Abstract
Previous studies demonstrated that an outer membrane c-type cytochrome, OmcB, was involved in Fe(III) reduction in Geobacter sulfurreducens. An OmcB-deficient mutant was greatly impaired in its ability to reduce both soluble and insoluble Fe(III). Reintroducing omcB restored the capacity for Fe(III) reduction at a level proportional to the level of OmcB production. Here, we report that the OmcB-deficient mutant gradually adapted to grow on soluble Fe(III) but not insoluble Fe(III). The adapted OmcB-deficient mutant reduced soluble Fe(III) at a rate comparable to that of the wild type, but the cell yield of the mutant was only ca. 60% of that of the wild type under steady-state culturing conditions. Analysis of proteins and transcript levels demonstrated that expression of several membrane-associated cytochromes was higher in the adapted mutant than in the wild type. Further comparison of transcript levels during steady-state growth on Fe(III) citrate with a whole-genome DNA microarray revealed a significant shift in gene expression in an apparent attempt to adapt metabolism to the impaired electron transport to Fe(III). These results demonstrate that, although there are many other membrane-bound c-type cytochromes in G. sulfurreducens, increased expression of these cytochromes cannot completely compensate for the loss of OmcB. The concept that outer membrane cytochromes are promiscuous reductases that are interchangeable in function appears to be incorrect. Furthermore, the results indicate that there may be different mechanisms for electron transfer to soluble Fe(III) and insoluble Fe(III) oxides in G. sulfurreducens, which emphasizes the importance of studying electron transport to the environmentally relevant Fe(III) oxides.
Microorganisms in the family of Geobacteraceae are the predominant Fe(III) reducers in many subsurface environments where dissimilatory Fe(III) reduction plays an important role in the degradation of organic matter or the bioremediation of organic or metal contaminants (12, 13). The mechanisms by which Fe(III)-reducing microorganisms reduce Fe(III) are poorly understood. It is known that phylogenetically distinct Fe(III) reducers reduce Fe(III) oxide with significantly different mechanisms. For example, Geothrix and Shewanella species produce soluble electron shuttles and/or Fe(III)-chelating compounds to alleviate the need for direct electron transfer from the cell surface to the surface of Fe(III) oxide (20-22, 26). However, current evidence suggests that Geobacter species require direct physical contact between cells and extracellular electron acceptors (19), which may be achieved with appendages such as flagella and pili (3).
Earlier biochemical studies suggested that c-type cytochromes are likely to be involved in Fe(III) reduction in Geobacter species (8, 9, 16). Further genetic studies, concentrating on Geobacter sulfurreducens, due to the availability of a genetic system (5) and the genome sequence (17), indicated that a periplasmic cytochrome, PpcA (11); an inner membrane-associated cytochrome, MacA (2); and an outer membrane cytochrome, OmcB (10), are important for Fe(III) reduction. Based on location, PpcA and MacA were proposed to be intermediary electron transfer components whereas the outer membrane localization of OmcB and the severe impact on Fe(III) reduction when omcB was deleted suggested that OmcB might be the terminal Fe(III) reductase (10).
However, sequencing of the G. sulfurreducens genome has demonstrated that this organism contains genes for over 100 putative c-type cytochromes (17). A potential reason for this unprecedented number of cytochromes is that this might provide the opportunity for G. sulfurreducens to form multiple routes for electron transfer to Fe(III) and thus better maximize rates of Fe(III) reduction and/or provide a high degree of flexibility to adapt to disruptions in electron transfer pathways. In order to investigate this potential for adaptation further, the long-term adaptation of the OmcB-deficient mutant was studied.
MATERIALS AND METHODS
Bacterial strains and culturing conditions.
Geobacter sulfurreducens strain DL1 (5) and DL6 (omcB::cam) (10) were routinely cultured anaerobically in NBAF (acetate/fumarate) or FWFC [acetate/Fe(III) citrate] medium at 30°C as previously described (5). Steady-state growth was investigated in chemostats under strict anaerobic conditions (N2-CO2 [80:20, vol/vol]) in a freshwater medium containing Fe(III) citrate (56 mM) as the electron acceptor and limited acetate (5.5 mM) as the electron donor as previously described (4, 6). The dilution rate was 0.05 h−1.
Detection of cytochromes in the membrane fraction.
The membrane fractions of G. sulfurreducens were isolated as described earlier (10). Proteins (50 μg) of membrane fractions were analyzed with Tris-Tricine denaturing polyacrylamide gel electrophoresis (1). c-type cytochromes were detected by staining with N,N,N′,N′-tetramethylbenzidine as previously described (7, 25). Molecular standard markers were purchased from Bio-Rad Laboratories (Hercules, CA) and were stained separately with Coomassie blue R-250.
Analytical techniques.
Protein concentration was determined with the bicinchoninic acid method with bovine serum albumin as a standard (24). Cells were counted using epifluorescence microscopy with acridine orange staining (15). Fe(II) concentrations were determined with the ferrozine assay as previously described (14). Acetate concentrations were measured with high-pressure liquid chromatography on a Hewlett-Packard series 1100 (Agilent Technologies, Inc., Albany, NY) with a Bio-Rad Aminex HPX-87H column (300 × 7.8 mm) and a mobile phase of 8 mM H2SO4.
DNA microarray hybridization experiments and data analysis.
Total RNA was isolated from two sets of identically treated chemostat cultures of both the wild type and the adapted OmcB-deficient mutant. A total of 11 replicate hybridizations were carried out: five replicate hybridizations were from one set of the biological samples, and six were from the other set. DNA microarray hybridization and data analyses were described previously (18). Briefly, total RNA (5 μg) that was isolated from steady-state continuous cultures of the wild type and the adapted OmcB-deficient mutant was used to synthesize cDNA labeled with Cy3-Cy5 fluorescent dyes, which was hybridized to gene arrays. The signal from each spot in the arrays served as a measure of the expression level of each gene and was used to calculate the expression ratio between the wild type and the mutant.
Quantitative reverse transcription-PCR (RT-PCR).
For cDNA synthesis, SuperScript III RNase H− reverse transcriptase (Invitrogen Co., Carlsbad, CA) was used according to the manufacturer's instructions with gene-specific antisense primers 8908-2 (10) and 8915 (4) for omcB and omcC, respectively.
