Abstract
Yersinia enterocolitica biovar 1B maintains three distinct type III secretion (TTS) systems, which independently operate to target proteins to extracellular sites. The Ysa and Ysc systems are prototypical contact-dependent TTS systems that translocate toxic effectors to the cytosols of targeted eukaryotic host cells during infection. The flagellar TTS system is utilized during the assembly of the flagellum and is required for secretion of the virulence-associated phospholipase YplA to the bacterial milieu. When ectopically produced, YplA is also a secretion substrate for the Ysa and Ysc TTS systems. In this study, we define elements that allow YplA recognition and export by the Ysa, Ysc, and flagellar TTS systems. Fusion of various amino-terminal regions of YplA to Escherichia coli alkaline phosphatase (PhoA) lacking its native secretion signal demonstrated that the first 20 amino acids or corresponding mRNA codons of YplA were sufficient for export of YplA-PhoA chimeras by each TTS system. Export of native YplA by each of the three TTS systems was also found to depend on the integrity of its amino terminus. Introduction of a frameshift mutation or deletion of yplA sequences encoding the amino-terminal 20 residues negatively impacted YplA secretion. Deletion of other yplA regions was tolerated, including that resulting in the removal of amino acid residues 30 through 40 of the polypeptide and removal of the 5′ untranslated region of the mRNA. This work supports a model in which independent and distantly related TTS systems of Y. enterocolitica recognize protein substrates by a similar mechanism.
Protein secretion plays an important role in the ability of Yersinia enterocolitica to evade the host immune system and cause disease. Like many gram-negative pathogens, Y. enterocolitica utilizes contact-dependent type III secretion (TTS) to localize proteins important for pathogenesis that function at sites outside the bacterium (11, 14, 22). Structurally, TTS systems consist of a heteromultimeric complex of proteins that span the bacterial cell envelope which, without modification, recognizes and directs the transport of substrates in a single step (14). Proteins targeted by TTS systems are commonly toxic effectors that are directed into a host cell during infection. The core components that make up the contact-dependent TTS apparatus are conserved among most bacteria and are homologous to the components that form the basal body of the bacterial flagellum (2). The flagellar basal body constitutes a base upon which the rest of the flagellum is assembled and functions as a bona fide TTS system to localize flagellar hook and filament-related proteins to the cell envelope during organelle biogenesis (2, 19). The flagellar TTS system also transports proteins not related to motility to extracellular sites (36).
Full pathogenesis of Y. enterocolitica requires three independent and distantly related TTS systems. The plasmid-encoded Ysc system, which secretes a set of proteins termed Yops (yersinia outer proteins), and the chromosomally encoded Ysa system, which secretes a set of proteins termed Ysps (yersinia-secreted proteins), are contact-dependent systems (8, 13). The third TTS system is an integral part of the flagellum and is required for the secretion of YplA, a secreted phospholipase that contributes to the survival of Y. enterocolitica in host tissues during infection (26, 27, 36). It was originally thought that each of these TTS systems targeted a dedicated set of proteins, but recent work has demonstrated that the Ysc TTS system substrates YopE, YopN, and YopP are also exported by the Ysa TTS system (33). Furthermore, both systems deliver YopP into macrophages, which results in YopP-dependent suppression of tumor necrosis factor alpha production (33). In addition, although YplA is an established substrate of the flagellar TTS system, recent work has demonstrated that YplA can additionally be secreted by the Ysa and Ysc TTS systems (34). These observations imply that despite their distant relationship, the Ysa, Ysc, and flagellar TTS systems share common mechanisms for protein targeting. To further test this hypothesis, we used YplA as a model TTS substrate and defined elements of it that allow recognition and export by the Ysc, Ysa, and flagellar TTS systems. The results revealed that each TTS system displays a similar requirement for a secretion signal located at the YplA amino terminus.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are described in Table 1. Strains of Escherichia coli were routinely grown at 37°C, and Y. enterocolitica strains were grown at 26°C in Luria broth (1% tryptone, 0.5% yeast extract, 90 mM NaCl) or on Luria agar (Difco). Media used for the examination of protein secretion by Y. enterocolitica were Luria broth base (L medium; 1% tryptone and 0.5% yeast extract) and phospholipase indicator agar (L medium supplemented with 1% Tween 80, 1 mM CaCl2, and 1.5% [wt/vol] agar). Antibiotics were used at the following concentrations: chloramphenicol at 25 μg/ml for E. coli and 12.5 μg/ml for Y. enterocolitica, kanamycin at 50 μg/ml for E. coli and 100 μg/ml for Y. enterocolitica, nalidixic acid at 20 μg/ml, streptomycin at 25 μg/ml, spectinomycin at 25 μg/ml, and tetracycline at 15 μg/ml for E. coli and 7.5 μg/ml for Y. enterocolitica.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or characteristics | Source or reference |
|---|---|---|
| Strains | ||
| Y. enterocolitica | ||
| JB580v | Serogroup O:8, Nalr, ΔyenR (r− m+) | 16 |
| GY4478 | pYV8081− | 33 |
| GY460 | ΔflhDC | 37 |
| GY4428 | ysaV::pEP185.2 | K. G. Venecia and G. M. Younga |
| GY4757 | ΔyplAB, pYV8081− | This study |
| GY4838 | ΔyplAB, pYV100 | This study |
| YEDS10 | yplA::pEP185.2 | 26 |
| GY4886 | ΔyopN | B. M. Young and G. M. Younga |
| E. coli | ||
| CC118 | araD139 Δ(ara leu)7697 ΔlacX74 phoAΔ20 galE galK thi rpsE rpoB argE(Am) recA1 | 21 |
| DH5α | F−endA1 hsdR17 supE44 thi-1 λ−recA1 gyrA96 relA1 Δ(lacZYA-argF)U169 deoR (φ80dlacZΔM15) | Laboratory stock |
| MC4100 | araD139 Δ(argF-lac)U169 e14−flhD5301 fruA25 relA1 rpsL150(Strr) rbsR22 deoC | 5 |
| S17-1 λpir | recA thi pro hsd (R− M+) RP4 2-Tc::Mu-Km::Tn7 λpir | 28 |
| HB101rif | HB101 spontaneous rifampin-resistant isolate | A. J. Darwin |
| Plasmids | ||
| pEP185.2 | mob+, ori R6K, Cmr | 16 |
