Graaf-Hess, A. de Kok, C. Veeger, A. J. Visser, and G. Voordouw. 1982. Pyruvate dehydrogenase complex from Azotobacter vinelandii. Ann. N. Y. Acad. Sci. 378:265-286. [DOI] [PubMed] [Google Scholar]
- 6.Burns, G., T. Brown, K. Hatter, J. M. Idriss, and J. R. Sokatch. 1988. Similarity of the E1 subunits of branched-chain-oxoacid dehydrogenase from Pseudomonas putida to the corresponding subunits of mammalian branched-chain-oxoacid and pyruvate dehydrogenases. Eur. J. Biochem. 176:311-317. [DOI] [PubMed] [Google Scholar]
- 7.Canback, B., S. G. E. Andersson, and C. G. Kurland. 2002. The global phylogeny of glycolytic enzymes. Proc. Natl. Acad. Sci. USA 99:6097-6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cocaign-Bousquet, M., A. Guyonvarch, and N. D. Lindley. 1996. Growth rate-dependent modulation of carbon flux through central metabolism and the kinetic consequences for glucose-limited chemostat cultures of Corynebacterium glutamicum. Appl. Environ. Microbiol. 62:429-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cocaign-Bousquet, M., and N. D. Lindley. 1995. Pyruvate overflow and carbon flux within the central metabolic pathways of Corynebacterium glutamicum during growth on lactate. Enzyme Microb. Technol. 17:260-267. [Google Scholar]
- 10.Costeas, P. A., and J. M. Chinsky. 1996. Effects of insulin on the regulation of branched-chain alpha-keto acid dehydrogenase E1 alpha subunit gene expression. Biochem. J. 318:85-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Kok, A., A. F. Hengeveld, A. Martin, and A. H. Westphal. 1998. The pyruvate dehydrogenase multi-enzyme complex from Gram-negative bacteria. Biochim. Biophys. Acta 1385:353-366. [DOI] [PubMed] [Google Scholar]
- 12.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eikmanns, B. J. 2005. Central metabolic pathways of Corynebacterium glutamicum: tricarboxylic cycle and anaplerotic reactions, p. 241-276. In L. Eggeling and M. Bott (ed.), Handbook of Corynebacterium glutamicum. CRC Press, Boca Raton, Fla.
- 14.Eikmanns, B. J., M. Metzger, D. Reinscheid, M. Kircher, and H. Sahm. 1991. Amplification of three threonine biosynthesis genes in Corynebacterium glutamicum and its influence on carbon flux in different strains. Appl. Microbiol. Biotechnol. 34:617-622. [DOI] [PubMed] [Google Scholar]
- 15.Eikmanns, B. J., N. Thum-Schmitz, L. Eggeling, K. U. Ludtke, and H. Sahm. 1994. Nucleotide sequence, expression and transcriptional analysis of the Corynebacterium glutamicum gltA gene encoding citrate synthase. Microbiology 140:1817-1828. [DOI] [PubMed] [Google Scholar]
- 16.Fast, N. M., and P. J. Keeling. 2001. Alpha and beta subunits of pyruvate dehydrogenase E1 from the microsporidian Nosema locustae: mitochondrion-derived carbon metabolism in microsporidia. Mol. Biochem. Parasitol. 117:201-209. [DOI] [PubMed] [Google Scholar]
- 17.Fries, M., H. J. Chauhan, G. J. Domingo, H. I. Jung, and R. N. Perham. 2003. Site-directed mutagenesis of a loop at the active site of E1 (alpha2beta2) of the pyruvate dehydrogenase complex. A possible common sequence motif. Eur. J. Biochem. 270:861-870. [DOI] [PubMed] [Google Scholar]
- 18.Ghosh, R., J. R. Guest, and K. Jeyaseelan. 1981. Regulatory properties of the pyruvate dehydrogenase complex in Pseudomonas aeruginosa. Biochim. Biophys. Acta 658:232-237. [DOI] [PubMed] [Google Scholar]
- 19.Guest, J. R., and I. T. Creaghan. 1974. Further studies with lipoamide dehydrogenase mutants of Escherichia coli K12. J. Gen. Microbiol. 81:237-245. [DOI] [PubMed] [Google Scholar]
- 20.Guest, J. R., S. J. Angier, and G. C. Russell. 1989. Structure, expression, and protein engineering of the pyruvate dehydrogenase complex of Escherichia coli. Ann. N. Y. Acad. Sci. 573:76-99. [DOI] [PubMed] [Google Scholar]
- 21.Hanahan, D. 1985. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 22.Heath, C., A. C. Jeffries, D. W. Hough, and M. J. Danson. 2004. Discovery of the catalytic function of a putative 2-oxoacid dehydrogenase multienzyme complex in the thermophilic archaeon Thermoplasma acidophilum. FEBS Lett. 577:523-527. [DOI] [PubMed] [Google Scholar]
- 23.Hederstedt, L. 1993. The Krebs citric acid cycle, p. 181-197. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. ASM Press, Washington, D.C.