The quantitative real-time PCR was carried out as previously described (4). Two specific primer sets, 8912/8908-2 and 8917/8915 (4), were used to determine levels of mRNA for omcB and omcC, respectively. The temperature profile was composed of an initial incubation step for 2 min at 50°C (activation of the polymerase) followed by a 10-min denaturation step at 95°C, 40 cycles of denaturation for 45 s at 95°C, annealing for 1 min at 58°C, elongation for 1 min at 72°C, and a final elongation step for 6 min at 72°C.
DNA and RNA manipulations.
PCR product purification was carried out using QIAGEN PCR purification kits (QIAGEN Inc., Valencia, CA). Probes for Northern blot analysis were labeled with [α-32P]dATP using a Strip-EZ DNA probe synthesis and removal kit (Ambion Inc., Austin, Texas). [α-32P]dATP was purchased from PerkinElmer Life and Analytical Sciences, Inc., Boston, MA. All primers used to amplify G. sulfurreducens sequences were designed using the G. sulfurreducens genome sequence (17). QIAGEN Taq DNA polymerase (QIAGEN Inc., Valencia, CA) was used for all PCR amplifications.
Total RNA was isolated from mid-log cultures using RNeasy Midi kits (QIAGEN Inc., Valencia, CA) followed by treatment with RNase-free DNase (Ambion Inc., Austin, Texas). Northern blot analyses were carried out with the NorthernMax-Gly system (Ambion Inc., Austin, Texas) according to the manufacturer's instructions. All probes for Northern blot analyses were purified PCR product. Primers for amplifying probes are listed in Table 1. The PCR amplification program was as follows: 96°C for 40 s followed by 25 cycles of 96°C for 40 s, 58°C for 1 min, and 72°C for 1 min and a final extension at 72°C for 10 min.
TABLE 1.
Primers used for Northern blot analyses for c-type cytochrome genes whose expression levels were significantly altered in the Fe(III)-adapted OmcB-deficient mutant
| Target gene | Primer | Sequences (5′-3′) | Length (bp) | Reference(s) |
|---|---|---|---|---|
| GSU0594 | 994F | CCACAGCAGTAAGGATATCG | 375 | This work |
| 994R | GCATCGATATGCAGATCCG | |||
| GSU2494 | 4120F | GCACTCAAGTTCAAGCTCAAG | 469 | This work |
| 4120R | GGATGATATCGCTCTTCAGC | |||
| GSU2495 | 4123F | GCTCATTCTTCACCTGTCGG | 574 | This work |
| 4123R | GATAGTGCAGCGCTTGCTTTC | |||
| GSU2503 (omcT) | 4140-1F | CCAACCAGTTCAGCTGCATC | 382 | This work |
| 4140-1R | GAAGGGGCCAAGGTTCTGATC | |||
| GSU2504 (omcS) | 4142F | ACGTTCGTGGTCTCAACAC | 775 | This work |
| 4142R | GATGGTCGTGAACTCGTATG | |||
| GSU2731 (omcC) | 8914 | GCCAGAGTGAGGCCCAGA | 543 | This work; 4 |
| 8915 | GGGTGTTGTGGTAGAAGGG | |||
| GSU2737 (omcB) | 8916 | GGACTGCGCACCATCAAGG | 435 | This work; 10 |
| 8908-2 | GGTCAGCAGGCCACCGG | |||
| GSU2808 | 4655F | GGTGACAGTAGGAGTACCTG | 354 | This work |
| 4655R | GGTGTGACAGAGGTAGCAGAC | |||
| GSU2811 | 4659F | GTGTGACCAGCTATGCTCC | 395 | This work |
| 4659R | GTCAAAGTCCAGGTCGAACC | |||
| GSU2813 | 4662F | CCATCCTCTGTGCAGTCGC | 599 | This work |
| 4662R | GGTCTGTTTTCCGTCAAGG | |||
| GSU2887 | 4789-1F | CCATACGAGCACCAATGAGC | 445 | This work |
| 4789-1R | GCTGTTGCTGTCAGAGGAGG | |||
| GSU3259 | 5426F | CGTCTATCCTTGGCTTCGAG | 426 | This work |
| 5426R | CGATATGCCAGTGGATACC |
RESULTS
Adaptation of OmcB-deficient mutant (DL6) to grow on soluble Fe(III).
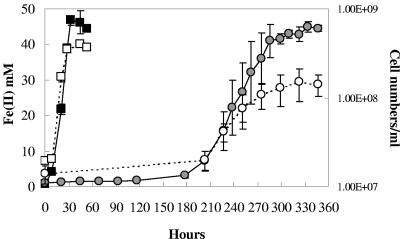
As previously reported (10), the OmcB-deficient mutant did not reduce Fe(III) citrate for periods of time well beyond those in which the wild-type cells had reduced all the Fe(III) (Fig. 1). However, after incubation for more than a week the OmcB-deficient mutant began to reduce Fe(III) (Fig. 1). The growth rate (doubling time, 35 h) was lower than that of the wild type (6 h), and the number of cells produced was only 38% of that of the wild type. The OmcB-deficient mutant never adapted to grow with Fe(III) oxide as the terminal electron acceptor (data not shown).
FIG. 1.
Adaptation of OmcB-deficient mutant (DL6) to grow with Fe(III) citrate as the sole electron acceptor. Mid-log (optical density at 600 nm, ∼0.3) fumarate-grown cells served as the inoculum. Symbols: filled or empty squares, wild-type Fe(II) or cell numbers, respectively; shaded or empty circles, OmcB-deficient mutant Fe(II) or cell numbers, respectively. Data are means ± standard deviations of triplicates.