| pTM100 | mob+, derivative of pACYC184, Cmr Tcr | 23 |
| pGY20 | Ptac-flhDC derivative of pVLT33, Kanr | 37 |
| pGY100 | Pcat-yplAB derivative of pTM100 constructed by cloning into the vector EcoRI site a 1.7-kb DNA fragment extending from 78 bp upstream of yplA to 40 bp downstream of yplB, Tcr | 34 |
| pRK2013 | mob+tra+, self-transmissible plasmid, Kanr | 10 |
| pWKS130 | oriPSC101, Kmr | 31 |
| pYV100 | mob+, mobilizable derivative of pYV8081, Str/Spcr | A. J. Darwin |
| pGY192 | pGY100 with TnphoA insertion at codon 105 of yplA | This study |
| pGY173 | pGY100 with TnphoA insertion at codon 132 of yplA | This study |
| pGY200 | pGY100 with TnphoA insertion at codon 182 of yplA | This study |
| pGY230 | pGY100 with TnphoA insertion at codon 194 of yplA | This study |
| pGY222 | pGY100 with TnphoA insertion at codon 204 of yplA | This study |
| pGY219 | pGY100 with TnphoA insertion at codon 224 of yplA | This study |
| pGY221 | pGY100 with TnphoA insertion at codon 238 of yplA | This study |
| pGY166 | pGY100 with TnphoA insertion at codon 264 of yplA | This study |
| pGY175 | pGY100 with TnphoA insertion at codon 302 of yplA | This study |
| pGY335 | BamHI-KpnI ′phoA fragment from TnphoA cloned into pGY100 | This study |
| pGY429 | Pcat-yplA1 upstream of ′phoA in pGY335 | This study |
| pGY353 | Pcat-yplA1-15 upstream of ′phoA in pGY335 | This study |
| pGY354 | Pcat-yplA1-20 upstream of ′phoA in pGY335 | This study |
| pGY378 | Pcat-yplA1-30 upstream of ′phoA in pGY335 | This study |
| pGY379 | Pcat-yplA1-40 upstream of ′phoA in pGY335 | This study |
| pGY366 | Pcat-yplA1-60 upstream of ′phoA in pGY335 | This study |
| pGY373 | Pcat-yplA1-75 upstream of ′phoA in pGY335 | This study |
| pGY363 | Pcat-yplA1-105 upstream of ′phoA in pGY335 | This study |
| pGY484 | Pcat-yplA cloned into pWKS130 | This study |
| pGY497 | Pcat-yplA2-30 cloned into pWKS130 | This study |
| pGY495 | Pcat-yplA2-10 cloned into pWKS130 | This study |
| pGY503 | Pcat-yplA10-20 cloned into pWKS130 | This study |
| pGY504 | Pcat-yplA20-30 cloned into pWKS130 | This study |
| pGY493 | Pcat-yplA30-40 cloned into pWKS130 | This study |
| pGY527 | Pcat-yplAΔUTR cloned into pWKS130 | This study |
| pGY528 | Pcat-yplA2-30 cloned into pWKS130 (deletion constructed with four-primer strategy) | This study |
| pGY549 | Pcat-yplA2-10 cloned into pWKS130 (deletion constructed with four-primer strategy) | This study |
| pGY548 | Pcat-yplA30-40 cloned into pWKS130 (deletion constructed with four-primer strategy) | This study |
| pGY556 | Pcat-yplAframeshift (Pcat-yplA with a frameshift of codons 2 through 6) cloned into pWKS130 | This study |
| pGY439 | ΔyplAB allele cloned into BglII site of pEP185.2 | This study |
Unpublished data.
Transposon mutagenesis.
E. coli strain CC118/pGY100 was mutagenized by transduction with λTnphoA under conditions described previously (21). In total, 305 separate transductions were plated on Luria agar containing kanamycin and tetracycline. To enrich for transposition events resulting in an insertion in the plasmid, colonies obtained following transduction were combined in 5 ml of L medium and incubated at 37°C for 2 h with mild aeration. Plasmids were isolated and reintroduced into E. coli strain CC118 by electroporation. Approximately 60,000 transformants were examined to identify strains that exhibited a phospholipase-negative phenotype (36). To identify insertions that had occurred in frame and in the appropriate orientation, phospholipase-negative candidates were examined by Western blotting. E. coli cells were grown overnight in Luria broth and subcultured 1:75 into fresh Luria medium. Following 6 h of growth at 37°C, supernatant proteins were harvested and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), as described previously (33, 34). After transfer to a nitrocellulose membrane, the membrane was probed with α-PhoA polyclonal antibody.
Characterization of transposon insertion sites and DNA sequencing.
Plasmid DNA was purified and analyzed by a restriction digest to confirm the orientation of the TnphoA insertion within yplA. The sequence of yplA immediately adjacent to the transposon was then determined using a primer that annealed near the 5′ end of TnphoA (primer phoA) (Table 2). DNA sequence was obtained using an Applied Biosystems DNA sequencing system and the BigDye terminator cycle sequencing kit according to the manufacturer's instructions (Applied Biosystems). A list of the fusions obtained is presented in Table 1.
TABLE 2.
Primers used in this study
| Primer name | Nucleotide sequence |
|---|---|
| phoA (for sequencing) | 5′-GGTGCAGTAATATCGC-3′ |
| yplA198 (for sequencing) | 5′-CCGCCTAATGAATGGCCG-3′ |
| SacII-Pcat | 5′-TCCCCGCGGGGAAAGTATCTTCCTGGC-3′ |
| BamHI-yplA-end | 5′-GGATCCTCATTCAGGCATACTGCCGC-3′ |
| phoA-KpnI | 5′-GGGGTACCCCATCTGCCATTAAGTCTG-3′ |
| BamHI-phoA | 5′-CGGGATCCCGCTGACTCTTATACAC-3′ |
| yplA-codon 1 | 5′-CGGGATCCCTCATGCCAACTCCTTGTCG-3′ |
| yplA-codon 9 | 5′-CGGGATCCCTCCGGGCTAAACTAAC-3′ |
| yplA-codon 15 | 5′-CGGGATCCCTGCCTGCTAATGGAACAGG-3′ |
| yplA-codon 20 | 5′-CGGGATCCCTCACCTGGTTCATCAAGCC-3′ |
| yplA-codon 30 | 5′-CGGGATCCCTGTTCTCCTTCTTGACCTG-3′ |
| yplA-codon 40 | 5′-CGGGATCCCTAAGCTGGTTCGAAGACCC-3′ |
| yplA-codon 60 | 5′-CGGGATCCCTCATGCTGTGTAATAAACATT-3′ |
| yplA-codon 75 | 5′-CGGGATCCCTGGCAGCTAAAACACGTTC-3′ |
| yplA-codon 105 | 5′-CGGGATCCCTAGCAGGAGCGTAAACATC-3′ |
| BamHI-yplA11 | 5′-AGGGATCCCGTTGGATTAGCAGGC-3′ |
| BamHI-yplA21 | 5′-AGGGATCCCCAACCAATAGAGCAG-3′ |
| BamHI-yplA31 | 5′-AGGGATCCCTCTACTCCTGTTGGG-3′ |
| BamHI-yplA41 | 5′-AGGGATCCCCAAAAAGAATCCGCC-3′ |
| YplAΔ2-30-2 | 5′-CCCAACAGGAGTAGACATGCCAACTCCTTGTCG-3′ |
| YplAΔ2-30-3 | 5′-GGCATGTCTACTCCTGTTGGG-3′ |
| YplAΔ2-10-2 | 5′-GCCTGCTAATGGAACCATGCCAACTCCTTGTCG-3′ |
| YplAΔ2-10-3 | 5′-GGCATGGTTCCATTAGCAGGC-3′ |
| YplAΔ31-40-2 | 5′-GGATTCTTTTTGAAGGTTCTCCTTCTTGACCTG-3′ |
| YplAΔ31-40-3 | 5′-GAGAACCTTCAAAAAGAATCC-3′ |
| UTR2 | 5′-ACTAACAGATATACTCATTTTAGCTTCCTTAGC-3′ |
| UTR3 | 5′-GCTAAGGAAGCTAAAATGAGTATATCTGTTAGT-3′ |
| Frameshift2 | 5′-AGGCCGGGCTAAAACTAACAGATATACCATTTTAGCTTC-3′ |
| Frameshift3 | 5′-GAAGCTAAAATGGTATATCTGTTAGTTTTAGCCCGGCCT-3′ |
| ΔyplAB1 | 5′-GGATCCCTGATCCAATTCGGTCAG-3′ |
| ΔyplAB2 | 5′-CGGGCTTCAGGATCAGCAAGATCTCATGCCAACTCCTTGTCG-3′ |
| ΔyplAB3 | 5′-AGATCTTGCTGATCCTGAAGCCCG-3′ |
| ΔyplAB4 | 5′-GGATCCCGTTCTGGTTGCAACTGA-3′ |
Amplification of DNA fragments by PCR.