- 24.Hemila, H., A. Palva, L. Paulin, S. Arvidson, and I. Palva. 1990. Secretory S complex of Bacillus subtilis: sequence analysis and identity to pyruvate dehydrogenase. J. Bacteriol. 172:5052-5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henning, U., G. Dennert, R. Hertel, and W. S. Shipp. 1966. Translation of the structural gene of the E. coli pyruvate dehydrogenase complex. Cold Spring Harbor Symp. Quant. Biol. 31:227-234. [DOI] [PubMed] [Google Scholar]
- 26.Ikeda, M., and S. Nakagawa. 2003. The Corynebacterium glutamicum genome: features and impacts on biotechnological processes. Appl. Microbiol. Biotechnol. 62:99-109. [DOI] [PubMed] [Google Scholar]
- 27.Jeyaseelan, K., and J. R. Guest. 1980. Isolation and properties of pyruvate dehydrogenase complex mutants of Pseudomonas aeruginosa PAO. J. Gen. Microbiol. 120:385-392. [DOI] [PubMed] [Google Scholar]
- 28.Johnston, M. L., M. H. Luethy, J. A. Miernyk, and D. D. Randall. 1997. Cloning and molecular analyses of the Arabidopsis thaliana plastid pyruvate dehydrogenase subunits. Biochim. Biophys. Acta 1321:200-206. [DOI] [PubMed] [Google Scholar]
- 29.Kalinowski, J., B. Bathe, D. Bartels, N. Bischoff, M. Bott, A. Burkovski, N. Dusch, L. Eggeling, B. J. Eikmanns, L. Gaigalat, A. Goesmann, M. Hartmann, K. Huthmacher, R. Kramer, B. Linke, A. C. McHardy, F. Meyer, B. Mockel, W. Pfefferle, A. Puhler, D. A. Rey, C. Ruckert, O. Rupp, H. Sahm, V. F. Wendisch, I. Wiegrabe, and A. Tauch. 2003. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J. Biotechnol. 104:5-25. [DOI] [PubMed] [Google Scholar]
- 30.Kellmann, J. W., E. Piechersky, and B. Piechulla. 1990. Analysis of the diurnal expression patterns of the tomato chlorophyll a/b binding protein genes. Influence of light and characterization of the gene family. Phytobiology 52:35-41. [DOI] [PubMed] [Google Scholar]
- 31.Kerscher, L., and D. Oesterhelt. 1981. Purification and properties of two 2-oxoacid:ferredoxin oxidoreductases from Halobacterium halobium. Eur. J. Biochem. 116:587-594. [DOI] [PubMed] [Google Scholar]
- 32.Kerscher, L., S. Nowitzki, and D. Oesterhelt. 1982. Thermoacidophilic archaebacteria contain bacterial-type ferredoxins acting as electron acceptors of 2-oxoacid:ferredoxin oxidoreductases. Eur. J. Biochem. 128:223-230. [DOI] [PubMed] [Google Scholar]
- 33.Langley, D., and J. R. Guest. 1974. Biochemical and genetic characteristics of deletion and other mutant strains of Salmonella typhimurium LT2 lacking α-keto acid dehydrogenase complex activities. J. Gen. Microbiol. 82:319-335. [DOI] [PubMed] [Google Scholar]
- 34.Langley, D., and J. R. Guest. 1977. Biochemical genetics of the α-keto acid dehydrogenase complexes of Escherichia coli K12: genetic characterization and regulatory properties of deletion mutants. J. Gen. Microbiol. 106:103-117. [DOI] [PubMed] [Google Scholar]
- 35.Liebl, W. 1991. The genus Corynebacterium—nonmedical, p. 1157-1171. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The Prokaryotes, vol. 2, Springer, New York, N.Y. [Google Scholar]
- 36.Nakai, T., N. Nakagawa, N. Maoka, R. Masui, S. Kuramitsu, and N. Kamiya. 2004. Ligand-induced conformational changes and a reaction intermediate in branched-chain 2-oxo acid dehydrogenase (E1) from Thermus thermophilus HB8, as revealed by X-ray crystallography. J. Mol. Biol. 337:1011-1033. [DOI] [PubMed] [Google Scholar]
- 37.Neveling, U., S. Bringer-Meyer, and H. Sahm. 1998. Gene and subunit organization of bacterial pyruvate dehydrogenase complexes. Biochim. Biophys. Acta 1385:367-372. [DOI] [PubMed] [Google Scholar]
- 38.Niebisch, A., and M. Bott. 2001. Molecular analysis of the cytochrome bc1-aa3 branch of the Corynebacterium glutamicum respiratory chain containing an unusual diheme cytochrome c1. Arch. Microbiol. 175:282-294. [DOI] [PubMed] [Google Scholar]
- 39.Oku, H., and T. Kaneda. 1988. Biosynthesis of branched-chain fatty acids in Bacillus subtilis. J. Biol. Chem. 263:18386-18396. [PubMed] [Google Scholar]