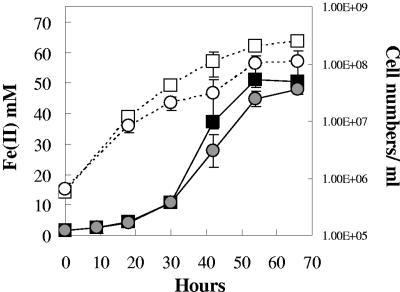
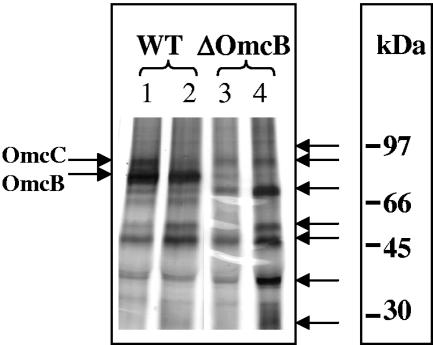
With two successive transfers of the adapted OmcB-deficient cells in Fe(III) citrate medium, the rate of Fe(III) reduction approached that of the wild type (Fig. 2). However, the final cell yield of OmcB-deficient mutant remained less than half of that of the wild type. The same pattern of adaptation was observed repeatedly when fumarate-grown cultures of the OmcB-deficient mutant were inoculated into Fe(III) citrate medium, suggesting that the adaptation was not due to spontaneous mutations; instead the growth may have resulted from a subpopulation of phase variants or cells with epigenetic modifications. Furthermore, when membrane fractions were isolated from fumarate or Fe(III) citrate-adapted cultures OmcB was not detected (Fig. 3, lanes 3 and 4). However, at least six other cytochromes, including OmcC, which is a close relative of OmcB but is not required for Fe(III) reduction (10), were present in higher concentrations in the OmcB-deficient mutant than in wild-type cells when membrane fractions were isolated from Fe(III) citrate cultures (Fig. 3, lane 4). The higher concentration of OmcC in the adapted OmcB-deficient mutant was consistent with the finding that levels of omcC transcripts as determined with quantitative RT-PCR were ca. 103 times higher in adapted, Fe(III)-grown cells than in the wild type.
FIG. 2.
Growth of the OmcB-deficient mutant after adaptation and two additional transfers on Fe(III) citrate medium. Symbols: filled or empty squares, wild-type Fe(II) or cell numbers, respectively; shaded or empty circles, OmcB-deficient mutant Fe(II) or cell numbers, respectively. Data are means ± standard deviations of triplicates.
FIG. 3.
Tricine-polyacrylamide gel electrophoresis and heme staining of membrane fractions prepared from wild type (WT) and the OmcB-deficient mutant. Membrane fractions were prepared from cultures grown with fumarate (lanes 1 and 3) or Fe(III) citrate (lanes 2 and 4). OmcB and OmcC are indicated with arrows on the left side of the gel. Other cytochromes which were highly expressed in the adapted Fe(III)-grown OmcB-deficient mutant are indicated with arrows on the right side of the gel.
Transcriptional profile comparison of the OmcB-deficient mutant and the wild type.
To further investigate potential differences in gene expression in the adapted OmcB-deficient mutant, transcript levels in continuous cultures of the adapted OmcB-deficient mutant grown with Fe(III) citrate as the electron acceptor were compared to those in wild-type cells with a whole-genome microarray (18). Although the OmcB-deficient mutant reduced Fe(III) at a rate comparable to that of the wild type in the chemostat [steady-state Fe(II) concentration, wild type versus mutant, 36.5 ± 1.2 mM versus 35.6 ± 2.9 mM, respectively; mean ± standard deviation; n = 3], the steady-state cell yield of the mutant was only 61% of that of the wild type (total protein of wild type versus mutant, 0.036 ± 0.003 versus 0.022 ± 0.007 mg/ml, respectively).
The adaptation of the OmcB-deficient mutant to grow on Fe(III) citrate was associated with changes in expression of a variety of genes involved in electron transfer and metabolism (Tables 2 and 3). A total of 83 genes had higher transcript levels in the OmcB-deficient mutant than in the wild type (Table 2), and 88 genes appeared to have lower transcript levels in the mutant (Table 3). Eighty-six genes had transcript levels that changed more than twofold in the OmcB-deficient mutant, with 47 genes up-regulated and 39 genes down-regulated.
TABLE 2.
Genes that were significantly up-regulated in the adapted OmcB-deficient mutant compared to the wild type grown in acetate-limiting Fe(III) citrate continuous culture
| Locus IDa | Common name | Role category | Log2 ratio ± SD |
|---|---|---|---|
| GSU0944 | Cystathionine beta-lyase | Amino acid biosynthesis | 0.884 ± 0.059 |
| GSU0945 | Cystathionine beta-lyase | 0.577 ± 0.095 | |
| GSU1906 | 2-Isopropylmalate synthase | 0.504 ± 0.125 | |
| GSU0265 | Putative membrane protein | Cell envelope | 0.855 ± 0.114 |
| GSU2078 | Rod shape-determining protein | 1.185 ± 0.264 | |
| GSU2497 | Putative lipoprotein | 1.618 ± 0.170 | |
| GSU2498 | Putative lipoprotein | 0.964 ± 0.095 | |
| GSU2526 | Putative membrane protein | 0.622 ± 0.121 | |
| GSU1140 | Methyl-accepting chemotaxis protein | Cellular processes | 0.873 ± 0.193 |
| GSU1899 | Virulence factor Mce family protein | 0.625 ± 0.145 | |
| GSU2236 | GTP pyrophosphokinase | 0.947 ± 0.195 | |
| GSU2502 | Spermine/spermidine synthase family protein | Central intermediary metabolism | 0.723 ± 0.121 |