DNA fragments used for cloning steps were amplified by PCR with Pfu-Turbo DNA polymerase (Stratagene). The DNA sequence of each fragment was determined through DNA sequencing as described above to be sure there were no unexpected mutations.
Construction of yplA′-′phoA fusions.
A ca. 1.5-kb ′phoA fragment encoding the carboxy-terminal activity domain was amplified by PCR using pGY192 as a template. This ′phoA fragment was designed to be flanked by the KpnI and BamHI sites using the oligonucleotide primers phoA-KpnI and BamHI-phoA (Table 2). This fragment was cloned into plasmid pCR-Blunt II TOPO using the manufacturer's instructions (Invitrogen). Subsequently, this DNA fragment was liberated with KpnI and BamHI and subcloned into the corresponding restriction sites of pWKS130 to give the resulting plasmid pGY335. Translational fusions between yplA and ′phoA were generated by cloning defined fragments of DNA, amplified by PCR from pGY100, into pGY335. The upstream primer used for each fragment anneals to a region 5′ to the cat promoter of pGY100 and contains a SacII linker (primer SacII-Pcat) (Table 2). Downstream primers were designed to anneal to particular codons of yplA followed by a BamHI linker (Table 2, primers yplA-codon X). Using this approach, each fusion is transcriptionally driven by the constitutively expressed Pcat promoter (35) and fusion of YplA to PhoA occurs via an Arg-Asp-Pro-Ala peptide linker. Eight Pcat-yplA-phoA translational fusions were constructed with this methodology: Pcat-yplA1-phoA (pGY429), Pcat-yplA1-15-phoA (pGY353), Pcat-yplA1-20-phoA (pGY354), Pcat-yplA1-30-phoA (pGY378), Pcat-yplA1-40-phoA (pGY379), Pcat-yplA1-60-phoA (pGY366), Pcat-yplA1-75-phoA (pGY373), and Pcat-yplA1-105-phoA (pGY363) (Table 1).
Construction of pGY484.
A Pcat-yplA fragment was PCR amplified using pGY100 as a template and primers SacII-Pcat and BamHI-yplA-end (Table 2). The resulting PCR product was cloned into pCR-Blunt II-TOPO and subcloned to pWKS130 to generate plasmid pGY484 (Table 1).
Construction of in-frame deletions of yplA.
Two methods were used to generate in-frame deletions of yplA: a linker method and a four-primer in-frame deletion strategy (32). Deletions generated with the linker method were constructed using the same methodology that was used to generate the yplA′-′phoA fusions (described above). Five in-frame deletions of yplA were generated in this way: deletions of codons 2 through 30 (Pcat-yplAΔ2-30, pGY497), 2 through 10 (Pcat-yplAΔ2-10, pGY495), 10 through 20 (Pcat-yplAΔ10-20, pGY503), 20 through 30 (Pcat-yplAΔ20-30, pGY504), and 30 through 40 (Pcat-yplAΔ30-40, pGY493) (Table 1). In each case, the codons removed by the deletion were replaced by the linker sequence 5′-AGGGATCCC-3′. Deletions were also generated by a four-primer in-frame deletion strategy. This method is a modified version of the PCR-based technique termed splicing by overlap extension (32). Four in-frame deletions were constructed this way: a deletion of codons 2 through 30 (Pcat-yplAΔ2-30, pGY528), a deletion of codons 2 through 10 (Pcat-yplAΔ2-10, pGY549), a deletion of codons 31 through 40 (Pcat-yplAΔ31-40, pGY548), and a deletion of the 5′ untranslated region (Pcat-yplAΔUTR, pGY527) (Table 1). In all cases, fragments of yplA were PCR amplified using pGY100 as a template with outside primers SacII-Pcat and BamHI-yplA-end and internal primers yplA-codonX and BamHI-yplAX or untranslated region 2 (UTR2) and UTR3 (Table 2). Each yplAdeletion fragment resides within the BamHI site of pWKS130 and is transcriptionally driven by the constitutively expressed Pcat promoter (35).
Construction of a frameshift mutation in the amino-terminal coding region of yplA.
A frameshift extending from codons 2 through 6 of yplA was generated using a four-primer strategy as described above with primers SacII-Pcat, BamHI-yplA-end, frameshift2, and frameshift3 (Table 2). This strategy introduced two compensatory point mutations in yplA. The first mutation introduces a −1 frameshift by the removal of adenine at position 1 of codon 2, and the second mutation introduces a +1 frameshift by the insertion of a thymine at position 3 of codon 6. This changes the original peptide sequence coded by this region from MSISVSLA to MYVLLVLA.
Deletion of the yplAB locus.
A four-primer PCR strategy was used to generate a deletion extending from ca. 1 kb upstream of yplAB to ca. 1 kb downstream of the region. Primers used for the reaction were ΔyplAB1, ΔyplAB2, ΔyplAB3, and ΔyplAB4 (Table 2). The fragment was initially cloned into plasmid pCR-Blunt II-TOPO and then subcloned into a BglII site of the suicide plasmid pEP185.2 to generate pGY439 (Table 1). The ΔyplAB mutation was introduced into the Y. enterocolitica GY4478 chromosome by allelic exchange as described previously (37). This new strain is designated GY4757, and the deletion was confirmed by PCR amplification of the yplAB locus of genomic DNA purified from the candidate strain using primers ΔyplAB1 and ΔyplAB4 (Table 2). GY4838 was constructed by conjugation of pYV100 into GY4757 by triparental mating with the E. coli strains HB101/pYV100 and DH5α/pRK2013.
Preparation of extracellular proteins, SDS-PAGE, and Western blot analysis.