- 40.Patek, M., B. J. Eikmanns, J. Patek, and H. Sahm. 1996. Promoters from Corynebacterium glutamicum: cloning, molecular analysis and search for a consensus motif. Microbiology 142:1297-1309. [DOI] [PubMed] [Google Scholar]
- 41.Patek, M., J. Nesvera, A. Guyonvarch, O. Reyes, and G. Leblon. 2003. Promoters of Corynebacterium glutamicum. J. Biotechnol. 104:311-323. [DOI] [PubMed] [Google Scholar]
- 42.Perham, R. N. 1991. Domains, motifs, and linkers in 2-oxo acid dehydrogenase multienzyme complexes: a paradigm in the design of a multifunctional protein. Biochemistry 30:8501-8512. [DOI] [PubMed] [Google Scholar]
- 43.Quail, M. A., Haydon, D. J., and J. R. Guest. 1994. The pdhR-aceEF-lpd operon of Escherichia coli expresses the pyruvate dehydrogenase complex. Mol. Microbiol. 12:95-104. [DOI] [PubMed] [Google Scholar]
- 44.Quail, M. A., and J. R. Guest. 1995. Purification, characterization and mode of action of PdhR, the transcriptional repressor of the pdhR-aceEF-lpd operon of Escherichia coli. Mol. Microbiol. 15:519-529. [DOI] [PubMed] [Google Scholar]
- 45.Reinscheid, D. J., S. Schnicke, D. Rittmann, U. Zahnow, H. Sahm, and B. J. Eikmanns. 1999. Cloning, sequence analysis, expression and inactivation of the Corynebacterium glutamicum pta-ack operon encoding phosphotransacetylase and acetate kinase. Microbiology 145:503-513. [DOI] [PubMed] [Google Scholar]
- 46.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 47.Sakuraba, H., S. Goda, and T. Ohshima. 2004. Unique sugar metabolism and novel enzymes of hyperthermophilic archaea. Chem. Rec. 3:281-287. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook, J., D. W. Russel, N. Irwin, and U. A. Janssen. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 49.Sauer, U., and B. J. Eikmanns. C3-carboxylation and C4-decarboxylation reactions: the anaplerotic node as a switchpoint for C-flux distribution. FEMS Microbiol. Rev., in press.
- 50.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the E. coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 51.Schnarrenberger, C., and W. Martin. 2002. Evolution of the enzymes of the citric acid cycle and the glyoxylate cycle of higher plants. A case study of endosymbiotic gene transfer. Eur. J. Biochem. 269:868-883. [DOI] [PubMed] [Google Scholar]
- 52.Schreiner, M. E., and B. J. Eikmanns. 2005. Pyruvate:quinone oxidoreductase from Corynebacterium glutamicum: purification and biochemical characterization. J. Bacteriol. 187:862-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schulze, E., A. H. Westphal, R. Hanemaaijer, and A. de Kok. 1990. The 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. 1. Molecular cloning and sequence analysis of the gene encoding the 2-oxoglutarate dehydrogenase component. Eur. J. Biochem. 187:229-234. [DOI] [PubMed] [Google Scholar]
- 54.Schwartz, E. R., and L. J. Reed. 1970. Regulation of the activity of the pyruvate dehydrogenase complex of Escherichia coli. Biochemistry 9:1434-1439. [DOI] [PubMed] [Google Scholar]
- 55.Schwinde, J. W., N. Thum-Schmitz, B. J. Eikmanns, and H. Sahm. 1993. Transcriptional analysis of the gap-pgk-tpi-ppc gene cluster of Corynebacterium glutamicum. J. Bacteriol. 175:3905-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schwinde, J. W., P. F. Hertz, H. Sahm, B. J. Eikmanns, and A. Guyonvarch. 2001. Lipoamide dehydrogenase from Corynebacterium glutamicum: molecular and physiological analysis of the lpd gene and characterization of the enzyme. Microbiology 147:2223-2231. [DOI] [PubMed] [Google Scholar]
- 57.Shaw, W. V. 1975. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 43:737-755. [DOI] [PubMed] [Google Scholar]
- 58.Shen, L. C., and D. E. Atkinson. 1970. Regulation of pyruvate dehydrogenase from Escherichia coli. J. Biol. Chem. 245:5974-5978. [PubMed] [Google Scholar]