| GSU2537 | Biosynthetic arginine decarboxylase | 0.638 ± 0.079 | |
| GSU3245 | Putative DNA polymerase II | DNA metabolism | 2.133 ± 0.199 |
| GSU0193 | l-Sorbosone dehydrogenase | Energy metabolism | 0.716 ± 0.111 |
| GSU0594 | Cytochrome c | 1.133 ± 0.271 | |
| GSU0782 | Nickel-dependent hydrogenase, small subunit | 2.165 ± 0.250 | |
| GSU0783 | Nickel-dependent hydrogenase, iron-sulfur cluster-binding protein | 2.547 ± 0.370 | |
| GSU0784 | Nickel-dependent hydrogenase, membrane protein | 2.605 ± 0.193 | |
| GSU0785 | Nickel-dependent hydrogenase, large subunit | 2.416 ± 0.306 | |
| GSU1640 | Cytochrome d ubiquinol oxidase, subunit I | 1.779 ± 0.776 | |
| GSU1641 | Cytochrome d ubiquinol oxidase, subunit II | 2.508 ± 0.427 | |
| GSU1707 | Group II decarboxylase | 0.664 ± 0.052 | |
| GSU1722 | Creatinine amidohydrolase | 0.570 ± 0.103 | |
| GSU1761 | Cytochrome c | 0.652 ± 0.164 | |
| GSU2018 | Glycine cleavage system H protein | 0.575 ± 0.112 | |
| GSU2098 | Carbon monoxide dehydrogenase subunit | 1.782 ± 0.286 | |
| GSU2494 | Cytochrome c | 1.863 ± 0.388 | |
| GSU2495 | Cytochrome c | 1.810 ± 0.294 | |
| GSU2501 | Cytochrome c | 0.870 ± 0.204 | |
| GSU2503 | Cytochrome c | 3.340 ± 0.154 | |
| GSU2504 | Cytochrome c | 3.676 ± 0.626 | |
| GSU2811 | Cytochrome c | 0.745 ± 0.196 | |
| GSU2812 | Glutaredoxin family protein | 0.815 ± 0.124 | |
| GSU2813 | Cytochrome c551 peroxidase | 0.839 ± 0.186 | |
| GSU3246 | Thioredoxin peroxidase | 1.049 ± 0.229 | |
| GSU1916 | Phosphatidate cytidylyltransferase | Fatty acid and phospholipid metabolism | 0.556 ± 0.146 |
| GSU0192 | Hypothetical protein | Hypothetical proteins | 1.433 ± 0.203 |
| GSU0208 | Hypothetical protein | 1.029 ± 0.354 | |
| GSU0593 | Hypothetical protein | 1.083 ± 0.217 | |
| GSU0647 | Hypothetical protein | 1.002 ± 0.139 | |
| GSU0788 | Hypothetical protein | 1.453 ± 0.329 | |
| GSU0919 | Hypothetical protein | 1.587 ± 0.326 | |
| GSU1160 | Hypothetical protein | 1.220 ± 0.134 | |
| GSU1309 | Hypothetical protein | 0.701 ± 0.216 | |
| GSU1770 | Hypothetical protein | 0.762 ± 0.117 | |
| GSU1947 | Hypothetical protein | 1.464 ± 0.098 | |
| GSU1948 | Hypothetical protein | 1.168 ± 0.162 | |
| GSU2496 | Hypothetical protein | 1.418 ± 0.374 | |
| GSU2499 | Hypothetical protein | 1.998 ± 0.238 | |
| GSU2500 | Hypothetical protein | 1.792 ± 0.532 | |
| GSU3309 | Hypothetical protein | 0.664 ± 0.091 | |
| GSU3310 | Hypothetical protein | 0.670 ± 0.160 | |
| GSU3403 | Hypothetical protein | 2.586 ± 0.168 | |
| GSU3409 | Hypothetical protein | 1.248 ± 0.215 | |
| GSU3410 | Hypothetical protein | 1.588 ± 0.170 | |
| GSU0786 | Hydrogenase maturation protease | Protein fate | 1.894 ± 0.178 |
| GSU0787 | TatA/E family twin-arginine translocation protein | 0.820 ± 0.155 | |
| GSU1159 | PfpI family intracellular protease | 1.185 ± 0.245 | |
| GSU0646 | tRNA (guanine-N1)-methyltransferase | Protein synthesis | 0.607 ± 0.129 |
| GSU0648 | Ribosomal protein L19 | 0.788 ± 0.185 | |
| GSU0537 | Sensory box/GGDEF family protein | Regulatory functions | 1.138 ± 0.081 |
| GSU2506 | Sigma-54-dependent DNA-binding response regulator | 2.264 ± 0.345 | |
| GSU2507 | Sensor histidine kinase | 2.797 ± 0.432 | |
| GSU3387 | AraC/XylS family transcriptional regulator | 0.561 ± 0.146 | |
| GSU0189 | ATP-dependent RNA helicase | Transcription | 1.097 ± 0.139 |
| GSU1161 | RND family efflux transporter | Transport and binding proteins | 0.949 ± 0.198 |
| GSU1346 | Sulfate ABC transporter, periplasmic sulfate-binding protein | 1.047 ± 0.224 | |
| GSU1723 | Mechanosensitive ion channel family protein | 0.812 ± 0.177 | |
| GSU3401 | Branched-chain amino acid ABC transporter, periplasmic amino acid-binding protein | 2.590 ± 0.255 | |
| GSU3404 | Amino acid ABC transporter, ATP-binding protein | 2.452 ± 0.341 | |
| GSU3405 | Amino acid ABC transporter, permease protein | 0.973 ± 0.187 | |
| GSU0500 | GTP-binding protein TypA | Unknown functions | 0.610 ± 0.086 |
| GSU0534 | RrF2 family protein | 0.814 ± 0.248 | |
| GSU0664 | GTP binding protein YchF | 0.546 ± 0.091 | |
| GSU0769 | RarD protein | 1.262 ± 0.177 | |
| GSU1643 | GGDEF/response regulator receiver domain protein | 2.053 ± 0.162 | |
| GSU1708 | Atz/Trz family chlorohydrolase | 0.599 ± 0.098 | |
| GSU1877 | 2-Nitropropane dioxygenase family oxidoreductase | 1.059 ± 0.287 | |
| GSU1945 | Fibronectin type III domain protein | 1.553 ± 0.177 | |
| GSU2493 | NHL repeat domain protein | 1.656 ± 0.166 | |
| GSU2505 | NHL repeat domain protein | 2.102 ± 0.573 | |
| GSU2527 | Nitrate/sulfite reductase domain protein | 1.200 ± 0.296 |
ID, identification.
TABLE 3.