Extracellular proteins were prepared as previously described (34). E. coli was grown overnight in Luria broth and subcultured 1:75 in 5 ml of fresh Luria broth. Y. enterocolitica was grown overnight in L medium and subcultured to an optical density at 600 nm (OD600) of 0.1 in 5 ml of appropriate medium to induce secretion of proteins by the Ysa, Ysc, or flagellar TTS systems. Induction of the Ysa TTS system was achieved by subculturing strains into L medium supplemented with 290 mM NaCl, followed by growth at 26°C for 6 h with mild aeration. The Ysc TTS system was induced by subculturing into L medium depleted of calcium by the addition of 20 mM sodium oxalate and 20 mM MgCl2, followed by incubation at 37°C for 6 h with mild aeration. To induce the flagellar TTS system, overnight cultures were grown in L medium at 26°C with mild aeration.
To examine secreted proteins, cultures grown under appropriate inducing conditions were collected. The OD600 of the culture was determined at the time of harvesting. Bacterial cells were removed by centrifugation in a microcentrifuge at 16,000 × g for 5 min. The upper two-thirds of the supernatant was removed and centrifuged again. The upper two-thirds of the supernatant was collected. Proteins were precipitated with 10% (wt/vol) ice-cold trichloroacetic acid and washed with ice-cold acetone. Proteins were suspended in sample buffer containing 2-mercaptoethanol (20), and the volume was adjusted according to the OD600 of the culture. Equivalent amounts of sample were analyzed by SDS-PAGE. Samples were heated to 95°C for 5 min and were then loaded onto 10% or 12.5% polyacrylamide gels. Proteins were transferred to nitrocellulose membranes for Western blot analysis. Detection of YplA and YplA-PhoA hybrids was completed with polyclonal anti-YplA antibody (26) or polyclonal anti-alkaline phosphatase antibody (Rockland Inc., Gilbertsville, PA), respectively. Western blots were visualized by chemiluminescence (enhanced chemiluminescence Western blotting detection reagents; Amersham Biosciences).
Enzymatic assay of YplA.
Y. enterocolitica was grown under conditions to induce the production of Fops or Ysps as described above. For experiments that assessed secretion by the Ysc TTS system, strains were derived from GY4886 (ΔyopN). This allowed Yop production to be induced in a medium that contained calcium, an essential cofactor of YplA. To induce Yop production, overnight cultures were subcultured to an OD600 of 0.1 in 5 ml of Luria broth and grown for 6 h at 37°C. Using a turbidimetric assay as described previously (30), 100 μl to 300 μl of culture supernatant was assayed for phospholipase activity at 37°C. The OD500 of each reaction was measured at several time intervals. Extracellular phospholipase activity (EPA) was calculated as a rate of turbidimetric change using the following formula: EPA = (1,000 × OD500)/(OD600 × V × T), where OD500 is the optical density of the assay solution, OD600 is the optical density of the Y. enterocolitica culture immediately before harvesting, V is the volume of supernatant used, and T is the time elapsed of the assay.
RESULTS
TnphoA mutagenesis of yplA reveals an amino-terminal secretion signal.
For other type III substrates, elements for substrate recognition and secretion have been determined by the construction of hybrid protein fusions (3, 4, 7, 15, 23-25, 29). These and other studies have led to two mechanistic models: one predicts that the type III secretion signal involves peptide-based elements of the secreted protein, and the other predicts mRNA-based elements. Our goal was to determine whether the distantly related flagellar Ysa and Ysc TTS systems share common requirements for the recognition of YplA with the expectation that the results may not ultimately favor one or the other secretion signal model. We set out to map the secretion signal of YplA by constructing a set of fusion proteins to E. coli alkaline phosphatase (PhoA) lacking its native secretion signal. Our initial experimental strategy was to generate a YplA-PhoA chimera by in vivo mutagenesis with λTnphoA (21). This strategy required that the experiments be completed in E. coli K-12 and that this bacterium could export YplA by its sole TTS pathway, the flagellar TTS system. To establish that YplA was indeed secreted, E. coli strain CC118 was transformed with the cloning vector pTM100 or pGY100, which carries a constitutively expressed copy of yplA. Strains were then cultivated under conditions that induce the production of flagella. Secreted proteins collected from the growth medium were examined by Western blotting using α-YplA polyclonal antibodies. YplA was secreted into the culture medium when pGY100 was present, indicating that E. coli can export this protein (Fig. 1A). In addition, when observed on phospholipase indicator agar medium, E. coli CC118 with pGY100 (yplA+), but not pTM100 (vector control), exhibited phospholipase activity indicative of YplA export (data not shown).
FIG. 1.
Secretion of YplA in E. coli. Secretion of YplA in E. coli was assessed by detection of the protein in culture supernatants from strains cultured in Luria medium at 37°C. Concentrated proteins from culture supernatants were separated on a 10% polyacrylamide gel, transferred to a nitrocellulose membrane, and probed with α-YplA antibody. (A) Secretion of YplA was initially examined in strain CC118, which is proficient for the production of flagella. Lanes: 1, E. coli CC118; 2, CC118/pTM100 (plasmid control); 3, CC118/pGY100 (yplA+). (B) The effect of flhD on YplA secretion by E. coli was tested in strain MC4100. Lanes: 1, E. coli MC4100/pTM100/pVLT33 (plasmid control); 2, MC4100/pTM100/pGY20 (flhD+); 3, MC4100/pGY100/pVLT33 (yplA+); 4, MC4100/pGY100/pGY20 (yplA+ flhD+).
To determine if YplA secretion by E. coli requires the flagellar TTS system, export of YplA was tested in E. coli strain MC4100 (flhD mutant), which carries a single mutation that blocks the production of flagella. Strains carrying vectors alone (pTM100 and pVLT33), pGY100 (yplA+), pGY20 (flhD+), or both pGY100 and pGY20 (yplA+, flhD+) were grown to induce the production of flagella (Fig. 1B) (1). Secreted proteins collected from the growth medium were examined by Western blotting using α-YplA polyclonal antibodies. YplA was not secreted by strains that carried only pGY100, suggesting that flhD is required for export of the phospholipase. Indeed, when the flhD mutation was complemented by the addition of pGY20 to MC4100/pGY100, YplA was secreted to the culture medium (Fig. 1B). These results indicate that E. coli secretes YplA and that secretion requires the flagellar TTS system.
To generate YplA-PhoA chimeras, E. coli strain CC118/pGY100 was subjected to transposon mutagenesis with λTnphoA as described in Material and Methods. Twenty-seven candidate E. coli strains carrying a plasmid-encoded gene fusion were examined for secretion of the corresponding YplA-PhoA chimera by screening for the presence of the fusion protein in culture supernatants. Nine independent YplA-PhoA hybrids were identified with ′phoA fused to codons 105, 132, 182, 194, 204, 224, 238, 265, and 302 of yplA, indicating that, in E. coli, the first 105 residues, or potentially codons, of YplA were sufficient for secretion when fused to PhoA (Fig. 2).
FIG. 2.