- 59.Shiio, I., Y. Toride, and S. Sugimoto. 1984. Production of lysine by pyruvate dehydrogenase mutants of Brevibacterium flavum. Agric. Biol. Chem. 48:3091-3098. [Google Scholar]
- 60.Skinner, D. D., M. R. Morgenstern, R. W. Fedechko, and C. D. Denoya. 1995. Cloning and sequencing of a cluster of genes encoding branched-chain alpha-keto acid dehydrogenase from Streptomyces avermitilis and the production of a functional E1 [alpha beta] component in Escherichia coli. J. Bacteriol. 177:183-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spencer, M. E., and J. R. Guest. 1985. Transcription analysis of the sucAB, aceEF and lpd genes of Escherichia coli. Mol. Gen. Genet. 200:145-154. [DOI] [PubMed] [Google Scholar]
- 62.Tauch, A., I. Homann, S. Mormann, S. Ruberg, A. Billault, B. Bathe, S. Brand, O. Brockmann-Gretza, C. Ruckert, N. Schischka, C. Wrenger, J. Hoheisel, B. Mockel, K. Huthmacher, W. Pfefferle, A. Puhler, and J. Kalinowski. 2002. Strategy to sequence the genome of Corynebacterium glutamicum ATCC 13032: use of a cosmid and a bacterial artificial chromosome library. J. Biotechnol. 95:25-38. [DOI] [PubMed] [Google Scholar]
- 63.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Uy, D., S. Delaunay, P. Germain, J. M. Engasser, and J. L. Goergen. 2003. Instability of glutamate production by Corynebacterium glutamicum 2262 in continuous culture using the temperature-triggered process. J. Biotechnol. 104:173-184. [DOI] [PubMed] [Google Scholar]
- 65.Van der Rest, M. E., C. Lange, and D. Molenaar. 1999. A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogenic plasmid DNA. Appl. Microbiol. Biotechnol. 52:541-545. [DOI] [PubMed] [Google Scholar]
- 66.Vasicova, P., Z. Abrhamova, J. Nesvera, M. Patek, H. Sahm, and B. Eikmanns. 1998. Integrative and autonomously replicating vectors for analysis of promoters in Corynebacterium glutamicum. Biotechnol. Tech. 12:743-746. [Google Scholar]
- 67.Venkova-Canova, T., M. Patek, and J. Nesvera. 2003. Control of rep gene expression in plasmid pGA1 from Corynebacterium glutamicum. J. Bacteriol. 185:2402-2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wallbrandt, P., V. Tegman, B. H. Jonsson, and A. Wieslander. 1992. Identification and analysis of the genes coding for the putative pyruvate dehydrogenase enzyme complex in Acholeplasma laidlawii. J. Bacteriol. 174:1388-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang, G. F., T. Kuriki, K. L. Roy, and T. Kaneda. 1993. The primary structure of branched-chain alpha-oxo acid dehydrogenase from Bacillus subtilis and its similarity to other alpha-oxo acid dehydrogenases. Eur. J. Biochem. 213:1091-1099. [DOI] [PubMed] [Google Scholar]
- 70.Wanner, C., and J. Soppa. 2002. Functional role for a 2-oxo acid dehydrogenase in the halophilic archaeon Haloferax volcanii. J. Bacteriol. 184:3114-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wendisch, V. F., M. Spies, D. J. Reinscheid, S. Schnicke, H. Sahm, and B. J. Eikmanns. 1997. Regulation of acetate metabolism in Corynebacterium glutamicum: transcriptional control of the isocitrate lyase and malate synthase genes. Arch. Microbiol. 168:262-269. [DOI] [PubMed] [Google Scholar]
- 72.Westphal, A. H., and A. de Kok. 1988. Lipoamide dehydrogenase from Azotobacter vinelandii. Molecular cloning, organization and sequence analysis of the gene. Eur. J. Biochem. 172:299-305. [DOI] [PubMed] [Google Scholar]
- 73.Westphal, A. H., and A. de Kok. 1990. The 2-oxoglutarate dehydrogenase complex from Azotobacter vinelandii. 2. Molecular cloning and sequence analysis of the gene encoding the succinyltransferase component. Eur. J. Biochem. 187:235-239. [DOI] [PubMed] [Google Scholar]
- 74.Zhu, P. P., and A. Peterkofsky. 1996. Sequence and organization of genes encoding enzymes involved in pyruvate metabolism in Mycoplasma capricolum. Protein Sci. 5:1719-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]