Genes that were significantly down-regulated in the adapted OmcB-deficient mutant compared to the wild type in acetate-limiting Fe(III) citrate-grown continuous culture
| Locus IDa | Common name | Role category | Log2 ratio ± SD |
|---|---|---|---|
| GSU0656 | Branched-chain amino acid aminotransferase | Amino acid biosynthesis | −0.765 ± 0.179 |
| GSU0862 | FolD bifunctional protein | Biosynthesis of cofactors, prosthetic groups and carriers | −0.881 ± 0.176 |
| GSU1577 | Cob(I) alamin adenosyltransferase | −0.635 ± 0.087 | |
| GSU0967 | Putative membrane protein | Cell envelope | −1.843 ± 0.242 |
| GSU1010 | Slt family transglycosylase | −0.809 ± 0.186 | |
| GSU1493 | Type IV pilus biogenesis protein PilC | −0.692 ± 0.128 | |
| GSU1013 | Putative chemotaxis MotB protein | Cellular processes | −0.673 ± 0.191 |
| GSU2707 | Acetate kinase | Central intermediary metabolism | −1.150 ± 0.162 |
| GSU0096 | Recombination protein RecR | DNA metabolism | −0.896 ± 0.098 |
| GSU0338 | NADH dehydrogenase I; A subunit | Energy metabolism | −0.980 ± 0.156 |
| GSU1538 | Putative methylamine utilization protein MauG | −1.708 ± 0.244 | |
| GSU2612 | Putative rubrerythrin/rubredoxin protein | −0.765 ± 0.098 | |
| GSU2706 | Phosphate acetyltransferase | −1.189 ± 0.271 | |
| GSU2737 | Cytochrome c (OmcB) | −0.940 ± 0.152 | |
| GSU2808 | Cytochrome c | −0.730 ± 0.145 | |
| GSU2887 | Cytochrome c | −1.515 ± 0.208 | |
| GSU3259 | Cytochrome c | −1.069 ± 0.150 | |
| GSU3444 | NADH dehydrogenase I B/C/D subunits | −0.669 ± 0.136 | |
| GSU3061 | Squalene-hopene cyclase | Fatty acids and phospholipid metabolism | −1.180 ± 0.349 |
| GSU0384 | Hypothetical protein | Hypothetical proteins | −0.785 ± 0.190 |
| GSU0712 | Hypothetical protein | −0.867 ± 0.115 | |
| GSU0875 | Hypothetical protein | −1.037 ± 0.259 | |
| GSU0964 | Hypothetical protein | −1.460 ± 0.196 | |
| GSU0965 | Hypothetical protein | −1.557 ± 0.543 | |
| GSU0966 | Hypothetical protein | −1.731 ± 0.304 | |
| GSU0976 | Hypothetical protein | −0.858 ± 0.254 | |
| GSU0977 | Hypothetical protein | −0.912 ± 0.254 | |
| GSU0980 | Hypothetical protein | −0.592 ± 0.176 | |
| GSU0981 | Hypothetical protein | −0.847 ± 0.231 | |
| GSU0982 | Hypothetical protein | −0.824 ± 0.217 | |
| GSU0988 | Hypothetical protein | −0.652 ± 0.291 | |
| GSU0990 | Hypothetical protein | −1.066 ± 0.296 | |
| GSU0996 | Hypothetical protein | −0.597 ± 0.116 | |
| GSU1339 | Hypothetical protein | −1.374 ± 0.412 | |
| GSU1943 | Hypothetical protein | −1.212 ± 0.184 | |
| GSU1994 | Hypothetical protein | −1.675 ± 0.211 | |
| GSU2353 | Hypothetical protein | −1.264 ± 0.296 | |
| GSU2780 | Hypothetical protein | −0.902 ± 0.223 | |
| GSU3079 | Hypothetical protein | −0.709 ± 0.171 | |
| GSU3080 | Hypothetical protein | −1.280 ± 0.150 | |
| GSU3081 | Hypothetical protein | −1.051 ± 0.198 | |
| GSU3084 | Hypothetical protein | −1.344 ± 0.144 | |
| GSU3085 | Hypothetical protein | −1.227 ± 0.246 | |
| GSU0555 | ISGsu7, transposase OrfA | Mobile and extrachromosomal element functions | −0.785 ± 0.188 |
| GSU0556 | ISGsu7, transposase OrfB | −1.069 ± 0.343 | |
| GSU0761 | ISGsu7, transposase OrfB | −0.844 ± 0.289 | |
| GSU0762 | ISGsu7, transposase OrfA | −0.742 ± 0.173 | |
| GSU0975 | Phage tail sheath protein | −0.866 ± 0.283 | |
| GSU0986 | Tail lysozyme | −0.767 ± 0.097 | |
| GSU1355 | ISGsu7, transposase OrfA | −0.882 ± 0.222 | |
| GSU1356 | ISGsu7, transposase OrfB | −1.029 ± 0.366 | |
| GSU1847 | ISGsu7, transposase OrfA | −0.856 ± 0.212 | |
| GSU1848 | ISGsu7, transposase OrfB | −1.057 ± 0.327 | |
| GSU2127 | ISGsu7, transposase OrfA | −0.784 ± 0.204 | |
| GSU2128 | ISGsu7, transposase OrfB | −0.969 ± 0.214 | |
| GSU2139 | ISGsu7, transposase OrfA | −0.734 ± 0.198 | |
| GSU2140 | ISGsu7, transposase OrfB | −1.009 ± 0.300 | |
| GSU2171 | ISGsu7, transposase OrfB | −0.993 ± 0.263 | |
| GSU2279 | ISGsu7, transposase OrfB | −0.827 ± 0.309 | |
| GSU2391 | ISGsu7, transposase OrfB | −1.136 ± 0.397 | |
| GSU2392 | ISGsu7, transposase OrfA | −0.833 ± 0.253 | |
| GSU3082 | ISGsu7, transposase OrfA | −0.946 ± 0.237 | |
| GSU3083 | ISGsu7, transposase OrfB | −1.249 ± 0.374 | |
| GSU0079 | Cro/CI family transcriptional regulator | Regulatory functions | −0.542 ± 0.075 |
| GSU1072 | IclR family transcriptional regulator | −1.200 ± 0.414 | |
| GSU1626 | GntR family transcriptional regulator | −0.626 ± 0.109 | |
| GSU2046 | DNA-binding response regulator | −0.728 ± 0.091 | |
| GSU2779 | MerR family transcriptional regulator | −0.966 ± 0.172 | |
| GSU1068 | Sodium/solute symporter family protein | Transport and binding proteins | −1.771 ± 0.167 |
| GSU1070 | Sodium/solute symporter family protein | −1.670 ± 0.231 | |
| GSU1261 | ABC transporter, ATP-binding protein | −0.534 ± 0.053 | |
| GSU1330 | Metal ion efflux outer membrane protein family protein | −1.142 ± 0.400 | |
| GSU1331 | RND family efflux transporter | −0.774 ± 0.144 | |
| GSU2005 | Branched-chain amino acid ABC transporter, periplasmic amino acid-binding protein | −1.938 ± 0.395 | |
| GSU2006 | Branched-chain amino acid ABC transporter, permease protein | −1.776 ± 0.312 | |
| GSU2007 | Branched-chain amino acid ABC transporter, permease protein | −1.540 ± 0.346 | |
| GSU2008 | Branched-chain amino acid ABC transporter, ATP-binding protein | −1.259 ± 0.133 | |
| GSU2352 | Sodium/solute symporter family protein | −1.508 ± 0.193 | |
| GSU2490 | Oxalate/formate antiporter | −1.957 ± 0.510 | |
| GSU2886 | TonB-dependent receptor | −1.501 ± 0.239 | |
| GSU0711 | Endonuclease/exonuclease/phosphatase family protein | Unknown functions | −0.745 ± 0.136 |
| GSU0930 | Sulfur transferase | −0.758 ± 0.159 | |
| GSU1338 | Heavy-metal-associated domain protein | −1.225 ± 0.341 | |
| GSU1398 | SCO1/SenC family protein | −0.774 ± 0.144 | |
| GSU2010 | CBS domain protein | −1.142 ± 0.215 | |
| GSU3059 | Radical SAM domain protein | −0.623 ± 0.114 | |
| GSU3157 | Alpha/beta fold family hydrolase | −0.725 ± 0.186 | |
| GSU3435 | Ankyrin repeat protein | −0.665 ± 0.105 |
ID, identification.