The amino terminus of YplA is sufficient for export of YplA-PhoA hybrids by E. coli. To test whether YplA-PhoA hybrids were secreted, E. coli strains carrying pGY100 derivatives with TnphoA insertions in yplA were grown in Luria medium at 37°C. Supernatant proteins were concentrated, separated on a 10% polyacrylamide gel, transferred to a nitrocellulose membrane, and probed with α-PhoA antibody. Lanes: 1, E. coli CC118/pTM100 (plasmid control); 2, CC118/pGY100 (yplA+); 3, CC118/pGY192; 4, CC118/pGY173; 5, CC118/pGY200; 6, CC118/pGY230; 7, CC118/pGY222; 8, CC118/pGY219; 9, CC118/pGY221; 10, CC118/pGY166; 11, CC118/pGY175. The fusion protein encoded by each plasmid is indicated below each lane.
The amino-terminal secretion signal directs the export of YplA-PhoA chimeras by Y. enterocolitica TTS systems.
To determine whether the secretion signal identified in E. coli also functioned in Y. enterocolitica, each of the plasmid-encoded YplA-PhoA chimeras was expressed in Y. enterocolitica GY4757 (ΔyplAB). When grown under conditions that induce the flagellar TTS system, each fusion protein was detected in culture supernatants, suggesting that the amino-terminal 105 residues of YplA contain elements of a secretion signal sufficient for export of YplA-PhoA fusions by the flagellar TTS system of Y. enterocolitica (data not shown).
To test if the amino terminus of YplA could direct hybrid proteins to the apparatus of either the Ysa or Ysc TTS systems, the same strains were grown under conditions that specifically induce each of these secretion pathways. Examination of culture supernatants by Western blotting revealed that the YplA-PhoA hybrids were each secreted under conditions inducing the Ysa and Ysc TTS systems (data not shown). From this initial analysis, we reasoned that a secretion signal is located in the amino-terminal region of YplA, or alternatively, in the 5′ region of yplA mRNA, that can direct proteins for export by the three distantly related TTS systems of Y. enterocolitica. We next focused our efforts on characterizing this amino-terminal secretion signal.
The first 20 residues are sufficient for the secretion of YplA-PhoA chimeras by the Ysc, Ysa, and flagellar TTS systems.
We further narrowed the location of the secretion signal within the amino terminus of YplA by generating additional plasmids encoding YplA-PhoA fusions using in vitro molecular cloning techniques. We began by cloning the -phoA coding sequence into the low-copy-number vector pWKS130. Translational fusions were generated by cloning specific PCR-generated Pcat-yplA cassettes upstream of ′phoA. When grown under conditions that induce type III secretion by the flagellar apparatus, export of fusion proteins consisting of 20 or more amino-terminal residues of YplA fused to PhoA was detected in culture supernatants (Fig. 3A). When these experiments were conducted under conditions that induce the Ysa or Ysc TTS systems, a minimum of 15 amino-terminal residues of YplA were required to direct the export of YplA-PhoA chimeras (Fig. 3B and C).
FIG. 3.
The first 20 residues of YplA can target PhoA to the flagellar, Ysa, and Ysc TTS systems of Y. enterocolitica. Strains expressing various portions of the amino terminus of YplA fused to PhoA were grown under conditions that induce the flagellar, Ysa, or Ysc TTS systems. Secreted proteins were examined as described in the legend to Fig. 3. (A) Secretion by the flagellar TTS system. Lanes: 1, GY4757/pGY335; 2, GY4757/pGY429; 3, GY4757/pGY353; 4, GY4757/pGY354;5, GY4757/pGY378; 6, GY4757/pGY379; 7, GY4757/pGY366; 8, GY4757/pGY373; 9, GY4757/pGY363. (B) Secretion by the Ysa TTS system. Lanes are the same as listed for panel A. (C) Secretion by the Ysc TTS system. Lanes: 1, GY4838/pGY335; 2, GY4838/pGY429; 3, GY4838/pGY353; 4, GY4838/pGY354; 5, GY4838/pGY378; 6, GY4838/pGY379; 7, GY4838/pGY366; 8, GY4838/pGY373; 9, GY4838/pGY363. The fusion protein encoded by each plasmid is indicated below each lane.
To confirm that protein secretion was indeed dependent upon a TTS pathway, the export of a representative YplA-PhoA fusion was examined in various mutants that are defective for either the Ysa, Ysc, or flagellar TTS system. The representative fusion contained the amino-terminal 30 amino acids of YplA fused to PhoA (YplA1-30-PhoA). Secretion of YplA30-PhoA under flagellar TTS-system-inducing conditions was affected by a mutation in flhDC, which is required for the expression of flagellar genes (37). This mutation did not affect the Ysa or Ysc TTS systems (Fig. 4A). Similarly, mutants defective for Ysa TTS (ysaV) or Ysc TTS (pYV8081−) were affected for YplA30-PhoA export under conditions that induce the respective pathway (Fig. 4B and C). We also examined whole-cell lysates for the presence of YplA1-30-PhoA, but despite several attempts, no protein was detected even for samples generated from the different TTS system mutants (data not shown). This may indicate that whole-cell-associated YplA1-30-PhoA is rapidly degraded or could indicate that YplA1-30-PhoA synthesis is coupled to secretion. In either case, these results confirm that YplA-PhoA hybrids are directed for type III secretion by any TTS system in Yersinia by a secretion signal within the amino terminus of YplA.
FIG. 4.

Analysis of YplA1-30-PhoA export by selected flagellar, Ysa, and Ysc TTS system mutants. The ability to secrete the YplA1-30-PhoA hybrid was tested in strains carrying mutations that block secretion by one of the three distinct TTS systems. Each mutant was grown under conditions that induce secretion by the flagellar, Ysa, or Ysc TTS system. Supernatant proteins were concentrated, separated on a 10% polyacrylamide gel, transferred to a nitrocellulose membrane, and probed with α-YplA antibody. In each panel, lanes are as follows: 1, JB580v(wild type)/pGY378; 2, GY4478(pYV8081−)/pGY378; 3, GY460(ΔflhDC)/pGY378; 4, GY4428(ysaV::pEP185.2)/pGY378. (A) Secretion by the flagellar TTS system; (B) secretion by the Ysa TTS system; (C) secretion by the Ysc TTS system.
The amino-terminal secretion signal is necessary for the export of native YplA by all Y. enterocolitica TTS systems.
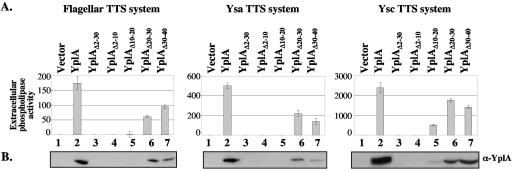
To test if the same YplA residues, or codons, shown to be important for the secretion of YplA-PhoA hybrids were also important for the secretion of native YplA, we constructed a series of mutations in yplA that alter specific regions of the protein. We began by cloning PCR-generated downstream portions of yplA (′yplA) into the low-copy-number vector pWKS130. Translational fusions were generated by cloning specific PCR-generated Pcat-yplA cassettes upstream of ′yplA. By this approach, YplA residues/codons 2 through 30, 2 through 10, 10 through 20, 20 through 30, and 30 through 40 were replaced with a short Arg-Asp-Pro peptide/9-bp nucleotide linker. Each mutant YplA was expressed in Y. enterocolitica GY4757. Supernatants of cultures grown under conditions that induce the flagellar, Ysa, or Ysc TTS systems of Yersinia were examined in two ways. First, proteins were precipitated and YplA was detected by Western blotting. Second, the relative amounts of secreted YplA were quantified by measurement of phospholipase activity. This analysis revealed that deletion of residues/codons 2 through 30 (YplAΔ2-30) or 2 through 10 (YplAΔ2-10) abolishes secretion of YplA (Fig. 5). Deletion of residues/codons 10 through 20 (YplAΔ10-20) abolished secretion through the Ysa and flagellar TTS systems and largely eliminated secretion by the Ysc TTS system. By contrast, YplA retained the capacity to be exported by each TTS system when residues/codons 20 through 30 (YplAΔ20-30) or 30 through 40 (YplAΔ30-40) were deleted. Taken together, these results indicate that the first 20 residues, or codons, are necessary for targeting YplA to the flagellar, Ysa, and Ysc TTS systems (Fig. 5).
FIG. 5.
The amino-terminal secretion signal is necessary for the export of native YplA by the flagellar, Ysa, and Ysc TTS systems of Y. enterocolitica. Secretion of YplA modified to replace specific regions with the tripeptide linker Arg-Asp-Pro was examined. These modifications included replacement of codons 2 through 30 (YplAΔ2-30), 2 through 10 (YplAΔ2-10), 10 through 20 (YplAΔ10-20), 20 through 30 (YplAΔ20-30), and 30 through 40 (YplAΔ30-40). Strains expressing each YplA variant were grown under conditions to induce the flagellar, Ysa, or Ysc TTS systems as indicated above each panel. (A) Culture supernatants were assayed for phospholipase activity to provide a quantitative assessment of the levels of extracellular YplA as described in Materials and Methods. (B) Supernatant proteins were concentrated, separated on a 10% polyacrylamide gel, transferred to a nitrocellulose membrane, and probed with α-YplA antibody. Columns/lanes for samples grown under conditions inducing the flagellar and Ysa TTS systems are as follows: 1, GY4757/pWKS130; 2, GY4757/pGY484; 3, GY4757/pGY497; 4, GY4757/pGY495; 5, GY4757/pGY503; 6, GY4757/pGY504; 7, GY4757/pGY493. Columns/lanes for samples grown under conditions inducing the Ysc TTS system are as follows: 1, GY4886/pWKS130; 2, GY4886/pGY484; 3, GY4886/pGY497; 4, GY4886/pGY495; 5, GY4886/pGY503; 6, GY4886/pGY504; 7, GY4886/pGY493.
To be sure that the effect upon secretion is a result of the removal of specific YplA residues and not due to the Arg-Asp-Pro tripeptide/9-bp nucleotide linker that was introduced during cloning, an additional representative set of mutations was generated. This set consisted of YplA mutants with a deletion of residues/codons 2 through 30, 2 through 10, or 30 through 40 and was constructed using a four-primer PCR strategy to prevent the addition of an extraneous peptide/nucleotide linker (Materials and Methods). Supernatants of cultures grown under conditions that induce the flagellar, Ysa, or Ysc TTS systems were examined by Western blotting, and secretion was quantified by phospholipase activity. The results of this analysis are consistent with those previously obtained: deletion of residues/codons 2 through 30 (YplAΔ2-30) or 2 through 10 (YplAΔ2-10) abolished YplA secretion by all TTS systems, while deletion of residues/codons 30 through 40 (YplAΔ30-40) did not affect export (Fig. 6). These data confirm that the amino-terminal residues of YplA, or 5′ mRNA, are necessary for the secretion of native YplA by the three TTS systems of Y. enterocolitica. We made several attempts to detect YplA associated with whole cells. When Y. enterocolitica cultures were grown to independently induce each of the three TTS systems, no YplA was detectable in whole-cell fractions when analyzed by Western blotting (data not shown). Similarly, when whole-cell fractions were analyzed by a liquid phospholipase assay, no phospholipase activity was detected (data not shown). This is similar to the results obtained when we examined YplA1-30-PhoA. It may indicate that cell-associated YplA, like YplA1-30-PhoA, is rapidly degraded or could indicate that polypeptide synthesis is coupled to secretion.
FIG. 6.
The effect on secretion of mutations altering the amino terminus of YplA or the untranslated region of yplA mRNA. The effect of mutations resulting in the deletion of the 5′ untranslated region (YplAΔUTR), a −1 frameshift of codons 2 through 6 (YplAframeshift), or an in-frame deletion within the amino-terminal amino acids (YplAΔ2-30, YplAΔ2-10, YplAΔ30-40) on YplA secretion was assessed under conditions that induce the flagellar, Ysa, and Ysc TTS systems. (A) Culture supernatants were assayed for phospholipase activity to provide a quantitative assessment of the levels of extracellular YplA as described in Materials and Methods. (B) Supernatant proteins were concentrated, separated on a 10% polyacrylamide gel, transferred to a nitrocellulose membrane, and probed with α-YplA antibody. Columns/lanes for samples grown under conditions inducing the flagellar and Ysa TTS systems are as follows: 1, GY4757/pWKS130; 2, GY4757/pGY484; 3, GY4757/pGY527; 4, GY4757/pGY556; 5, GY4757/pGY528; 6, GY4757/pGY549; 7, GY4757/pGY548. Columns/lanes for samples grown under conditions inducing the Ysc TTS system are as follows: 1, GY4886/pWKS130; 2, GY4886/pGY484; 3, GY4886/pGY527; 4, GY4886/pGY556; 5, GY4886/pGY528; 6, GY4886/pGY549; 7, GY4886/pGY548.
The 5′ untranslated region is dispensable for YplA export.
To determine if the 5′ untranslated region of yplA mRNA influences secretion, this region was removed (YplAΔUTR) and replaced by the untranslated region of cat, the gene encoding the cytoplasmic protein chloramphenicol acetyltranferase. YplAΔUTR secreted by Y. enterocolitica cultivated under conditions that individually induce each of the three TTS system was examined by Western blotting and liquid phospholipase assay (Fig. 6). In each case, YplAΔUTR was secreted at levels that exceeded the wild-type control, suggesting that the native untranslated region of yplA mRNA has no particular properties that are essential for YplA targeting and secretion by the TTS systems (Fig. 6). These results further indicate that the amino-terminal secretion signal for YplA is limited to residues/codons 1 through 20.
A frameshift mutation in the secretion signal affects YplA export.