The greatest increase in transcript levels was for GSU2504 and GSU2503 (Tables 2 and 4), which are cotranscribed and are predicted to encode two outer membrane c-type cytochromes, OmcS and OmcT, with high similarity to each other (D. Holmes et al., submitted for publication; T. Mehta et al., submitted for publication). OmcS, the cytochrome encoded by GSU2504, is readily sheared off from the outer surface of G. sulfurreducens and is involved in insoluble Fe(III) reduction and electricity production (T. Mehta, submitted for publication). The protein encoded by GSU2503, OmcT, has yet to be detected in any proteomic analysis of G. sulfurreducens (R. Ding, unpublished data). Transcripts of omcS and omcT were detected in wild-type cells during growth on insoluble Fe(III) oxide but not on soluble Fe(III) citrate (T. Mehta et al., submitted for publication). However, transcripts of both omcS and omcT were detected in the adapted OmcB-deficient mutant during growth on Fe(III) citrate (Table 2; see also Fig. 5). Thus, it is likely that the heme-staining protein recovered in higher levels from the adapted OmcB-deficient mutant (Fig. 3) with a molecular mass of ca. 45 kDa was OmcS.
TABLE 4.
Genes coding for c-type cytochromes whose expression levels were altered significantly (ca. twofold) in the OmcB-deficient mutant versus the wild type via microarray assaya
| Locus ID | Predicted location | Microarray log2 ratio | Predicted mol mass (kDa) | Northern blot result |
|---|---|---|---|---|
| GSU0594 | IM | 1.133 | 38.2 | Up |
| GSU2494 | IM | 1.863 | 47.7 | Up |
| GSU2495 | P | 1.810 | 69.7 | Up |
| GSU2503 | OM | 3.340 | 45.5 | Up |
| GSU2504 | OM | 3.676 | 45.4 | Up |
| GSU2811 | OM | 0.745 | 52.7 | Up |
| GSU2813 | P | 0.839 | 36.9 | Up |
| GSU2731 (OmcC) | OM | NA | 77.2 | Up |
| GSU2737 (OmcB) | OM | −1.189 | 77.1 | Down |
| GSU2808 | OM | −0.730 | 20.4 | Down |
| GSU2887 | OM | −1.515 | 89.7 | Down |
| GSU3259 | IM | −1.069 | 57.4 | Down |
Abbreviations: ID, identification; IM, inner membrane; P, periplasm; OM, outer membrane; NA, not applicable; mol, molecular.
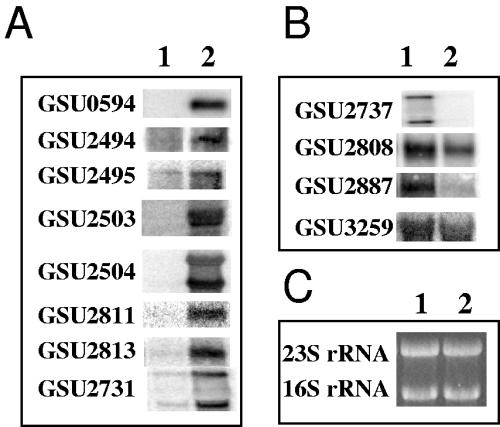
FIG. 5.
Northern blot analyses of up- or down-regulated c-type cytochrome genes. Total RNA was isolated from mid-log Fe(III) citrate-grown cultures of wild type (lane 1) or OmcB-deficient mutant (lane 2). (A) c-type cytochrome genes that were identified to be up-regulated by a ≥2-fold change in the adapted OmcB-deficient mutant. (B) c-type cytochrome genes that were identified to be down-regulated in the adapted OmcB-deficient mutant. (C) A replicate gel was run and stained with ethidium bromide, revealing the 16S and 23S rRNA, as a loading control.
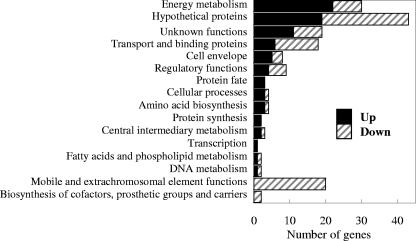
Genes with significant changes in their transcript levels could be categorized into 16 different groups according to their functions (Fig. 4). The largest functional groups that had higher levels of transcripts in the OmcB-deficient mutant than in the wild type were those related to energy metabolism and genes for hypothetical proteins. Other than omcS and omcT, the other genes related to energy metabolism which had transcript levels over twofold higher in the adapted mutant were those associated with a nickel-dependent hydrogenase, a cytochrome d ubiquinol oxidase, and a carbon monoxide dehydrogenase (Table 2). The transcript of a relA homolog (GSU2236), whose protein product is predicted to be involved in a stringent response during stress or nutrient depletion, was ca. twofold up-regulated in the mutant. The largest functional groups that had lower levels of transcripts in the OmcB-deficient mutant than in the wild type were those coding for hypothetical proteins, mobile and extrachromosomal element functions, and transport and binding proteins (Fig. 4). Four c-type cytochrome genes including the omcB gene were among the genes with lower transcript levels than those of the wild type (Tables 3 and 4). Genes coding for acetate metabolism (acetate kinase, GSU2707, and phosphate acetyltransferase, GSU2706) were among the groups of genes with over twofold-lower transcript levels.
FIG. 4.
Functional groups of up- or down-regulated genes from the adapted OmcB-deficient mutant. The up- or down-regulated genes of the OmcB-deficient mutant were identified by comparing expression patterns with that of the wild type as described in Materials and Methods. Functional classes are determined using the Geobacter sulfurreducens genome page from TIGR (http://www.tigr.org/tigr-scripts/CMR2/GenomePage3.spl?database=ggs). Functional groups of up-regulated genes from OmcB-deficient mutant are represented by black bars, and functional groups of down-regulated genes are shown by gray-striped bars. The number of genes represented in each bar is indicated in the x axis.