Previous studies of proteins exported by TTS systems have shown that frameshift mutations are tolerated in some instances, but not others (3, 4, 6, 17). We therefore determined if the amino-terminal secretion signal of YplA could tolerate a frameshift mutation. Due to the presence of stop codons in the alternate reading frames of yplA downstream of codon 7, we chose to frameshift codons 2 through 6. Removal of 1 nucleotide at codon 2 (−1 frameshift) was followed by a compensatory insertion at codon 7 (+1 frameshift). This set of mutations changed the amino terminus of YplA from an amphipathic series of residues (MSISVSLA) to a series that is essentially nonpolar (MYVLLVLA) while leaving the rest of the amino acid sequence intact. This alteration significantly decreased YplA export by the Ysa, Ysc, and flagellar TTS systems (Fig. 6), indicating that the signal required for the type III secretion of YplA cannot tolerate a frameshift mutation.
DISCUSSION
We set out to define elements of YplA that are necessary for its export by the flagellar, Ysa, and Ycs TTS systems. Previously, it had been shown that substrate recognition by the Ysc system, and other contact-dependent systems, requires a secretion signal located in the amino terminus of the target substrate or, alternatively, the 5′ end of the coding mRNA (9). Since YplA can be recognized by the flagellar TTS system and the distantly related contact-dependent Ysa and Ysc TTS systems (29), we initially took a reductionist approach to locate the region(s) of YplA important for substrate recognition. This analysis revealed that the reporter protein PhoA could be exported by each TTS pathway when fused to the amino terminus of YplA. Assuming that PhoA does not contribute to the process, this indicates that neither the carboxy terminus of YplA nor the yplA mRNA regions 3′ to the fusion site are necessary for targeting the protein to a TTS apparatus. This observation allowed us to then take a more directed approach. We constructed an additional series of YplA-PhoA chimeras to define the region sufficient for export. Our analysis of Y. enterocolitica culture supernatants shows that a hybrid consisting of the first 20 residues of YplA fused to PhoA is recognized and exported by the flagellar, Ysa, and Ysc TTS systems. From this observation, we concluded that a secretion signal common to all three pathways is present within the first 20 residues, or codons, of YplA.
To gain some insight on the role of this region of YplA during export of the native protein, a systematic functional survey of the amino terminus/5′ mRNA region was conducted. Removal of residues/codons 2 through 30 (YplAΔ2-30) and 2 through 10 (YplAΔ2-10) essentially blocked the secretion of YplA by all three TTS pathways, reducing the level of export by at least two orders of magnitude. Conversely, alteration of other regions, such as the removal of residues/codons 20 through 30 (YplAΔ20-30) or 30 through 40 (YplAΔ30-40), had only a minor effect on the export of YplA regardless of the TTS system utilized. In these cases, YplA secretion was reduced by two- to threefold. Interestingly, we routinely observed that YplA is secreted by Y. enterocolitica grown to induce the Ysc TTS system at levels fivefold more than that which occurs when YplA is exported by the flagellar TTS system and twofold more when compared to conditions inducing the Ysa TTS system. The basis for this difference is not clear, but it is not likely due to differences in the levels of yplA transcription since the gene was under control of the constitutive Pcat promoter in all three cases. An alternative explanation is that there may be variability in the number of complete export apparatuses. Possibly, there are more Ysc TTS apparatuses than Ysa or flagellar TTS apparatuses on a per cell basis.
Previous work in Y. enterocolitica has shown that individual type III substrates can be recognized by multiple TTS systems (33). This has suggested that the elements required for peptide targeting and export by each TTS system are conserved. Consistent with this idea, we have defined the amino terminus/5′ mRNA coding sequence of YplA as an important element, or secretion signal, affecting recognition of this protein by the flagellar, Ysa, and Ysc TTS systems. We have also shown that this secretion signal is sufficient for the export of YplA-PhoA chimeras by E. coli K-12 whose only TTS pathway is the flagellar TTS system. Further, YplA export by E. coli K-12 is dependent upon flhD, the master regulator of the flagellar regulon, supporting the conclusion that secretion depends on the flagellar TTS system. Recognition of the YplA secretion signal by four TTS systems in two bacterial species is consistent with the hypothesis that at least some aspects of substrate recognition are conserved despite the distant evolutionary relationship these TTS systems share (12).
The possibility that substrate recognition by TTS systems involves both peptide and mRNA-based signals has been a subject of much debate. In the case of YplA, we have not unambiguously discerned the nature of the secretion signal, but the data favor the peptide-based secretion signal model. First, we have shown that the presence of the untranslated region of yplA mRNA is not essential for YplA secretion and its removal can actually enhance the level of YplA exported by the cell. This indicates that an important signal required for YplA recognition by a TTS system is limited to the first 20 residues/codons. Second, a hallmark of the mRNA-based hypothesis is that a frameshift at the amino terminus, which radically affects the polypeptide residues by minor changes to the nucleotide sequence, does not affect export of the protein. In the case of YplA, a frameshift at the amino terminus greatly reduced secretion of YplA by all three TTS systems. This frameshift mutation changed the amino-terminal amino acids from a series that is described as amphipathic (MSISVSLA) to a more nonpolar series of amino acids (MYVLLVLA) while leaving the rest of the sequence intact. Interestingly, using the predictions made by Lloyd and coworkers (18), this mutation changes the amino terminus from one that contains a peptide-type secretion signal predicted to result in secretion to one that is predicted to be not functional. However, we cannot eliminate the possibility that the particular point mutations we used to form the frameshift coincidentally affect the critical nucleotides of an mRNA-based secretion signal. In addition, under no circumstance did we detect cytoplasmically located YplA despite efforts to measure the accumulation of this polypeptide under a wide range of conditions, including those which induce the flagellar, Ysa, or Ysc TTS systems. We also did not detect the cytoplasmic accumulation of YplA in secretion-defective mutants. These negative results may indicate that the secretion of YplA is tightly correlated with its translation, an observation typically associated with substrates hypothesized to utilize an mRNA secretion signal (4). Alternatively, cytoplasmic YplA may be especially unstable and consequently, steady-state levels of this protein in the cell are below the level of detection. While our observations do not rule out a role for an mRNA signal for YplA export by the flagellar, Ysa, or Ysc TTS systems, the data do support a hypothesis in which the nature of the amino acids is the critical aspect of the secretion signal recognized by TTS systems.
It has been shown that Yersinia can target virulence substrates to multiple TTS apparatuses (33). Here, we show that this action is accomplished through the use of a common secretion signal within the amino terminus. This analysis suggests that Y. enterocolitica has developed a strategy for sharing secreted substrates among distinct TTS systems. It also points to the importance of alternative mechanisms for the regulation of protein export, including those that control gene transcription. Such regulatory mechanisms would be necessary to properly deploy virulence factors as needed during infection.
Acknowledgments
We thank Briana Young for experimental contributions and members of the Young laboratory for critical reading of the manuscript.
This work was supported by a grant from the National Institutes of Health, R21 AI156042 (G.M.Y.).