Validation by Northern blotting of expression ratios from transcriptional profiling.
Transcript levels for cytochrome genes which were significantly different (ca. twofold) between the adapted mutant and the wild type from the whole-genome microarray analysis were evaluated with Northern blot analyses (Fig. 5). The results from the Northern analysis compared well with those from the microarray analysis. The only exception was that omcC was not detected as up-regulated in the microarray, but transcript levels of omcC were significantly higher in the Northern analysis, consistent with quantitative real-time RT-PCR results mentioned above. The predicted molecular weights of those cytochromes whose transcript levels were significantly higher (ca. twofold) than those of the wild type from both microarray and Northern analyses (Table 4) are in agreement with those protein bands that were identified from the membrane fractions of the OmcB-deficient mutant (Fig. 3, lane 4).
DISCUSSION
The results demonstrate that even though G. sulfurreducens has genes for ca. 30 outer membrane c-type cytochromes, including the closely related OmcC, it cannot fully adapt to the loss of OmcB, which is essential for the most effective growth on soluble Fe(III) as well as for any growth on Fe(III) oxide, the most abundant form of Fe(III) in most environments. As detailed below, these results suggest that outer membrane cytochromes, which are frequently viewed as rather promiscuous agents for electron transfer to a variety of extracellular electron acceptors, may in fact have quite specific functions that cannot be replicated by other cytochromes.
Adaptation of OmcB-deficient mutant for growth on soluble Fe(III).
Previous short-term studies identified OmcB as an important component in electron transfer based on the finding that deleting omcB inhibited Fe(III) reduction and reintroducing omcB restored the capacity for Fe(III) reduction at a level proportional to the level of OmcB production (10). Furthermore, there is a direct correlation between transcript levels of omcB and rates of Fe(III) reduction in continuous cultures (4). However, as shown here, after extended incubation the OmcB-deficient mutant is able to adapt and grow with soluble Fe(III) as the sole electron acceptor.
One of the more surprising aspects of this adaptation is that, during growth, adapted OmcB-deficient cultures reduced soluble Fe(III) at rates comparable to the wild type, but cell yields were substantially lower, indicating that there is less energy conservation from the pathway for Fe(III) reduction in the adapted mutant. It is unlikely that OmcB, an outer membrane protein, plays a direct role in coupling the flow of electron transport to the generation of a proton motive force, because it is expected that this is primarily established from proton pumping across the inner membrane. However, it is possible that the electron transport pathway from inner membrane electron transport components to OmcB is tightly coupled and that alternative electron transfer pathways to Fe(III) reduction, in the absence of OmcB, require electron transfer from alternative inner membrane electron transfer components that yield less proton pumping. Alternatively, the apparent need for increased biosynthesis of alternative c-type cytochromes, and possibly other proteins, may increase energy consumption and decrease growth yields.
The increased production of other outer membrane c-type cytochromes associated with adaptation in the OmcB-deficient mutant may account for the ability of the adapted mutant to reduce Fe(III) by providing an alternative route(s) for electron transfer to soluble Fe(III).
Five of the nine up-regulated c-type cytochrome genes with higher levels of transcripts in the OmcB-deficient mutant (GSU594, 2495, 2503 [OmcT], 2811, and 2813) were also found to have higher transcript levels when G. sulfurreducens was grown under electron-acceptor limiting conditions, rather than with the electron donor limiting growth (A. Esteve-Nunez, unpublished data). Thus, the physiological state associated with the disruption of the electron transport pathway in the OmcB-deficient mutant may mimic the physiological state under electron acceptor-limiting conditions, and this, as well as possible other physiological signals, may account for the increased production of outer membrane cytochromes.
In addition to affecting cytochrome production, the disruption of electron flow in the OmcB-deficient mutant appeared to have an impact on other aspects of metabolism. This was evident from the microarray studies which suggested that there was a decrease in transcription of several genes involved in acetate metabolism and an increase in transcript levels for genes such as a nickel-dependent hydrogenase (GSU0782 to -0787), a cytochrome d ubiquinol oxidase (GSU1640-1641), and a carbon monoxide dehydrogenase (GSU2098), which presumably played a role in maintaining the appropriate balance of reducing equivalents in the OmcB-deficient mutant (23, 27). Evidence that the mutant was under stress includes the apparent up-regulation of a relA homolog (GSU2236) which has been shown to be important in regulating levels of ppGpp and stress responses in G. sulfurreducens (L. DiDonato et al., submitted for publication), as well as higher transcript levels for other stress response proteins, such as thioredoxin peroxidase (GSU3246), glutaredoxin family protein (GSU2812), and proteins involved in cell envelope biosynthesis (GSU2078, 2497), etc.
Significance of failure to adapt for Fe(III) oxide reduction.
The fact that the OmcB-deficient mutant was never able to adapt to grow on Fe(III) oxide emphasizes the central role that OmcB plays in the reduction of this most environmentally relevant form of Fe(III). Although it was initially considered that OmcB might function as the terminal Fe(III) reductase because of its location in the outer membrane (10), subsequent studies have identified other proteins, such as the outer membrane c-type cytochrome OmcS (T. Mehta et al., submitted for publication) and pili (23a), that are more exposed on the outside of the cell and are specifically required for the reduction of Fe(III) oxide, but not soluble Fe(III). Thus, the critical role of OmcB in Fe(III) oxide reduction may be as an intermediary electron transfer component that establishes electron transfer to the terminal Fe(III) oxide reductase (13). The finding that the mutant deficient in OmcB can reduce soluble Fe(III), but not Fe(III) oxides, demonstrates that G. sulfurreducens can reduce soluble Fe(III) via mechanisms different than those required for Fe(III) oxide reduction. This is an important consideration when extrapolating results from pure cultures to Fe(III) reduction in sedimentary environments where Fe(III) oxides are the predominant Fe(III) form readily available for microbial reduction and concentrations of soluble Fe(III) are likely to be low (13, 21).
Acknowledgments
This research was supported by the Office of Science (BER), U.S. Department of Energy, cooperative agreement no. DE-FC02-02ER63446.
We are grateful for the excellent technical support from Betsy Blunt.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1999. Current protocols in molecular biology, vol. 2. John Wiley & Sons, Inc., New York, N.Y.