REFERENCES
- 1.Adler, J., and B. Templeton. 1967. The effect of environmental conditions on the motility of Escherichia coli. J. Gen. Microbiol. 46:175-184. [DOI] [PubMed] [Google Scholar]
- 2.Aizawa, S. 2001. Bacterial flagella and type III secretion systems. FEMS Microbiol. Lett. 202:157-164. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, D. M., and O. Schneewind. 1997. A mRNA signal for the type III secretion of Yop proteins by Yersinia enterocolitica. Science 278:1140-1143. [DOI] [PubMed] [Google Scholar]
- 4.Anderson, D. M., and O. Schneewind. 1999. Yersinia enterocolitica type III secretion: an mRNA signal that couples translation and secretion of YopQ. Mol. Microbiol. 31:1139-1148. [DOI] [PubMed] [Google Scholar]
- 5.Casadaban, M. J. 1975. Fusion of the Escherichia coli lac genes to the ara promoter: a general technique using bacteriophage Mu-1 insertions. Proc. Natl. Acad. Sci. USA 72:809-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng, L. W., D. M. Anderson, and O. Schneewind. 1997. Two independent type III secretion mechanisms for YopE in Yersinia enterocolitica. Mol. Microbiol. 24:757-765. [DOI] [PubMed] [Google Scholar]
- 7.Cheng, L. W., O. Kay, and O. Schneewind. 2001. Regulated secretion of YopN by the type III machinery of Yersinia enterocolitica. J. Bacteriol. 183:5293-5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cornelis, G. R., A. Boland, A. P. Boyd, C. Geujin, M. Iriarte, C. Neyt, M. P. Sory, and I. Stainier. 1998. The virulence plasmid of Yersinia, an antihost genome. Microbiol. Mol. Biol Rev. 62:1315-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornelis, G. R., and F. Van Gijsegem. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54:735-774. [DOI] [PubMed] [Google Scholar]
- 10.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghosh, P. 2004. Process of protein transport by the type III secretion system. Microbiol. Mol. Biol. Rev. 68:771-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gophna, U., E. Z. Ron, and D. Graur. 2003. Bacterial type III secretion systems are ancient and evolved by multiple horizontal-transfer events. Gene 312:151-163. [DOI] [PubMed] [Google Scholar]
- 13.Haller, J. C., S. Carlson, K. J. Pederson, and D. E. Pierson. 2000. A chromosomally encoded type III secretion pathway in Yersinia enterocolitica is important in virulence. Mol. Microbiol. 36:1436-1446. [DOI] [PubMed] [Google Scholar]
- 14.Hueck, C. J. 1998. Type III secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacobi, C. A., A. Roggenkamp, A. Rakin, R. Zumbihl, L. Leitritz, and J. Heesemann. 1998. In vitro and in vivo expression studies of yopE from Yersinia enterocolitica using the gfp reporter gene. Mol. Microbiol. 30:865-882. [DOI] [PubMed] [Google Scholar]
- 16.Kinder, S. A., J. L. Badger, G. O. Bryant, J. C. Pepe, and V. L. Miller. 1993. Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O:8 and construction of a transformable R-M+ mutant. Gene 136:271-275. [DOI] [PubMed] [Google Scholar]
- 17.Lloyd, S. A., M. Norman, R. Rosqvist, and H. Wolf-Watz. 2001. Yersinia YopE is targeted for type III secretion by N-terminal, not mRNA, signals. Mol. Microbiol. 39:520-531. [DOI] [PubMed] [Google Scholar]
- 18.Lloyd, S. A., M. Sjostrom, S. Andersson, and H. Wolf-Watz. 2002. Molecular characterization of type III secretion signals via analysis of synthetic N-terminal amino acid sequences. Mol. Microbiol. 43:51-59. [DOI] [PubMed] [Google Scholar]
- 19.Macnab, R. M. 1999. The bacterial flagellum: reversible rotary propellor and type III export apparatus. J. Bacteriol. 181:7149-7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Manoil, C., and J. Beckwith. 1985. TnphoA: a transposon probe for protein export signals. Proc. Natl. Acad. Sci. USA 82:8129-8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marenne, M. N., L. J. Mota, and G. R. Cornelis. 2004. The pYV plasmid and the Ysc-Yop type III secretion system, p. 319-348. In E. Carniel and B. J. Hinnebusch (ed.), Yersinia: molecular and cellular biology. Horizon Bioscience, Norfolk, United Kingdom.
- 23.Michiels, T., and G. R. Cornelis. 1991. Secretion of hybrid proteins by the Yersinia Yop export system. J. Bacteriol. 173:1677-1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russmann, H., T. Kubori, J. Sauer, and J. E. Galan. 2002. Molecular and functional analysis of the type III secretion signal of the Salmonella enterica InvJ protein. Mol. Microbiol. 46:769-779. [DOI] [PubMed] [Google Scholar]
- 25.Schesser, K., E. Frithz-Lindsten, and H. Wolf-Watz. 1996. Delineation and mutational analysis of the Yersinia pseudotuberculosis YopE domains which mediate translocation across bacterial and eukaryotic cellular membranes. J. Bacteriol. 178:7227-7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmiel, D. H., E. Wagar, L. Karamanou, D. Weeks, and V. L. Miller. 1998. Phospholipase A of Yersinia enterocolitica contributes to pathogenesis in a mouse model. Infect. Immun. 66:3941-3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmiel, D. H., G. M. Young, and V. L. Miller. 2000. The Yersinia enterocolitica phospholipase gene yplA is part of the flagellar regulon. J. Bacteriol. 182:2314-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negtive bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 29.Sory, M. P., A. Boland, I. Lambermont, and G. R. Cornelis. 1995. Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc. Natl. Acad. Sci. USA 92:11998-12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.von Tigerstrom, R. G., and S. Stelmaschuk. 1988. The use of Tween 20 in a sensitive turbidimetric assay of lipolytic enzymes. Can. J. Microbiol. 35:511-514. [DOI] [PubMed] [Google Scholar]
- 31.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 32.Warrens, A. N., M. D. Jones, and R. I. Lechler. 1997. Splicing by overlap extension by PCR using asymmetric amplification. Gene 186:29-35. [DOI] [PubMed] [Google Scholar]
- 33.Young, B. M., and G. M. Young. 2002. Evidence for targeting of Yop effectors by the chromosomally encoded Ysa type III secretion system of Yersinia enterocolitica. J. Bacteriol. 184:5563-5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Young, B. M., and G. M. Young. 2002. YplA is exported by the Ysc, Ysa, and flagellar type III secretion systems of Yersinia enterocolitica. J. Bacteriol. 184:1324-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Young, G. M., and V. L. Miller. 1997. Identification of novel chromosomal loci affecting Yersinia enterocolitica pathogenesis. Mol. Microbiol. 25:319-328. [DOI] [PubMed] [Google Scholar]
- 36.Young, G. M., D. H. Schmiel, and V. L. Miller. 1999. A new pathway for the secretion of virulence factors by bacteria: the flagellar export apparatus functions as a protein secretion system. Proc. Natl. Acad. Sci. USA 96:6456-6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young, G. M., M. J. Smith, S. A. Minnich, and V. L. Miller. 1998. The Yersinia enterocolitica motility master regulatory operon, flhDC, is required for flagellin production, swimming motility, and swarming motility. J. Bacteriol. 181:2823-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]