- 2.Butler, J. E., F. Kaufmann, M. V. Coppi, C. Nunez, and D. R. Lovley. 2004. MacA, a diheme c-type cytochrome involved in Fe(III) reduction by Geobacter sulfurreducens. J. Bacteriol. 186:4042-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Childers, S. E., S. Ciufo, and D. R. Lovley. 2002. Geobacter metallireducens accesses insoluble Fe(III) oxide by chemotaxis. Nature 416:767-769. [DOI] [PubMed] [Google Scholar]
- 4.Chin, K.-J., A. Esteve-Núñez, C. Leang, and D. R. Lovley. 2004. Direct correlation between rates of anaerobic respiration and levels of mRNA for key respiratory genes in Geobacter sulfurreducens. Appl. Environ. Microbiol. 70:5183-5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coppi, M. V., C. Leang, S. J. Sandler, and D. R. Lovley. 2001. Development of a genetic system for Geobacter sulfurreducens. Appl. Environ. Microbiol. 67:3180-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esteve-Nunez, A., M. Rothermich, M. Sharma, and D. Lovley. 2005. Growth of Geobacter sulfurreducens under nutrient-limiting conditions in continuous culture. Environ. Microbiol. 7:641-648. [DOI] [PubMed] [Google Scholar]
- 7.Francis, R. T., Jr., and R. R. Becker. 1984. Specific indication of hemoproteins in polyacrylamide gels using a double-staining process. Anal. Biochem. 136:509-514. [DOI] [PubMed] [Google Scholar]
- 8.Gaspard, S., F. Vazquez, and C. Holliger. 1998. Localization and solubilization of the iron(III) reductase of Geobacter sulfurreducens. Appl. Environ. Microbiol. 64:3188-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorby, Y. A., and D. R. Lovley. 1991. Electron transport in the dissimilatory iron reducer, GS-15. Appl. Environ. Microbiol. 57:867-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leang, C., M. V. Coppi, and D. R. Lovley. 2003. OmcB, a c-type polyheme cytochrome, involved in Fe(III) reduction in Geobacter sulfurreducens. J. Bacteriol. 185:2096-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lloyd, J. R., C. Leang, A. L. Hodges Myerson, M. V. Coppi, S. Cuifo, B. Methe, S. J. Sandler, and D. R. Lovley. 2003. Biochemical and genetic characterization of PpcA, a periplasmic c-type cytochrome in Geobacter sulfurreducens. Biochem. J. 369:153-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lovley, D. R. 2003. Cleaning up with genomics: applying molecular biology to bioremediation. Nat. Rev. Microbiol. 1:35-44. [DOI] [PubMed] [Google Scholar]
- 13.Lovley, D. R., D. E. Holmes, and K. P. Nevin. 2004. Dissimilatory Fe(III) and Mn(IV) reduction. Adv. Microb. Physiol. 49:219-286. [DOI] [PubMed] [Google Scholar]
- 14.Lovley, D. R., and E. J. P. Phillips. 1986. Organic matter mineralization with the reduction of ferric iron in anaerobic sediments. Appl. Environ. Microbiol. 51:683-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lovley, D. R., and E. J. P. Phillips. 1988. Novel mode of microbial energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl. Environ. Microbiol. 54:1472-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Magnuson, T. S., A. L. Hodges-Myerson, and D. R. Lovley. 2000. Characterization of a membrane-bound NADH-dependent Fe3+ reductase from the dissimilatory Fe3+-reducing bacterium Geobacter sulfurreducens. FEMS Microbiol. Lett. 185:205-211. [DOI] [PubMed] [Google Scholar]
- 17.Methé, B. A., K. E. Nelson, J. A. Eisen, I. T. Paulsen, W. Nelson, J. F. Heidelberg, D. Wu, M. Wu, N. Ward, M. J. Beanan, R. J. Dodson, R. Madupu, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, A. S. Durkin, M. Gwinn, J. F. Kolonay, S. A. Sullivan, D. H. Haft, J. Selengut, T. M. Davidsen, N. Zafar, O. White, B. Tran, C. Romero, H. A. Forberger, J. Weidman, H. Khouri, T. V. Feldblyum, T. R. Utterback, S. E. Van Aken, D. R. Lovley, and C. M. Fraser. 2003. Genome of Geobacter sulfurreducens: metal reduction in subsurface environments. Science 302:1967-1969. [DOI] [PubMed] [Google Scholar]
- 18.Methe, B. A., J. Webster, K. Nevin, J. Butler, and D. R. Lovley. 2005. DNA microarray analysis of nitrogen fixation and Fe(III) reduction in Geobacter sulfurreducens. Appl. Environ. Microbiol. 71:2530-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nevin, K. P., and D. R. Lovley. 2000. Lack of production of electron-shuttling compounds or solubilization of Fe(III) during reduction of insoluble Fe(III) oxide by Geobacter metallireducens. Appl. Environ. Microbiol. 66:2248-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nevin, K. P., and D. R. Lovley. 2002. Mechanisms for accessing insoluble Fe(III) oxide during dissimilatory Fe(III) reduction by Geothrix fermentans. Appl. Environ. Microbiol. 68:2294-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nevin, K. P., and D. R. Lovley. 2002. Mechanisms for Fe(III) oxide reduction in sedimentary environments. Geomicrobiol. J. 19:141-159. [Google Scholar]
- 22.Newman, D. K., and R. Kolter. 2000. A role for excreted quinones in extracellular electron transfer. Nature 405:94-97. [DOI] [PubMed] [Google Scholar]
- 23.Rees, D. C. 2002. Great metalloclusters in enzymology. Annu. Rev. Biochem. 71:221-246. [DOI] [PubMed] [Google Scholar]
- 23a.Reguera, G., K. D. McCarthy, T. Mehta, J. S. Nicoll, M. T. Tuominen, and D. R. Lovley. 2005. Extracellular electron transfer via microbial nanowires. Nature 435:1098-1101. [DOI] [PubMed] [Google Scholar]
- 24.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 25.Thomas, P. E., D. Ryan, and W. Levin. 1976. An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal. Biochem. 75:168-176. [DOI] [PubMed] [Google Scholar]
- 26.Turick, C. E., L. S. Tisa, and F. Caccavo, Jr. 2002. Melanin production and use as a soluble electron shuttle for Fe(III) oxide reduction and as a terminal electron acceptor by Shewanella algae BrY. Appl. Environ. Microbiol. 68:2436-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White, D. 2000. The physiology and biochemistry of prokaryotes, 2nd ed. Oxford University Press, New York, N.Y.